Abstract
An intricate network of hormone signals regulates plant development and responses to biotic and abiotic stress. Salicylic acid (SA), derived from the shikimate/isochorismate pathway, is a key hormone in resistance to biotrophic pathogens. Several SA derivatives and associated modifying enzymes have been identified and implicated in the storage and channeling of benzoic acid intermediates or as bioactive molecules. However, the range and modes of action of SA-related metabolites remain elusive. In Arabidopsis, Enhanced Disease Susceptibility 1 (EDS1) promotes SA-dependent and SA-independent responses in resistance against pathogens. Here, we used metabolite profiling of Arabidopsis wild type and eds1 mutant leaf extracts to identify molecules, other than SA, whose accumulation requires EDS1 signaling. Nuclear magnetic resonance and mass spectrometry of isolated and purified compounds revealed 2,3-dihydroxybenzoic acid (2,3-DHBA) as an isochorismate-derived secondary metabolite whose accumulation depends on EDS1 in resistance responses and during ageing of plants. 2,3-DHBA exists predominantly as a xylose-conjugated form (2-hydroxy-3-β-O-d-xylopyranosyloxy benzoic acid) that is structurally distinct from known SA-glucose conjugates. Analysis of DHBA accumulation profiles in various Arabidopsis mutants suggests an enzymatic route to 2,3-DHBA synthesis that is under the control of EDS1. We propose that components of the EDS1 pathway direct the generation or stabilization of 2,3-DHBA, which as a potentially bioactive molecule is sequestered as a xylose conjugate.
Keywords: Arabidopsis, Innate Immunity, Metabolomics, Pathogen-associated Molecular Pattern (PAMP), Plant, Plant Immunity, Plant Senescence, Salicylic Acid
Introduction
Plants possess a multi-layered innate immune system that protects local tissues from infection by the majority of pathogens and primes systemic tissues against subsequent attack. The structural range of plant secondary metabolites produced during defense activation is vast, although the functions and biosynthetic origins have only been established for a few molecules (1). A complex interaction between plant hormone pathways influences the accumulation and activities of particular pathogen-induced metabolites (2). Molecules such as reactive oxygen intermediates and fatty acid derivatives, which are produced as part of a plant immune response, in turn modulate local and systemic resistance (3). Also, glucosinolates with established roles in insect defense are now known to act as anti-microbial and signaling molecules in plant resistance to pathogens (4, 5).
The phenolic hormone salicylic acid (SA)4 is an important defense signal in plant resistance to biotrophic and hemi-biotrophic pathogens (2). Pathogen-induced accumulation of SA derived from isochorismate in the chloroplast is essential for basal resistance to virulent pathogens and for establishment of systemic acquired resistance. The precise mode of SA action is unclear, although it has been shown to interact in a complex manner with reactive oxygen intermediate (6–8) and to modulate cellular redox homeostasis leading to changes in transcription factor activities and defense gene activation (9). Many SA-induced responses are regulated by the transcriptional co-activator NPR1, which moves into the nucleus after SA application to modulate the expression of defense and secretory pathway genes (10, 11).
SA synthesized endogenously after pathogen challenge or applied exogenously is rapidly metabolized to the SA 2-O-β-glucoside, which has been reported to be a predominant storage form in various plant species including tobacco (12) and Arabidopsis (13). A number of SA derivatives and genes regulating SA modification steps also appear to be important for plant resistance (14). For example, methyl-SA acts as a systemic defense signal in tobacco (15). Studies in Arabidopsis suggest that other molecules are also needed for the establishment of systemic acquired resistance (16). Mutations in PBS3 (AvrPphB Susceptible3) encoding an acyl-adenylate/thioester-forming (GH3) enzyme cause enhanced susceptibility to bacterial and oomycete pathogens, indicating a role for this enzyme in resistance (17–19). In in vitro assays, PBS3 conjugates amino acids preferentially to 4-substituted benzoates and is inhibited by SA (20). Thus, PBS3 may control the direction of a chorismate flux, leading to SA synthesis.
In Arabidopsis, the nucleo-cytoplasmic protein EDS1 and its interactor PAD4 regulate SA accumulation in basal resistance to virulent pathogens and in effector-triggered immunity mediated by intracellular Toll/interleukin 1 receptor-nucleotide-binding leucine-rich repeat (NB-LRR) receptors (21, 22). A direct role for EDS1 or PAD4 in pathogen-inducible SA synthesis can be excluded because the corresponding null mutants display intact resistance and SA accumulation triggered by NB-LRR receptors containing a coiled-coil domain (23, 24). Although EDS1 and PAD4 are dispensable for local resistance and cell death triggered by the coiled-coil NB-LRR receptor RPM1 (recognizing Pseudomonas syringae effectors AvrRpm1 and AvrB), they are needed for the release of signals from infection sites leading to systemic acquired resistance (16, 25). Several components of an SA-independent branch of EDS1 signaling in RPM1 resistance were identified from analysis of Arabidopsis wild type and mutant gene expression profiles (26). For example, FMO1 encoding a flavin-dependent mono-oxygenase acts as a positive modulator, and a cytosolic nudix hydrolase (NUDT7) acts as a negative component of EDS1-regulated resistance (26). FMO1 promotes resistance responses through oxygenase catalysis (26–28), but its precise biochemical activity is not known. NUDT7 has a role in limiting reactive oxygen intermediate-induced cellular stress (29, 30), and nudt7 mutants, which display deregulated defense, cell death, and SA accumulation (26, 29), were reported to have a more oxidizing cellular redox status compared with wild type (29). The above phenotypes point to the importance of EDS1-regulated processes in plant stress signaling that do not involve SA accumulation per se.
Here we exploited the intact local Arabidopsis RPM1 resistance response of Arabidopsis eds1 pathway mutants (26) to characterize secondary metabolites that may play a role in EDS1 stress signal relay and explain an SA-independent component of the plant immune response. We have identified 2,3-dihydroxybenzoic acid xyloside (2,3-DHBX) as a metabolite that is reduced in Arabidopsis eds1 compared with wild type in resistance responses to pathogens and in senescing plants. Analysis of Arabidopsis mutants in EDS1 signaling and senescence reveals that an increase in 2,3-DHBX content correlates with the onset of plant senescence. Our data suggest that 2,3-DHBA is generated enzymatically from isochorismate or isochorismate-derived SA. Conjugation of 2,3-DHBA to xylose may represent a mechanism for controlling the availability of free, bioactive SA-derivatives in pathogen-infected and senescing plant tissues.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
All of the Arabidopsis thaliana genotypes used are in the Columbia (Col-0) background and have been previously described: Col eds1-2, fmo1-1, nudt7-1 (26), sid2-1 (31), pad2-1 (32), vtc1-1 (33), and Col/pPR1:GUS (34). The plants were grown on soil in controlled environment chambers under a regime of 10 h of light at 180 μE m−2 s−1, 22 °C, and 60% relative humidity unless otherwise stated. The DEX-inducible transgenic line AvrRpm1-HA has been described (35).
Pathogen Inoculations and Application of SA and 2,3-DHBA
P. syringae pv. tomato (Pst) strain DC3000 expressing avrRpm1 was inoculated by vacuum infiltration into leaves, and Hyaloperonospora arabidopsidis conidiospores were spray-inoculated onto leaves as previously described (21). Arabidopsis sterile seedlings were grown in microtiter plate wells containing Murashige and Skoog medium with 0.5% sucrose, as described (36). After 2 weeks the medium was replaced by fresh Murashige and Skoog medium supplemented with SA (Duchefa) or 2,3-DHBA (Sigma-Aldrich) at the indicated concentrations, and incubation was continued for 24 h. Seedling extracts were prepared, and fluorimetric β-glucuronidase (GUS) activities relative to protein concentration were performed as previously described (36).
