Abstract
The density of hepatitis C virus (HCV) particles circulating in the blood of chronically infected patients and of cell-culture produced HCV is heterogeneous. Specific infectivity and fusion of low density particles are higher than those of high density particles. We recently characterized hybrid particles produced by Caco-2 colon or Huh-7.5 liver cells transduced with HCV E1 and E2 envelope glycoproteins. Caco-2-derived particles, called empty lipo-viral particles (eLVP), are composed of triglyceride-rich lipoproteins positive for apolipoproteins B (i.e. apoB100 and apoB48) and contain HCV E1 and E2. Here we aimed at characterizing the morphology and in vitro fusion properties of eLVP using electron microscopy and fluorescence spectroscopy. They displayed the aspect of β-lipoproteins, and immunogold labeling confirmed the presence of apoB and HCV E1 and E2 at their surface. These particles are able to fuse with lipid bilayers (liposomes) in a fusion process leading to the coalescence of internal contents of triglyceride-rich lipoproteins particles and liposomes. Fusion was pH-dependent and could be inhibited by either Z-fFG, a peptide known to inhibit viral fusion, or by monoclonal antibodies directed against HCV E2 or the apolipoprotein moiety of the hybrid particle. Interestingly, particles derived from Huh-7.5 cells failed to display equivalent efficient fusion. Optimal fusion activity is, thus, observed when HCV envelope proteins are associated to apoB-positive hybrid particles. Our results, therefore, point to a crucial role of the E1 and E2 proteins in HCV fusion with a subtle interplay with the apolipoprotein part of eLVP.
Keywords: Apolipoproteins, Electron Microscopy (EM), Fluorescence, Hepatitis Virus, Lipoprotein, Membrane Fusion, Cryo Electron Microscopy, Fusion Inhibition, Hepatitis C Virus
Introduction
Hepatitis C virus (HCV)3 is an important public health concern worldwide as it is a major cause of chronic hepatitis, cirrhosis and hepatocellular carcinoma and infects an estimated 3% of the world population (1). HCV is a member of the Hepacivirus genus of the Flaviviridae family (2), to which belong the Flavivirus and Pestivirus genera as well. Based on sequence comparison, patient HCV isolates are classified into seven genotypes, differing in their nucleotide sequence by 30–35% (3–6). HCV only infects humans and chimpanzees, which sets this virus apart from other flaviviruses. Another peculiarity of HCV is the exceptional low density of the virus particles resulting from the association of the virus with lipoproteins (7). Indeed the majority of HCV circulating in blood was found associated with β-lipoproteins, very low and low density lipoproteins ((VLDL, LDL (7–9)), and the LDL receptor has been reported as a receptor for HCV (10–13). Interestingly, serum-derived HCV displays a highly heterogeneous density, of which low density particles are more infectious for chimpanzees than viruses with higher density (14). A transmission case of hepatitis C suggests that low density viral particles are also infectious in humans (15). Similarly, cell culture-grown HCV particles (HCVcc) with low density (1.09–1.10 g/ml) display the highest specific infectivity (16). Serum viral particles in density fractions below 1.06 g/ml are associated with triglyceride-rich lipoproteins (TRLs) bearing apolipoprotein B (apoB), i.e. the low, intermediate and very low density lipoproteins (low, intermediate, and very low density lipoproteins, respectively) and chylomicrons (9, 11, 17, 18). Taken together, these data suggest a key role of lipids and/or lipoprotein-associated lipids for productive infection by HCV, which may be related to facilitated virus binding, entry, and/or fusion. These highly infectious low density HCV particles were termed lipo-viral particles (LVPs). As lipoprotein-like particles, they are thought to be delimited by a phospholipid monolayer surrounding the core enriched in triglycerides and cholesterol esters. They are recognized by host antibodies and contain apolipoproteins B, CII, CIII, and E but not the high density lipoprotein-associated apoA and also contain HCV RNA, core protein, and envelope glycoproteins E1 and E2 (9, 11). E1 and E2 appear to be exposed on the surface of purified LVPs as they are recognized by anti-envelope antibodies under non-denaturing conditions (17). To better understand how these LVPs could be formed in vivo, we developed in vitro cellular systems where E1 and E2 proteins were stably expressed in cell lines supporting the production of apoB-positive lipoproteins, namely differentiated human intestinal Caco-2 cells and the human hepatic cell lines HepG2 and Huh-7.5. Using these models, relevant to HCV assembly and maturation, we recently showed that HCV glycoproteins were secreted only when the synthesis and secretion pathways of TRL were functional (19). The resulting hybrid particles are apoB-positive, harbor E1 and E2 glycoproteins at their surface, and display densities ≤1.05 g/ml. Interestingly our most recent data demonstrated that HCVcc of densities ≤1.06 g/ml exhibited the highest infectivity toward cultured hepatocyte Huh-7.5 cells together with the highest fusogenicity toward liposomal membranes (20). Low density fractions of HCVcc would contain apoB and apoE (21); however, the exact lipid nature of low density HCVcc is unknown. Lipid elements associated to or co-floating with HCVcc are, therefore, strongly suspected to play a prominent role in HCV infection and notably in membrane fusion, a key step in the HCV lifecycle leading to the delivery of its genetic material into the cytosol. Because a number of molecular details regarding the HCV fusion process are still missing, the E1-E2/apoB-positive hybrid particles obtained from producer cells offer the unique opportunity to further improve our comprehension of HCV fusion. In this study we, therefore, aimed at characterizing the morphology and in vitro fusion properties of TRL hybrid particles (further called empty LVP (eLVP)) by using electron microscopy and the fusion assays we previously designed (20, 22). These particles resembled β-lipoproteins, and immunogold labeling revealed the presence of HCV E1-E2 and apoB at their surface. We also report that these particles are able to fuse with lipid bilayers (liposomes). Interestingly, only particles secreted from E1-E2-transduced Caco-2 cells displayed efficient fusion. Because they contain HCV glycoproteins together with both apoB isoforms (apoB 100 and apoB 48), our results, therefore, point to a crucial role of the E1-E2 proteins in HCV fusion with a subtle interplay with the apolipoprotein part of the triglyceride-rich lipoprotein hybrid particle.
