Abstract
Ca2+ signals through store-operated Ca2+ (SOC) channels, activated by the depletion of Ca2+ from the endoplasmic reticulum, regulate various physiological events. Orai1 is the pore-forming subunit of the Ca2+ release-activated Ca2+ (CRAC) channel, the best characterized SOC channel. Orai1 is activated by stromal interaction molecule (STIM) 1, a Ca2+ sensor located in the endoplasmic reticulum. Orai1 and STIM1 are crucial for SOC channel activation, but the molecular mechanisms regulating Orai1 function are not fully understood. In this study, we demonstrate that protein kinase C (PKC) suppresses store-operated Ca2+ entry (SOCE) by phosphorylation of Orai1. PKC inhibitors and knockdown of PKCβ both resulted in increased Ca2+ influx. Orai1 is strongly phosphorylated by PKC in vitro and in vivo at N-terminal Ser-27 and Ser-30 residues. Consistent with these results, substitution of endogenous Orai1 with an Orai1 S27A/S30A mutant resulted in increased SOCE and CRAC channel currents. We propose that PKC suppresses SOCE and CRAC channel function by phosphorylation of Orai1 at N-terminal serine residues Ser-27 and Ser-30.
Keywords: Calcium/Channels, Channels/Calcium, Enzymes/Kinase, Lipid/Diacylglycerol, Membrane/Channels, Signal Transduction/Calcium, Signal Transduction/Phorbol Esters, Signal Transduction/Protein Kinases/Serine/Threonine
Introduction
Ca2+ signals regulate various physiological events such as cell growth, motility, secretion, and gene expression (1). Activation of receptors in the plasma membrane induces Ca2+ influx. Store-operated Ca2+ (SOC)3 channels are activated by depletion of Ca2+ from the endoplasmic reticulum (ER) allowing Ca2+ influx from the extracellular space. Ca2+ entry through SOC channels was proposed in the 1980s (2) and has been characterized extensively in patch clamp experiments. The best defined SOC channel is the Ca2+ release-activated Ca2+ (CRAC) channel in immune cells (3), which is characterized by voltage-independent activation and an inwardly rectifying, Ca2+-selective current. An increase in the intracellular Ca2+ concentration following SOCE activates several types of Ca2+-dependent enzymes, such as calcineurin and calmodulin kinase, which collectively regulate gene expression and influence cell fate decisions (4).
Some protein kinase C (PKC) subtypes are activated by Ca2+ in the presence of diacylglycerol (DAG). The PKC family consists of at least 10 subtypes and is divided into three subfamilies based on primary structure: conventional PKC (cPKC; α, βI, βII, and γ), novel PKC (nPKC; δ, ϵ, η, and θ), and atypical PKC (aPKC; ζ and ι/κ) (5, 6). cPKC is activated by Ca2+ and DAG, nPKC is activated by DAG but not Ca2+, whereas aPKC is independent of both DAG and Ca2+. Activator selectivity of PKC subfamilies is mediated by a domain in their regulatory region. Ca2+-dependent activation of cPKC depends on the C2 domain, which binds to Ca2+ and anionic lipids, such as phosphatidylserine (7). DAG and its structural analog, TPA, activate PKC by binding to the C1 domain, cysteine-rich zinc finger-like subdomain (8). DAG and Ca2+ required for cPKC activation are generated by the engagement of G protein-coupled receptors or receptors associated with protein-tyrosine kinases, which activates phospholipase C and induces subsequent production of inositol trisphosphate (IP3) and DAG from phosphatidylinositol 4,5-bisphosphate. IP3 binds to IP3 receptors in the ER to induce Ca2+ store depletion leading to SOC/CRAC channel activation. Increases in the intracellular Ca2+ concentration via SOCE and production of DAG induce activation and translocation of PKC to the plasma membrane (5, 6). Although PKC is activated by Ca2+, conversely PKC has been shown to regulate SOC channel activation in Jurkat T cells (9), RBL-2H3 cells (10), HL60 cells (11), and Xenopus oocytes (12). The molecular mechanism of PKC-mediated regulation of SOC channels, however, is not understood.
Although SOCE and CRAC channel currents were first reported approximately two decades ago, the molecular identity of the channel-forming proteins was revealed only recently. Orai1 (or CRACM1) was identified in 2006 (13–15) and was suggested to be the pore-forming subunit of the CRAC channel (16–18). Orai1 belongs to a novel class of Ca2+ channels with a tetraspanning membrane topology and intracellular N and C termini required for CRAC channel activation. In addition to Orai1, two other subtypes, Orai2 and Orai3, were identified that can mediate SOCE and Ca2+ currents when ectopically expressed (19). Orai1 is activated by stromal interaction molecule (STIM)-1, which is located in the ER membrane and senses the Ca2+ concentration in the ER (20, 21). Two STIM genes have been reported in the mammalian genome (STIM1 and STIM2), which have distinct but overlapping functional roles (20, 22). In addition to Orai1, several types of Ca2+-permeable, non-selective transient receptor potential cation (TRPC) channels have also been proposed to be regulated by store depletion and STIM1 (23). Whereas TRPC channels were reported to be regulated by PKC (24–26), regulation of Orai1 by PKC has not been analyzed. In this study, we focused on SOC channels formed by Orai proteins, and found that PKC suppresses SOC channel activity by phosphorylation of Orai1 at serine residues Ser-27 and Ser-30.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293 cells were cultured in minimum essential medium (Amersham Biosciences) and Orai1 deficient fibroblasts (13) in RPMI 1640 (Amersham Biosciences) at 37 °C/5% CO2. The medium contained 25 mm glucose, buffered with 44 mm NaHCO3, and was supplemented with 10% fetal bovine serum, non-essential amino acids, penicillin (100 units/ml), and streptomycin (100 μg/ml).
Plasmid Construction and Protein Expression
Orai1, Orai2, and Orai3 were subcloned from HEK293 cells. PKC phosphorylation sites were predicted using NetPhos2.0 Server (Technical University of Denmark) and substitutions of Ser to Ala at predicted phosphorylation sites were introduced using PRIMEstar (Takara, Japan). To purify intracellular regions of Orai1, N-terminal (1–71 aa), intracellular loop (141–173 aa), and C-terminal (257–301 aa) regions were amplified by PCR and cloned into the pGEX-6P vector at EcoR1/BamH1 sites. EcoR1 sites were replaced with BamH1 sites to delete the serine residue encoded by the EcoR1 sequence. BL21 pLys Escherichia coli were transformed with expression plasmids for the production of recombinant protein. Expression of Orai1 intracellular domains was induced by 0.1 mm isopropyl 1-thio-β-d-galactoside at 37 °C for 4 h. E. coli were harvested and lysed in a buffer containing 20 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mm dithiothreitol, 250 mm sucrose, 20 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride. GST fusion proteins were captured on glutathione-Sepharose 4B resin (Amersham Biosciences), and the GST region was excluded by incubation with PreScission protease (Amersham Biosciences) overnight at 4 °C. A GST-tagged N-terminal (1–88 aa) region was attached to FLAG peptide sequence at the C-terminal to avoid degradation. The GST fusion protein was lysed and harvested in a buffer containing 20 mm Tris-HCl (pH 6.5), 1 mm EDTA, 1 mm dithiothreitol, 250 mm sucrose, 20 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride, and GST tag was eliminated as described above. For purification of recombinant PKCβI, Sf9 cells were infected with baculovirus encoding GST-tagged PKCβI and were cultured in EX-CELLTM 420 (JRH Biosciences) containing 10% fetal bovine serum and 0.5% antibiotic/antimycotic in a temperature-controlled orbital shaker operating at 150 rpm at 27.5 °C. PKCβI-expressing baculovirus was a kind gift of Dr. Mukai (Kobe University). Cells were lysed in a buffer containing 20 mm Tris-HCl (pH 8.0), 1 mm EDTA, 62 μm MgCl2, 1% Triton, 1 mm dithiothreitol, 250 mm sucrose, 20 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride, and GST-tagged PKCβI was purified using glutathione-Sepharose 4B resin. Cloning and purification of the Orai1 activating CCb9 domain in STIM1 (339–446 aa) was performed as previously described (27). For RNA interference experiments, double-stranded oligonucleotides were subcloned into the short hairpin RNA (shRNA) expression vector pSuper.gfp/neo (OligoEngine) containing the H1 small nuclear RNA promoter at its BglII/HindIII sites. The GFP coding region in pSuper.gfp/neo was replaced with mRFP. Target sequences for knockdown were as follows: Orai1 target, AAGAGAAATTTCTGCACTT; PKCα target, AAAGGCTGAGGTTGCTGAT (28); PKCβ target, GGAAGCTGTGGCCATCTGC (29); and luciferase (control) target, AACATAAAGAAAGGCCCGG. Orai1 rescue experiments were performed using pSuper.mrfp/neo expressing both Orai1 shRNA targeting a sequence in the 3′-untranslated region and the Orai1 coding sequence. Non-tagged wild-type or mutant Orai1 was cloned between AgeI/AflII sites replacing mRFP. The target sequence for Orai1 knockdown is located in the 3′-untranslated region of Orai1 mRNA; the Orai1 expression sequence does not contain a 3′-untranslated region to avoid knockdown of ectopically expressed Orai1. Retroviral transduction of fibroblasts with IRES-GFP containing bicistronic vectors containing Orai1 was done as previously described (13).
