Abstract
Transforming growth factor-β-activated kinase 1 (TAK1), an MAP3K, is a key player in processing a multitude of inflammatory stimuli. TAK1 autoactivation involves the interplay with TAK1-binding proteins (TAB), e.g. TAB1 and TAB2, and phosphorylation of several activation segment residues. However, the TAK1 autoactivation is not yet fully understood on the molecular level due to the static nature of available x-ray structural data and the complexity of cellular systems applied for investigation. Here, we established a bacterial expression system to generate recombinant mammalian TAK1 complexes. Co-expression of TAK1 and TAB1, but not TAB2, resulted in a functional and active TAK1-TAB1 complex capable of directly activating full-length heterotrimeric mammalian AMP-activated protein kinase (AMPK) in vitro. TAK1-dependent AMPK activation was mediated via hydrophobic residues of the AMPK kinase domain αG-helix as observed in vitro and in transfected cell culture. Co-immunoprecipitation of differently epitope-tagged TAK1 from transfected cells and mutation of hydrophobic αG-helix residues in TAK1 point to an intermolecular mechanism of TAB1-induced TAK1 autoactivation, as TAK1 autophosphorylation of the activation segment was impaired in these mutants. TAB1 phosphorylation was enhanced in a subset of these mutants, indicating a critical role of αG-helix residues in this process. Analyses of phosphorylation site mutants of the activation segment indicate that autophosphorylation of Ser-192 precedes TAB1 phosphorylation and is followed by sequential phosphorylation of Thr-178, Thr-187, and finally Thr-184. Finally, we present a model for the chronological order of events governing TAB1-induced TAK1 autoactivation.
Keywords: AMP-activated Kinase (AMPK), MAPKs, Protein Phosphorylation, Protein-Protein Interactions, Serine/Threonine Protein Kinase, TAK1-binding Protein 1 (TAB1), Transforming Growth Factor-beta-activated Kinase 1 (TAK1), Bacterial Co-expression, Kinase Domain alphaG-helix, Mitogen-activated Protein Kinase Kinase Kinase (MAP3K)
Introduction
Transforming growth factor-β-activated kinase 1 (TAK1) was first identified as an MAP3K4 involved in transforming growth factor-β and bone morphogenetic protein-mediated signaling (1). Because it transduces a multitude of extracellular stimuli, such as those from interleukin-1, tumor necrosis factor-α, and lipopolysaccharides, the serine/threonine kinase TAK1 represents a key activator of pathways involving IκB kinase, c-Jun NH2-terminal kinase (JNK), and p38 (2–9). Furthermore, a regulatory function of TAK1 has also been described in the Wnt signaling pathway (10, 11).
TAK1 activity is regulated by association of TAK1-binding proteins, namely TAB1, TAB2, TAB3, and TAB4 (12–17). TAB1, a “pseudophosphatase” that shows close structural similarity with the Mg2+- or Mn2+-dependent protein phosphatase family member protein phosphatase 2Cα (PP2Cα) (18), was originally found in a yeast two-hybrid screen as a protein, which directly triggers TAK1 activity (13). Both structural and functional studies revealed that an evolutionarily conserved motif in the carboxyl-terminal region of TAB1 (residues 480–504) is sufficient to bind to the catalytic domain of TAK1. However, the presence of a slightly extended TAB1 carboxyl-terminal domain (residues 437–504) is indispensable for full activation of TAK1 in HeLa cells (19–22). Purification and co-immunoprecipitation experiments from cultured cells revealed the co-existence of TAB1 and TAB2 in TAK1-containing complexes (3, 15). The functional integrity and interplay of both TAK1-binding proteins in these complexes is not fully understood yet. The TAB2 carboxyl terminus interacts with a carboxyl-terminal region of TAK1 (13, 14). TAB2 activates TAK1 by recruiting the MAP3K to tumor necrosis factor-α receptor-associated factor family members in a polyubiquitin-dependent manner (3, 14, 23, 24). Also, nonconjugated, free Lys-63 polyubiquitin chains are capable of directly activating TAK1 via TAB2 (25). TAB3 shares 48% amino acid identity with TAB2, similarly binds ubiquitin, and interacts and activates TAK1 (15, 16). Thus, TAB2 and TAB3 were suggested to function redundantly as mediators in TAK1 activation (15, 16, 26). Recently, TAB4, a type 2A phosphatase-interacting protein, was described as the fourth TAK1-binding protein capable of triggering TAK1 activity (17).
It has been shown for many protein kinases that phosphorylation or dephosphorylation of critical serine and threonine residues within the kinase domain activation segment is essential for regulation of kinase activity (27–29). Phosphorylation of these crucial residues can be mediated by upstream kinases and autoactivation events, respectively, and can often be efficiently mimicked by substitution of the respective site with acidic amino acids (30–32). Replacement of Thr-187 and Ser-192 with acidic residues did not render TAK1 constitutively active and instead rather inactivated the kinase (19, 21, 33, 34), whereas TAK1 mutants carrying acidic amino acids at the Thr-178 and Thr-184 sites at least partially mimicked the activated wild type, although still requiring co-expression of TAB1 in cultured cells (21, 35). TAB1, upon association with TAK1, is suggested to get phosphorylated at a carboxyl-terminal Ser/Thr rich domain, which is part of the TAK1 autoactivation process (19).
Recently, TAK1 was found to phosphorylate and activate AMP-activated protein kinase (AMPK), a key player in sensing and regulating energy metabolism on the cellular and whole body level (36–39). Momcilovic et al. (36) showed TAK1-dependent phosphorylation of sucrose nonfermenting kinase 1 (SNF1), the yeast ortholog of the AMPK α-subunit, and the recombinant AMPK catalytic domain in vitro, as well as AMPK activation elicited by co-transfection of TAK1 and TAB1 in HeLa cells. Xie et al. (37) confirmed that TAK1 plays an important role in AMPK signaling in heart but suggested a rather indirect mechanism of AMPK activation, involving an alternative upstream kinase of AMPK, the tumor suppressor kinase LKB1.
Mammalian AMPK is a highly conserved heterotrimeric serine/threonine protein kinase, which consists of a catalytic α-subunit and regulatory β- and γ-subunits, each existing in different isoforms (α1, α2, β1, β2, γ1, γ2, and γ3) (40). Direct phosphorylation of Thr-172 in the activation segment of the AMPK kinase domain by Ca2+/calmodulin-dependent protein kinase kinase-2 (CamKK2) (41) or the tumor suppressor kinase complex LKB1-MO25-STRAD (42) is the crucial step in AMPK activation, which then leads to autophosphorylation of multiple sites on its α- and β-subunits (43). Recently, we described a distal recognition site for the upstream kinases residues of the amphipathic kinase domain αG-helix of AMPK, namely Val-219 and Phe-223 (44).
Here, we established the expression of TAK1 subunits, complexes, and mutants in bacteria. The direct functional interplay of TAK1 and AMPK was investigated in cultured cells and in cell-free assays. A two-step protocol was developed, involving TAK1-dependent AMPK activation followed by quantification of AMPK activity, which allowed for accurate TAK1 activity determination. TAK1-TAB1 complexes were found functional and active, whereas TAK1-TAB2 complexes were inactive after isolation from bacterial lysates. TAK1-TAB1 complexes were further studied by probing a series of mutant enzyme complexes. Based on the findings, we discuss a model of the autoactivation process. Our analyses point to a sequential bimolecular autophosphorylation mechanism requiring the TAK1 kinase domain αG-helix and the TAK1-binding protein TAB1.
EXPERIMENTAL PROCEDURES
Plasmids, Enzymes, and Inhibitors
Bacterial expression plasmids coding for mammalian nontagged AMPK α1β1γ1 (WT-AMPK) (45, 46), nontagged α1(V219E,F223E)β1γ1 (VEFE-AMPK) (44), and mammalian expression constructs coding for Myc-tagged AMPK α1 and α1(V219E,F223E) subunits, respectively (44, 47), were described earlier. The bacterial expression plasmid encoding nontagged kinase-deficient mutant α1(D157A)β1γ1 (DA-AMPK) was generated analogous to WT-AMPK (45, 46). The His-tagged TAK1-TAB1 fusion protein encodes the human TAK1 amino acids 1–303 fused to human TAB1 amino acids 437-end. The purified active TAK1-TAB1 fusion protein was purchased from Upstate (catalog no. 14-600). The resorcylic acid lactone (5Z)-7-oxozeaenol, an irreversible inhibitor of selected kinases (TAK1, VEGFR, PDGFR, MKNK1, MEK1, KIT, GAK, and FLT1), was prepared as described previously (48–51).
Cloning
The bacterial expression vectors pOLY3.TEV and pOLY1 are derived from pET14bx and pET3ax vectors, respectively (45), which were modified to encode additional unique restriction enzyme recognition sites and a tobacco etch virus protease cleavage site instead of thrombin (to be described in detail elsewhere). The cDNAs coding for human TAK1, TAB1, and TAB2 (GenBankTM accession numbers NM_003188, NM_006116, and NM_015093, respectively) were subcloned by PCR and inserted in the NdeI/SpeI sites of the pOLY vectors as listed in Table 1. The following primers were used for subcloning: TAK1-pOLY-fwd (5′-GTCATATGTCTACAGCCTCTGCCGCCTCCTCC-3′); TAK1-pOLY-rev (5′-GTACTAGTCATGAAGTGCCTTGTCGTTTCTGCTG-3′); TAB1-pOLY-fwd (5′-CTCATATGGCGGCGCAGAGGAGGAGCTTGCTGC-3′); TAB1-pOLY-rev (5′-GTACTAGTCTACGGTGCTGTCACCACGCTCTGCTC-3′); TAB2-pOLY-fwd (5′-GTACATATGGCCCAAGGAAGCCACCAAATTG-3′); and TAB2-pOLY-rev (5′-GTGGATCCACTAGTCAGAAATGCCTTGGCATCTCACACTG-3′). Polycistronic bacterial expression plasmids were generated by successive restriction of individual cistrons from donor plasmids by XbaI/BlpI and ligation of the latter into the complementary SpeI/BlpI sites of the corresponding acceptor plasmid (Table 1 and supplemental Fig. S1) (45). The cytomegalovirus promoter-based mammalian expression vectors 3×FLAG pCMV5b and 3×HA pCMV5b code for amino-terminal FLAG and HA tags, respectively, and were kindly provided by Sabine Werner (ETH Zurich, Zurich, Switzerland). Prior to ligation into the XbaI/BamHI sites, the human TAK1 and TAB1 cDNAs were amplified by PCR utilizing the following primer pairs (restriction sites are underlined): TAK1-CMV-fwd (5′-CAGTTCTAGAATGTCTACAGCCTCTGCCGCC-3′); TAK1-CMV-rev (5′-ACTGGGATCCTCATGAAGTGCCTTGTCGTTTCTG-3′); TAB1-CMV-fwd (5′-CAGTTCTAGAATGGCGGCGCAGAGGAGGAGC-3′); and TAB1-CMV-rev (5′- ACTGGGATCCCTACGGTGCTGTCACCACGC-3′). The sequences of all plasmids were verified (Microsynth AG, Balgach, Switzerland).