MATERIALS AND METHODS
Chemicals and Reagents
Phosphatidylcholine from egg yolk (99% pure), cholesterol (99% pure), and Triton X-100 were from Avanti Polar Lipids and Sigma. Octadecylrhodamine B chloride (R18) was from Invitrogen. Terbium (Tb) chloride, dipicolinic acid (DPA, 2,6 pyridine-dicarboxylic acid), fatty acid-free bovine serum albumin (BSA), and the goat anti-mouse secondary antibody labeled with 5 nm-colloidal gold particles were purchased from Sigma. The rabbit anti-goat secondary antibody labeled with 15 nm-colloidal gold particles was purchased from EMS (Hatfield, PA). Protein A-superparamagnetic beads (Bio-Adembeads) were purchased from Ademtech (Pessac, France).
Monoclonal antibodies 1D1 and 5E11 (isotype IgG1) were from the Heart Institute (University of Ottawa, Ontario, Canada). The monoclonal antibody to measles H protein (clone 55) was an isotype IgG2b. Monoclonal antibodies against HCV E1(A4) and E2 (H48) were kind gifts from Jean Dubuisson (IBL, Lille, France). Goat antibody K45253G to human apoB 48 and 100 was from Biodesign International (Meridian Life Science, Saco, ME). Cell culture reagents were obtained from Invitrogen, and FCS (fetal calf serum) was from PAN-Biotech GmbH (Aidenbach, Germany). Microporous polyethylene terephthalate membrane inserts were from BD Biosciences (4.2 cm2, 1-mm pore size).
Lentiviral Particles Production and Cell Transduction
A lentiviral vector E1E2-HIV-SIN for the transduction of the HCV glycoproteins E1 and E2 was constructed. The gene coding the proteins E1 and E2 from HCV genotype 1a H77 strain (AJ 318514, nucleotides 733–2579) were introduced at the BsrgI and BamHI restriction sites of the HIV-1 self-inactivating vector backbone pRRL-SIN18-cPPT-hPGK-EGFP-WPRE (pHIV-hPGK-GFPW+ (23)) under the control of the internal human phosphoglycerate kinase promoter. Retrovirus carrying the E1E2-HIV-SIN sequence was produced by co-transfection of the Gag-Pol packaging construct pCMVdeltaR8.91 (24) and glycoprotein expression construct pCMV-VSV-G (AJ 318514) in 293T cells (ATCC CRL-1573). 2.4 × 106 293T cells per 55-cm2 culture dish (Corning) were seeded 24 h before transfection in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mm glutamine, 100 IU/ml penicillin, 100 mg/ml streptomycin, and 10% heat-inactivated FCS in a 95% humidified incubator containing 5% CO2 in air at 37 °C. For the production of E1E2-HIV particles, 13.2 mg of pCMV-VSV-G envelope plasmid, 10.2 mg of pCMVdeltaR8.91 packaging plasmid, and 13.2 mg of E1E2-HIV-SIN-derived vector plasmid were co-transfected by calcium phosphate precipitation. Medium was replaced with 0.1 ml/cm2 fresh culture medium without serum 14–16 h post-transfection. Viral supernatants were harvested 24 h later, cleared by low speed centrifugation, and filtered through a 0.45-μm low-binding protein filter (Millex filter unit, Millipore) before concentration. High titer viral stocks were prepared by concentrating viral supernatants ∼100-fold by ultracentrifugation for 2 h in a Beckman LE 70 ultracentrifuge at 26,000 rpm at 4 °C in an SW28 rotor. Viral stocks were aliquoted in PBS supplemented with 1% glycerol and stored at −80 °C. 7 ng of p24 antigen (HIV P24 II Vidas, Vidas apparatus, BioMérieux) with 8 mg/ml Polybrene in culture medium was used to infect target cells overnight, plated at 100,000 cells in a 2-cm2 well the day before.