Quantitative Real-time PCR
Knockdown efficiency and specificity were quantified by quantitative real-time PCR analysis. RNA was obtained by purification kit (Promega, Madison, WI), and random nucleotides were primed for synthesis of first-strand cDNA (Invitrogen). PCR primers (5′/3′), were as follows: Orai1, TATGCCTAGGCCCATGTGGTCT/AATCCTCTTCCCTCCATGCTCT; Orai2, TTGCGTTCAGAGAAGGATTGC/GCCTTCGTGTCATCACTGTCA; Orai3, CCTTGGTGGCACACAAGACAGA/GGTGGCTAACACCAGTCTCACA; and ribosomal protein L13a (RPL14A), CCTGGAGGAGAAGAGGAAAGAGA/TTGAGGACCTCTGTGTATTTGTCAA. The amplified DNA was normalized to RPL14A expression.
Measurement of Intracellular Ca2+
HEK293 cells were plated onto a 96-well CC3 black plate (Nunc) 1 day before measurement at a rate of 1.8 × 103 cells/well. Cells were loaded with 2.5 μm Fura2/AM and 1% Pluronic 100 (both from Dojin, Japan) for 20 min in Ringer's solution (155 mm NaCl, 4.5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm glucose, and 5 mm HEPES (pH 7.4)). Subsequently, cells were washed and pretreated with 1 μm thapsigargin (TG) in Ca2+ free Ringer's solution. At the indicated time points, Ringer's solution containing 2 mm CaCl2 was added. Fura2 intensity was monitored using a Mithras LB940 microplate reader (Berthold Technologies, Germany). Emission was measured at 535 ± 20 nm following excitation at 340 ± 10 nm and 380 ± 10 nm. Before calculation of the Fura2 ratio, background Fura2 intensity was subtracted after quenching by addition of 10 mm MnCl2.
In Vitro PKC Phosphorylation Assay
Purified Orai proteins were incubated with 100 ng of PKCβI and the following buffers: activation buffer, 20 mm Tris (pH 7.4), 0.5 m CaCl2, 10 μm ATP, 0.5 mCi of [γ-32P]ATP, 25 μg/ml phosphatidylserine (PS), and 50 ng/ml (4 μm) 4α-phorbol 12-myristate 13-acetate (TPA), or 1.6 ng/ml (4 μm) 1,2-didecanoylglycerol (10:1, DG); inactivation buffer, 20 mm Tris (pH 7.4), 0.5 m EGTA, 10 μm ATP, 0.5 mCi of [γ-32P]ATP in a 25-μl final volume. The samples were incubated at 30 °C for 15 min, and the reactions were stopped by the addition of SDS sample buffer. The phosphorylated proteins were separated by SDS-PAGE and were visualized and quantitated by measurement of the intensity of photostimulated luminescence with a Bio-imaging Analyzer (Fuji).
In Vivo PKC Phosphorylation Assay
HEK293 cells were transfected with pFLAG-CMV10 vector containing wild-type Orai1 or phosphorylation site mutants of Orai1 using FuGENE6, according to the manufacturer's instructions. To avoid the suppressive effect on SOCE caused by overexpression of wild-type Orai1 (30, 31), the transfected cells were maintained for 7 days before experiments, because we found that longer cultivation of cells after transfection decreased the suppressive effect of Orai1 on SOCE (data not shown). Cells were plated on 100-mm dishes and labeled with Ringer's solution containing [32P]monophosphate (50 Ci/ml) and 10% fetal bovine serum for 3 h at 37 °C. Following TG stimulation for 120 s in Ringer's solution at 37 °C, cells were washed with ice-cold buffer containing 150 mm NaCl, 10 mm EGTA, 2 mm EDTA, and 10 mm HEPES (pH 7.4). Cells were collected and resuspended in a homogenization buffer containing 150 mm NaCl, 10 mm EGTA, 2 mm EDTA, 10 mm HEPES (pH 7.4), 1 mm phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, 50 mm NaF, 1 mm vanadate, and 25 mm β-glycerolphosphate. Samples were centrifuged for 15 min at 20,000 × g to collect the membrane fraction as pellets, and pellets were resuspended in homogenization buffer containing 1% Triton X-100. Following centrifugation at 20,000 × g for 15 min, the supernatants were collected and incubated with anti-FLAG-agarose resin for 3 h at 4 °C. Precipitated proteins were boiled in SDS sample buffer and separated by SDS-PAGE. The phosphorylated proteins were visualized and quantitated by measuring the intensity of photostimulated luminescence with a Bio-imaging Analyzer (Fuji). For calculation of relative phosphorylation level, densitometries of autoradiography were normalized by maximum value in each experiment, and averages relative to phosphorylation in three independent experiments are shown in the bar graph.
Pulldown Assay
Antibodies against Orai1 were described previously (32). Pulldown of Orai1 with the Orai1 activation domain of STIM1 (CCb9) was performed as previously described (27). Briefly, HEK293T cells were transfected with FLAG-tagged Orai1. Two days after transfection, membrane fractions of HEK293T cells were solubilized in buffer containing 150 mm NaCl, 25 mm Tris, pH 7.2, 10 mm EGTA, 2 mm EDTA, 0.5% Tween 20. The purified GST-tagged CCb9 domain or control GST protein were incubated with membrane fractions for 3 h together with glutathione-Sepharose 4B resin. Bound proteins were detected by Western blotting analysis using anti-FLAG antibody.