TABLE 1.
Overview of plasmids generated for this study (encoding wild-type protein)
| Plasmid | Insert/Cloning strategy |
|---|---|
| pHis-TAK1 | TAK1 subcloned into the NdeI/SpeI sites of pOLY3.TEV |
| pTAB1 | TAB1 subcloned into the NdeI/SpeI sites of pOLY1 |
| pTAB2 | TAB2 subcloned into the NdeI/SpeI sites of pOLY1 |
| pHis-TAK1-TAB1 | pHis-TAK1 digested with SpeI/BlpI and ligated to XbaI/BlpI fragment of pTAB1 |
| pHis-TAK1-TAB2 | pHis-TAK1 digested with SpeI/BlpI and ligated to XbaI/BlpI fragment of pTAB2 |
| pHis-TAK1-TAB1-TAB2 | pHis-TAK1-TAB1 digested with SpeI/BlpI and ligated to XbaI/BlpI fragment of pTAB2 |
Mutagenesis
Kinase-deficient (K63W), kinase domain αG helix (A236E, A236K, F237E, F237K, I239E, I239K, W241E, W241K, V243E, and V243K), and activation segment phosphorylation site mutants (T178A, T178E, T184A, T184E, T187A, T187E, S192A, and S192E) of TAK1 were generated by site-directed mutagenesis (QuikChange, Stratagene) using the respective mono- or dicistronic bacterial expression plasmid as a template. The sequences of the applied primers are available upon request. The sequences of all expression plasmids were verified (Microsynth AG, Balgach, Switzerland).
Bacterial Expression and Purification
Recombinant TAK1, TAB1, and TAB2 proteins and complexes, as well as heterotrimeric AMPK were expressed in Rosetta 2 (DE3) Escherichia coli cells (Novagen). Usually, 2 liters of autoinduction medium (52) were inoculated, and bacteria were grown at 37 °C for 3 h before cultures were shifted to room temperature with continuous shaking at 220 rpm for 22 h. Cells were harvested and proteins were extracted as previously described (46). Proteins were purified by application of a two-dimensional purification protocol as reported earlier (44). Briefly, the soluble protein fraction was subjected to batch affinity purification for 60–90 min at room temperature using 1 ml of nickel-Sepharose HP (GE Healthcare). Eluted proteins were further purified by size exclusion chromatography with a Superdex 200 16/60 column (GE Healthcare) equilibrated in 50 mm Tris-Cl, 200 mm NaCl, 4 mm MgCl2, 8 mm EDTA, 0.02% NaN3, pH 8.0, at 7 °C. Peak fractions were pooled and concentrated using a Vivaspin 20 centrifugal concentrator with a 10,000 molecular weight cutoff (Sartorius AG, Goettingen, Germany). Afterward, proteins were supplemented with 2 mm Tris(2-carboxyethyl)phosphine hydrochloride and stored either at 4 °C or in 50% glycerol at −20 °C until usage.
SDS-PAGE and Western Blotting
SDS-PAGE and Western blot analysis were performed as reported previously (53). AMPKα (catalog no. 2532), AMPK-Thr(P)-172 (catalog no. 2535), TAK1 (catalog no. 4505), TAK1-Thr(P)-184 (catalog no. 4537S), TAK1-Thr(P)-187 (catalog no. 4536S), TAK1-Thr(P)-184/187 (catalog no. 4531S), TAB1 (catalog no. 3225), and TAB2 (catalog no. 3744S) antibodies were from Cell Signaling Inc. Penta-His (catalog no. 34660) antibody was purchased from Qiagen, FLAG® M2 (catalog no. F1804) and HA (catalog no. H3663) antibodies were from Sigma. Signal intensities were densitometrically quantified using the software Quantity One, version 4.6.3.088 (Bio-Rad).
TAK1 Autoactivation, AMPK Activation by TAK1, and Determination of Kinase Activity
TAK1 autoactivation was carried out with 150 μg/ml bacterially expressed and purified His-TAK1 or His-TAK1 complexes. For determination of TAK1 activity, 75 μg/ml heterotrimeric AMPK was added to the reaction mixture. If not otherwise specified in the figure legends, all activation assays were incubated for 20–25 min at 30 °C. Accurate comparison of TAK1 kinase activities of different complexes or mutants was facilitated by expressing and purifying all respective proteins in parallel. Additionally, the activity data were normalized to the amount of TAK1 protein, which was determined by Western blotting and subsequent densitometric analysis. AMPK phosphorylation by TAK1 was monitored by incorporation of [γ-32P]ATP (Hartmann Analytic, Braunschweig, Germany) by autoradiography using Hyperfilm MP (GE Healthcare). AMPK activity was determined by Western blot analysis of AMPKα Thr-172 phosphorylation and activity assay using the artificial peptide substrate with the sequence HMRSAMSGLHLVKRR as reported previously (53). Activity of immunoprecipitated AMPK from HeLa cell lysates was assayed as described (44).
Cell Culture Methods
Immortalized AMPKα subunit double knock-out (α1−/− and α2−/−) mouse embryo fibroblasts (MEF) (54) and HeLa cells were cultured under standard conditions in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 4.5 g/liter glucose, 2 mm l-glutamine, and 10% fetal calf serum (Bioconcept, Allschwil, Switzerland). Transfection and lysis of cells were performed as reported previously (44). To activate AMPK by TAK1 in HeLa cells, 24 h post-transfection cells were serum-starved for 4 h prior to treatment with 1 mm H2O2 for 15 min and subsequent lysis (36).
Co-immunoprecipitation
Anti-HA antibodies (1 μg) coupled to GammaBindTM Plus-SepharoseTM matrix (GE Healthcare) (15 μl) were used to immunoprecipitate recombinant protein from 1 mg of (α1−/− and α2−/−) MEF soluble and precleared lysate. After overnight incubation with gentle agitation at 4 °C, the beads were washed three times in lysis buffer. Laemmli buffer was added; samples were heated, and the supernatants were subjected to SDS-PAGE and Western blot analysis.
Statistical Analysis
Background signal subtraction and normalization of the data were performed as described in the corresponding figure legends. Results are depicted as means ± S.D.
RESULTS
Bacterial Expression of Recombinant TAK1 and TAK1-containing Complexes
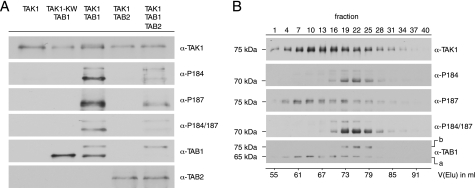
Based on earlier work from our group (45), we generated mono- and polycistronic vectors for the bacterial expression of hexahistidine-tagged TAK1 (TAK1) alone and in combination with its binding proteins TAB1 and TAB2 (Table 1 and supplemental Fig. S1). Because of the absence of interfering eukaryotic protein kinases, the bacterial system allowed for the more precise investigation of TAK1 autophosphorylation in the presence or absence of TAB1 and/or TAB2. This approach contrasts to earlier studies, where eukaryotic cell systems were used, thus potentially impacting on the TAK1 phosphorylation pattern prior to analysis. TAK1, TAK1-TAB1, TAK1-TAB2, and TAK1-TAB1-TAB2 were successfully expressed in E. coli, as revealed by Western blotting using antibodies specifically recognizing the respective proteins (supplemental Fig. S2). Although soluble levels of the recombinant proteins were generally low, enrichment of sufficient protein amounts by immobilized metal affinity chromatography allowed for second step purification by size exclusion chromatography. Subsequently, individual fractions of the purified proteins were pooled, concentrated, and subjected to Western blot analysis, which included the probing with phospho-specific antibodies recognizing the established TAK1 activation segment phosphorylation sites Thr-184 and Thr-187 (Fig. 1A). Direct interaction of His-TAK1 with TAB1 and His-TAK1 with TAB2 could be confirmed by co-purification of corresponding proteins. Strong phosphorylation of the activation segment sites Thr-184 and Thr-187 was observed when wild-type TAK1 (WT-TAK1) was co-expressed with TAB1. In addition, TAB1 shifted partly to lower electrophoretic mobility upon co-expression with WT-TAK1, thus indicating autophosphorylation of TAB1 as suggested previously (19). Autophosphorylation is TAK1-dependent, because it only appeared in WT-TAK1-TAB1 complexes but not in kinase-deficient TAK1(K63W)-TAB1 (KW-TAK1-TAB1) complexes. TAK1 expressed alone, KW-TAK1-TAB1, and also WT-TAK1 co-expressed with TAB2 did not show detectable autophosphorylation of the Thr-184 and Thr-187 sites. After co-expression of WT-TAK1, TAB1, and TAB2, the purified WT-TAK1-TAB1-TAB2 complex contained only few TAB1s, and this species exhibited a reduced electrophoretic mobility. WT-TAK1-TAB1-TAB2 complexes exhibited reduced TAK1 autophosphorylation at the Thr-184 and Thr-187 sites compared with WT-TAK1-TAB1, which could be interpreted as inability of TAB2 to directly induce TAK1 autophosphorylation (13, 14), and it could reflect impaired binding of TAB1 due to the presence of TAB2.
FIGURE 1.
Analysis of TAK1 containing complexes after two-dimensional purification. A, Western blot analysis of bacterially expressed His-TAK1 containing complexes subsequent to two-dimensional purification. Anti-TAK1, anti-TAB1, anti-TAB2, and phospho-specific anti-TAK1-Thr(P)-184 (α-P184), anti-TAK1-Thr-187 (α-P187), and anti-TAK1-Thr(P)-184/187 (α-P184/187) antibodies were used for detection. KW-TAK1 represents the kinase-deficient mutant His-TAK1(K63W). For better comparability of activation segment phosphorylation and protein complex composition, the amount of analyzed protein was adjusted to equal His-TAK1 content. B, Western blot analysis of bacterially expressed and purified His-TAK1-TAB1 subjected to size exclusion chromatography using antibodies as in A. 1-ml fractions were collected, and every third fraction was analyzed. The corresponding elution volume V(Elu) is specified. The given molecular weights correspond to the electrophoretic mobility estimated from SDS-PAGE. Line a, unmodified TAB1 species; line b, shifted TAB1 species.
Western blot analysis of WT-TAK1-TAB1 elution fractions from size exclusion chromatography using anti-His, anti-TAK1, and anti-TAB1 antibodies again verified interaction of TAK1 and TAB1, resulting in co-elution of both proteins (Fig. 1B). Although phosphorylation at Thr-187 was detected in a broad elution peak, phosphorylation of Thr-184 was restricted to a smaller subset of elution fractions, which accordingly also showed double phosphorylation of the Thr-184 and Thr-187 sites. These data show that TAK1 autophosphorylation at Thr-187 can occur in the absence of the Thr-184 modification, thus indicating that autophosphorylation at Thr-187 could precede phosphorylation of the Thr-184 site. The doubly phosphorylated species also co-eluted with shifted phosphorylated TAB1.