Cell Culture
Caco-2 cells (ATCC HTB-37) were plated on inserts at a density of 250,000 cells/4.2 cm2 transwell and grown to confluence (6 days post-seeding) in DMEM containing 25 mm glucose and 2 mm glutamine supplemented with penicillin (100 IU/ml) and streptomycin (100 mg/ml), 1% nonessential amino acids, and 20% (v/v) heat-inactivated FCS. Cells were then cultured for an additional week under asymmetrical conditions with medium containing FCS in the lower compartment and medium without FCS in the upper compartment. Fresh medium without FCS and phenol red was added to the lower and upper compartment 48 h before ultracentrifugation. Huh-7.5 (from Charles Rice, Rockefeller University, New York) were plated in 9.5 cm2 wells at a density of 400,000 cells/well and grown for 24 or 48 h (until 80% confluence) in DMEM containing 25 mm glucose and 2 mm glutamine supplemented with penicillin (100 IU/ml), streptomycin (100 mg/ml), 1% nonessential amino acids, and 10% (v/v) heat-inactivated FCS before subsequent experiments. Fresh medium without FCS and phenol red was added to the wells 48 h before ultracentrifugation.
Particle Purification
E1-E2 hybrid particles secreted into cell culture medium were immediately purified by iodixanol gradients prepared as described by Nielsen et al. (9). Isopycnic linear density gradients were prepared from 6% (w/v) (1.7 ml of 60% (w/v) iodixanol, 0.34 ml of 0.5 m Tris-HCl, pH 8.0, 0.34 ml of 0.1 m EDTA, pH 8.0, and 14.6 ml 0.25 m sucrose) and 56.4% (w/v) (16.0 ml of 60% iodixanol, 0.34 ml of 0.5 m Tris-HCl, pH 8.0, 0.34 ml of 0.1 m EDTA, pH 8.0, and 0.34 ml 0.25 m sucrose) iodixanol solutions in thin-wall centrifuge tubes (14 × 89 mm, Beckman Instruments) using a two-chamber gradient maker. A sample of 2 ml of lower compartment medium (for Caco-2 cells) or 2 ml of supernatant (for Huh-7.5 cells) was applied to the top of 6–56% iodixanol gradients and centrifuged for 10 h in a Beckman Optima L100 XP ultracentrifuge at 41,000 rpm and 4 °C in an SW41 rotor. The gradient was harvested by tube puncture from the bottom, and 11 fractions of 1 ml each were collected. The density of each fraction was determined by measuring the mass of 100 μl and by assaying apoB concentration by ELISA, as described in Icard et al. 19. Upper fractions (density ≤1.06 g/ml) were pooled before subsequent analyses.
Conventional Electron Microscopy
Particles were deposited onto glow-discharged Formvar/carbon copper grids and negatively stained with 1% uranyl acetate. In some experiments Formvar-coated nickel grids were floated onto a drop of the anti-HCV E2 H48 antibody for 5 min followed by washes in Tris-HCl, pH 7.4. Grids were then floated onto a drop of particle suspension for 10 min followed by washes in Tris-HCl, pH 7.4, and by negative staining with 1% uranyl acetate. Other experiments consisted in depositing a drop of hybrid particles on grids followed by blocking for 1 h at room temperature in Tris-HCl, pH 7.4, containing 2% fatty acid-free BSA; grids were then incubated with the anti-HCV E1 A4 antibody. After extensive washes in Tris-HCl, 2% BSA at pH 7.4 then at pH 8.2, grids were floated for 1 h at room temperature on top of an anti-mouse secondary antibody labeled with 5-nm colloidal gold particles and washed in Tris-HCl, pH 8.2 then pH 7.4, and finally in distilled water. Negative staining was then achieved with 1% uranyl acetate. ApoB immunolabeling of hybrid particles was achieved after capture on copper/Formvar H48-coated grids of particles positive for HCV envelopes; grids were then floated onto a drop of the polyclonal goat anti-human apoB 48 and 100 antibody (K45253G, from Biodesign International, Saco, ME) and after extensive washes reacted with a rabbit anti-goat secondary antibody labeled with 15 nm-colloidal gold particles. Negative staining was then achieved with 1% uranyl acetate.
Cryo-electron Microscopy
Protein A-superparamagnetic beads were washed with a large volume of working buffer (NaCl 145 mm, 1 mm EDTA, 2.5 mm Hepes pH 7.4) before use. Five μl of beads were incubated with H48 in a final volume of 100 μl at 4 °C for 30 min. The magnetic bead-protein A·H48 complex was sedimented with a magnet. The pellet was washed three times to remove antibody. H48-coated beads were then incubated with eLVP at room temperature for 30 min (final volume 100 μl). After sedimentation with the magnet, the pellet was washed to remove unbound particles. For cryo-electron microscopy, a 5-μl sample was deposited onto a holey carbon-coated copper grid; the excess was blotted with filter paper, and the grid was plunged into a liquid ethane bath cooled with liquid nitrogen (Leica EM CPC). Specimens were maintained at a temperature of approximately −170 °C using a cryo-holder (Gatan) and were observed with a FEI Tecnai F20 electron microscope operating at 200 kV and at a nominal magnification of 50,000× under low dose conditions. Images were recorded with a 2Kx2K Ultrascan 1000 CCD camera (Gatan).