HPLC Analysis of Metabolites
Leaves of plants were harvested and ground to a fine powder in liquid nitrogen. 200 mg of plant material/sample were extracted in 80% methanol, as described (37). For quantification of total SA, 2,3-DHBA, and 2,5-DHBA contents, leaf extracts were hydrolyzed with β-glucosidase (EC 3.2.1.21; Sigma-Aldrich), resulting in the release of free phenolics from their glycon residues (glucose or xylose). The phenolics were then extracted as described for SA analysis (38). HPLC analyses were preformed on an Agilent 1100 HPLC system. SA was detected at a 415-nm wavelength using fluorescence detection, whereas 2,3-DHBA and 2,5-DHBA were quantified using a photodiode array detector at 254 and 320 nm, respectively. The identified peaks were quantified by comparison with the pure standards SA (Duchefa), 2,3-DHBA, and 2,5-DHBA (Sigma-Aldrich). In this method, β-glucosidase treatment of the plant extracts to hydrolyze sugar-conjugated metabolites was used to determine the total levels of particular benzoic acid derivatives. The free amounts were subtracted from the total to determine the amounts of the corresponding sugar-conjugated form. Detection limits for SA, 2,3-DHBA, and 2,5-DHBA were 0.05, 0.1, and 0.05 μg/g of fresh weight, respectively.
NMR Spectroscopy
1H NMR, 1H-1H COSY, HSQC, and heteronuclear multiple bond correlation (HMBC) spectra were recorded on an Avance 500 NMR spectrometer (Bruker-Biospin, Karlsruhe, Germany) at 300 K using a 5-mm TCI CryoProbeTM. The sample was dissolved in methanol-d4 (120 μl) and measured in a 2.5-mm NMR tube. Chemical shift values (δ) are given relative to tetramethylsilane as an internal standard, and coupling constants are in Hertz. The 13C NMR chemical shift values were extracted from the HSQC and HMBC spectra.
Ultra Performance Liquid Chromatography Electrospray Ionization-Quantitative Time-of-Flight-MS
Chromatographic separations were performed on an Acquity Ultra Performance Liquid Chromatography system (Waters) equipped with a modified C18 column (HSS T3, 1.0 × 100 mm; particle size, 1.8 μm; Waters) applying the following binary gradient at a flow rate of 150 μl min−1: 0–1 min, linear from 95% A (water, 0.1% formic acid) to 85% A; 1–7 min, linear from 85% A to 55% A; 7–7.5 min, linear from 55% A to 45% A; 7.5–8 min, linear from 45% A to 5% A; 8–11 min, isocratic 95% B (acetonitrile, 0.1% formic acid); and 11.5–14 min, isocratic 95% A. The injection volume was 2 μl. Eluted compounds were detected from m/z 100–1000 by a MicrOTOF-Q hybrid quadruple time-of-flight mass spectrometer (Bruker Daltonics) equipped with an Apollo II electrospray ion source in negative ion mode using following instrument settings: nebulizer gas, nitrogen, 1.6 bar; dry gas, nitrogen, 6 liters/min, 190 °C; collision gas, argon; capillary, −4000 V; end plate offset, −500 V; collision RF 200 Vpp (50%)/400 Vpp (50%), transfer time, 70 μs; prepulse storage, 5 μs; pulser frequency, 5 kHz; and spectra rate, 3 Hz. Mass calibration was achieved by infusion of 10 mm lithium formiate in isopropanol/water, 1/1 (v/v). For the acquisition of collision-induced fragmentation spectra, the ions were isolated with a width of ± 4 Da. Collision energy was chosen in the range of 10–17 eV according to the substance in question. The raw data files were analyzed by instrumental software (Compass 1.3 for MicrOTOF).
Deuterium [2H]-Labeled SA Application and Metabolite Analysis
4-week-old soil-grown AvrRpm1-HA plants were sprayed with 0.2 μm DEX or a mock solution of sterile water containing 0.01% Tween 20. After 30 min, the leaves were infiltrated with 500 μm [2H]SA (2-hydroxybenzoic acid-d6; Dr. Ehrenstorfer GmbH) in sterile 10 mm MgCl2. Infiltrated leaves were harvested at 16 hpi and analyzed by LC-MS.
RT-PCR Analysis
Total RNA was extracted using TRI reagent (Sigma). RT reactions were performed with 1 μg of total RNA and 0.5 μg of oligo(dT)18 primer at 42 °C using reverse transcriptase and RNase inhibitor RNasin (both from Promega) in a 20-μl reaction volume. Aliquots of 1-μl RT reaction product were subsequently used for quantitative RT-PCR analysis with iQ SYBR Green Supermix on a iQCycler5 system (Bio-Rad) using the following gene-specific primers: SAG13 forward, 5′-CATGACCGCTCTTGTCACTG-3′; SAG13 reverse, 5′-TCGCAGACAGAAGTGGTGAC-3′; Actin2 forward, 5′-GGTAACATTGTGCTCAGTGGTGG-3′; and Actin2 reverse, 5′-AACGACCTTAATCTTCATGCTGC-3′. The values were normalized to those for the Actin2 reference gene and expressed relative to the Col-0 sample at 5 weeks. The experiment was repeated twice with RNA from independent biological samples, and RT-PCRs were preformed in triplicate.
RESULTS
Transcriptional Co-regulation of EDS1 Pathway Components with ICS1
In leaf extracts of eds1, pad4, and fmo1 null mutants, SA accumulates to levels seen in wild type plants after triggering coiled-coil NB-LRR (RPM1) resistance, consistent with regulatory rather than SA biosynthetic functions of these defense components (24, 26). Meta-analysis of Arabidopsis gene co-regulation using the ACT (39) and ATTED-II databases identified FMO1, EDS1, and PAD4 as genes co-expressed (Pearson correlation coefficient (r) of 0.71, 0.61 and 0.82, respectively) with ICS1 (isochorismate synthase 1) encoding a key enzyme in pathogen-inducible SA biosynthesis (supplemental Fig. S1). Also, PBS3 encoding a potential benzoate-amino acid conjugating enzyme (see Introduction) is closely co-regulated with FMO1 (r value of 0.86) (supplemental Fig. S1). These data prompted us to investigate by metabolic profiling whether EDS1 pathway components affect metabolism of SA or SA-related benzoic acids.
Identification of EDS1-dependent Phenolic Metabolites
Leaf material was harvested from wild type and eds1 plants grown in soil under a short day light regime (inoculations were done 2 h after the start of the light period). Four-week-old plants were infiltrated with 10 mm MgCl2 or with 5 × 106 colony-forming units/ml Pst DC3000 avrRpm1 in 10 mm MgCl2, and the samples were prepared for HPLC analysis. Initially, a crude methanol extraction was used (37). Then, to increase the likelihood of identifying individual phenolic molecules, a methanol extraction with or without β-glucosidase treatment, followed by extraction with ethyl acetate (which is routinely used for SA analysis (38), was also applied (see “Experimental Procedures”).
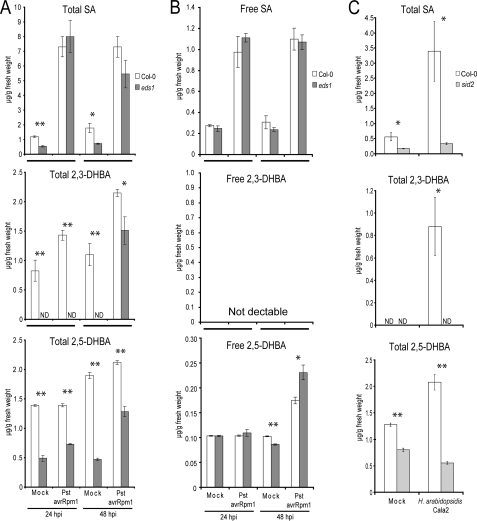
The amounts of total SA were lower in mock inoculated eds1 leaves compared with wild type Col-0 but increased to similar high levels 24 and 48 h after infiltration with Pst DC3000 avrRpm1 (Fig. 1A), consistent with EDS1 being dispensable for RPM1-triggered SA accumulation. HPLC analysis of nonhydrolyzed crude methanol leaf extracts revealed several peaks with UV absorption spectra resembling SA whose accumulation was absent or strongly reduced in eds1 compared with wild type plants. Detailed examination of chromatograms of β-glucosidase-treated samples revealed two compounds with SA-like spectral properties present in extracts from wild type plants and reduced in abundance in eds1 samples. By comparing column retention times and UV absorption spectra of the respective peaks with standards of various SA derivatives, we tentatively identified the differentially accumulating compounds as 2,3-dihydroxybenzoic acid (2,3-DHBA) and 2,5-DHBA (gentisic acid; supplemental Fig. S2). Conjugates of SA-related compounds, such as 2,5-dihydroxybenzoic acid glucoside, have been identified previously in Arabidopsis (13). Comparison of free and hydrolyzed leaf extracts indicated that both 2,3- and 2,5-DHBA accumulate in pathogen-inoculated Arabidopsis leaves of 4-week-old plants predominantly as a conjugated form, although 2,5-DHBA can be detected in its free form at low concentrations (Fig. 1, A and B).