In conclusion, bacterially expressed TAK1 forms complexes with TAB1 and TAB2 if co-expressed in bacteria, but only the WT-TAK1-TAB1 complex showed autophosphorylation of the two critical activation segment residues Thr-184 and Thr-187 that existed only in conjunction with phosphorylation of TAB1.
TAK1 Directly Activates Heterotrimeric AMPK in Vitro
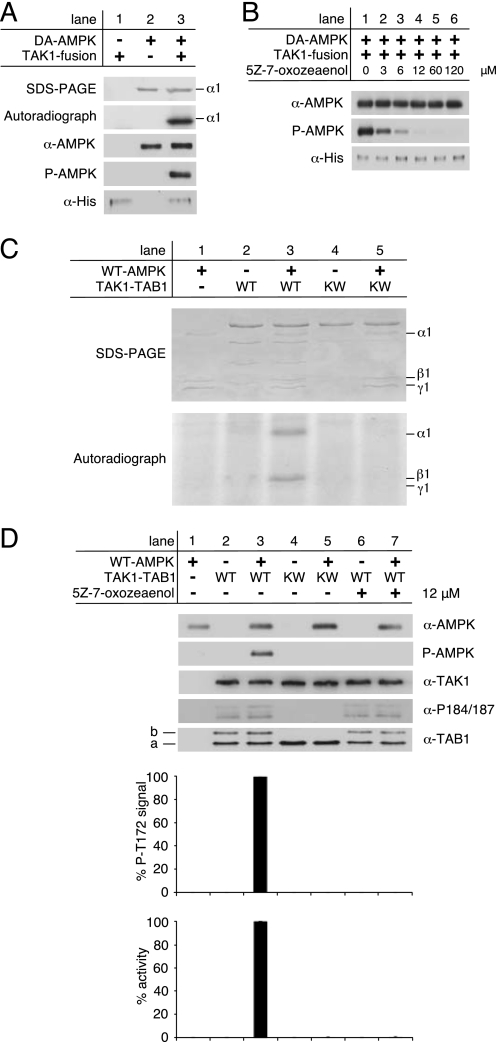
Autophosphorylation of the Thr-184 and Thr-187 sites is considered important for TAK1 kinase activity. Thus, we wanted to determine kinase activity of bacterially expressed TAK1-TAB1 complexes. Recently, it was reported that TAK1 can activate the isolated recombinant AMPK catalytic domain at Thr-172 in vitro (36). These studies employed a commercially available, recombinant TAK1-TAB1 fusion protein that was obtained by baculoviral expression and encodes the isolated TAK1 kinase domain fused to a small carboxyl-terminal fragment of TAB1, thus exhibiting constitutive activity. Incubation of a kinase-deficient mutant of full-length heterotrimeric AMPK, α1(D157A)β1γ1 (DA-AMPK), with TAK1-TAB1 fusion protein in the presence of [γ-32P]ATP revealed incorporation of radioactivity into the AMPK α-subunit as shown by autoradiography (Fig. 2A). Moreover, Western blot analysis confirmed that Thr-172, the residue critical for AMPK activation, was phosphorylated by the TAK1-TAB1 fusion protein in vitro. To show TAK1 dependence of this phosphorylation, we additionally applied the TAK1-selective inhibitor (5Z)-7-oxozeaenol (Fig. 2B). Indeed, preincubation with increasing concentrations of (5Z)-7-oxozeaenol dose-dependently decreased Thr-172 phosphorylation of DA-AMPK. TAK1-dependent phosphorylation of AMPK was completely abolished at an inhibitor concentration of 12 μm. Here, we proceeded to investigate the functional integrity of the bacterially expressed TAK1-TAB1 complex using recombinant heterotrimeric AMPK. Incubation of wild-type AMPK (WT-AMPK) with WT-TAK1-TAB1 in the presence of [γ-32P]ATP led to incorporation of 32P into the AMPK α- and β-subunits, but incubation with KW-TAK1-TAB1 showed no incorporation (Fig. 2C). Labeling of the AMPK β-subunit upon activation of WT-AMPK has been reported previously and reflects autophosphorylation of this subunit in response to activation (43, 53). The putative direct AMPK activation by full-length bacterially expressed TAK1-TAB1 was further evaluated by Western blot analysis of Thr-172 phosphorylation and AMPK activity determination using an artificial peptide substrate of AMPK, the SAMS peptide (SAMS, synthetic peptide HMRSAMSGLHLVKRR), and the HPLC-based detection method (53). In contrast to the kinase-deficient mutant TAK1 complex (KW-TAK1-TAB1), WT-TAK1-TAB1 efficiently phosphorylated AMPK at Thr-172, as revealed by Western blotting and activity assay (Fig. 2D, compare lanes 3 and 5). Furthermore, preincubation of WT-TAK1-TAB1 with 12 μm of the TAK1-specific inhibitor (5Z)-7-oxozeaenol completely blocked TAK1-mediated activation of AMPK (Fig. 2D, lane 7).
FIGURE 2.
Activation of AMPK by active TAK1 in vitro. A, AMPK kinase-dead mutant α1(D157A)β1γ1 (DA-AMPK) was incubated with commercially available, active His-tagged TAK1-TAB1 fusion protein from baculoviral expression (TAK1 fusion) in the presence of [γ-32P]ATP for 20 min at 37 °C. Phosphorylation of DA-AMPK was detected by incorporation of 32P followed by autoradiography and Western blot analysis using anti-AMPKα and phospho-specific anti-AMPKα Thr-172 antibodies (P-AMPK). The input of TAK1-fusion protein was probed with anti-His antibody. B, 25 ng of TAK1 fusion was preincubated with increasing concentrations of the TAK1-selective inhibitor (5Z)-7-oxozeaenol in 10 mm Hepes, pH 7.5, for 15 min at 37 °C before adding DA-AMPK. C, phosphorylation of recombinant WT-AMPK by the bacterially expressed recombinant wild-type TAK1-TAB1 (WT) and the kinase-deficient mutant TAK1(K63W)-TAB1 (KW) in the presence of [γ-32P]ATP was monitored by autoradiography. The corresponding Coomassie Blue-stained SDS-polyacrylamide gel is shown on the top. D, in vitro activation of recombinant WT-AMPK by bacterially expressed and purified recombinant WT-TAK1-TAB1 and KW-TAK1-TAB1. TAK1-TAB1 proteins were preincubated for 15 min at 37 °C in the presence or absence of 12 μm (5Z)-7-oxozeaenol. Western blot analysis was performed with anti-AMPKα, anti-TAK1, anti-TAB1, and phospho-specific anti-AMPKα Thr-172 (P-AMPK) and anti-TAK1 Thr(P)-184/187 (α-P184/187) antibodies. To quantify AMPKα Thr-172 phosphorylation as a consequence of TAK1 activity, the respective background signals of WT-AMPK (lane 1) and TAK1-TAB1 complexes (lanes 2 and 4) were subtracted, and the AMPKα phospho-Thr172 signals were normalized to AMPKα and TAK1 signal intensities. AMPK activity was determined, and respective background activities were subtracted. All activities were standardized to corresponding TAK1 signal intensities as revealed from densitometric Western blot analysis to determine TAK1 activity in relation to TAK1 amount. All data were normalized relative to WT-AMPK activation by WT-TAK1-TAB1 (n = 3; S.D.). Line a, unmodified TAB1 species; line b, shifted TAB1 species.
These results strongly suggest that bacterial co-expression of TAK1 and TAB1 yielded functional and active TAK1-TAB1 complex. The observed TAK1-dependent phosphorylation and activation of heterotrimeric AMPK also enable the quantification of TAK1 activity in a two-step assay involving preincubation of heterotrimeric AMPK with TAK1 followed by activity determination of AMPK.
TAK1-TAB1-mediated AMPK Activation Requires Hydrophobic Residues of the AMPK Kinase Domain αG-helix
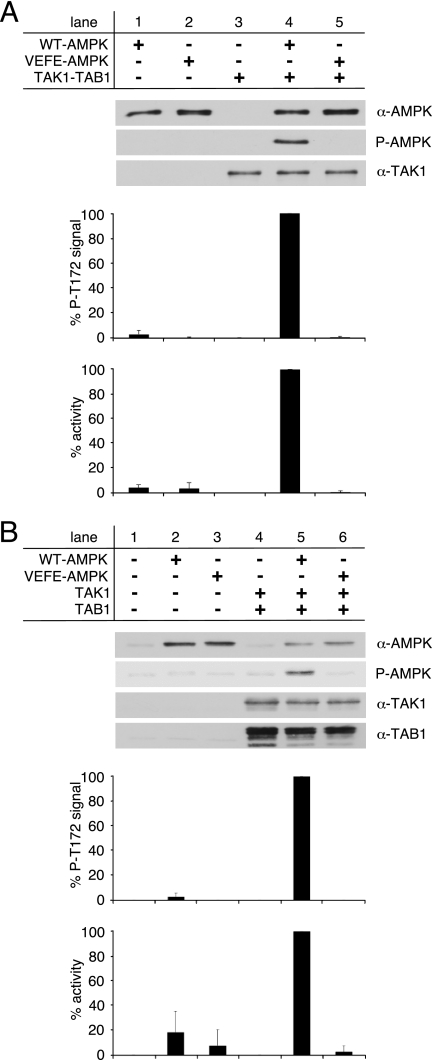
Recently, we showed that the hydrophobic residues Val-219 and Phe-223 of the AMPK kinase domain αG-helix constitute an important distal recognition motif for the AMPK upstream kinases CamKK2 and LKB1 (44). Replacement of both residues, Val-219 and Phe-223, by glutamate strongly impaired activation of AMPK by its upstream kinases. Because TAK1 was suggested as an alternative AMPK upstream kinase (36), we consequently tested whether TAK1-dependent AMPK activation requires the same distal recognition motif. Thus, bacterially expressed WT-TAK1-TAB1 was incubated with either WT-AMPK or the αG-helix-mutant AMPK, α1(V219E,F223E)β1γ1 (VEFE-AMPK), in presence of Mg-ATP. Subsequently, AMPK activation was determined by Western blot analysis of Thr-172 phosphorylation, as well as AMPK activity assay (Fig. 3A). WT-TAK1-TAB1 was completely unable to phosphorylate Thr-172 of VEFE-AMPK and thus did not activate the mutant, whereas Thr-172 phosphorylation and activation of WT-AMPK by WT-TAK1-TAB1 were evident (Fig. 3A, lanes 4 and 5).
FIGURE 3.