Fusion Assays
Preparation of Liposomes
All liposomes were large unilamellar vesicles (100 nm) consisting of (phosphatidylcholine:cholesterol (70:30 mol %). R18-labeled liposomes were obtained by mixing R18 and lipids as ethanol and chloroform solutions, respectively (5 mol % R18 final), and liposomes were prepared as previously described (22). R18 was used for liposome labeling due to its photostability and the relative insensitivity of its fluorescence to pH variations.
Lipid Mixing Assay
Lipid mixing was assessed essentially as described (22) and monitored as the dequenching of R18. Briefly, hybrid particles from the upper fractions of the iodixanol gradient were added to a cuvette containing R18-labeled liposomes (final lipid concentration, 15 μm). After temperature equilibration at pH 7.4 for 2 min, fusion was initiated by adding an appropriate volume of diluted HCl to the cuvette, and kinetics were recorded using an SLM Aminco 8000 fluorimeter at excitation and emission wavelengths of 560 and 590 nm, respectively. Maximal R18 dequenching was measured after the addition of 0.1% Triton X-100 (final concentration) to the cuvette. Initial rates of fusion were taken as the value of the slope of the tangent, drawn to the steepest part of the fusion kinetics. Final extents of lipid mixing were the value obtained when fluorescence has reached a plateau.
When monoclonal antibodies were used, hybrid particles from the upper fractions of the iodixanol gradient were incubated for 20 min in the presence of 25 μg/ml of antibodies (except for the irrelevant antibody to measles hemagglutinin, used at 60 μg/ml) at pH 7.4. Liposomes were then added, and lipid mixing was measured as described above. For some experiments, hybrid particles produced in Caco-2 cells and positive for HCV E1-E2 were incubated for 20 min on ice at neutral or acidic pH in the presence of 25 μg/ml monoclonal antibodies against HCV E2 (H48), apoB 100 and apoB 48 (1D1), apoB 100 (5E11), or with 60 μg/ml of the irrelevant antibody against measles virus H protein. The Ab/particles mixture was then added to a 37 °C-thermostated cuvette containing R18-labeled liposomes in a buffer at the same pH as that of the preincubation, and lipid mixing was recorded.
Contents Mixing Assay
Tb-loaded and DPA-loaded liposomes were prepared as described (25, 26). Free compounds were discarded by gel filtration on PD-10 columns (Amersham Biosciences) equilibrated with 10 mm Hepes, 150 mm NaCl, 1 mm EDTA, pH 7.4. Contents mixing was performed in that buffer preheated to 37 °C, and hybrid particles (350 ng/ml apoB 100, final concentration) were added to the cuvette containing Tb-loaded and DPA-loaded liposomes (1:1 mixture, 50 μm lipid final). Tb·DPA chelation complex formation, as a measure of contents mixing, was performed at excitation and emission wavelengths of 276 and 545 nm, respectively.
RESULTS
Electron Microscopy of eLVP
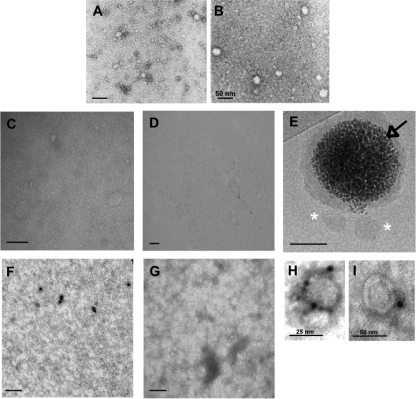
Electron microscopy was performed with purified eLVP obtained from E1-E2-transduced Caco-2 producer cells. At first, particles were directly observed after negative staining and mainly revealed two populations composed of spherical structures and centered around diameters of 25 and 50 nm (Fig. 1A). Larger objects were occasionally seen (data not shown). Conversely, TRL obtained from nontransduced Caco-2 cells appeared as a homogeneous layer of small structures (∼15 nm) covered with larger objects centered around 25 nm (Fig. 1B). Then we designed two strategies to sort eLVPs positive for HCV glycoproteins. The first one consisted of coating TEM grids with H48, an E2-specific monoclonal antibody (27), and “fishing” hybrid particles containing E2. As shown in Fig. 1C, the sample became enriched with ∼50-nm structures, whereas only a smooth layer of very small objects remained from TRL isolated from Caco-2 nontransduced cells (Fig. 1D). The second one relied on the use of magnetic beads covered with protein A and coated with H48. After incubation of these H48-coated beads with biological samples and collection of the magnetic beads with a magnet, the hybrid particles sorted through this strategy were observed by cryo-TEM. A representative image is shown in Fig. 1E, and eLVP appear again as ∼50-nm faintly contrasted objects (asterisks). No particle was retained from TRL particle samples obtained from untransduced Caco-2 cells (data not shown). Next we sought to identify the presence of E1 and/or E2 at the surface of the eLVP. This was achieved through immunogold labeling of particles directly deposited on TEM grids (see “Materials and Methods” for details). In samples of TRL hybrid particles, a few objects were immunolabeled with the anti-E1 specific monoclonal antibody A4 (Fig. 1F), whereas no labeling could be seen in samples of TRL particles from untransduced cells (Fig. 1G). Similar conclusions could be reached when using the H48 antibody instead of A4 (data not shown). Moreover, no labeling was observed when the primary antibodies were omitted (data not shown). A final strategy was performed as follows; grids coated with H48 were incubated with biological samples, then immunogold labeling was performed using an antibody directed against apoB 48 and 100 (Ac K45253G). Representative objects at high magnification are presented in Fig. 1, H and I, revealing spherical structures of ∼25 and 50 nm, respectively. Taken together these data demonstrate the concomitant presence of HCV E1 and E2 and of apoB 100 or 48 at the surface of the eLVP produced from E1-E2-transduced Caco-2 cells.