FIGURE 1.
Accumulation of SA, 2,3-DHBA, and 2,5-DHBA after challenge with avirulent pathogens. The data represent the averages ± S.D. of three replicate samples. Metabolite levels were significantly different in Col-0 samples compared with corresponding samples from the mutants at p < 0.05 (*) and p < 0.01 (**). The experiments were repeated twice with similar results. Total (A) and free (B) levels of phenolic metabolites were measured in leaves of 4-week-old plants that were syringe-infiltrated with 10 mm MgCl2 (mock) or with 5 × 106 colony-forming units/ml Pst DC3000 avrRpm1 (Pst avrRpm1). C, accumulation of total phenolic metabolites was determined in 3-week-old plants 4 days after treatment with water (mock) or spray inoculation with 105 spores/ml of avirulent H. arabidopsidis isolate Cala2 (recognized by R gene RPP2 in Col-0).
Total amounts of 2,3-DHBA and to a lesser extent 2,5-DHBA increased in Arabidopsis wild type leaves after inoculation with Pst DC3000 avrRpm1 (Fig. 1A) or an avirulent isolate of the oomycete pathogen, H. arabidopsidis (Fig. 1C). Accumulation of both compounds was low compared with that of total SA at 24 and 48 hpi with Pst DC3000 avrRpm1 (Fig. 1, A and B). Notably, 2,3-DHBA was also below the level of detection in mock treated or Pst DC3000 avrRpm1-infiltrated eds1 leaf samples 24 hpi but was identified in eds1 samples collected at 48 hpi (Fig. 1A). By contrast, the levels of 2,5-DHBA were only moderately reduced in both mock and pathogen-treated eds1 samples compared with wild type (Fig. 1, A and B). We concluded that mutation of EDS1 impacts more severely on the generation or stability of 2,3-DHBA than on 2,5-DHBA. This prompted us to characterize structurally the 2,3-DHBA-conjugated form.
Structural Analysis of the 2,3-DHBA Conjugate
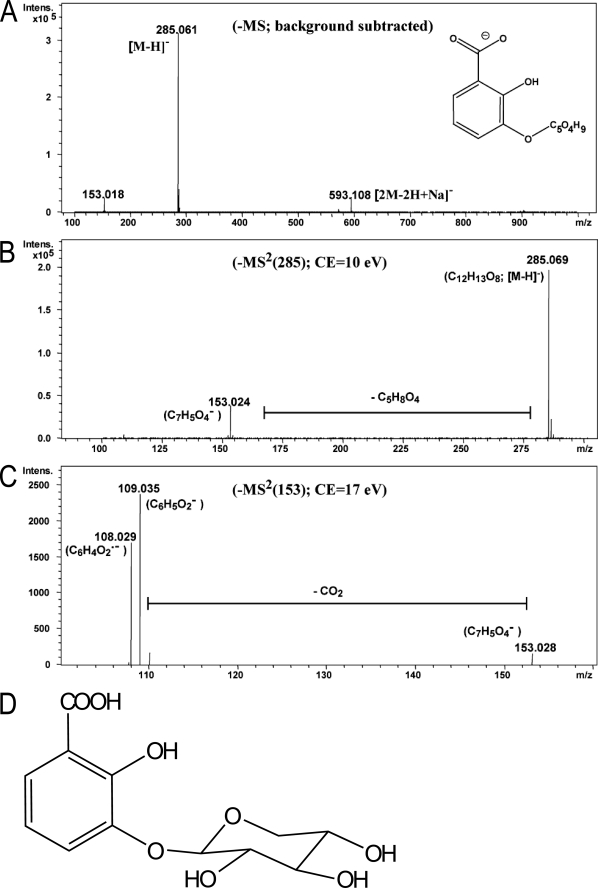
NMR and MS analysis of the putative 2,3-DHBA compound purified from crude Arabidopsis leaf extracts revealed its structure to be 2-hydroxy-3-β-O-d-xylopyranosyloxy benzoic acid, otherwise denoted 2,3-DHBA 3-O-β-d-xyloside (2,3-DHB3X) (Fig. 2). Detailed LC-MS (Fig. 2, A–C) and NMR data (supplemental Fig. S3) confirmed 2,3-DHBA as the aglycone present in the native conjugate. In addition to aglycone signals, the 1H NMR and 1H-1H COSY spectra of 2,3-DHB3X showed a carbohydrate spin system comprising a typical doublet of a β-configured proton H-1′ (δ 4.88, J = 7.3 Hz) assignable to the anomeric center and another five-well resolved one-proton signal. These data indicated a glycosidic structure in which the carbohydrate part must be a pentose and not glucose or another hexose. Consistent with the chair conformation of β-xylopyranose, a signal of the axial proton of the methylene group H-5′ax appeared at relatively high field (δ 3.32), and large coupling constants indicated axial configuration of the four sugar methine protons. 13C chemical shifts were obtained from the HSQC and HMBC spectra and confirmed the structure of the β-xylopyranosyl unit. Finally, the HMBC correlation of H-1′ with C-3 indicated that the xylose is attached to 3-OH of 2,3-DHBA. The xylose substitution at position 3 of the benzene ring identified here differs from other forms reported in Arabidopsis for SA and 2,5-DHBA conjugates in which β-d-glucose was linked to the respective aglycones via an ester or glucosidic bond at position 2 or 5 of the benzene ring (13).
FIGURE 2.
Structural elucidation of 2,3-DHB3X by LC-MS. The molecular structure was determined by NMR spectroscopy (supplemental Fig. S3) and confirmed by LC-MS. A, LC-electrospray ionization-MS analysis of the peak representing 2,3-DHB3X. LC-electrospray ionization-MS analysis in negative ion mode revealed an ion of m/z 285.061 as [M-H], suggesting a molecular formula of M = C12H14O8. B and C, the insource fragment at m/z 153.018 and subsequent CID experiments of the parent ion (285.06) (B) and the daughter ion (153.02) (C) showed a neutral loss of a pentose moiety indicating that 2,3-DHBA is linked to the pentose xylose. D, structure of 2,3-DHB3X.
The Bulk of 2,3- and 2,5-DHBA Is Synthesized via Isochorismate
To our knowledge, 2,3-DHBA has not been identified previously in Arabidopsis. 2,3-DHBA levels increased in Catharanthus roseus cell suspension and shoot cultures in response to elicitors (40). Also 2,5-DHBA accumulation was reported in tomato plants upon viral infection (41). In plants, both 2,3-DHBA and 2,5-DHBA are thought to be derived from the shikimate pathway via isochorismate, as established in bacteria (14, 42, 43). We therefore measured accumulation of the DHBAs in the SA biosynthetic mutant sid2-1 (salicylic acid induction deficient 2-1) encoding chloroplast localized ICS1 (31, 44). 2,3-DHBA was undetectable in mock and pathogen-inoculated sid2-1 leaves, whereas basal levels of 2,5-DHBA were present in this mutant but significantly reduced compared with wild type (Fig. 1C). These findings are consistent with DHBAs being derived substantially but not exclusively from the chloroplastic SA precursor isochorismate.