Activation of WT- and VEFE-AMPK by active TAK1. A, in vitro activation of recombinant WT- and VEFE-AMPK by bacterially expressed and purified TAK1-TAB1. Western blot analysis was performed using anti-TAK1, anti-AMPKα, and phospho-specific anti-AMPKα Thr-172 (P-AMPK) antibodies. TAK1 background signal (lane 3) was subtracted, and AMPKα phospho-Thr-172 signal intensities were standardized to AMPKα signals. Activity was determined by a HPLC- and SAMS-peptide-based activity assay. Background signal of TAK1 was subtracted, and data were normalized to WT-AMPK activation by TAK1-TAB1 (n = 3; S.D.). B, HeLa cells were transfected with plasmids encoding Myc-AMPKα1 wild-type and VEFE mutant, FLAG-TAK1, and HA-TAB1. Cells were treated with 1 mm hydrogen peroxide for 15 min before lysis. HeLa cell lysates were subjected to Western blot analysis using anti-TAK1, anti-TAB1, anti-AMPKα, and phospho-specific anti-AMPKα Thr-172 (P-AMPK) antibodies. Subsequent to immunoprecipitation using anti-AMPKα1 antibodies, AMPK activity was determined. Respective background signals from mock- and TAK1/TAB1-double transfected cells were subtracted, and data were normalized to the WT-AMPK/TAK1/TAB1 triple-transfected cells (n = 3; S.D.).
It was reported previously that co-transfection of TAK1 and TAB1 in HeLa cells promoted AMPK activation upon stimulation with H2O2 (36). HeLa cells naturally lack LKB1 and only express CamKK2 as an established AMPK upstream kinase. Therefore, HeLa cells represent a relevant system to further evaluate the AMPK αG-helix as a potential recognition site for the TAK1-TAB1 complex. Accordingly, HeLa cells were transfected with combinations of expression constructs encoding FLAG-tagged TAK1 (FLAG-TAK1), HA-tagged TAB1 (HA-TAB1), and Myc-tagged wild-type or (V219E,F223E) AMPK α1-subunit (WT- or VEFE-AMPK). Cells were stimulated with H2O2 and, subsequently, AMPK activation was evaluated by Western blot analysis of Thr-172 phosphorylation and activity determination (Fig. 3B). Single transfection of constructs encoding Myc-tagged VEFE-AMPK α1-subunit in HeLa cells showed a slightly decreased Thr-172 phosphorylation and AMPK activity compared with single transfection of a plasmid encoding Myc-tagged WT-AMPK α1-subunit (Fig. 3B, lanes 2 and 3). This difference in AMPK activity and activation remained statistically insignificant, which likely reflects the observation that CamKK2 is less sensitive to the AMPK αG-helix mutation (44). Co-transfection of WT-AMPK with FLAG-TAK1 and HA-TAB1 led to a strongly increased Thr-172 phosphorylation and AMPK activity (Fig. 3B, lane 5). Intriguingly, co-transfection of VEFE-AMPK with FLAG-TAK1 and HA-TAB1 completely failed to activate AMPK (Fig. 3B, lane 6), which fully correlates with the situation observed in vitro (Fig. 3A). These data suggest that direct activation of AMPK by TAK1-TAB1 in vitro, and presumably also direct activation in cultured cells, is mediated by the hydrophobic residues Val-219 and Phe-223 of the AMPK kinase domain αG-helix.
TAB1 but Not TAB2 Induces Autoactivation of TAK1 in E. coli
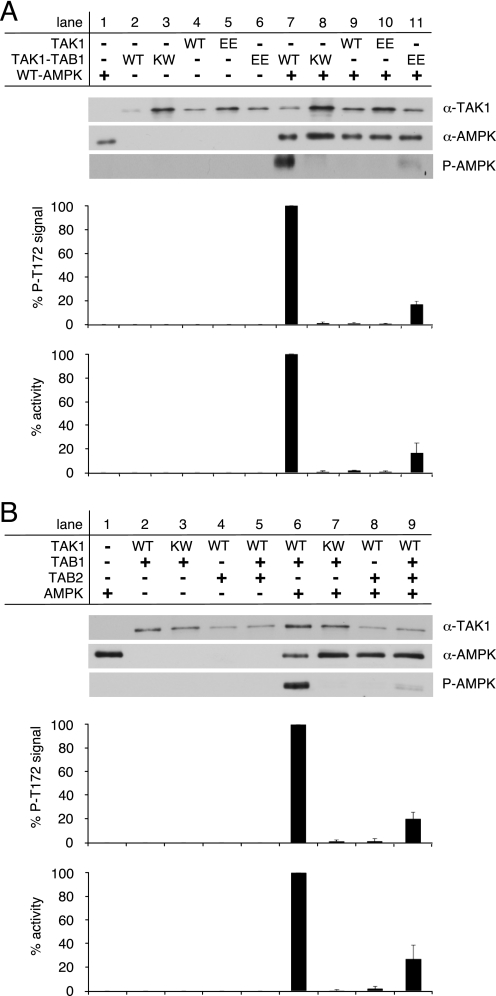
Because we established direct targeting of heterotrimeric AMPK by TAK1-TAB1 in vitro, we employed TAK1-dependent AMPK activation as a tool to evaluate the activity of bacterially expressed and purified TAK1. First, we investigated the particular role of TAB1 in TAK1 autoactivation. Only TAK1-TAB1 but not TAK1 alone was capable of activating WT-AMPK (Fig. 4A, lanes 7 and 9, respectively). Expectedly, the kinase-deficient mutant KW-TAK1-TAB1 did not activate AMPK (Fig. 4A, lane 8). Substitution of the activation segment residues Thr-178 and Thr-184 by glutamate was reported to mimic auto-activated wild-type TAK1 when co-overexpressed with TAB1 in cultured cells (35). Hence, bacterially expressed and purified TAK1(T178E,T184E) and TAK1(T178E,T184E)-TAB1 were analyzed similar to their wild-type counterparts by their ability to phosphorylate and activate AMPK. Substitution of Thr-178 and Thr-184 by glutamate to potentially mimic autophosphorylation of these sites had no effect on TAK1 activity when TAB1 was absent (Fig. 4A, lane 10). However, when co-expressed with TAB1, TAK1(T178E,T184E) conducted AMPK activation even though activity was reduced by 80% compared with WT-TAK1-TAB1 (Fig. 4A, compare lanes 11 and 7, respectively).
FIGURE 4.
Activation of TAK1 requires co-expression of TAB1. A, recombinant WT-AMPK was activated by bacterially expressed TAK1 and TAK1 co-expressed with TAB1 (TAK1-TAB1). Here, the activities of wild-type TAK1 proteins were additionally compared with the activities of TAK1 proteins carrying the phosphorylation site mimicking mutation T178E,T184E (EE) within the TAK1 activation segment. B, activity of recombinant TAK1 bacterially co-expressed with TAB1 and/or TAB2. The kinase-deficient mutant TAK1(K63W)-TAB1 (KW) served as a control. Western blot analysis utilized anti-TAK1, anti-AMPKα, and phospho-specific anti-AMPKα Thr-172 (P-AMPK) antibodies. The AMPKα phospho-Thr-172 signals were quantified after subtraction of respective WT-AMPK (lane 1) and TAK1 background signals (lanes 2–6) and normalization to AMPKα and TAK1 signals. Activity was determined, and respective background activities were subtracted. All activities were standardized to corresponding TAK1 signal intensities as revealed from densitometric Western blot analysis. The data were normalized relative to WT-AMPK activation by WT-TAK1-TAB1 (n = 3; S.D.).
Furthermore, we compared the roles of TAB1 and TAB2 in TAK1 autoactivation (Fig. 4B). Similar to KW-TAK1-TAB1, the WT-TAK1-TAB2 complex also entirely failed to phosphorylate and thus activate AMPK (Fig. 4B, lanes 7 and 8, respectively). If TAK1 was co-expressed with TAB1 and TAB2, AMPK was activated but to a much lesser extend as if WT-TAK1-TAB1 complexes were used (Fig. 4B, compare lanes 6 and 9, respectively).
These results suggest that direct binding of TAB1 is essential to induce significant TAK1 kinase activity, whereas TAB2 alone failed to induce TAK1 autoactivation. These data are also reminiscent of the diverging capability of TAB1 and TAB2 to induce TAK1 activation segment autophosphorylation (Fig. 1A).
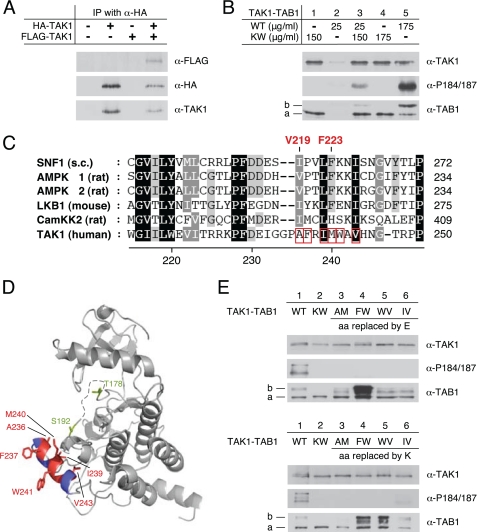
TAK1 Kinase Domain αG-helix Residues Mediate TAB1 Phosphorylation and Intermolecular TAK1 Autophosphorylation
The TAB1-induced TAK1 autoactivation has been reported as an intramolecular (33) and, contradictorily, an intermolecular process (25, 34). As a prerequisite for an intermolecular mechanism, one would predict stable or at least transient TAK1 self-association. To clarify this aspect, we transfected AMPKα subunit double knock-out (α1−/−, α2−/−) MEFs with plasmids encoding HA-TAK1 and FLAG- TAK1, and expression of respective proteins was confirmed by Western blot analysis of the total cell lysates (supplemental Fig. S3). As expected, only HA-TAK1 but not FLAG-TAK1 could be detected in immunoprecipitates from the respective single transfected cells when using an anti-HA antibody for immunoprecipitation (Fig. 5A). Importantly, FLAG-TAK1 clearly co-immunoprecipitated with HA-TAK1 from co-transfected cells, thus suggesting a physical interaction of TAK1 in cultured cells and rather pointing to an intermolecular autoactivation mode. TAK1 autoactivation was further analyzed by incubation of small amounts of bacterially expressed active TAK1-TAB1 with an excess of inactive KW-TAK1-TAB1 in the presence of Mg-ATP (Fig. 5B). Consistent with the results shown in Fig. 1A, KW-TAK1-TAB1 lacked phosphorylated (shifted) TAB1 as well as Thr-184/Thr-187 double phosphorylation (Fig. 5B, lanes 1 and 4). Low amounts of WT-TAK1-TAB1 neither revealed shifted TAB1 signals nor Thr-184/Thr-187 phosphorylation, but at higher loading the expected signals were apparent for both (Fig. 5B, compare lane 2 with 5). Consequently, the clear increase in phosphorylation of Thr-184 and Thr-187 upon incubation of low amounts of WT-TAK1-TAB1 with higher amounts of KW-TAK1-TAB1 reflects intermolecular activation segment phosphorylation (Fig. 5B, lane 3). Additionally, a slight increase in phosphorylated TAB1 species was detected, suggesting also that TAB1 phosphorylation could involve at least partly a bimolecular mechanism (Fig. 5B, lane 3). The intermolecular nature of TAK1 autoactivation raised the question whether the kinase domain αG-helix could also mediate TAK1 self-recognition and thus autoactivation. We previously showed that the hydrophobic AMPK kinase domain αG-helix residues Val-219 and Phe-223 promote AMPK homo-oligomerization (44). The homologous residues are conserved or conservatively replaced in yeast and mammalian AMPK, as well as in LKB1, but less well conserved in CamKK2 and TAK1 (Fig. 5C). In the TAK1 αG-helix region, the corresponding residues Ala-236 and Met-240 are surrounded by hydrophobic residues Phe-237, Ile-239, Trp-241, and Val-243. Focused analysis of the human TAK1 kinase domain structure (Protein Data Bank code 2eva) showed that Phe-237 and Trp-241, but also Ala-236 and Met-240, are well surface-exposed, whereas Ile-239 and Val-243 are situated closer to the core of the large kinase lobe (Fig. 5D). Interestingly, the activation segment phosphorylation site Ser-192 is located in close proximity to the rather hydrophobic αG-helix. The solvent accessibility of the hydrophobic residues of the αG-helix suggests their involvement in protein-protein interaction, and thus these residues could also play a role in intermolecular TAK1 autoactivation. To test this hypothesis, pairs of TAK1 αG-helix residues were either replaced by glutamate or lysine, both of which were expected to disrupt the recognition site as follows: Ala-236/Met-240 (AEME, AKMK), Phe-237/Trp-241 (FEWE, FKWK), Trp-241/Val-243 (WEVE, WKVK), and Ile-239/Val-243 (IEVE, IKVK). The respective TAK1-TAB1 mutant complexes were bacterially expressed, and TAK1 autoactivation was evaluated by Western blot analysis using antibodies recognizing TAK1, TAB1, and double phosphorylation at Thr-184 and Thr-187 (Fig. 5E). Intriguingly, replacement of αG-helix residues by either glutamate or lysine completely abolished phosphorylation of activation segment residues, thus further confirming intermolecular autophosphorylation of the Thr-184 and Thr-187 sites. Only IKVK-TAK1-TAB1 showed a weak TAK1 phosphorylation, but this was strongly reduced if compared with wild type (Fig. 5E, lower panel, lane 6), thus indicating replacement of these residues with lysine does not completely disrupt the αG-helix interface. Accordingly, the IKVK mutant also revealed a shifted TAB1 species, whereas the observed TAB1 shift was markedly different from WT in all other TAK1-TAB1 mutant complexes. More precisely, the phosphorylation-dependent shift of TAB1 was completely absent in kinase-deficient (KW) and AKMK mutants (Fig. 5E, upper panel, lane 2, lower panel, lanes 2 and 3). The overall signal intensity corresponding to the TAB1 protein, including the shifted species, was much stronger in the FEWE, FKWK, and WKVK mutants if compared with WT (Fig. 5E, upper panel, lane 4, lower panel, lanes 4 and 5, versus lane 1 of both panels). These results indicate involvement of the αG-helix in TAB1 association and phosphorylation. Notably, the TAB1 protein showed an intermediate, partly shifted species in AEME, WEVE, and IEVE mutant TAK1-TAB1 complexes (Fig. 5E, upper panel, lane 3, 5, and 6, lower panel, lane 3), which could indicate that TAB1 autophosphorylation is a sequential process involving multiple distinct sites.