Patch Clamp Measurements of CRAC Channel Currents
Patch clamp experiments were performed as described (27). Briefly, HEK293 cells were transfected with Cherry-STIM1 (a kind gift of R. S. Lewis, Stanford University) and Orai1-IRES-GFP plasmids. CRAC currents were measured in whole cell configuration at 21–25 °C. Patch pipettes were pulled from Kwik-Fil glass capillaries (World Precision Instruments) on a DMZ-Universal Puller (Dagan, Minneapolis, MN) and had resistances of 3–4 MΩ. Data were acquired with Clampex 8.2 software (Axon Instruments) controlling an Axopatch 200B amplifier (Axon Instruments). Voltage ramps from −130 to +100 mV were delivered at a rate of 0.5 Hz from a holding potential of 0 mV. The external bath solution contained (in mm): 120 NaCl, 10 CsCl, 2.8 KCl, 2 MgCl2, 10 CaCl2, 10 TEA-Cl, 10 HEPES, and 10 glucose. The standard internal solution contained (in mm): 120 cesium glutamate, 3 MgCl2, 8 NaCl, 2 ATP, 10 HEPES, and 10 EGTA. To induce store depletion, cells expressing Orai1 and STIM1 were pretreated with 1 μm thapsigargin before break-in; alternatively, 20 μm IP3 was included in the internal pipette solution. Voltages were corrected for liquid-junction potential; currents were filtered at 1 kHz and digitized at 200-μs intervals. Data were analyzed using Clampfit 9.2 (Axon Instruments), and IgorPro (WaveMetrics, Lake Oswego, OR). Data points for inward current amplitudes were obtained at −80 mV. Currents were normalized to cell size in picofarads. For current-voltage relationships (IV), leak was corrected by subtracting currents at 200 s after break-in (Fig. 7C) or currents prior to development of ICRAC (Fig. 7D) from peak currents. Statistical errors of averaged data are given as means ± S.E.
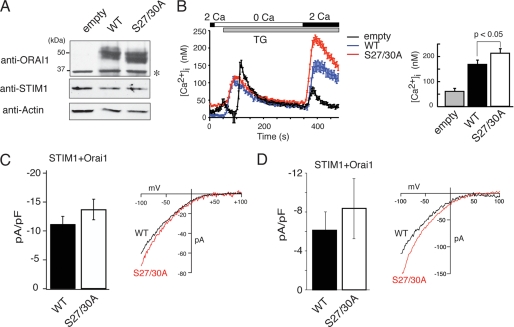
FIGURE 7.
Phosphorylation of Orai1 on Ser-27 and Ser-30 inhibits SOCE and CRAC channel current (ICRAC). A and B, wild-type Orai1 or mutant Orai1-S27A/S30A were expressed in SOCE-deficient fibroblasts from a patient with Orai1-R91W mutation by lentiviral transduction. A, equal expression of transduced wild-type and mutant Orai1 was verified by Western blotting using an anti-Orai1 antibody. Note that endogenous Orai1 levels are too low for detection; *, nonspecific band. B, Ca2+ measurements were conducted in transduced fibroblasts loaded with Fura-2. Cells were stimulated with thapsigargin (TG) in the absence of Ca2+ followed by addition of 2 mm Ca2+ to induce SOCE. Traces represent averages of Ca2+ responses in >30 GFP+ cells analyzed per experiment. GFP expression was from an IRES-GFP cassette in the lentiviral expression vector. Bar graphs show the averages of peak [Ca2+]i obtained from 6–8 independent experiments (empty vector, n = 6; WT, n = 8; S27A/S30A, n = 8). Error bars represent ±S.E. C and D, for measurements of ICRAC, HEK293 cells were transfected with Cherry-STIM1 and either wild-type Orai1 or Orai1-S27A/S30A vectors expressing GFP from an IRES site. Cherry+/GFP+ cells were selected for whole cell recordings. C, cells were stimulated with thapsigargin (1 μm) in 10 mm Ca2+. Shown are averages of current amplitudes at −80 mV measured 100 s after break-in. Orai1-WT (n = 10), Orai1-S27A/S30A (n = 7); error bars represent ±S.E. D, cells were stimulated with 20 μm IP3 in the patch pipette to deplete ER Ca2+ stores. Shown are current amplitudes at −80 mV measured 50 s after break-in. Orai1-WT (n = 6), Orai1-S27A/S30A (n = 6); error bars represent ±S.E. Representative current-voltage (I-V) relationships of Ca2+ currents recorded immediately (C) and 50 s (D) after break-in, respectively, are shown on the right.
RESULTS
PKC Activation Suppresses SOCE
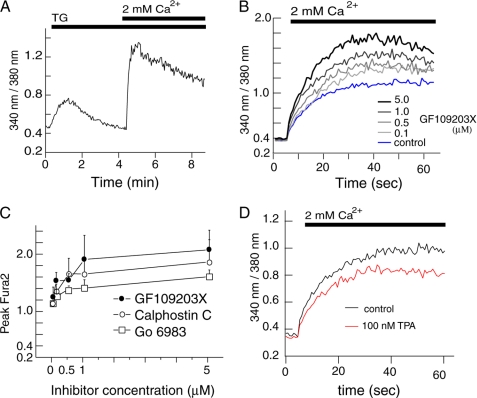
To study the functional role of PKC in the regulation of SOCE, we monitored changes in intracellular Ca2+ concentrations in HEK293 cells using Fura2. HEK293 cells were used as a model as they express all three subtypes of Orai proteins, Orai1, -2, and -3 (33). We confirmed the expression of Orai mRNA in HEK293 cells by reverse transcription-PCR (data not shown). To measure intracellular Ca2+ entry through SOC channels, HEK293 cells were pretreated with TG in the absence of extracellular Ca2+ followed by re-addition of 2 mm Ca2+ at the indicated time points (Fig. 1A). The first rise in intracellular Ca2+ shown in Fig. 1A represents Ca2+ release from the ER store, and the second peak represents Ca2+ influx through SOC channels. Focusing on Ca2+ influx, TG-treated HEK293 cells were incubated with the a PKC inhibitor GF109203X (GFX) at various concentrations. GFX increased Ca2+ entry in a dose-dependent manner (Fig. 1B). Similar increases of Ca2+ influx were observed after treatment with several other PKC inhibitors such as calphostin C and Go6983 (Fig. 1C and supplemental Fig. S1). Conversely, treatment with 100 nm TPA, a potent and specific PKC activator, suppressed TG-induced Ca2+ entry (Fig. 1D). Peak values of the Fura2 ratio were 1.1 ± 0.03 and 0.94 ± 0.01 in the absence and presence of TPA, respectively (p < 0.05). These results suggest that SOCE is suppressed by PKC activation in HEK293 cells.
FIGURE 1.
PKC inhibition enhances SOCE in HEK293 cells. Intracellular Ca2+ concentrations were measured in HEK293 cells loaded with the Ca2+ indicator Fura2. A, to induce SOCE, ER Ca2+ stores were depleted with 1 μm thapsigargin (TG) in the absence of extracellular Ca2+. Subsequently, Ca2+ was added to a final concentration of 2 mm. Ca2+ levels are shown as the emission ratio of Fura2 following excitation at 340 and 380 nm. B, HEK293 cells were pretreated with different concentrations of GF109203X after depletion of Ca2+ stores with TG. Fura2 emission ratios were monitored for 60 s after addition of 2 mm Ca2+. C, peak values of Fura2 emission ratios obtained after preincubation of TG-treated cells with different concentrations of PKC inhibitors GFX (B), calphostin C (supplemental Fig. S1A) and Go 6983 (supplemental Fig. S1B) are shown as mean ± S.D. (n = 3). D, Fura2 emission ratios of TG-stimulated cells pretreated with 100 nm TPA followed by addition of 2 mm Ca2+.