Total 2,3-DHBA Levels Increase in Ageing Wild type but Not eds1 Leaves
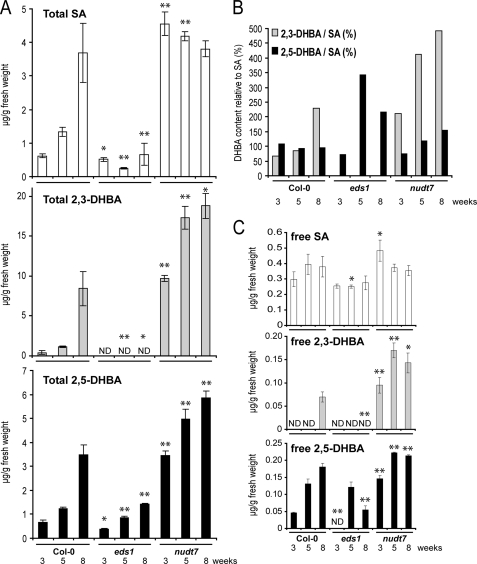
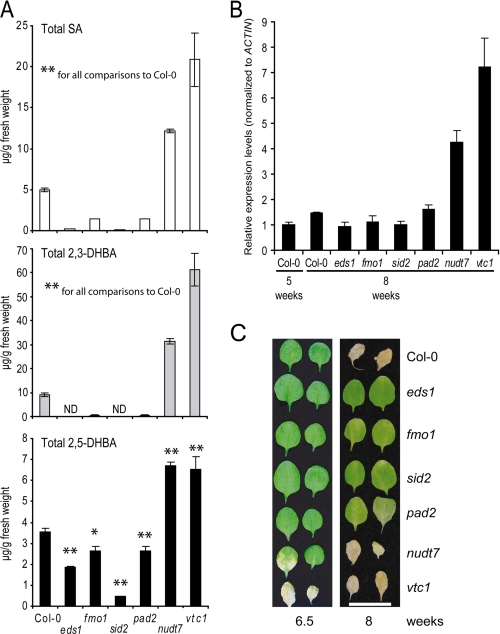
2,3-DHBA was detected in extracts from mock treated 4-week-old plants (Fig. 1A) but not 3-week-old plants (Fig. 1C), suggesting that 2,3-DHBA may accumulate with plant age, as shown previously for SA (45). We therefore determined the content of phenolic metabolites in 3-, 5-, and 8-week-old wild type and eds1 leaves and in the SA hyperaccumulating nudt7 mutant (26). Whole rosettes were harvested to avoid variation caused by different leaf ages. Total amounts of SA, 2,3-DHBA, and 2,5-DHBA increased with the age of wild type plants (Fig. 3A). By contrast, SA levels did not increase with age in eds1 (Fig. 3A). Also, the high SA levels seen in 3-week-old nudt7 plants did not rise further in samples from older plants (Fig. 3A). Levels of free 2,3- and 2,5-DHBA increased with plant age, although they remained low compared with the high amounts of free SA, which did not increase significantly with age (Fig. 3C). In 8-week-old wild type plants the absolute concentration of total 2,3-DHBA was higher than that of total SA, reaching 228% of SA levels (Fig. 3B). Strikingly, although SA and 2,5-DHBA were present in eds1, 2,3-DHBA was not detectable in this mutant at any plant age (Fig. 3, A and B). These data support our conclusion that accumulation of 2,3-DHBA, in particular, depends on functional EDS1. Analysis of Arabidopsis gene expression microarray data from young and senescing leaves (46) showed that EDS1 expression does not increase with leaf age, but expression of the EDS1-related genes FMO1, SID2, and PBS3 does (supplemental Fig. S4).
FIGURE 3.
2,3-DHBA accumulation increases with age in wild type but not eds1 mutant plants. A, total phenolic metabolite content was determined in healthy plants at the indicated age in weeks. B, relative total levels of 2,3-DHBA and 2,5-DHBA compared with total SA levels (%) were calculated from the data in Fig. 3A. C, content of free SA, 2,3-DHBA, and 2,5-DHBA. The data points are the averages ± S.D. of three replicate samples. The metabolite levels were significantly different in Col-0 samples compared with corresponding samples from the mutants at p < 0.05 (*) and p < 0.01 (**).
Comparison of the total leaf contents of 2,3-DHBA and 2,5-DHBA relative to SA revealed that their ratios varied depending on plant age and genetic background (Fig. 3B). The much larger increase in total 2,3-DHBA amounts with plant age compared with that of 2,5-DHBA is consistent with DHBAs being generated enzymatically rather than by nonenzymatic reaction of OH* with SA. This interpretation is only valid if the DHBAs are stable end products and not further metabolized by the plant. We therefore measured amounts of free and conjugated SA and 2,3- and 2,5-DHBA during development. All of the compounds were present mainly as their sugar conjugates (Fig. 3, A and C), supporting the idea that the DHBAs identified here are stable metabolites that are probably conjugated to a sugar to allow vacuolar storage, as has been shown for the SA glycoside (SAG) (12).
Exogenously Supplied SA Is Transformed to a SA Hexoside and 2,3-DHB3X
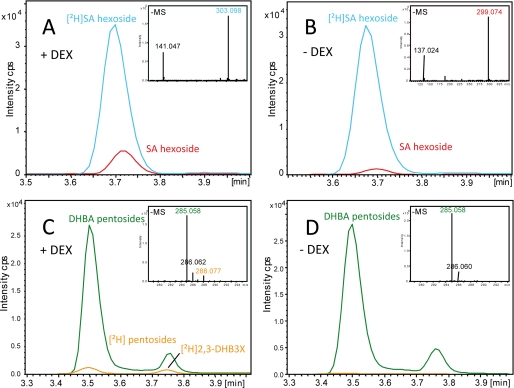
The above results show that 2,3- and 2,5-DHBA accumulation is strongly dependent on ICS1, and therefore isochorismate is an intermediate in DHBA synthesis. Because isochorismate is also a precursor for pathogen-induced SA, we determined whether SA can be converted to 2,3- and 2,5-DHBA during the resistance response. For this, deuterium 2H-labeled SA was infiltrated into leaves of a transgenic Arabidopsis line expressing avrRpm1 under the DEX-inducible promoter (AvrRpm1-HA). Leaves sprayed with either DEX or a mock solution followed by infiltration with [2H]SA in MgCl2 buffer were harvested at 16 hpi. LC-MS analysis of the leaf samples revealed that the majority of [2H]SA was conjugated to a [2H]SA hexoside (most likely SAG) in DEX-induced and mock treated tissues (Fig. 4, A and B). However, a small amount of [2H] was incorporated into two distinct [2H]DHBA pentosides. One was identified as 2,3-DHB3X, which was detectable after the DEX treatment (Fig. 4C), indicating that SA can be directly transformed into 2,3-DHBA.
FIGURE 4.
Exogenously applied deuteriated SA ([2H]SA) is transformed into [2H]SAG and [2H]DHBAs including [2H]2,3-DHB3X in planta. A and B, DEX-treated (A) and mock treated (B) plants of the DEX-inducible transgenic line AvrRpm1-HA infiltrated with [2H]SA show a high level of a [2H]SA-hexoside m/z 303.101 (calculated) (blue line). An increase of ∼3-fold was evident for the nondeuteriated natural form of the SA-hexoside at m/z 299.076 (calculated) (red line) in DEX-triggered (A) compared with mock treated plants (B). Infiltration of [2H]SA into DEX-elicited (C) Arabidopsis produced low amounts of [2H]DHBA pentosides m/z 288.079 (calculated) (orange line) compared with high concentrations of the natural DHBA pentosides m/z 285.060 (calculated) (green line). D, [2H]DHBA conjugates were not detected in mock treated leaves. Using a biosynthetic standard, the second peak at a retention time of 3.75 min was identified as [2H]2,3-DHB3X.