FIGURE 5.
Involvement of TAK1 αG-helix residues in TAB1 and intermolecular TAK1 phosphorylation. A, AMPKα subunit double knock-out (α1−/−, α2−/−) MEFs were transfected with plasmids encoding HA-TAK1 and FLAG-TAK1. Immunoprecipitation (IP) from cell lysates was performed using an anti-HA antibody. Immunoprecipitates were analyzed by Western blot with anti-FLAG, anti-HA, and anti-TAK1 antibodies. B, six times excess of bacterially expressed kinase-deficient KW-TAK1-TAB1 (150 μg/ml) was incubated with WT-TAK1-TAB1 (25 μg/ml). Intermolecular TAK1 activation segment phosphorylation was monitored by Western blot analysis using anti-TAK1, anti-TAB1, and phospho-specific anti-TAK1-Thr(P)-184/187 (α-P184/187) antibodies. 175 μg/ml KW-TAK1-TAB1 and WT-TAK1-TAB1 were used as a control. C, alignment of the αG-helix region sequences of Snf1 from Saccharomyces cerevisiae, AMPK α1 and α2 subunits, and CamKK2 from Rattus norvegicus, LKB1 from Mus musculus, and TAK1 from human (GenBankTM accession numbers P06782, AAC52355, CAA82620, NP_112628, NP_035622, and O43318). Residues highlighted in black show conservation in all sequences, whereas residues highlighted in gray show partial conservation. The position of the hydrophobic AMPK kinase domain αG-helix residues Val-219 and Phe-223 is specified. Hydrophobic TAK1 kinase domain αG-helix residues Ala-236, Phe-237, Ile-239, Met-240, Trp-241, and Val-243 are boxed in red. D, structural analysis of the TAK1 kinase domain (Protein Data Bank code 2eva). Surface-exposed location of the αG-helix with hydrophobic residues highlighted in red (hydrophobicity scale >0.2 (64)) and rather hydrophilic residues highlighted in blue (hydrophobicity scale < 0.0 (64)) is shown. Structurally unresolved activation segment residues 179–190 are represented by a dashed line. Activation segment phosphorylation sites Ser-192 and Thr-178 are shown in green. E, upper panel, Western blot analysis of bacterially expressed WT- and KW-TAK1-TAB1 as well as TAK1-TAB1 mutants with hydrophobic αG-helix residues Ala-236, Phe-237, Ile-239, Met-240, Trp-241, and Val-243 replaced by glutamate-TAK1(A236E, M240E)-TAB1 (AM), TAK1(F237E, W241E)-TAB1 (FW), TAK1(W241E, V243E)-TAB1 (WV), and TAK1(I239E, V243E)-TAB1 (IV). Lower panel as in upper panel, but the corresponding αG-helix residues are substituted by lysine instead of glutamate. Line a, unmodified TAB1 species; line b, shifted TAB1 species.
To restore αG-helix-mediated TAK1 self-recognition, we incubated the glutamate-substituted αG-helix mutants with the corresponding lysine-substituted counterpart, thus creating electrostatic attraction between the two different mutants. A similar approach successfully restored recognition of AMPK by LKB1 (44), but failed to rescue TAK1 autoactivation (data not shown). Initially, we assumed that this failure was due to the complete lack of TAK1 activity in both mutants. Because substitution of Thr-178 and Thr-184 with glutamate resulted in some fraction of wild-type activity in TAK1-TAB1 complexes (Fig. 4A), we introduced the same mutations into the activation segment of the lysine-substituted αG-helix mutants, thus hoping to create sufficient basal activity for TAK1 autoactivation. The FKWK-TAK1(T178E,T184E)-TAB1, WKVK-TAK1(T178E,T184E)-TAB1, and IKVK-TAK1(T178E,T184E)-TAB1 complexes again showed significant TAB1 autophosphorylation, whereas TAB1 shifted to a lesser extent in AKMK-TAK1(T178E,T184E)-TAB1 complexes (supplemental Fig. S4A). These data are consistent with results shown in Fig. 5E. However, also all of the lysine-substituted αG-helix mutant TAK1(T178E,T184E)-TAB1 complexes failed to phosphorylate Thr-184/Thr-187 of their corresponding glutamate-substituted αG-helix mutants AEME-TAK1-TAB1, FEWE-TAK1-TAB1, WEVE-TAK1-TAB1, and IEVE-TAK1-TAB1, respectively (supplemental Fig. S4B). Thus, we were unable to create a functional rescue based on electrostatic attraction of the TAK1 αG-helix residues, potentially indicating that additional functions important for TAK1 autoactivation apart from TAK1 self-recognition are fulfilled by the αG-helix. Taken together, specific hydrophobic residues of the TAK1 kinase domain αG-helix likely mediate the intermolecular autophosphorylation of Thr-184 and Thr-187 and play a crucial role in TAB1 phosphorylation during the TAK1 autoactivation process.
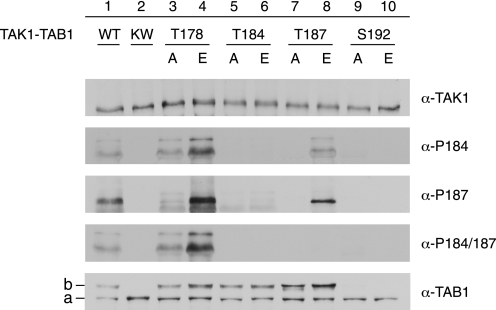
Sequential Phosphorylation Events in TAB1-induced TAK1 Autoactivation
The TAK1 activation segment residues Thr-178, Thr-184, Thr-187, and Ser-192 have been identified as important autophosphorylation sites during TAB1-induced TAK1 autoactivation (19, 33–35), whereas their specific roles and functional interplay in this process remained unclear. Therefore, each residue was individually replaced by either alanine or glutamate (T178A; T178E; T184A; T184E; T187A; T187E; S192A; S192E). The respective mutant TAK1-TAB1 complexes were bacterially expressed and purified and then subjected to Western blot analysis using antibodies recognizing TAK1, TAB1, single phosphorylation at Thr-184 or Thr-187, as well as double phosphorylation at Thr-184/Thr-187 (Fig. 6). Most activation segment residue mutants, except the Ser-192 mutants, performed TAB1 phosphorylation similar to WT-TAK1, and this was apparently independent from the detected phosphorylation status at Thr-184 or Thr-187. Thus, TAB1 phosphorylation may precede phosphorylation of Thr-178, Thr-184, and Thr-187. The S192A and S192E mutations precluded both the phosphorylation of Thr-184 or Thr-187 and the TAB1 shift (Fig. 6, lanes 9 and 10). Possibly, the replacement of Ser-192 with glutamate failed to effectively mimic phosphorylation, but this residue is evidently critical for mediation of TAB1 phosphorylation and, by this or other means, could affect TAK1 autophosphorylation. Furthermore, the T178E-TAK1-TAB1 mutant complex revealed elevated Thr-187 phosphorylation, whereas replacement of Thr-178 by alanine reduced phosphorylation of Thr-187 compared with WT-TAK1-TAB1 (Fig. 6, lanes 3 and 4). Hence, we assume that Thr-178 phosphorylation predates autophosphorylation of Thr-187. Furthermore, unlike T187A-TAK1-TAB1, which apart from phosphorylation at Thr-187 also lacked phosphorylation at Thr-184, the T187E-TAK1-TAB1 was able to autophosphorylate at Thr-184 (Fig. 6, lanes 7 and 8). Thus, phosphorylation at Thr-187 may precede autophosphorylation at Thr-184. The anti-TAK1-Thr(P)-187 antibody but not the anti-TAK1-Thr(P)-184/187 antibody recognized their respective epitope when Thr-187 was substituted by glutamate. Such impaired epitope recognition might also explain why both mutants with Thr-184 replaced by either alanine or glutamate did not show any phosphorylation (Fig. 6, lanes 5 and 6).