PKCβI Translocates to the Plasma Membrane in Response to Ca2+ Influx and Regulates SOCE
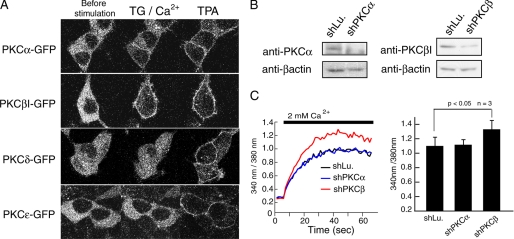
PKC subtypes expressed in HEK293 cells were studied by Western blot analysis revealing predominant expression of PKCα, -βI, -δ, and -ϵ among the cPKC and nPKC subfamilies (data not shown). Activation of these PKC subtypes was examined by monitoring the translocation of GFP-tagged PKC in HEK293 cells. PKCα and PKCβI translocated to the plasma membrane of TG-treated cells following the addition of Ca2+, whereas PKCδ and PKCϵ did not. By contrast, TPA induced membrane translocation of all PKC subtypes (Fig. 2A). Interestingly, PKCβI stably translocated to the plasma membrane after TG stimulation, whereas PKCα translocated only partially. Interestingly, almost all PKCβI translocated to the plasma membrane TG stimulation, whereas PKCα translocated only partially. These results suggested that PKCβI, and to a lesser degree PKCα, could be activated by SOCE in HEK293 cells. To determine which PKC subtype participates in the regulation of SOC channels, we measured SOCE in HEK293 in which PKC expression was attenuated by shRNA-mediated knockdown of PKCα (28) and PKCβ (29). Knockdown of PKCα and PKCβI was verified by Western blot analysis showing a decrease of PKCβI and PKCα protein expression (Fig. 2B). The knockdown of endogenous PKCβI, but not that of PKCα, resulted in increased TG-induced Ca2+ influx (Fig. 2C) consistent with results obtained with PKC inhibitors (Fig. 1). Together with the evident translocation of PKCβI induced by Ca2+ entry (Fig. 2A), we conclude that PKCβI participates in SOC channel regulation in HEK293 cells.
FIGURE 2.
PKCβI but not PKCα negatively regulates SOCE. A, HEK293 cells were transfected with GFP-tagged PKC isoforms α, βI, δ, and ϵ. For confocal microscopy, cells were imaged before stimulation (left), after addition of TG in the presence of 2 mm Ca2+ (middle) and following stimulation with 500 μm TPA. B, for knockdown of PKCα and PKCβ, HEK293 cells were transfected with shPKCα and shPKCβ, respectively, and expression levels of PKCα and PKCβI were detected by Western blotting. Knockdown of luciferase (shLu) was used as negative control. β-Actin expression levels were used for normalization of protein expression. C, Ca2+ entry after pretreatment with TG was measured in HEK293 cells transfected with control and knockdown vectors as described in Fig. 1. Averages of peak Fura2 emission ratios after addition of 2 mm Ca2+ are shown (n = 3). Error bars, ±S.D.
Orai1 Is Phosphorylated by PKCβI in Vitro and in Vivo
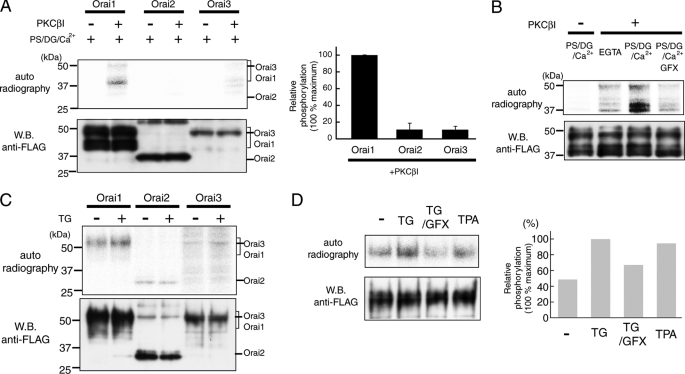
The Orai family consists of three members, Orai1, -2, and -3, which are capable of forming SOC channels. When ectopically expressed, the three Orai isoforms have different current properties (34), and the contribution of each isoform to SOCE may differ from cell type to cell type. Knockdown of Orai1 in HEK293 cells suppressed SOCE, whereas knockdown of Orai2 and Orai3 had no effect (33). To determine which SOCE-mediating Orai subtype is phosphorylated by PKCβI, we performed in vitro and in vivo phosphorylation experiments. For in vitro phosphorylation, FLAG-tagged Orai proteins were purified with anti-FLAG agarose resin from COS7 cells transfected with Orai expression vectors. Purified Orai proteins were incubated with [γ-32P]ATP with or without recombinant PKCβI in the presence of phosphatidylserine (PS), didecanoylglycerol (DG), and Ca2+ to activate PKC function. Under these conditions, Orai1 was highly phosphorylated by PKCβI, whereas no or little phosphorylation of Orai2 and Orai3 was observed (Fig. 3A). Although purified Orai1 was detected as multiple bands by Western blotting representing glycosylated (∼50 kDa) and non-glycosylated (∼37 kDa) forms of the protein, we detected phosphorylation predominantly of non-glycosylated Orai1 (lower band), whereas phosphorylation of the upper band was faint in the in vitro experiments. The phosphorylation of Orai1 was dependent on PKC activation, because the presence of PKC activators PS/DG and Ca2+ significantly enhanced phosphorylation, whereas addition of the PKC inhibitor GFX abolished phosphorylation (Fig. 3B).
FIGURE 3.
Orai1 is phosphorylated by PKCβI in vitro and in vivo. A and B, in vitro phosphorylation of Orai1. A, FLAG-tagged Orai proteins (Orai1, -2, and -3) were purified and incubated with or without recombinant PKCβI in the presence of PKC activator (PS/DG/Ca2+) and [γ-32P]ATP. Phosphorylated proteins were detected by autoradiography, and protein expression was determined by Western blotting using anti-FLAG antibody. The bar graph shows relative phosphorylation levels normalized to the amount of total protein (n = 3). B, purified Orai1 was incubated with PKCβI and PS/DG/Ca2+ in the presence or absence of PKC inhibitor GFX (5 μm). C and D, in vivo phosphorylation of Orai1. C, HEK293 cells expressing FLAG-tagged Orai proteins (Orai1, -2, and -3) were incubated with 32P-monosodium phosphate and stimulated with TG for 120 s in the presence of 2 mm extracellular Ca2+. Phosphorylated proteins were detected by autoradiography. D, cells expressing FLAG-tagged Orai1 were stimulated with TG in the presence or absence of GFX (1 μm), and with TPA (500 nm). Relative phosphorylation levels for each experimental condition were normalized to the strongest phosphorylation signal following TG stimulation (set as 100%). Averages of Orai1 phosphorylation from two independent experiments are shown.
Furthermore, we examined in vivo PKC phosphorylation of each Orai isoform. FLAG-tagged Orai proteins were expressed in HEK293 cells and, after incubation with 32P monosodium phosphate, cells were stimulated with TG in the presence of extracellular Ca2+. TG is expected to activate Ca2+/DAG-dependent PKC isoforms through mobilization of Ca2+ from ER stores and the extracellular space and activation of Ca2+-dependent phospholipases resulting in production of DAG. We observed that Orai1 phosphorylation was enhanced in response to TG stimulation (Fig. 3C). When the relative phosphorylation levels of Orai1, Orai2, and Orai3 were normalized to the expression levels of each protein, the levels of Orai2 and Orai3 phosphorylation were less than half of that observed for Orai1 (Fig. 3C and data not shown). Furthermore, TG-induced phosphorylation of Orai1 in vivo was reduced by GFX (Fig. 3D), whereas TPA stimulation increased the phosphorylation of Orai1 to a similar extent as TG. Collectively, results from in vitro and in vivo phosphorylation experiments show that Orai1 phosphorylation in HEK293 cells is mediated by PKCβI and is dependent on Ca2+ influx.