Total 2,3-DHBA Accumulation Correlates with Onset of Senescence in Arabidopsis Mutants
We have shown that the levels of SA and DHBAs depend on plant genotype, with eds1 and sid2 being depleted in 2,3-DHBA and nudt7 accumulating more 2,3-DHBA (Fig. 3). To understand more about the genetic control of accumulation of these phenolic compounds, we measured SA and DHBA levels in a further set of Arabidopsis mutants. Eight-week-old plants were analyzed because at this stage the level of 2,3-DHBA is particularly high in wild type (Fig. 3A). We included fmo1 in the analysis because FMO1 contributes to EDS1 defense independently of SA accumulation in younger plants (26). We also tested the pad2-1 (phytoalexin deficient 2) mutant, which accumulates 22% of wild type glutathione content because of a defect in γ-glutamylcysteine synthetase, a key enzyme in glutathione synthesis (32), because it was reported that SA and glutathione levels are positively correlated (7). Further, we tested the vtc1 (vitamin c1) mutant, which has a mutation in a GDP-Man pyrophosphorylase gene leading to greatly reduced ascorbate content (33). vtc1 has constitutively high levels of SA and displays an early senescence phenotype (47). The SA-depleted sid2-1 mutant was used as a negative control (low SA levels), and nudt7-1 was used as a positive control (high SA) in these assays.
Eight-week-old plants of eds1, fmo1, and sid2 mutants accumulated lower amounts of total SA than wild type, and this correlated with reduced accumulation of total 2,3-DHBA (Fig. 5A). Levels of 2,5-DHBA were comparably less affected in these mutants, indicating that 2,3-DHBA and 2,5-DHBA display distinct accumulation patterns (Fig. 5A). The nudt7 and vtc1 mutants with high constitutive SA also displayed high amounts of both 2,3-DHBA and 2,5-DHBA. Because 2,3-DHBA levels increased strongly with plant age in wild type, we examined whether the different amounts of total 2,3-DHBA in the mutants correlate with the onset of senescence. Symptoms of senescence including cell death and induction of senescence marker genes were reported to be delayed in pad4 (45). A role in senescence was also found for the signaling and interacting partner of EDS1, SAG101 (48), and an early senescence phenotype of vtc1 has been shown (47). We found that the high levels of SA and 2,3-DHBA in vtc1 and nudt7 positively correlated with an early senescence phenotype, as reflected by higher transcript levels of the senescence-associated gene, SAG13 (49) (Fig. 5B), and accelerated onset of senescence-induced cell death compared with wild type (Fig. 5C). The 2,3-DHBA depleted mutants eds1, fmo1, sid2, and pad2 displayed delayed senescence (Fig. 5C) but not significantly reduced SAG13 expression (Fig. 5B).
FIGURE 5.
2,3-DHBA levels correlate with the onset of senescence in different genetic backgrounds. A, total levels of phenolic compounds were measured in 8-week-old, untreated wild type and mutant plants, as indicated. The data represent the averages ± S.D. of three replicate samples. The metabolite levels were significantly different in Col-0 samples compared with corresponding samples from the mutants at p < 0.05 (*) and p < 0.01 (**). B, relative transcript levels of SAG13 determined by quantitative real time PCR and displayed after normalization to the internal control ACTIN and relative to the expression in Col-0 5-week-old sample (set at 1). The data bars represent the mean levels of transcripts ± S.D. C, leaves (third and fourth emerging after the cotyledons) from mutants displaying different degrees of senescence-induced leaf necrosis. The experiment was repeated twice with similar results, and representative leaves are shown. The bar represents 2 cm.
Because pad2 and vtc1 have altered levels of redox-active compounds (32, 33) as well as SA and DHBA (Fig. 5A), we tested whether a changed redox status in eds1, fmo1, and nudt7 mutants could account for their distorted SA and DHBA contents and composition. However, there was no correlation between the accumulation of DHBAs (Fig. 5A) and the levels of the major cellular redox buffers glutathione and ascorbate (supplemental Fig. S5).
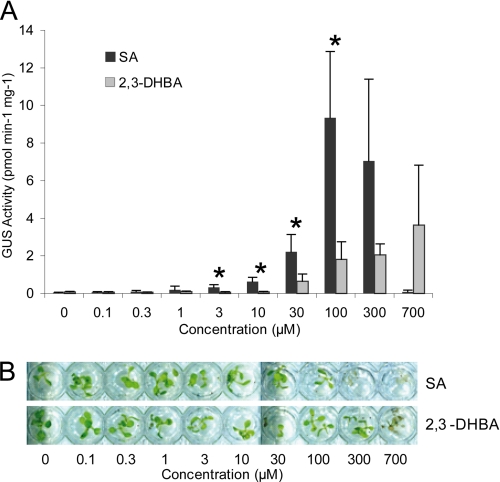
2,3-DHBA Is a Weaker Inducer of PR1 Expression than SA
SA induces expression of many plant defense-related genes, and PR1 (pathogenesis-related gene 1) is a commonly used marker of SA responses (11, 50). To compare the PR1 inducing activity of SA with 2,3-DHBA, we applied increasing concentrations of each substance to a transgenic Col-0 line expressing the GUS gene under the control of the PR1 promoter (34) and measured GUS activity. Free 2,3-DHBA displayed markedly weaker PR1 inducing activity (Fig. 6A) and toxicity (Fig. 6B) than free SA when applied to sterile liquid-grown seedlings or after spraying onto leaves of older soil-grown plants (supplemental Fig. S6).
FIGURE 6.
Exogenously applied 2,3-DHBA is a poor inducer of PR1 expression compared with SA. A, Arabidopsis seedlings harboring the SA-responsive PR1p::GUS reporter gene, grown for 2 weeks in liquid culture, were treated with increasing concentrations of SA or 2,3-DHBA for 24 h. GUS activity was determined in total extracts. The data represent the averages ± S.D. of four replicate samples normalized to total extractable protein. GUS activity was significantly different between SA and 2,3-DHBA at p < 0.05 (*). B, appearance of seedlings 24 h after treatment with SA or 2,3-DHBA. SA concentrations exceeding 100 μm are toxic (visible as tissue bleaching), whereas detrimental effects of 2,3-DHBA are apparent only at concentrations exceeding 300 μm.
DISCUSSION
SA is a key signaling molecule in resistance to biotrophic pathogens (2) and helps to protect cells against photo-oxidative stress (7, 51). Conversion of bioactive free SA to SAG and its transport to the vacuole are seen as a deactivation mechanism to limit the adverse effects of SA (12, 38). A number of recent publications point to SA modifications (such as methylation or amino acid conjugation) as potentially important reactions for modulating SA mobilization and activities (14, 15). Through metabolite profiling of Arabidopsis wild type and mutant plants, we have identified 2,3-DHBA and its xylose conjugate 2,3-DHB3X as isochorismate-derived metabolites whose accumulation in pathogen-challenged and ageing leaves requires a functional EDS1 pathway. The reduced 2,3-DHBA levels in pathogen-treated (Fig. 1) and senescing (Fig. 3) eds1 leaves when measured against SA and 2,5-DHBA suggest that EDS1 may help to direct the formation or stabilization of benzoic acid derivatives other than SA. Based on our results and those of previous studies, we discuss the biosynthesis of 2,3-DHBA and its potential role in plants (see also scheme in Fig. 7).
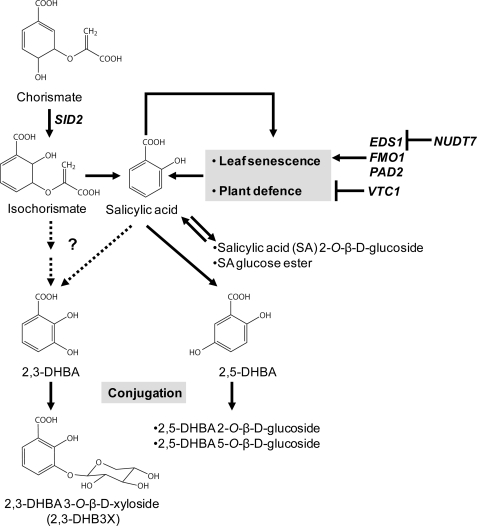
FIGURE 7.