FIGURE 1.
Electron microscopy of TRL hybrid particles. Panels A and B, negative staining of TRL hybrid particles produced from E1-E2-transduced Caco-2 cells or nontransduced cells, respectively, is shown. The bar is 100 nm unless otherwise indicated. Panels C and D, TEM grids were coated with the anti-HCV E2 H48 antibody and rinsed, and particles produced from E1-E2-transduced (C) or untransduced Caco-2 cells (D) were added. Panel E, magnetic beads (black arrow) covered with a layer of protein A coated with H48 were incubated with TRL hybrid particles, rinsed, and observed by cryo-TEM; asterisks, TRL and E2-positive particles retained on the magnetic bead. Panels F and G, immunogold labeling of TRL particles from E1-E2-transduced (F) or untransduced Caco-2 cells (G) are shown; particles deposited on TEM grids were immunoassayed for the presence of HCV E1 with the A4 antibody, and a gold-labeled secondary antibody was added. Panels H and I, TEM grids were coated with H48, rinsed, then incubated with E117-E2-positive particles and immunoassayed for human apoB 48 and 100 with the polyclonal antibody K45253G; a gold-labeled secondary antibody was then added. Images show representative objects at high magnification.
eLVP Fuse with Liposomes in an E1-E2-, Particle Dose-, and pH-dependent Manner
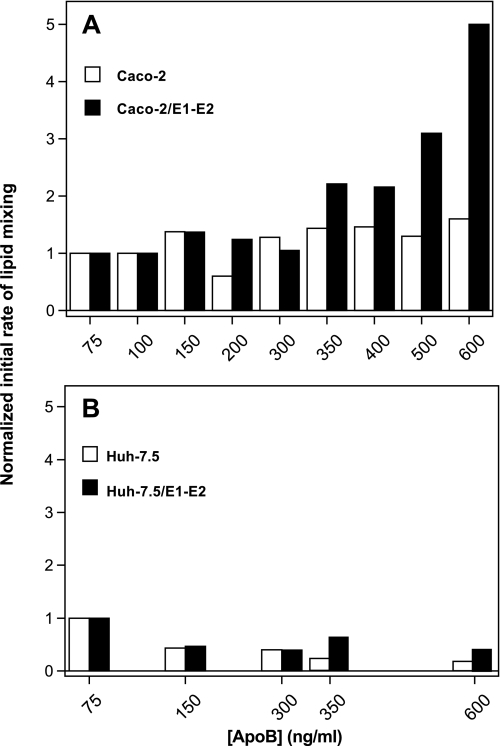
TRL particles collected from cells transduced or not with the HCV E1 and E2 glycoproteins were analyzed for their fusion properties using the R18 fusion assay we successfully applied to the study of HCV pseudoparticle- and HCVcc-mediated fusion (20, 22, 28). The initial rate of lipid mixing was the chosen parameter to evaluate and compare the fusion capacities of each particle type. Results obtained with particles collected from Caco-2 and Huh-7.5 cells are presented in Fig. 2, A and B, respectively. For particles from Caco-2 cells, efficient lipid mixing could not be measured below a certain threshold amount of apoB (350 ng/ml) and, therefore, a threshold of absolute number of particles present in the cuvette, as each particle is decorated by one molecule of apoB (19, 29). Starting from 350 ng/ml apoB, increasing the amount of apoB per fusion assay increased the initial rate of lipid mixing (solid bars). Indeed, a less than 2-fold increase in apoB concentration resulted in a more than 2-fold increase in lipid mixing rate. This is most likely related to a parallel increase in the amount of E1-E2 proteins at the surface of the particles considering that a similar increase in apoB amount is not paralleled by an increase in lipid mixing for particles devoid of HCV glycoproteins, collected from untransduced Caco-2 cells (open bars). For particles from Huh-7.5 cells (Fig. 2B), lipid mixing could not be related either to the amount of apoB per assay (compare solid bars) or to the presence or absence of HCV E1 and E2 proteins (compare solid and open bars) or only at high apoB concentrations. This agrees well with our previous observations and hypotheses (19) where E1-E2 eLVP produced from Huh-7.5 cells would lie in two distinct populations that could not likely merge in single hybrid particles. This could, therefore, explain their relative inability to fuse with liposomes under the conditions of our test.