FIGURE 6.
Sequence of phosphorylation events within the TAK1 kinase domain activation segment. Western blot analysis of bacterially expressed WT-TAK1-TAB1, KW-TAK1-TAB1 TAK1-TAB1 mutants carrying single amino acid mutations at phosphorylation sites within the TAK1 kinase domain activation segment: TAK1(T178A)-TAB1, TAK1(T178E)-TAB1, TAK1(T184A)-TAB1, TAK1(T184E)-TAB1, TAK1(T187A)-TAB1, TAK1(T187E)-TAB1, TAK1(S192A)-TAB1, and TAK1(S192E)-TAB1. Anti-TAK1, anti-TAB1, and phospho-specific anti-TAK1-Thr(P)-184 (α-P184), anti-TAK1-Thr(P)-187 (α-P187), and anti-TAK1-Thr(P)-184/187 (α-P184/187) antibodies were applied. Line a, unmodified TAB1 species; line b, shifted TAB1 species.
Taken together, TAB1-induced TAK1 autoactivation involves sequential phosphorylation of the four activation segment residues. According to our data, autoactivation starts with phosphorylation of Ser-192, which is also required for TAB1 autophosphorylation, and continues with Thr-178, which augments autoactivation, and finally allows for phosphorylation at Thr-187 and Thr-184.
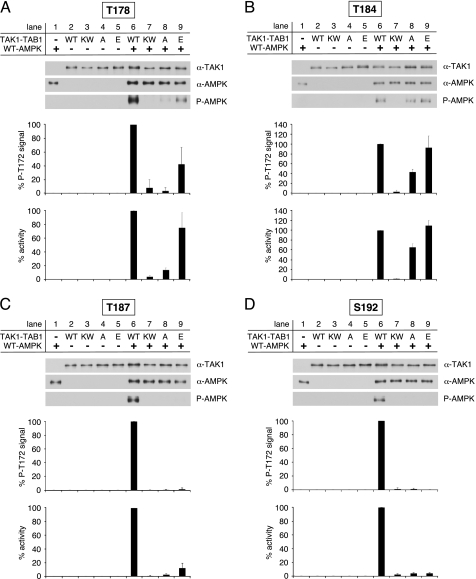
Role of Thr-178, Thr-184, Thr-187, and Ser-192 Autophosphorylation in TAK1 Autoactivation
To assess the activity of bacterially expressed TAK1-TAB1 mutants carrying single amino acid substitutions in the activation segment residues Thr-178, Thr-184, Thr-187, and Ser-192, we analyzed their ability to phosphorylate and activate WT-AMPK (Fig. 7, A–D). In agreement with the observed disability to promote the TAB1 shift, as well as Thr-184 and Thr-187 autophosphorylation, the replacement of Ser-192 with either alanine or glutamate prevented detectable TAK1 activity (Fig. 7D, lanes 6, 8, and 9). Furthermore, the substitution of Thr-178 and Thr-187 by alanine also resulted in a complete loss of TAK1 activity comparable with KW-TAK1-TAB1 (Fig. 7, A and C, lanes 6 and 8). Hence, besides Ser-192 phosphorylation, the phosphorylation of Thr-178 and Thr-187 was indispensable for TAB1-induced TAK1 autoactivation. Interestingly, T184A-TAK1-TAB1 was still capable of activating AMPK, albeit TAK1 activity of the mutant was lower if compared with the WT-TAK1-TAB1 complex (Fig. 7B, lanes 6–8). Substitution of Thr-184 by glutamate led to increased TAK1 activity similar to WT (Fig. 7B, lanes 6 and 9). Hence, these data suggest that phosphorylation of Thr-184 is not essential for TAK1 activation, although phosphorylation of this residue further enhances TAK1 activity. Replacement of Thr-187 by glutamate was sufficient to promote Thr-184 phosphorylation (Fig. 6, lane 8) but was apparently unable to efficiently mimic Thr-187 phosphorylation, as mutant TAK1 activity was much lower than WT (Fig. 7C, lane 9). In contrast, the T178E-TAK1-TAB1 complex was capable of activating AMPK similar to WT (Fig. 7A, lane 9).
FIGURE 7.
Activity of TAK1-TAB1 activation segment mutants. A, in vitro activation of recombinant WT-AMPK by bacterially expressed WT- and KW-TAK1-TAB1 as well as TAK1(T178A)-TAB1 and TAK1(T178E)-TAB1. B–D as in A but analyzing TAK1(T184A)-TAB1 and TAK1(T184E)-TAB1, TAK1(T187A)-TAB1 and TAK1(T187E)-TAB1, and TAK1(S192A)-TAB1 and TAK1(S192E)-TAB1, respectively. Western blot analysis was performed using anti-TAK1, anti-AMPKα, and phospho-specific anti-AMPKα Thr-172 (P-AMPK) antibodies. To quantify AMPKα Thr-172 phosphorylation, the respective background signals from nonactivated AMPK (lane 1) and TAK1-TAB1 (lane 2–5) were subtracted, and phospho-Thr-172 signal intensities were normalized to AMPKα and TAK1 signals. Activity was determined, and respective background activities (lane 1–5) were subtracted. All activities were standardized to TAK1 signal intensities as revealed from densitometric Western blot analysis. Data were normalized relative to WT-AMPK activation by WT-TAK1-TAB1 (n = 3; S.D.).
In summary, TAB1-induced TAK1 autoactivation requires phosphorylation of activation segment sites Thr-178, Thr-187, and Ser-192. Although clearly promoting TAK1 activity, phosphorylation of Thr-184 appears not to be mandatory.
DISCUSSION
In this study, we established a bacterial expression system for TAK1-containing complexes and a quantitative TAK1 activity assay that is based on the ability of TAK1 to activate AMPK. Using the cell-free and thus defined in vitro system, we shed new light on the TAK1 autoactivation mechanism with the focus on the following: (i) the TAK1-binding proteins TAB1 and TAB2, (ii) the TAK1 kinase domain αG-helix, and (iii) the autophosphorylation of the activation segment residues Thr-178, Thr-184, Thr-187, as well as Ser-192.
First, TAK1 co-purified with either TAB1 or TAB2 when co-expressed in E. coli. The capability of TAB1 to promote TAK1 autoactivation in cells is well established (13, 19, 33). However, these results mainly resulted from co-expression experiments in yeast, mammalian cells, or baculovirus, and therefore the cellular milieu of eukaryotes potentially impacted on the results prior to analysis. We show here the bacterial co-expression of TAK1 and TAB1 was essential and sufficient for TAK1 autoactivation. However, this does not exclude the possibility that other unknown cellular components such as phosphatases could suppress the activity of endogenous TAK1-TAB1 complexes, as TAK1 in association with TAB1 remained nonphosphorylated and inactive in the absence of interleukin-1 stimulation (33). Bacterial co-expression of TAK1 with TAB2 resulted in TAK1 complexes lacking both kinase activity and autophosphorylation of the TAK1 activation segment residues Thr-184 and Thr-187, which is in accordance with earlier studies in yeast (13, 14). Further cellular components, e.g. to tumor necrosis factor-α receptor-associated factor family members or nonconjugated free Lys-63 polyubiquitin chains, are involved in TAB2-dependent TAK1 autoactivation in vivo (3, 14, 23, 24, 25). In contrast, our in vitro setting lacked respective factors that could explain the absence of kinase activity and autophosphorylation of bacterially expressed TAK1-TAB2 complexes, thus underlining the importance of corresponding cellular components in TAB2-mediated TAK1 activation. TAB2 and TAB1 reportedly bind at distinct sites of TAK1, the catalytic and carboxyl-terminal domain, respectively (13). When co-expressed with both TAK1-binding proteins TAB1 and TAB2 in bacteria, the resulting TAK1 complex contained TAB2 but only few TAB1 and accordingly showed low activity, which requires further elucidation. In addition, we show here self-association of TAK1 as revealed by co-immunoprecipitation of HA-TAK1 and FLAG-TAK1 from transfected cell culture. In support of this notion, TAK1 autoactivation involved intermolecular transphosphorylation of the kinase activation segment (Fig. 5B), thus implying TAK1 self-association. Notably, apoptosis signal-regulating kinase 1 (ASK1) and mixed lineage kinase-3 (MLK-3) auto-activate intermolecularly (55, 56), suggesting similarities in activation of the different MAP3Ks.
Second, we identified a new regulatory element of TAB1-induced TAK1 autoactivation, namely the TAK1 kinase domain αG-helix located in subdomain X within the large kinase lobe. As evidenced by the molecular structure of TAK1, the αG-helix possesses a large solvent-accessible surface with mainly hydrophobic residues Ala-236, Phe-237, Ile-239, Met-240, Trp-241, and Val-243, thus being available for putative protein-protein interactions. Pairwise substitution of these residues by either glutamate or lysine (Ala-236/Met-240 (AEME, AKMK), Phe-237/Trp-241 (FEWE, FKWK), Trp-241/Val-243 (WEVE, WKVK), and Ile-239/Val-243 (IEVE, IKVK)) abolished Thr-184 and Thr-187 phosphorylation of the corresponding bacterially expressed TAK1-TAB1 mutants, except IKVK-TAK1-TAB1, which showed reduced Thr-184/Thr-187 phosphorylation. These data substantiate a crucial role for the αG-helix in mediating TAK1 autophosphorylation and further emphasize the intermolecular nature of this process. Analysis of the available structural data of the human TAK1 kinase domain revealed so far unrecognized crystal contacts between symmetry-related monomers, mainly involving the hydrophobic residues Phe-237, Trp-241, and Met-240 (supplemental Fig. S5). Although the physiological significance of this interface is questionable, it further indicates the potential of αG-helix residues to provide a docking site for intermolecular TAK1 autophosphorylation. Furthermore, the critical involvement of subdomain X residues in signaling and autophosphorylation has been shown in two other MAP3Ks, namely MEKK1 and MEKK2 (57–59), which points to conserved functions of the αG-helix within this family of kinases. Additionally, we provide evidence that the TAK1 kinase domain αG-helix also mediates the interplay between TAK1 and its regulatory binding protein TAB1, because pairwise replacement of Phe-237/Trp-241 by glutamate and of Trp-241/Val-243 by either lysine or glutamate increased TAB1 phosphorylation relative to the wild-type TAK-TAB1 complex (gain-of-function). In contrast, AEME-TAK1-TAB1, WEVE-TAK1-TAB1, IEVE-TAK1-TAB1, and AKMK-TAK1-TAB1 showed impaired TAB1 phosphorylation (loss-of-function). These observed differences in TAB1 phosphorylation levels did not correlate with TAK1 autophosphorylation, suggesting a direct involvement of the αG-helix in TAB1 positioning via electrostatic interactions prior to TAK1-dependent phosphorylation. Indeed, a charged residue, Arg-238, is situated right between Phe-237 and Trp-241 on the outer face of the αG-helix.