PKCβI Predominantly Phosphorylates the N Terminus of Orai1 in Vitro
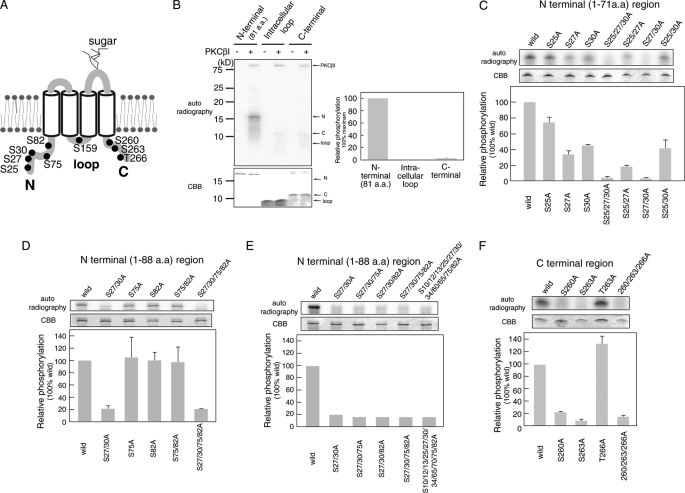
Orai1 is a tetraspanning plasma membrane protein (13) and thus contains three intracellular regions that can potentially be phosphorylated by PKC: the N terminus, an intracellular loop between transmembrane domains II and III, and the C terminus (Fig. 4A). Each intracellular region contains one or more predicted PKC phosphorylation sites (N terminus: Ser-25, Ser-27, Ser-30, Ser-75, and Ser-82; intracellular II and III loop: Ser-159; C terminus: Ser-260, Ser-263, and Thr-266). To study PKC-mediated phosphorylation of these sites, recombinant proteins encompassing the three regions were purified from E. coli and used for in vitro phosphorylation assays. Expression of all three Orai1 regions was confirmed by Coomassie Brilliant Blue staining (Fig. 4B). Importantly, both the N terminus (1–88 aa) and C terminus of Orai1 were phosphorylated by PKCβI in vitro, but phosphorylation of the N-terminal region was ∼20 times higher than that of the C terminus. Phosphorylation of the intracellular II and III loop was not detected.
FIGURE 4.
Analysis of PKC phosphorylation sites in intracellular regions of Orai1. A, the predicted PKC phosphorylation sites in Orai1 are shown. B–F, in vitro phosphorylation of the Orai1 N terminus (aa 1–81), intracellular loop (aa 141–173), and C terminus (aa 257–301). Intracellular Orai1 protein regions were expressed in E. coli, purified, and incubated with [γ-32P]ATP and recombinant PKCβI. Phosphorylated proteins were separated by SDS-PAGE and detected by autoradiography. Expression levels of recombinant Orai1 protein regions were detected by Coomassie Brilliant Blue (CBB) staining. Densitometry results of autoradiographs are shown in the bar graphs. Phosphorylation levels were normalized to the strongest phosphorylation signal (B) or phosphorylation of wild-type Orai1 (C–F), which were set to 100%. B, in vitro phosphorylation of the Orai1 N terminus (80 pmol), intracellular loop (400 pmol), and C terminus (240 pmol). C–F, in vitro phosphorylation of the Orai1 N terminus, intracellular loop, and C terminus containing the indicated mutations of predicted serine phosphorylation sites. Bars represent mean ± S.D. from three independent experiments.
PKCβI Phosphorylates Serine Residues Ser-27 and Ser-30 in Orai1 in Vitro
To identify the phosphorylation sites within the N and C termini of Orai1, we generated recombinant mutant proteins in which Ser or Thr residues were replaced with Ala. These proteins were subjected to in vitro phosphorylation assays. Among the five predicted phosphorylation sites in the N terminus (1–88 aa), we first examined the effect of substitution of three Ser residues (Ser-25, Ser-27, and Ser-30) on the phosphorylation of an N-terminal fragment (1–71 aa). Using various combinations of substitutions, we observed that all single substitutions impaired phosphorylation of the N-terminal fragment. Mutation of Ser-25 resulted in a moderate reduction while the S27A and S30A mutations strongly suppressed phosphorylation (Fig. 4C). No phosphorylation of the N-terminal Orai1 fragment was detectable with a triple mutant (S25A/S27A/S30A) and a double mutant (S27A/S30A). These results suggest that the Ser residues Ser-27 and Ser-30 are PKC phosphorylation sites but that Ser-25 is not.
Next, we examined the in vitro phosphorylation of Ser-75 and Ser-82 in the N-terminal region by using a longer recombinant peptide (1–88 aa) of the N terminus (Fig. 4, D and E). Neither single nor double substitutions of Ser-75 and Ser-82 with Ala affected the phosphorylation of Orai1 1–88 (Fig. 4D). The low level phosphorylation observed in the S27A/S30A mutant of Orai1 1–88 (Fig. 4, D and E) most likely represents nonspecific background, as a similar level of phosphorylation was detected in an Orai1 1–88 mutant in which almost all Ser residues (Ser-10, Ser-12, Ser-13, Ser-25, Ser-27, Ser-30, Ser-34, Ser-60, Ser-65, Ser-75, and Ser-82) in the N-terminal region had been replaced (Fig. 4E). The C-terminal region of Orai1 contains three predicted phosphorylation sites (Ser-260, Ser-263, and Thr-266), which were substituted with Ala to assess site-specific phosphorylation (Fig. 4F). Mutations at Ser-260 and Ser-263 eliminated phosphorylation of the C terminus, whereas replacement of Ser-266 with Ala had no effect.
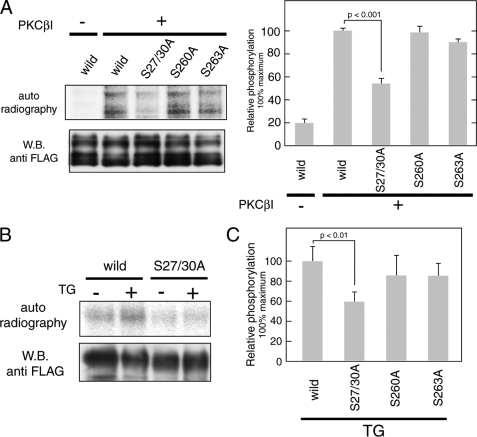
PKC Phosphorylates Orai1 on Serine Residues Ser-27 and Ser-30 in Vitro and in Vivo
To confirm the predicted PKC phosphorylation sites in the context of full-length Orai1, FLAG-tagged full-length wild-type and mutant Orai1 proteins were expressed in COS7 cells and purified with anti-FLAG agarose resin (Fig. 5A). Purified proteins were subjected to in vitro phosphorylation assays with PKCβI. Phosphorylation of the S27A/S30A mutant of Orai1 was significantly lower than that of wild-type Orai1. The S263A Orai1 mutant showed slightly reduced phosphorylation compared with wild-type Orai1, whereas mutation of Ser-260 to Ala had no effect on phosphorylation in contrast to results obtained with the isolated C terminus of Orai1. These findings suggested that, in the context of full-length Orai1, Ser-27 and Ser-30 are the predominant PKC phosphorylation sites.
FIGURE 5.