A model for SA-metabolizing pathways leading to 2,3- and 2,5-DHBA and its various sugar conjugates in Arabidopsis. SA synthesis is promoted during leaf senescence and plant defense, forming a positive feedback loop. This feedback loop is under the positive control of EDS1, FMO1, and PAD2 and under the negative control of both NUDT7 and VTC1. Free cellular SA is removed by SA conjugation with glucose and by SA hydroxylation leading to 2,5-DHBA and subsequent glucose conjugation to 2,5-DHBA glucosides. SA hydroxylation to 2,3-DHBA and subsequent xylose conjugation to 2,3-DHB3X is a newly discovered pathway, representing a detoxification mechanism for SA or 2,3-DHBA or a means of channeling bioactive benzoic acid molecules in cells and tissues (see “Discussion”). The dotted arrows represent uncertainty regarding whether 2,3-DHBA is derived from SA or from isochorismate under normal physiological conditions.
Biosynthetic Origin of 2,3-DHBA/2,3-DHB3X
Arabidopsis mutant sid2 is defective in the chloroplastic enzyme isochorismate synthetase, which converts chorismate to isochorismate, the main precursor of pathogen-induced SA (31, 44). We found that sid2 is also deficient in accumulation of 2,3-DHBA, indicating that, as for the majority of SA, 2,3-DHBA is derived from the shikimate/isochorimate pathway and not from phenylalanine. In contrast, the basal level of 2,5-DHBA is reduced but detectable in sid2-1 (Fig. 1C and Fig. 5A) and might either be derived from phenylalanine or generated by the SID2 homologue isochorismate synthase2 (52). The question of whether 2,3-DHBA is synthesized from isochorismate, as demonstrated in some bacteria (53) or is generated by hydroxylation from SA as found in in vitro assays with hydroxyl radicals and during degradation of hydroxybenzoic acids by various microorganisms remains to be answered (54, 55) (Fig. 7). SA was shown to be a precursor of 2,3-DHBA in experiments in which radioactive labeled SA was fed to leaves and converted to 2,3-DHBA and 2,5-DHBA in varying ratios depending on the plant species examined (56). We also found that [2H]SA infiltrated into avrRpm1-triggered tissue is converted into two distinct DHBA-pentosides (Fig. 4). One peak was identified as 2,3-DHB3X (Fig. 4). The other is unknown, although it is likely to be 2,5-DHBA-xyloside based on previous findings in pathogen-induced tomato (57). Thus, exogenously applied SA can be directly transformed into 2,3-DHBA without isochorismate as an intermediate. However, the very low amount of [2H]2,3-DHB3X formed suggests that SA conversion to 2,3-DHBA is inefficient, and it remains to be seen whether exogenous SA application reflects the metabolism of pathogen-induced endogenous SA.
If SA serves as a direct DHBA precursor, its hydroxylation in plant cells could be catalyzed by enzymes, as found in some microorganisms that use hydroxylation as a first step for metabolizing hydroxybenzoic acids (54). An alternative nonenzymatic route would be through reaction of SA with highly reactive OH* radicals. In in vitro assays, SA reacted with OH* to produce approximately equal amounts of 2,3-DHBA and 2,5-DHBA (55, 58). The different ratios of 2,3-DHBA and 2,5-DHBA measured in Arabidopsis mutants in our study (Fig. 3B) suggest that 2,3-DHBA is generated by enzymes that may change in activity depending on the genetic background and developmental stage. Although hydrogen peroxide can form OH* in the presence of free Fe2+ by the Fenton reaction, we found that the total 2,3-DHBA levels were not altered in atrbohD or atrbohF mutants (supplemental Fig. S7), which are defective in pathogen-inducible hydrogen peroxide (59). This lends further support to an enzymatic origin for 2,3-DHBA from SA or isochorismate.
Hydroxylation of SA at position 3 of the phenyl ring to 2,3-DHBA would allow subsequent xylose conjugation at this hydroxyl group by sugar-conjugating enzymes. A candidate enzyme for 2,3-DHBA xylosylation is encoded by Arabidopsis At4g01070, which has been characterized as glycosyltransferase UGT72B1 catalyzing glycosylation of 2,3-DHBA to 2,3-DHB3G in vitro (60). The same enzyme was reported to conjugate the phenolic pollutant 2,4,5-trichlorophenol with UDP-xylose in vivo, thus behaving as a xylosyltransferase (61). Our metabolic and phenotypic characterization of corresponding Arabidopsis T-DNA insertion mutants (UGT72B1 SALK_049597_and SAIL_611_E04 obtained from the Nottingham Arabidopsis Stock Centre), however, suggests that UGT72B1 is not involved in 2,3-DHBA conjugation to xylose unless genetic redundancy with other xylosylation enzymes masks its activity.5 Given that the majority of 2,3-DHBA accumulates as the xylose-conjugated form 2,3-DHB3X (Figs. 1, 3, and 7) and is therefore not likely to be an unstable intermediate in the formation of another metabolite, we propose that generation of 2,3-DHB3X serves as a reservoir of nonconjugated 2,3-DHBA, as has been shown for SAG, which can be remobilized to SA in tobacco (62).
It is possible that 2,3-DHBA, as a much less potent defense-inducing molecule (Fig. 6), can then be dehydroxylated to the more active SA, although, to our knowledge, dehydroxylation of an aromatic ring has not been described in plant secondary metabolism. Another scenario is that free 2,3-DHBA itself has a defense-modulating role that is distinct from that of SA and not associated with PR1 activation.
2,3-DHBA Accumulation Varies with Arabidopsis Genotype and Age
SA was reported to accumulate with increasing age of Arabidopsis leaves (45). The age-related accumulation of SA was greatly reduced in defense mutants eds1, fmo1, pad2, and sid2, whereas SA levels were constitutively elevated in nudt7 and vtc1 mutant lines (Fig. 5A). 2,3-DHBA accumulation broadly correlated with the levels of SA produced during plant development (Fig. 3). Reduced amounts of 2,3-DHBA in sid2 are thus likely caused by precursor deficiency in the shikimate/isochorismate pathway, at the level of isochorismate and/or SA (Fig. 7). However, SA and 2,3-DHBA accumulation is less well correlated in mutant responses to Pst DC3000 avrRpm1 because eds1 mutants displayed disproportionately low 2,3-DHBA accumulation compared with SA and 2,5-DHBA (Fig. 1A). Moreover, fmo1 mutant plants that are defective in an SA-independent branch of the EDS1 pathway (26) accumulated wild type levels of SA but undetectable 2,3-DHBA 24 h after Pst DC3000 avrRpm1 infiltration (26) (supplemental Fig. S8). Therefore, EDS1 and/or FMO1 may regulate enzymes responsible for 2,3-DHBA synthesis. In a separate experiment examining early time points (9 and 16 hpi) after infiltration with Pst DC3000 avrRpm1, we could not detect 2,3-DHBA but observed slightly reduced SA levels in eds1 compared with wild type (supplemental Fig. S9). If SA is the direct precursor of 2,3-DHBA, it is possible that it does not reach a threshold that would activate 2,3-DHBA synthesis in eds1 and fmo1. Alternatively, the defense regulators FMO1 and EDS1 could be more directly involved in 2,3-DHBA biosynthesis. However, a typical FMO would not hydroxylate SA because FMO enzymes tend to catalyze the oxidation of nucleophilic, heteroatom-containing (e.g. nitrogen or sulfur) substrates and not carbon atoms (63). Also, the eds1 mutant is able to accumulate 2,3-DHBA at late stages of the RPM1-triggered response (Fig. 1A). We therefore favor a regulatory role for EDS1 and FMO1, which may help to mobilize distinct benzoic acid derivatives depending on the status of the cells and tissues.
Is 2,3-DHBA a Deactivated Form of SA?