Third, our data define a chronological sequence of the molecular events governing TAB1-induced TAK1 autoactivation. As discussed above, TAB1 binding to TAK1 is required and sufficient to cause TAK1 autoactivation. In the absence of detectable Thr-184/Thr-187 double phosphorylation, the mutant complexes FKWK-TAK1-TAB1, FEWE-TAK1-TAB1, and WKVK-TAK1-TAB1 showed increased TAB1 phosphorylation if compared with wild type (Fig. 5E), suggesting that phosphorylation of TAB1 precedes phosphorylation of the activation segment residues Thr-184 and Thr-187. Furthermore, the replacement of Thr-178, Thr-184, and Thr-187 by alanine or glutamate did not affect TAB1 phosphorylation (Fig. 6). Hence, phosphorylation of TAB1 was independent of the phosphorylation status within the activation segment of the respective TAK1-TAB1 mutant. Importantly, the exchange of Ser-192 to either alanine or glutamate totally abolished the presence of phosphorylated TAB1, indicating that TAB1 phosphorylation requires Ser-192 but readily proceeds in the absence of other activation segment phosphorylations. In addition, the data allude to glutamate not sufficiently mimicking phosphorylation at Ser-192, as observed earlier (33). The Ser-192 residue was suggested to be an autophosphorylation site (33) and, notably, is located in very close proximity to the TAK1 kinase domain αG-helix (Fig. 5D). As discussed above, the αG-helix mediates TAK1-induced TAB1 phosphorylation, and therefore, phosphorylation at Ser-192 will rather likely impact on the position of the αG-helix within the TAK1-TAB1 complex, which potentially could facilitate interactions between TAK1 and TAB1 and promote TAK1-mediated TAB1 phosphorylation. Substitution of the activation segment residues Thr-178, Thr-184, and Thr-187 by glutamate or alanine revealed a complex pattern of either inhibition or promotion of phosphorylation at the remaining sites that were amenable to probing by Western blotting (Fig. 6). These results suggest sequential autophosphorylation of the activation segment residues. Such turning on by sequentially ordered autophosphorylation of multiple activation segment residues has been studied, e.g. for FGF-receptor-1 kinase (60), thus suggesting a more widespread mechanism among kinases.
Taking together the results (Figs. 6 and 7), we propose that autophosphorylation at Thr-178 precedes Thr-187. Thereafter, TAK1 activity may be further stimulated by phosphate incorporation at Thr-184. However, not all data may appear fully in accord with this proposed scheme.
First, as shown in Fig. 6 (lanes 1, 3, and 4), alanine substitution of Thr-178 is suppressing Thr-187 phosphorylation if compared with the T178E and WT, which indicates Thr-178 phosphorylation could stimulate the modification at Thr-187. The T178A mutation also suppresses Thr-184 phosphorylation if compared with the T178E mutant. Only the T178A mutant is incapable of inhibiting phosphorylation at Thr-184 in relation to WT. Thus, Thr-178 phosphorylation is permissive for modification at Thr-187 and Thr-184, but the lack of phosphate at Thr-178 does not preclude phosphorylation at Thr-184/Thr-187. Therefore, the suggested order of events might not be strict.
Second, the T187A mutation completely abolishes phosphorylation at Thr-184, whereas the T187E mutant shows detectable anti-TAK1-Thr(P)-184 signal levels, thus implying that Thr-187 phosphorylation could finally facilitate autophosphorylation of Thr-184. Also, the T184A mutant retains considerable activity, whereas the other alanine substitutions almost completely abolished TAK1 activity (Fig. 7, A–D, lanes 8). At least these data indicate that Thr-184 phosphorylation is not mandatory for antecedent parts of the autoactivation process, which may immediately put it in ultimate position. However, if the final step is indeed phosphorylation at Thr-184, we would expect to see phosphorylation signals at Thr-187 in the Thr-184 mutants. The best answer we can give to this apparent discrepancy is the unknown properties of the anti-TAK1-Thr(P)-187 antibody. We presume the absence of any Thr-187 phosphorylation signal for the T184A and T184E mutants might be due to the altered protein sequence in close proximity to Thr-187, which could impair epitope recognition by the anti-TAK1-Thr(P)-187 antibodies. The notion that Thr-184 phosphorylation is occurring at last is also partly backed by Western blot analyses of the size exclusion chromatography elution fractions of WT-TAK1-TAB1 complexes (Fig. 1B), which show phosphorylation of Thr-184 only in conjunction with Thr-187, whereas Thr-187 phosphorylation was observed in a broader range of elution fractions. Furthermore, the final phosphorylation at Thr-184 is in line with earlier observations from co-transfection experiments in cultured cells, where substitution of Thr-178, Thr-187, and Ser-192 by alanine resulted in a complete loss of TAK1 activity (33–35), again in full accordance with our data (Fig. 7, A, C and D). Hence, Thr-184 phosphorylation clearly enhances TAK1 activity but might not be essential for TAB1-induced TAK1 activation.
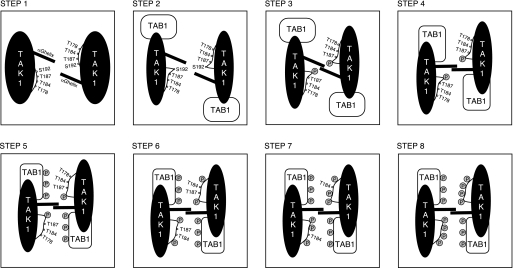
According to our own and published data, the following model for TAB1-induced TAK1 autoactivation emerges (Fig. 8): TAB1 binds to TAK1, thus facilitating a conformational change of TAK1 and subsequent phosphorylation of Ser-192. Consequently, the αG-helix repositions, thereby promoting phosphorylation of TAB1. Next, the TAK1 activation segment liberates, permitting intermolecular autophosphorylation, which is facilitated by hydrophobic interaction of αG-helix residues. Sequential phosphorylation of the remaining activation segment phosphorylation sites occurs in the following order: 1) Thr-178, 2) Thr-187, and 3) Thr-184.
FIGURE 8.
Model of sequential TAB1-induced TAK1 autoactivation. Step 1, TAK1 activation segment autophosphorylation sites Thr-178, Thr-184, Thr-187, and Ser-192 are not accessible in the absence of TAB1. Step 2, upon TAB1 binding to the TAK1 catalytic domain, a conformational change is induced in TAK1, leading to improved accessibility of Ser-192. Step 3, Ser-192 autophosphorylation is mediated by an intermolecular mechanism, which involves hydrophobic residues of the TAK1 kinase domain αG-helix. Step 4, Ser-192 phosphorylation re-positions the αG-helix, thus facilitating the further functional interplay between TAB1 and TAK1. Step 5, TAB1 gets phosphorylated, which results in improved accessibility of the remaining activation segment autophosphorylation sites Thr-178, Thr-184, and Thr-187. Steps 6–8, completion of TAB1-induced TAK1 autoactivation by αG-helix-mediated sequential intermolecular autophosphorylation of Thr-178, Thr-187, and finally Thr-184.
In this study, we took advantage of TAK1 as an upstream kinase of AMPK. Momcilovic et al. (36) identified TAK1 as an SNF1-activating kinase in a yeast genetic screen and went on to show that a highly truncated constitutively active TAK1-TAB1 fusion protein resulting from baculoviral expression was capable of phosphorylating the isolated recombinant AMPK catalytic domain at Thr-172 in vitro. Here, we show activation of full-length heterotrimeric mammalian AMPK by full-length mammalian TAK1-TAB1 in vitro and exploit the findings to determine TAK1 activity quantitatively (Fig. 2). As we reported recently, Val-219 and Phe-223 of the AMPK kinase domain αG-helix are critically involved in AMPK activation by providing a distal recognition site for its AMPK upstream kinases, i.e. LKB1 and CamKK2 (44). As shown here, direct AMPK activation by TAK1 is also mediated via Val-219 and Phe-223 of the AMPK kinase domain αG-helix in vitro and in cultured cells (Fig. 3). Important evidence for the physiological role of TAK1-dependent AMPK activation was presented in a recent study showing that activation of AMPK by tumor necrosis factor-related apoptosis-inducing ligand was refractory to depletion of LKB1 and CamKK2 and dependent on TAK1 (61). Taking these findings together, we consider TAK1 as a genuine third AMPK upstream kinase, in addition to LKB1 and CamKK2.
In summary, we elucidated molecular details of the TAK1 autoactivation mechanism, enabling us to present a model for the sequence of events causing TAK1 to fully activate. Our studies also underline the importance of the kinase domain αG-helix that provides a docking site for self-association of TAK1 as shown in this study. Work by us and others have demonstrated essential roles of the αG-helix for only few kinases of the Ca2+/calmodulin-dependent kinase, cAMP-dependent protein kinase/protein kinase G/protein kinase C, and sterile kinase group (44, 62, 63), which could be studied in a wider selection of kinases, because interference with this docking motif potentially offers a new therapeutic perspective.
Supplementary Material
Acknowledgments
We thank all members of our laboratories for help and S. Werner (ETH Zurich) for critical reading of the manuscript. D. G. Hardie (Dundee, United Kingdom) is acknowledged for the provision of antibodies and B. Viollet (Paris, France) for providing immortalized (α1−/− and α2−/−) MEF cells.
This work was supported in part by the Swiss National Science Foundation Grant 3100A0-114137, the European Union FP6 Contract LSHM-CT-2004-005272 (EXGENESIS), and Graduate Training Fellowships ETHIIRA 36/05-3 and 32/05-3 (for R. S. and R. F. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- MAP3K
- mitogen-activated protein kinase kinase kinase
- AMPK
- AMP-activated protein kinase
- CamKK2
- Ca2+/calmodulin-dependent kinase kinase 2
- HPLC
- high pressure liquid chromatography
- MEF
- mouse embryo fibroblast
- MEKK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase
- STRAD
- STE20-related adaptor protein
- TAB
- TAK1-binding protein
- WT
- wild type
- HA
- hemagglutinin
- fwd
- forward
- rev
- reverse.