FIGURE 2.
Dose-dependent lipid mixing of eLVP produced from Caco-2 cells (A) or Huh-7.5 cells (B). TRL particles from untransduced (open bars) or E1-E2-transduced cells (solid bars) were added at indicated apoB100 concentrations to a cuvette containing phosphatidylcholine:cholesterol:R18 liposomes (see “Materials and Methods”), and lipid mixing was recorded at pH 5.0 over a 10-min period. Initial rates of lipid mixing were calculated from the tangent at the steepest part of the fusion curves and results normalized, with 1 representing the basal lipid mixing rate measured for the lowest concentration of apoB tested (75 ng/ml).
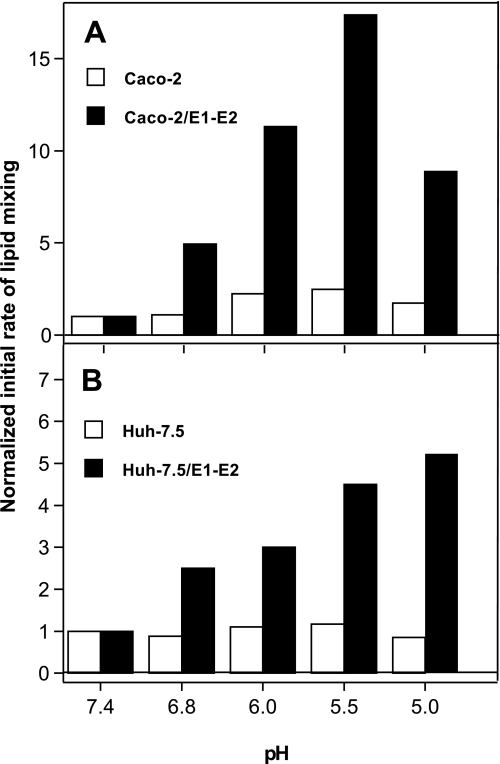
We then studied the dependence upon pH of the fusion mediated by these hybrid particles at a nominal apoB concentration of 350 ng/ml. As shown in Fig. 3A, solid bars, eLVP from Caco-2 cells displayed a strong pH dependence for their lipid mixing, with an optimum at pH 5.5 corresponding to a 17-fold increase in rate as compared with that measured at pH 7.4. Lipid mixing rate then decreased for lower pH values, although it was still 10-fold higher at pH 5.0 than at pH 7.4. Conversely, TRL particles devoid of HCV E1-E2 exhibited low to negligible lipid mixing at any pH tested, with only a 2-fold increase in initial rate between pH 7.4 and 5.5 (Fig. 3A, open bars). This again emphasizes the crucial role played by the HCV envelope glycoproteins in the fusion process of these TRL particles. Strikingly, hybrid particles produced by Huh-7.5 cells displayed a pH-dependent lipid mixing (Fig. 3B), although lower than that observed for Caco-2-produced particles. At this apoB concentration, lipid mixing was dependent upon the presence of E1-E2 in the samples (compare solid with open bars) for each pH tested. However, contrary to what was observed with eLVP from Caco-2 cells, lipid mixing rate continuously increased, whereas pH decreased. Under the conditions of our fusion assay, the rate optimum was reached at pH 4.0 (data not shown) and corresponded to a 7-fold increase as compared with that measured at pH 7.4. This reveals that a reduction in the free energy of activation for fusion of the Huh-7.5-produced hybrid particles is achieved only at low pH values and is in line with our recent data on HCVcc membrane fusion (Ref 20 and see “Discussion”).