PKC mediates phosphorylation of full-length Orai1 protein at Ser-27 and Ser-30. A, in vitro kinase assay using full-length Orai1. FLAG-tagged wild-type and mutant (S27A/S30A, S260A, and S263A) Orai1 was expressed in COS7 cells and purified by anti-FLAG-agarose resin. Full-length proteins were incubated with [γ-32P]ATP and recombinant PKCβI. Phosphorylated proteins were separated by SDS-PAGE and detected by autoradiography. Total amounts of protein were determined by Western blotting using anti-FLAG antibody. Bar graphs represent the averages of densitometry results ±S.D. (n = 3); phosphorylation levels for wild-type Orai1 in the presence of PKCβI were set to 100%. B, in vivo phosphorylation of Orai1 in HEK293 cells. Cells transfected with FLAG-tagged wild-type and S27A/S30A Orai1 mutants were left unstimulated or stimulated with TG after incubation with 32P-monosodium phosphate. FLAG-tagged Orai1 was precipitated and separated by SDS-PAGE. Phosphorylated Orai proteins were detected by autoradiography. Total amounts of protein were determined by Western blotting using anti-FLAG antibody. C, phosphorylation levels of FLAG-tagged Orai1 mutants (S27A/S30A, S260A, and S263A) after TG stimulation were normalized to phosphorylation of wild-type Orai1 (set to 100%). Bars represent averages ± S.D. (n = 3).
To examine whether serine residues Ser-27, Ser-30, Ser-260, and Ser-263 are phosphorylated in vivo, FLAG-tagged wild-type and mutant Orai1 proteins were expressed in HEK293 cells. Cells were stimulated with TG in the presence of extracellular Ca2+ after incubation with 32P monosodium phosphate. Expression of the Orai1 S27A/S30A mutant suppressed background phosphorylation levels, and abolished the increase in phosphorylation after TG stimulation observed with wild-type Orai1 (Fig. 5B). By contrast, mutating Ser-260 or Ser-263 in the C terminus of Orai1 did not result in significantly decreased phosphorylation (Fig. 5C). Collectively, these data show that serine residues Ser-27 and Ser-30 are the main PKC phosphorylation sites in Orai1.
PKC-mediated Phosphorylation of Orai1 at Serine Residues Ser-27 and Ser-30 Suppresses SOCE
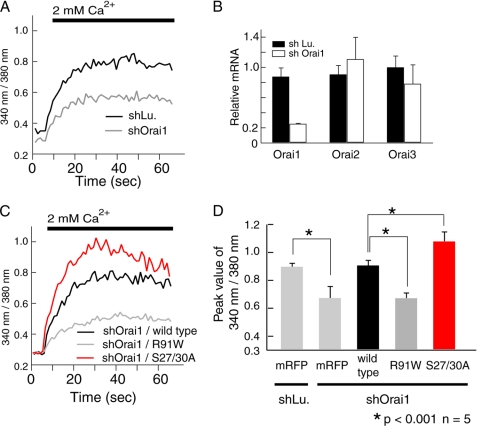
To study the role of PKC-mediated phosphorylation of Orai1 for SOCE, we substituted wild-type Orai1 with the Orai1 S27A/S30A mutant by knocking down endogenous Orai1 using shRNA while simultaneously expressing a knockdown-resistant form of Orai1 S27A/S30A. shOrai1 specifically reduced Orai1 mRNA levels by ∼80%, but had no effect on Orai2 and Orai3 expression (Fig. 6B). Knockdown of Orai1 suppressed SOCE to approximately half of the Ca2+ levels observed in cells treated with control shRNA (differential between peak and baseline [Ca2+]i 0.60 ± 0.02 in cells treated with control shRNA versus 0.37 ± 0.08 in cells treated with shOrai1; p < 0.05) (Fig. 6, A and D). Ca2+ entry in cells transfected with shOrai1 was rescued completely following expression of wild-type Orai1, but not using a non-functional mutant form of Orai1 (Orai1-R91W) (Fig. 6, C and D). Substituting endogenous Orai1 with Orai1 S27A/S30A resulted in enhanced SOCE compared with cells ectopically expressing wild-type Orai1. Comparable levels of all ectopically expressed Orai1 proteins were confirmed by Western blot analysis (supplemental Fig. S2); by contrast, endogenous Orai1 was undetectable. Localization of ectopically expressed wild-type and mutant Orai1 proteins on the plasma membrane was confirmed by immunofluorescence by using N-terminally as well as C-terminally FLAG-tagged Orai1 constructs (data not shown). Similar results, that is more efficient rescue of SOCE by Orai1-S27A/S30A than by wild-type Orai1, were obtained in fibroblasts from patients lacking SOCE (30), which were retrovirally transduced with wild-type or mutant Orai1-S27A/S30A (Fig. 7, A and B). Under these conditions, no functional endogenous Orai1 was present that could be phosphorylated by PKC interfering with the regulation of SOCE. Taken together, our results suggested that PKC-mediated phosphorylation of serine residues Ser-27 and Ser-30 in the N terminus of Orai1 negatively regulates SOCE by modulating the CRAC channel function of Orai1.
FIGURE 6.
Phosphorylation of Orai1 at S27A/S30A suppresses SOCE. A, SOCE was measured in HEK293 cells transfected with shOrai1 (targeting the 3′-untranslated region of Orai1) and shLu (Lu, luciferase) as negative control. B, knockdown efficiency and specificity were determined by quantitative real-time PCR. Bar graphs show results from three independent experiments as mean ± S.D. C, SOCE was measured in HEK293 cells transfected with shOrai1 and the coding sequences of wild-type Orai1, non-functional Orai1-R91W and Orai1-S27A/S30A (for protein expression levels see supplemental Fig. S2). D, averages of peak Fura2 ratio values obtained from five independent experiments similar to those in A and C are shown as means ± S.D.
To directly show that PKC negatively regulates CRAC channel activity by phosphorylating Orai1 and does not act on SOCE by modulating other mechanisms involved in Ca2+ homeostasis, we measured CRAC channel currents (ICRAC) in HEK293 cells co-transfected with STIM1 and either wild-type or mutant (S27A/S30A) Orai1. Co-expression of STIM1 and Orai1 was shown to dramatically enhance CRAC channel currents while maintaining most if not all properties characteristic of native CRAC channels (30, 35). Expression of mutant Orai1-S27A/S30A in HEK293 cells followed by TG stimulation resulted in large Ca2+ currents, which showed the same inwardly rectifying current-voltage relationship obtained by expression of wild-type Orai1, suggesting that gross CRAC channel properties were not affected by the mutation (Fig. 7C). The amplitude of CRAC channel currents measured 100 s after break-in was moderately increased in cells expressing Orai1-S27A/S30A compared with cells expressing wild-type Orai1 (Fig. 7C). These findings are consistent with enhanced SOCE in TG-stimulated HEK293 cells (Fig. 6, C and D) and fibroblasts (Fig. 7, A and B) expressing the Orai1-S27A/S30A mutant. A similar increase in CRAC channel currents in cells expressing the Orai1-S27A/S30A mutant compared with those expressing wild-type Orai1 was observed when ER Ca2+ stores were depleted with IP3 in the patch-pipette (Fig. 7D). As stated earlier, the increase in [Ca2+]i induced by either TG or IP3 results in activation of Ca2+-dependent phospholipases, production of DAG, and subsequently activation of Ca2+/DAG-dependent PKC isoforms. Collectively, these data show that PKC-mediated phosphorylation of Orai1 at Ser-27 and Ser-30 inhibits CRAC channel function directly rather than indirectly by modulating other proteins involved in Ca2+ homeostasis.