REFERENCES
- 1.Yamaguchi K., Shirakabe K., Shibuya H., Irie K., Oishi I., Ueno N., Taniguchi T., Nishida E., Matsumoto K. (1995) Science 270, 2008–2011 [DOI] [PubMed] [Google Scholar]
- 2.Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. (1999) Nature 398, 252–256 [DOI] [PubMed] [Google Scholar]
- 3.Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 4.Takaesu G., Surabhi R. M., Park K. J., Ninomiya-Tsuji J., Matsumoto K., Gaynor R. B. (2003) J. Mol. Biol. 326, 105–115 [DOI] [PubMed] [Google Scholar]
- 5.Lee J., Mira-Arbibe L., Ulevitch R. J. (2000) J. Leukocyte Biol. 68, 909–915 [PubMed] [Google Scholar]
- 6.Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 1087–1095 [DOI] [PubMed] [Google Scholar]
- 7.Moriguchi T., Kuroyanagi N., Yamaguchi K., Gotoh Y., Irie K., Kano T., Shirakabe K., Muro Y., Shibuya H., Matsumoto K., Nishida E., Hagiwara M. (1996) J. Biol. Chem. 271, 13675–13679 [DOI] [PubMed] [Google Scholar]
- 8.Sakurai H., Miyoshi H., Toriumi W., Sugita T. (1999) J. Biol. Chem. 274, 10641–10648 [DOI] [PubMed] [Google Scholar]
- 9.Shirakabe K., Yamaguchi K., Shibuya H., Irie K., Matsuda S., Moriguchi T., Gotoh Y., Matsumoto K., Nishida E. (1997) J. Biol. Chem. 272, 8141–8144 [DOI] [PubMed] [Google Scholar]
- 10.Meneghini M. D., Ishitani T., Carter J. C., Hisamoto N., Ninomiya-Tsuji J., Thorpe C. J., Hamill D. R., Matsumoto K., Bowerman B. (1999) Nature 399, 793–797 [DOI] [PubMed] [Google Scholar]
- 11.Ishitani T., Ninomiya-Tsuji J., Nagai S., Nishita M., Meneghini M., Barker N., Waterman M., Bowerman B., Clevers H., Shibuya H., Matsumoto K. (1999) Nature 399, 798–802 [DOI] [PubMed] [Google Scholar]
- 12.Inagaki M., Omori E., Kim J. Y., Komatsu Y., Scott G., Ray M. K., Yamada G., Matsumoto K., Mishina Y., Ninomiya-Tsuji J. (2008) J. Biol. Chem. 283, 33080–33086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibuya H., Yamaguchi K., Shirakabe K., Tonegawa A., Gotoh Y., Ueno N., Irie K., Nishida E., Matsumoto K. (1996) Science 272, 1179–1182 [DOI] [PubMed] [Google Scholar]
- 14.Takaesu G., Kishida S., Hiyama A., Yamaguchi K., Shibuya H., Irie K., Ninomiya-Tsuji J., Matsumoto K. (2000) Mol. Cell 5, 649–658 [DOI] [PubMed] [Google Scholar]
- 15.Cheung P. C., Nebreda A. R., Cohen P. (2004) Biochem. J. 378, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishitani T., Takaesu G., Ninomiya-Tsuji J., Shibuya H., Gaynor R. B., Matsumoto K. (2003) EMBO J. 22, 6277–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prickett T. D., Ninomiya-Tsuji J., Broglie P., Muratore-Schroeder T. L., Shabanowitz J., Hunt D. F., Brautigan D. L. (2008) J. Biol. Chem. 283, 19245–19254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conner S. H., Kular G., Peggie M., Shepherd S., Schüttelkopf A. W., Cohen P., Van Aalten D. M. (2006) Biochem. J. 399, 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakurai H., Miyoshi H., Mizukami J., Sugita T. (2000) FEBS Lett. 474, 141–145 [DOI] [PubMed] [Google Scholar]
- 20.Ono K., Ohtomo T., Sato S., Sugamata Y., Suzuki M., Hisamoto N., Ninomiya-Tsuji J., Tsuchiya M., Matsumoto K. (2001) J. Biol. Chem. 276, 24396–24400 [DOI] [PubMed] [Google Scholar]
- 21.Sakurai H., Nishi A., Sato N., Mizukami J., Miyoshi H., Sugita T. (2002) Biochem. Biophys. Res. Commun. 297, 1277–1281 [DOI] [PubMed] [Google Scholar]
- 22.Brown K., Vial S. C., Dedi N., Long J. M., Dunster N. J., Cheetham G. M. (2005) J. Mol. Biol. 354, 1013–1020 [DOI] [PubMed] [Google Scholar]
- 23.Takaesu G., Ninomiya-Tsuji J., Kishida S., Li X., Stark G. R., Matsumoto K. (2001) Mol. Cell. Biol. 21, 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishida S., Sanjo H., Akira S., Matsumoto K., Ninomiya-Tsuji J. (2005) Genes Cells 10, 447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Z. P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., Chen Z. J. (2009) Nature 461, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besse A., Lamothe B., Campos A. D., Webster W. K., Maddineni U., Lin S. C., Wu H., Darnay B. G. (2007) J. Biol. Chem. 282, 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoji S., Titani K., Demaille J. G., Fischer E. H. (1979) J. Biol. Chem. 254, 6211–6214 [PubMed] [Google Scholar]
- 28.Alessi D. R., Saito Y., Campbell D. G., Cohen P., Sithanandam G., Rapp U., Ashworth A., Marshall C. J., Cowley S. (1994) EMBO J. 13, 1610–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson L. N., Noble M. E., Owen D. J. (1996) Cell 85, 149–158 [DOI] [PubMed] [Google Scholar]
- 30.Raingeaud J., Whitmarsh A. J., Barrett T., Dérijard B., Davis R. J. (1996) Mol. Cell. Biol. 16, 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein S. C., Woods A., Jones N. A., Davison M. D., Carling D. (2000) Biochem. J. 345, 437–443 [PMC free article] [PubMed] [Google Scholar]
- 32.Pike A. C., Rellos P., Niesen F. H., Turnbull A., Oliver A. W., Parker S. A., Turk B. E., Pearl L. H., Knapp S. (2008) EMBO J. 27, 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishimoto K., Matsumoto K., Ninomiya-Tsuji J. (2000) J. Biol. Chem. 275, 7359–7364 [DOI] [PubMed] [Google Scholar]
- 34.Singhirunnusorn P., Suzuki S., Kawasaki N., Saiki I., Sakurai H. (2005) J. Biol. Chem. 280, 7359–7368 [DOI] [PubMed] [Google Scholar]
- 35.Yu Y., Ge N., Xie M., Sun W., Burlingame S., Pass A. K., Nuchtern J. G., Zhang D., Fu S., Schneider M. D., Fan J., Yang J. (2008) J. Biol. Chem. 283, 24497–24505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momcilovic M., Hong S. P., Carlson M. (2006) J. Biol. Chem. 281, 25336–25343 [DOI] [PubMed] [Google Scholar]
- 37.Xie M., Zhang D., Dyck J. R., Li Y., Zhang H., Morishima M., Mann D. L., Taffet G. E., Baldini A., Khoury D. S., Schneider M. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17378–17383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann D., Schlattner U., Wallimann T. (2003) Biochem. Soc. Trans. 31, 169–174 [DOI] [PubMed] [Google Scholar]
- 39.Hardie D. G., Hawley S. A., Scott J. W. (2006) J. Physiol. 574, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kahn B. B., Alquier T., Carling D., Hardie D. G. (2005) Cell Metab. 1, 15–25 [DOI] [PubMed] [Google Scholar]
- 41.Woods A., Dickerson K., Heath R., Hong S. P., Momcilovic M., Johnstone S. R., Carlson M., Carling D. (2005) Cell Metab. 2, 21–33 [DOI] [PubMed] [Google Scholar]
- 42.Hawley S. A., Boudeau J., Reid J. L., Mustard K. J., Udd L., Mäkelä T. P., Alessi D. R., Hardie D. G. (2003) J. Biol. 2, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tuerk R. D., Auchli Y., Thali R. F., Scholz R., Wallimann T., Brunisholz R. A., Neumann D. (2009) Anal. Biochem. 390, 141–148 [DOI] [PubMed] [Google Scholar]
- 44.Scholz R., Suter M., Weimann T., Polge C., Konarev P. V., Thali R. F., Tuerk R. D., Viollet B., Wallimann T., Schlattner U., Neumann D. (2009) J. Biol. Chem. 284, 27425–27437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neumann D., Woods A., Carling D., Wallimann T., Schlattner U. (2003) Protein Expr. Purif. 30, 230–237 [DOI] [PubMed] [Google Scholar]
- 46.Riek U., Scholz R., Konarev P., Rufer A., Suter M., Nazabal A., Ringler P., Chami M., Müller S. A., Neumann D., Forstner M., Hennig M., Zenobi R., Engel A., Svergun D., Schlattner U., Wallimann T. (2008) J. Biol. Chem. 283, 18331–18343 [DOI] [PubMed] [Google Scholar]
- 47.Woods A., Salt I., Scott J., Hardie D. G., Carling D. (1996) FEBS Lett. 397, 347–351 [DOI] [PubMed] [Google Scholar]
- 48.Ninomiya-Tsuji J., Kajino T., Ono K., Ohtomo T., Matsumoto M., Shiina M., Mihara M., Tsuchiya M., Matsumoto K. (2003) J. Biol. Chem. 278, 18485–18490 [DOI] [PubMed] [Google Scholar]
- 49.Jogireddy R., Dakas P. Y., Valot G., Barluenga S., Winssinger N. (2009) Chemistry 15, 11498–11506 [DOI] [PubMed] [Google Scholar]
- 50.Dakas P. Y., Jogireddy R., Valot G., Barluenga S., Winssinger N. (2009) Chemistry 15, 11490–11497 [DOI] [PubMed] [Google Scholar]
- 51.Dakas P. Y., Barluenga S., Totzke F., Zirrgiebel U., Winssinger N. (2007) Angew. Chem. Int. Ed. Engl. 46, 6899–6902 [DOI] [PubMed] [Google Scholar]
- 52.Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 53.Suter M., Riek U., Tuerk R., Schlattner U., Wallimann T., Neumann D. (2006) J. Biol. Chem. 281, 32207–32216 [DOI] [PubMed] [Google Scholar]
- 54.Laderoute K. R., Amin K., Calaoagan J. M., Knapp M., Le T., Orduna J., Foretz M., Viollet B. (2006) Mol. Cell. Biol. 26, 5336–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tobiume K., Saitoh M., Ichijo H. (2002) J. Cell. Physiol. 191, 95–104 [DOI] [PubMed] [Google Scholar]
- 56.Leung I. W., Lassam N. (2001) J. Biol. Chem. 276, 1961–1967 [DOI] [PubMed] [Google Scholar]
- 57.Tu Z., Mooney S. M., Lee F. S. (2003) Cell. Signal. 15, 65–77 [DOI] [PubMed] [Google Scholar]
- 58.Huang J., Tu Z., Lee F. S. (2003) Biochem. Biophys. Res. Commun. 303, 532–540 [DOI] [PubMed] [Google Scholar]
- 59.Deak J. C., Templeton D. J. (1997) Biochem. J. 322, 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furdui C. M., Lew E. D., Schlessinger J., Anderson K. S. (2006) Mol. Cell 21, 711–717 [DOI] [PubMed] [Google Scholar]
- 61.Herrero-Martín G., Høyer-Hansen M., García-García C., Fumarola C., Farkas T., López-Rivas A., Jäättelä M. (2009) EMBO J. 28, 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lei M., Lu W., Meng W., Parrini M. C., Eck M. J., Mayer B. J., Harrison S. C. (2000) Cell 102, 387–397 [DOI] [PubMed] [Google Scholar]
- 63.Taylor S. S., Kim C., Cheng C. Y., Brown S. H., Wu J., Kannan N. (2008) Biochim. Biophys. Acta 1784, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eisenberg D., Weiss R. M., Terwilliger T. C., Wilcox W. (1982) Faraday Symp. Chem. Soc. 17, 109–120 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.