FIGURE 3.
pH-dependent lipid mixing of eLVP produced from Caco-2 cells (A) or Huh-7.5 cells (B). TRL particles (350 ng/ml apoB concentration) from untransduced (open bars) or E1-E2-transduced cells (solid bars) were added to a cuvette containing phosphatidylcholine:cholesterol:R18 liposomes, and lipid mixing was recorded at the indicated pH values. Initial rates of lipid mixing were then normalized, with 1 corresponding to the rate measured at pH 7.4.
eLVP Fusion Process Is Complete
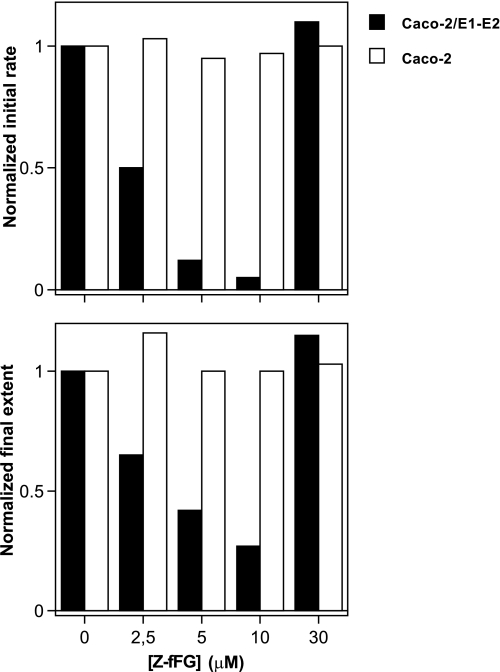
In the following we only focused on particles produced from Caco-2 cells. To further characterize their fusion properties, we first determined whether the fusion of eLVP could be inhibited by the viral fusion peptide inhibitor carbobenzoxy-d-phenyl-l-phenylglycine (Z-fFG), known to act as a membrane stabilizer (30, 31). Increasing concentrations of this tripeptide decreased both the initial rate and final extent of lipid mixing between eLVP and liposomes (Fig. 4, closed bars), whereas no effect was observed when E1-E2 proteins were absent from the particles (open bars). The effect of diluted acetic acid alone (the solvent into which Z-fFG had been diluted) on final pH and on lipid mixing remained negligible (data not shown). Maximal fusion inhibition occurred at 10 μm Z-fFG, in agreement with literature and the respective proportions of lipid and proteins present in the assays (30). Interestingly, lipid mixing returned to a normal rate and extent when 30 μm Z-fFG were added; this “rebound” effect could likely be attributed to a lytic membrane activity of Z-fFG in the conditions of our assay, leading to artifactual R18 fluorescence dequenching due to membrane rupture.
FIGURE 4.
Inhibition of eLVP lipid mixing by Z-fFG. Lipid mixing of TRL particles from Caco-2 cells transduced (solid bars) or not (open bars) with HCV E1-E2 was measured as described above in the presence of indicated concentrations of Z-fFG. Initial rates and final extents of lipid mixing were measured as described under “Materials and Methods” and normalized (1 is the value of the considered parameter obtained in the absence of Z-fFG).
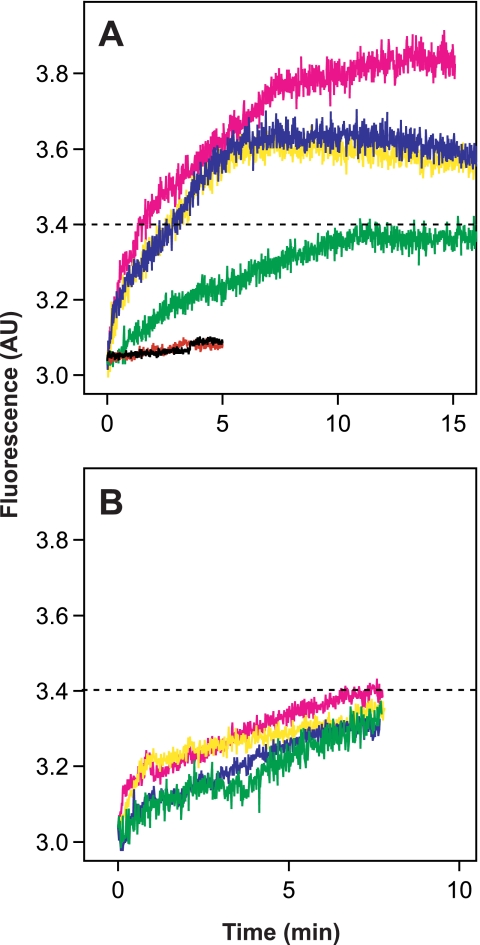
Second, we wanted to assess if fusion between eLVP and liposomes could lead or not lead to coalescence of vesicle contents (contents mixing) and, therefore, to complete fusion or not. A convenient contents mixing assay relies on the use of terbium chloride (TbCl3) and DPA, which eventually form a highly fluorescent chelation complex when in contact (25, 26). Because hybrid particles cannot be filled up, TbCl3 was encapsulated in one population of liposomes and DPA in another, and eLVP acted as the fusion “trigger.. This was performed in buffer containing EDTA to avoid formation of the Tb·DPA complex in the external medium (see “Materials and Methods”). As shown in Fig. 5A, this way of performing the assay was successful, i.e. formation of the Tb·DPA chelation complex occurred only when eLVP were added to liposomes (compare black and red curves with other curves). The increase in Tb·DPA fluorescence reflects the mixing of internal content of Tb-loaded liposomes with that of DPA-loaded vesicles, significantly induced only by the addition of HCV E1-E2-positive TRL particles (compare panel B with A, where the dotted line indicates threshold of significance). This most likely reflects the mixing of the luminal content of eLVP with that of both liposome populations. Moreover, contents mixing, as already observed for lipid mixing, was pH-dependent and increased proportionally with medium acidity (Fig. 5A). Conversely, increasing acidification did not modify the residual and base-line fusion activity of TRL particles devoid of HCV E1-E2 (Fig. 5B). This result indicates that eLVP-mediated fusion is dependent on pH and HCV E1-E2 and leads to both lipid and content mixings.