Free 2,3-DHBA applied to liquid-grown Arabidopsis seedlings or sprayed onto leaves of soil grown plants had much weaker PR1 inducing activity and phytotoxic effects than free SA (Fig. 6 and supplemental Fig. S6) in accordance with a previous report that 2,3-DHBA is a poor inducer of PR-b1 protein in Nicotiana tabacum (64). Therefore, 2,3-DHBA accumulation may serve to limit the availability of SA after pathogen challenge and, as the plant ages, as part of a deactivation mechanism. SA conversion to 2,3-DHBA could limit the adverse effects of SA such as phytotoxicity (65) or the fitness costs of constitutive activation of plant defenses (66). The relatively low increase and absolute amounts of total 2,3-DHBA after pathogen inoculation (∼2-fold; 1.4 μg/g of fresh weight at 24 hpi with Pst DC3000 avrRpm1 over mock treatment) compared with SA (7-fold, 7.2 μg/g of fresh weight at 24 hpi) (Fig. 1A) do not support a role for 2,3-DHBA as an activator of plant defense responses. Also, when SA was fed to non-pathogen-challenged 2-week-old Arabidopsis seedlings grown in liquid culture, we did not observe 2,3-DHB3X accumulation,6 in accordance with Dean and Delaney (13), who found that radioactively labeled SA fed to detached Arabidopsis leaves is transformed to SA- and 2,5-DHBA glucose conjugates. We conclude that 2,3-DHBA formation is unlikely to be a general detoxification step for exogenously applied SA in Arabidopsis. However, this does not exclude the transformation of endogenously produced SA to 2,3-DHBA as a physiologically important modification.
If synthesis of 2,3-DHBA is a means to limit the availability of bioactive SA, an important attribute of EDS1 and FMO1 regulation (Figs. 3 and 4) would be to dampen SA signaling as part of their defense-promoting roles. Evidence from analysis of nudt7 mutants alone or in combination with eds1 or sid2 reinforces a role for EDS1 in promoting SA accumulation as a mechanism to limit cell death (8). Hence, control of SA metabolism may be an important strategy to cope with environmental stress during development.
2,3-DHBA as a Biologically Active Metabolite
It is significant that the majority of 2,3-DHBA in pathogen-responding and ageing leaves is present as a xylose conjugate (Fig. 3). By contrast, free SA is rapidly conjugated with glucose, which is associated with deactivation and transport into the plant vacuole for storage (12). Thus, the plant may use a similar but apparently not identical sugar conjugation process to remove active free 2,3-DHBA from the cytoplasm. Although 2,3-DHBA is a weak inducer of PR1 gene expression (Fig. 6), we cannot exclude the possibility that it has the capacity to induce different sets of genes or cellular processes important for pathogen resistance or the progression to senescence. In this respect, age-related resistance to pathogens in Arabidopsis is dependent on EDS1 signaling and associated more with the mobilization of SA to intercellular spaces between cells than with conventional SA-mediated NPR1-dependent intracellular changes in defense gene activation (67). At high concentrations, 2,3-DHBA was found to possess antifungal activity by disrupting cellular glutathione homeostasis in in vitro assays (68, 69). At lower concentrations, 2,3-DHBA might protect the plant cell from oxidative stress because this is a function of 2,3-DHBA produced by bacterial pathogens infecting mammals. By acting as an iron siderophore, 2,3-DHBA is thought either to prevent the formation of the reactive hydroxyl radical generated in the presence of free iron and hydrogen peroxide (the Fenton reaction) that is harmful to the pathogen or to help acquire limited iron resources from the host cell and thus facilitate pathogen growth (70–72). The 2,3-DHBA we detect in planta upon increasing plant age might thus also serve as a protectant against oxidative stress associated with senescence (73).
Supplementary Material
Acknowledgments
We thank Allan D. Shapiro for Col/pPR1:GUS seeds, Jeff Dangl for AvrRpm1-HA seeds, and Jaqueline Bautor for technical assistance. We are grateful to Gerald Bannenberg for assistance with the figures.
This work was supported by the Max Planck Society, Bundesministerium für Bildung und Forschung GABI DILEMA, and Deutsche Forschungsgemeinschaft Schwerpunkt programm 1212 (Plant-Microbe) grants (to J. E. P. and M. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
M. Bartsch and P. Bednarek, unpublished observations.
P. Bednarek, unpublished observations.
- SA
- salicylic acid
- EDS1
- Enhanced Disease Susceptibility 1
- DHBA
- dihydroxybenzoic acid
- DHB3X
- DHBA 3-O-β-d-xyloside
- OH*
- hydroxyl radical
- SAG
- salicylic acid glycoside
- NB-LRR
- nucleotide-binding leucine-rich repeat
- DEX
- dexamethasone
- HA
- hemagglutinin
- GUS
- β-glucuronidase
- HPLC
- high pressure liquid chromatography
- HSQC
- heteronuclear single quantum coherence
- MS
- mass spectrometry
- LC
- liquid chromatography
- hpi
- hours post-inoculation.
REFERENCES
- 1.Pieterse C., Leon-Reyes A., Van der Ent S., Van Wees S. (2009) Nat. Chem. Biol. 5, 308–316 [DOI] [PubMed] [Google Scholar]
- 2.Glazebrook J. (2005) Annu. Rev. Phytopathol. 43, 205–227 [DOI] [PubMed] [Google Scholar]
- 3.Shah J. (2009) Curr. Opin. Plant Biol. 12, 459–464 [DOI] [PubMed] [Google Scholar]
- 4.Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P. (2009) Science 323, 101–106 [DOI] [PubMed] [Google Scholar]
- 5.Clay N., Adio A., Denoux C., Jander G., Ausubel F. (2009) Science STKE 323, 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres M. A., Jones J. D., Dangl J. L. (2005) Nat. Genet. 37, 1130–1134 [DOI] [PubMed] [Google Scholar]
- 7.Mateo A., Funck D., Mühlenbock P., Kular B., Mullineaux P. M., Karpinski S. (2006) J. Exp. Bot. 57, 1795–1807 [DOI] [PubMed] [Google Scholar]
- 8.Straus M. R., Rietz S., van Themaat E. V., Bartsch M., Parker J. E. (2010) Plant J. 62, 628–640 [DOI] [PubMed] [Google Scholar]
- 9.Fobert P. R., Després C. (2005) Curr. Opin. Plant Biol. 8, 378–382 [DOI] [PubMed] [Google Scholar]
- 10.Mou Z., Fan W., Dong X. (2003) Cell 113, 935–944 [DOI] [PubMed] [Google Scholar]
- 11.Wang D., Weaver N. D., Kesarwani M., Dong X. (2005) Science 308, 1036–1040 [DOI] [PubMed] [Google Scholar]
- 12.Dean J. V., Mohammed L. A., Fitzpatrick T. (2005) Planta. 221, 287–296 [DOI] [PubMed] [Google Scholar]
- 13.Dean J. V., Delaney S. P. (2008) Physiol. Plant. 132, 417–425 [DOI] [PubMed] [Google Scholar]
- 14.Wildermuth M. C. (2006) Curr. Opin. Plant Biol. 9, 288–296 [DOI] [PubMed] [Google Scholar]
- 15.Park S. W., Kaimoyo E., Kumar D., Mosher S., Klessig D. F. (2007) Science 318, 113–116 [DOI] [PubMed] [Google Scholar]
- 16.Attaran E., Zeier T. E., Griebel T., Zeier J. (2009) Plant Cell 21, 954–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Biezen E. A., Freddie C. T., Kahn K., Parker J. E., Jones J. D. (2002) Plant J. 29, 439–451 [DOI] [PubMed] [Google Scholar]
- 18.Jagadeeswaran G., Raina S., Acharya B. R., Maqbool S. B., Mosher S. L., Appel H. M., Schultz J. C., Klessig D. F., Raina R. (2007) Plant J. 51, 234–246 [DOI] [PubMed] [Google Scholar]