DISCUSSION
In the present study, we investigated whether phosphorylation of Orai1 is implicated in the regulation of store-operated calcium entry. Using HEK293 cells as a model system, we found that knockdown of endogenous Orai1 suppressed Ca2+ entry through SOC channel (Fig. 6A) as previously reported (14, 33), suggesting that Orai1 is an important SOC channel in this cell type. We identified a potentially important role for PKC-mediated negative regulation of the SOC channel activity of Orai1, because Ca2+ entry was enhanced by knockdown of PKC and in the presence of PKC inhibitors, whereas the PKC activator TPA suppressed SOCE. Consistent with these results we found that Orai1 was strongly phosphorylated by PKC in vitro and in vivo (Fig. 3). Studying the phosphorylation of three distinct intracellular Orai1 domains, we found that Orai1 is predominantly phosphorylated at its N terminus but much less at its C terminus or its intracellular II and III loop (Figs. 4 and 5). We identified Ser-27 and Ser-30 as the main phosphorylation sites in Orai1 that are targeted by PKCβI. It is noteworthy that the phosphorylation sites Ser-27 and Ser-30 are conserved throughout evolution in all mammalian Orai1 proteins, suggesting that PKC-mediated phosphorylation of Orai1 is an important mechanism involved in the modulation of SOCE. By contrast, Ser-27 and Ser-30 are not present in Orai2 and Orai3 (supplemental Fig. S3), consistent with a lack of significant Orai2 and Orai3 phosphorylation in our studies (Fig. 3).
Mutation of both Ser-27 and Ser-30 not only strongly impair Orai1 phosphorylation in vitro and in vivo but also enhanced store-operated Ca2+ entry in TG-stimulated HEK293 cells (Fig. 6) and SOCE-deficient fibroblasts (Fig. 7B) expressing the mutant Orai1 protein. Although our results do not exclude the possibility that the intracellular Ca2+ concentration is also modulated by PKC-mediated phosphorylation of other molecules such as the IP3 receptor, TRPC channels, SERCA pumps, or plasma membrane Ca2+ ATPases, they provide strong evidence that PKC regulates SOCE by direct phosphorylation of Orai1. Direct measurements of CRAC channel currents in cells overexpressing STIM1 together with either wild-type Orai1 or the Orai1-S27A/S30A mutant show that Ca2+ currents resulting from the non-phosphorylatable mutant Orai1 channel are increased compared with wild-type Orai1 (Fig. 7, C and D). Under the conditions tested, Ca2+-dependent PKC activation occurred in response to SOCE induced by either TG or IP3 resulting in phosphorylation of wild-type but not mutant Orai1. These findings suggest that phosphorylation of Orai1 at S27A/S30A inhibits CRAC channel function directly rather than indirectly by modulating other proteins involved in Ca2+ homeostasis.
How exactly PKC phosphorylation regulates Orai1 function remains unclear. Phosphorylation of Orai1 is unlikely to modulate Ca2+ permeation through the ion channel pore, because Ser-27 and Ser-30 are ∼60 aa residues away from the pore-forming first transmembrane domain of Orai1 (16–18, 36). Importantly, Ca2+ currents in cells expressing Orai1 S27A/S30A together with STIM1 show the same inwardly rectifying I-V relationship observed in cells expressing wild-type Orai1, indicating that the mutant Orai1 S27A/S30A retains its high Ca2+ selectivity and that the channel pore is unlikely to be affected by Orai1 phosphorylation. A more likely possibility is that phosphorylation of S27A/S30A in the N terminus of Orai1 regulates SOC channel gating. Although deletion of the Orai1 N terminus did not affect Orai1 clustering and association with STIM1, it resulted in loss of channel activation (37, 38). This is consistent with our finding that the Orai1 S27A/S30A mutant is able to bind to the minimal CRAC channel activation domain (CCb9, CAD, SOAR, or OASF) in STIM1 (supplemental Fig. S4) recently identified by several laboratories, including ours (27, 39–41). Phosphorylation of Orai1 at its N terminus is therefore unlikely to interfere with Orai1 binding to STIM1, although we cannot exclude the possibility at this point that binding of full-length STIM1, in contrast to the shorter CCb9 fragment, to Orai1 may be modulated by phosphorylation.
The function of PKC in the regulation of SOCE has been investigated in various cell types, with the observed effects of PKC on SOC channel function being very variable (9–12). This may be due to the different PKC subtypes (5, 6) and potentially different SOC channels expressed in the cell types studied. In the present study, we show that members of the Ca2+-activated cPKC subfamily are responsible for Orai1 regulation (Fig. 2). It is noteworthy that PKCβ more effectively phosphorylated Orai1 than PKCα and that knockdown of PKCβI but not PKCα enhanced SOCE. Although both PKCs are activated by Ca2+, the specific effect of PKCβ on Orai1 phosphorylation after TG stimulation may be explained by the higher affinity of the C2 domain in PKCβ for Ca2+ and lipid binding compared with the C2 domain of PKCα (42, 43). Although the substrate specificity of each PKC subtype seems to be quite low in vitro, there are several reports showing PKC subtype-specific regulation of functional proteins under physiological conditions (44).
The inconsistent results regarding PKC regulation of SOCE may also be explained by variations in the molecular composition of SOC channels in different cell types. The Orai1 paralogues Orai2 and Orai3 can form SOC channels, at least when ectopically expressed together with STIM1 (31, 34). Furthermore, several TRPC channels have been suggested to function as SOC channels. Studies using knock-out mice and siRNA demonstrated that TRPC1, TRPC4, and TRPC6 may be activated in a store-dependent manner (45, 46) and TRPC1 (24), TRPC3 (25), and TRPC6 (26) were shown to be regulated by PKC mediated phosphorylation. Taken together, the regulation of SOCE by PKC is likely to depend on the PKC isoforms and types of SOC channels expressed in any given cell type.
In this study, we demonstrate that SOCE and CRAC channel function in HEK293 cells is negatively regulated by PKC-mediated phosphorylation of Orai1 on serine residues Ser-27 and Ser-30. Serines Ser-27 and Ser-30 are unique to Orai1 and are not conserved in Orai2 and -3. The physiological importance of this regulatory mechanism for cell functions remains unclear, but because Orai1 is the predominant SOC channel in immune cells (47), phosphorylation of Orai1 by PKC might have an important role in the regulation of immune responses.
Supplementary Material
Acknowledgment
We thank W. Coetzee for his generous support with patch clamp equipment. We also thank Dr. Y. Shirai for helpful discussions of our work.
This work was supported by a grant-in-aid for scientific research from the Global Center of Excellence (COE) Program of the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to N. S.), grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to N. S.), the Hyogo Science and Technology Association (to N. S.), the Takeda Science Foundation (to N. S.), and an Uehara Foundation postdoctoral fellowship (to T. K.). This work was also supported in part by National Institutes of Health Grant AI066128 (to S. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- SOC
- store-operated Ca2+
- SOCE
- store-operated Ca2+ entry
- ER
- endoplasmic reticulum
- CRAC
- Ca2+ release-activated Ca2+
- PKC
- protein kinase C
- DAG
- diacylglycerol
- cPKC
- conventional PKC
- nPKC
- novel PKC
- aPKC
- atypical PKC
- STIM
- stromal interaction molecule
- aa
- amino acid(s)
- GST
- glutathione S-transferase
- shRNA
- short hairpin RNA
- GFP
- green fluorescent protein
- TG
- thapsigargin
- PS
- phosphatidylserine
- DG
- 1,2-didecanoylglycerol
- GFX
- GF109203X
- TPA
- 4α-phorbol 12-myristate 13-acetate
- IP3
- inositol 1,4,5-triphosphate
- TRPC
- transient receptor potential cation
- mRFP
- monomer red fluorescent protein
- IRES
- internal ribosome entry site
- SERCA
- sarco/endoplasmic reticulum Ca2+-ATPase.