- 19.Nobuta K., Okrent R. A., Stoutemyer M., Rodibaugh N., Kempema L., Wildermuth M. C., Innes R. W. (2007) Plant Physiol. 144, 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okrent R. A., Brooks M. D., Wildermuth M. C. (2009) J. Biol. Chem. 284, 9742–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feys B. J., Wiermer M., Bhat R. A., Moisan L. J., Medina-Escobar N., Neu C., Cabral A., Parker J. E. (2005) Plant Cell 17, 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiermer M., Feys B. J., Parker J. E. (2005) Curr. Opin. Plant Biol. 8, 383–389 [DOI] [PubMed] [Google Scholar]
- 23.Zhou N., Tootle T. L., Tsui F., Klessig D. F., Glazebrook J. (1998) Plant Cell 10, 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feys B. J., Moisan L. J., Newman M. A., Parker J. E. (2001) EMBO J. 20, 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truman W., Bennett M. H., Kubigsteltig I., Turnbull C., Grant M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J. L., Bautor J., Parker J. E. (2006) Plant Cell 18, 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishina T. E., Zeier J. (2006) Plant Physiol. 141, 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch M., Vorwerk S., Masur C., Sharifi-Sirchi G., Olivieri N., Schlaich N. L. (2006) Plant J. 47, 629–639 [DOI] [PubMed] [Google Scholar]
- 29.Ge X., Li G. J., Wang S. B., Zhu H., Zhu T., Wang X., Xia Y. (2007) Plant Physiol. 145, 204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishikawa K., Ogawa T., Hirosue E., Nakayama Y., Harada K., Fukusaki E., Yoshimura K., Shigeoka S. (2009) Plant Physiol. 151, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M. (2001) Nature 414, 562–565 [DOI] [PubMed] [Google Scholar]
- 32.Parisy V., Poinssot B., Owsianowski L., Buchala A., Glazebrook J., Mauch F. (2007) Plant J. 49, 159–172 [DOI] [PubMed] [Google Scholar]
- 33.Conklin P. L., Norris S. R., Wheeler G. L., Williams E. H., Smirnoff N., Last R. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro A. D., Zhang C. (2001) Plant Physiol. 127, 1089–1101 [PMC free article] [PubMed] [Google Scholar]
- 35.Mackey D., Holt B. F., 3rd, Wiig A., Dangl J. L. (2002) Cell 108, 743–754 [DOI] [PubMed] [Google Scholar]
- 36.Kienow L., Schneider K., Bartsch M., Stuible H. P., Weng H., Miersch O., Wasternack C., Kombrink E. (2008) J. Exp. Bot. 59, 403–419 [DOI] [PubMed] [Google Scholar]
- 37.Bednarek P., Schneider B., Svatos A., Oldham N. J., Hahlbrock K. (2005) Plant Physiol. 138, 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H., Raskin I. (1998) Phytopathology 88, 692–697 [DOI] [PubMed] [Google Scholar]
- 39.Jen C., Manfield I., Michalopoulos I., Pinney J., Willats W., Gilmartin P., Westhead D. (2006) Plant J. 46, 336–348 [DOI] [PubMed] [Google Scholar]
- 40.Moreno P. R., Heijden R., Verpoorte R. (1994) Plant Cell Rep. 14, 188–191 [DOI] [PubMed] [Google Scholar]
- 41.Belles J. M., Garro R., Fayos J., Navarro P., Primo J., Conejero V. (1999) Mol. Plant-Microbe Interact. 12, 227–235 [Google Scholar]
- 42.Muljono R. A., Scheffer J. J., Verpoorte R. (2002) Plant Physiol. Biochem. 40, 231–234 [Google Scholar]
- 43.Mustafa N. R., Kim H. K., Choi Y. H., Erkelens C., Lefeber A. W., Spijksma G., van der Heijden R., Verpoorte R. (2009) Phytochemistry 70, 532–539 [DOI] [PubMed] [Google Scholar]
- 44.Strawn M. A., Marr S. K., Inoue K., Inada N., Zubieta C., Wildermuth M. C. (2007) J. Biol. Chem. 282, 5919–5933 [DOI] [PubMed] [Google Scholar]
- 45.Morris K., MacKerness S. A., Page T., John C. F., Murphy A. M., Carr J. P., Buchanan-Wollaston V. (2000) Plant J. 23, 677–685 [DOI] [PubMed] [Google Scholar]
- 46.Wagstaff C., Yang T. J., Stead A. D., Buchanan-Wollaston V., Roberts J. A. (2009) Plant J. 57, 690–705 [DOI] [PubMed] [Google Scholar]
- 47.Barth C., Moeder W., Klessig D. F., Conklin P. L. (2004) Plant Physiol. 134, 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He Y., Gan S. (2002) Plant Cell 14, 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver L. M., Gan S., Quirino B., Amasino R. M. (1998) Plant Mol. Biol. 37, 455–469 [DOI] [PubMed] [Google Scholar]
- 50.Uknes S., Mauch-Mani B., Moyer M., Potter S., Williams S., Dincher S., Chandler D., Slusarenko A., Ward E., Ryals J. (1992) Plant Cell 4, 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y., Qi M., Mei C. (2004) Plant J. 40, 909–919 [DOI] [PubMed] [Google Scholar]
- 52.Garcion C., Lohmann A., Lamodière E., Catinot J., Buchala A., Doermann P., Métraux J. P. (2008) Plant Physiol. 147, 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young I. G., Batterham T., Gibson F. (1968) Biochim. Biophys. Acta 165, 567–568 [DOI] [PubMed] [Google Scholar]
- 54.Karegoudar T. B., Kim C. K. (2000) J. Microbiol. 38, 53–61 [Google Scholar]
- 55.Floyd R. A., Watson J. J., Wong P. K. (1984) J. Biochem. Biophys. Methods 10, 221–235 [DOI] [PubMed] [Google Scholar]
- 56.Ibrahim R. K., Towers G. H. (1959) Nature 184, 180313811171 [Google Scholar]
- 57.Fayos J., Bellés J. M., López-Gresa M. P., Primo J., Conejero V. (2006) Phytochemistry 67, 142–148 [DOI] [PubMed] [Google Scholar]
- 58.Grootveld M., Halliwell B. (1986) Biochem. J. 237, 499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres M. A., Dangl J. L., Jones J. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim E. K., Doucet C. J., Li Y., Elias L., Worrall D., Spencer S. P., Ross J., Bowles D. J. (2002) J. Biol. Chem. 277, 586–592 [DOI] [PubMed] [Google Scholar]
- 61.Brazier-Hicks M., Edwards R. (2005) Plant J. 42, 556–566 [DOI] [PubMed] [Google Scholar]
- 62.Hennig J., Malamy J., Grynkiewicz G., Indulski J., Klessig D. F. (1993) Plant J. 4, 593–600 [DOI] [PubMed] [Google Scholar]
- 63.Schlaich N. L. (2007) Trends Plant Sci. 12, 412–418 [DOI] [PubMed] [Google Scholar]
- 64.Abad P., Marais A., Cardin L., Poupet A., Ponchet M. (1988) Antiviral Res. 9, 315–327 [DOI] [PubMed] [Google Scholar]
- 65.van Leeuwen H., Kliebenstein D. J., West M. A., Kim K., van Poecke R., Katagiri F., Michelmore R. W., Doerge R. W., St Clair D. A. (2007) Plant Cell 19, 2099–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Hulten M., Pelser M., van Loon L. C., Pieterse C. M., Ton J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carviel J. L., Al-Daoud F., Neumann M., Mohammad A., Provart N. J., Moeder W., Yoshioka K., Cameron R. K. (2009) Mol. Plant Pathol. 10, 621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amborabé B. E., Fleurat-Lessard P., Chollet J. F., Roblin G. (2002) Plant Physiol. Biochem. 40, 1051–1060 [Google Scholar]
- 69.Kim J. H., Mahoney N., Chan K. L., Molyneux R. J., May G. S., Campbell B. C. (2008) FEMS Microbiol. Lett. 281, 64–72 [DOI] [PubMed] [Google Scholar]
- 70.Bellaire B. H., Elzer P. H., Baldwin C. L., Roop R. M., 2nd (2003) Infection Immunity 71, 2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leonard B. A., Lopez-Goni I., Baldwin C. L. (1997) Vet. Res. 28, 87–92 [PubMed] [Google Scholar]
- 72.Parent M. A., Bellaire B. H., Murphy E. A., Roop R. M., 2nd, Elzer P. H., Baldwin C. L. (2002) Microb. Pathog. 32, 239–248 [DOI] [PubMed] [Google Scholar]
- 73.Zimmermann P., Zentgraf U. (2005) Cell. Mol. Biol. Lett. 10, 515–534 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.