REFERENCES
- 1.Berridge M. J., Lipp P., Bootman M. D. (2000) Nat. Rev. Mol. Cell Biol. 1, 11–21 [DOI] [PubMed] [Google Scholar]
- 2.Putney J. W., Jr. (1986) Cell Calcium 7, 1–12 [DOI] [PubMed] [Google Scholar]
- 3.Macian F. (2005) Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 4.Malviya A. N., Rogue P. J. (1998) Cell 92, 17–23 [DOI] [PubMed] [Google Scholar]
- 5.Nishizuka Y. (1988) Nature 334, 661–665 [DOI] [PubMed] [Google Scholar]
- 6.Nishizuka Y. (1992) Science 258, 607–614 [DOI] [PubMed] [Google Scholar]
- 7.Murray D., Honig B. (2002) Mol. Cell 9, 145–154 [DOI] [PubMed] [Google Scholar]
- 8.Zhang G., Kazanietz M. G., Blumberg P. M., Hurley J. H. (1995) Cell 81, 917–924 [DOI] [PubMed] [Google Scholar]
- 9.Haverstick D. M., Dicus M., Resnick M. S., Sando J. J., Gray L. S. (1997) J. Biol. Chem. 272, 15426–15433 [DOI] [PubMed] [Google Scholar]
- 10.Parekh A. B., Penner R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7907–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H., Suh B. C., Kim K. T. (1997) J. Biol. Chem. 272, 21831–21838 [DOI] [PubMed] [Google Scholar]
- 12.Petersen C. C., Berridge M. J. (1994) J. Biol. Chem. 269, 32246–32253 [PubMed] [Google Scholar]
- 13.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.-H., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. (2006) 441, 179–185 [DOI] [PubMed] [Google Scholar]
- 14.Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S. L., Yeromin A. V., Zhang X. H., Yu Y., Safrina O., Penna A., Roos J., Stauderman K. A., Cahalan M. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9357–9362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., Hogan P. G. (2006) Nature 443, 230–233 [DOI] [PubMed] [Google Scholar]
- 17.Vig M., Beck A., Billingsley J. M., Lis A., Parvez S., Peinelt C., Koomoa D. L., Soboloff J., Gill D. L., Fleig A., Kinet J. P., Penner R. (2006) Curr. Biol. 16, 2073–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeromin A. V., Zhang S. L., Jiang W., Yu Y., Safrina O., Cahalan M. D. (2006) Nature 443, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeHaven W. I., Smyth J. T., Boyles R. R., Putney J. W., Jr. (2007) J. Biol. Chem. 282, 17548–17556 [DOI] [PubMed] [Google Scholar]
- 20.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandman O., Liou J., Park W. S., Meyer T. (2007) Cell 131, 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan J. P., Kim M. S., Zeng W., Shin D. M., Huang G., Worley P. F., Muallem S. (2009) Channels 3, 221–225 [DOI] [PubMed] [Google Scholar]
- 24.Albert A. P., Large W. A. (2003) Cell Calcium 33, 345–356 [DOI] [PubMed] [Google Scholar]
- 25.Venkatachalam K., Zheng F., Gill D. L. (2003) J. Biol. Chem. 278, 29031–29040 [DOI] [PubMed] [Google Scholar]
- 26.Kim J. Y., Saffen D. (2005) J. Biol. Chem. 280, 32035–32047 [DOI] [PubMed] [Google Scholar]
- 27.Kawasaki T., Lange I., Feske S. (2009) Biochem. Biophys. Res. Commun. 385, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X., Yu Y., Zhao H., Zhang Y., Manela J., Tonetti D. A. (2006) Carcinogenesis 27, 1538–1546 [DOI] [PubMed] [Google Scholar]
- 29.Cejas P. J., Carlson L. M., Kolonias D., Zhang J., Lindner I., Billadeau D. D., Boise L. H., Lee K. P. (2005) Mol. Cell Biol. 25, 7900–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peinelt C., Vig M., Koomoa D. L., Beck A., Nadler M. J., Koblan-Huberson M., Lis A., Fleig A., Penner R., Kinet J. P. (2006) Nat. Cell Biol. 8, 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer J. C., Dehaven W. I., Smyth J. T., Wedel B., Boyles R. R., Bird G. S., Putney J. W., Jr. (2006) J. Biol. Chem. 281, 24979–24990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarl C. A., Picard C., Khalil S., Kawasaki T., Röther J., Papolos A., Kutok J., Hivroz C., Ledeist F., Plogmann K., Ehl S., Notheis G., Albert M. H., Belohradsky B. H., Kirschner J., Rao A., Fischer A., Feske S. (2009) J. Allergy Clin. Immunol. 124, 1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gwack Y., Srikanth S., Feske S., Cruz-Guilloty F., Oh-hora M., Neems D. S., Hogan P. G., Rao A. (2007) J. Biol. Chem. 282, 16232–16243 [DOI] [PubMed] [Google Scholar]
- 34.Lis A., Peinelt C., Beck A., Parvez S., Monteilh-Zoller M., Fleig A., Penner R. (2007) Curr. Biol. 17, 794–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soboloff J., Spassova M. A., Tang X. D., Hewavitharana T., Xu W., Gill D. L. (2006) J. Biol. Chem. 281, 20661–20665 [DOI] [PubMed] [Google Scholar]
- 36.McNally B. A., Yamashita M., Engh A., Prakriya M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22516–22521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muik M., Frischauf I., Derler I., Fahrner M., Bergsmann J., Eder P., Schindl R., Hesch C., Polzinger B., Fritsch R., Kahr H., Madl J., Gruber H., Groschner K., Romanin C. (2008) J. Biol. Chem. 283, 8014–8022 [DOI] [PubMed] [Google Scholar]
- 38.Li Z., Lu J., Xu P., Xie X., Chen L., Xu T. (2007) J. Biol. Chem. 282, 29448–29456 [DOI] [PubMed] [Google Scholar]
- 39.Yuan J. P., Zeng W., Dorwart M. R., Choi Y. J., Worley P. F., Muallem S. (2009) Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keranen L. M., Newton A. C. (1997) J. Biol. Chem. 272, 25959–25967 [DOI] [PubMed] [Google Scholar]
- 43.Kohout S. C., Corbalán-García S., Torrecillas A., Goméz-Fernandez J. C., Falke J. J. (2002) Biochemistry 41, 11411–11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchino M., Sakai N., Kashiwagi K., Shirai Y., Shinohara Y., Hirose K., Iino M., Yamamura T., Saito N. (2004) J. Biol. Chem. 279, 2254–2261 [DOI] [PubMed] [Google Scholar]
- 45.Freichel M., Suh S. H., Pfeifer A., Schweig U., Trost C., Weissgerber P., Biel M., Philipp S., Freise D., Droogmans G., Hofmann F., Flockerzi V., Nilius B. (2001) Nat. Cell Biol. 3, 121–127 [DOI] [PubMed] [Google Scholar]
- 46.Dietrich A., Mederos Y., Schnitzler M., Gollasch M., Gross V., Storch U., Dubrovska G., Obst M., Yildirim E., Salanova B., Kalwa H., Essin K., Pinkenburg O., Luft F. C., Gudermann T., Birnbaumer L. (2005) Mol. Cell Biol. 25, 6980–6989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feske S. (2007) Nat. Rev. Immunol 7, 690–702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.