FIGURE 5.
Internal contents mixing of eLVP. Tb-loaded and DPA-loaded liposomes were prepared as described under “Materials and Methods.” A 1:1 mixture of liposomes was added to 10 mm Hepes, 150 mm NaCl, 1 mm EDTA buffer, pH 7.4, at 37 °C, and TRL particles were added (final apoB concentration 350 ng/ml). Formation of the Tb·DPA chelation complex, as a measure of internal contents mixing, leads to a fluorescence increase (λexc 276 nm; λem 545 nm). Panel A, contents mixing of TRL particles produced from HCV E1-E2-transduced Caco-2 cells is shown; green, pH 7.4; blue, pH 6.5; yellow, pH 5.5; magenta, pH 4.5; black and red curves, Tb- and DPA-loaded liposomes without particles, at pH 7.4 and 5.0, respectively. Panel B, contents mixing of TRL particles devoid of HCV glycoproteins. The color code is as described in panel A. AU, absorbance units.
eLVP Fusion Is Partially Abolished by Antibodies Directed against HCV E2 and ApoB 100/ApoB 48
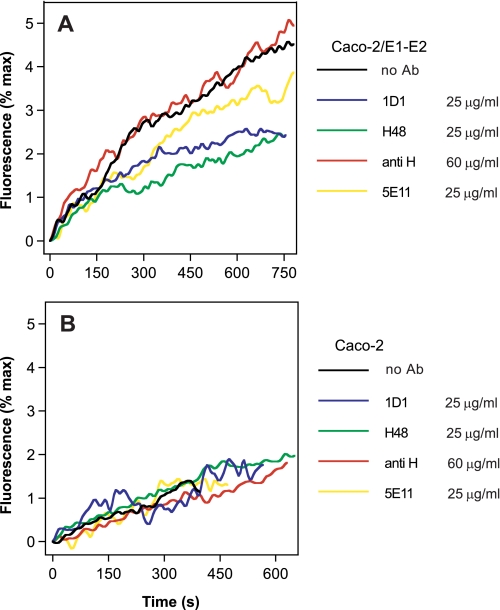
We further investigated the role played by each component of the eLVP in their membrane fusion process by incubating these particles with a series of monoclonal antibodies directed against either HCV E2 (H48) or apoB100 and apoB48 (1D1) or apoB100 only, targeting an LDL receptor binding site (5E11) (32, 33). As shown in Fig. 6A, targeting of specific regions in E2 with H48 led to a 2-fold decrease in eLVP-mediated fusion as compared with that observed in the absence of Ab (green versus black curves). This decrease was visible on both the initial rate and final extent of lipid mixing. Fusion was reduced down to a level comparable with that induced by particles devoid of HCV E1-E2 proteins (Fig. 6B, black curve). Surprisingly, blocking both apoB isoforms at the surface of hybrid particles with Ab 1D1 led to a similar reduction in fusion rate and extent (Fig. 6A, blue versus black), which suggests that the apolipoprotein part of the TRL particle could play a role in particle fusion in addition to HCV envelope proteins. Further analysis was performed using the 5E11 Ab, which displayed an intermediate behavior between the conditions without Ab and with H48 or 1D1 (Fig. 6A, yellow). This lends further support to our hypothesis that the apolipoprotein component of the TRL particle would play a key role in the fusion process together with HCV glycoproteins. Moreover, if this hypothesis were valid, this suggests that both isoforms of apoB would be required for optimal fusion. The presence of a high amount of an irrelevant antibody directed against the hemagglutinin of the measles virus (anti-H) did not affect fusion between eLVP and liposomes (Fig. 6A, red versus black curves). Also, the (very low) fusion of particles devoid of HCV proteins remained unaffected by the presence of any antibody (Fig. 6B).
FIGURE 6.
eLVP lipid mixing in the presence of monoclonal antibodies. TRL particles produced in Caco-2 cells and positive for HCV E1-E2 (panel A) or not (panel B) were incubated for 20 min on ice in buffer at pH 7.4, with the indicated amounts of monoclonal antibodies against apoB 100 and apoB 48 (1D1), HCV E2 (H48), apoB 100 (5E11), or with an irrelevant (but isotype-matched) antibody against measles virus hemagglutinin (anti H). The Ab/particles mixture was then added to a 37 °C-thermostated cuvette containing R18-labeled liposomes, and lipid mixing was recorded after acidification as described above.
Inhibition of eLVP Fusion by Antibodies Is Dependent upon pH
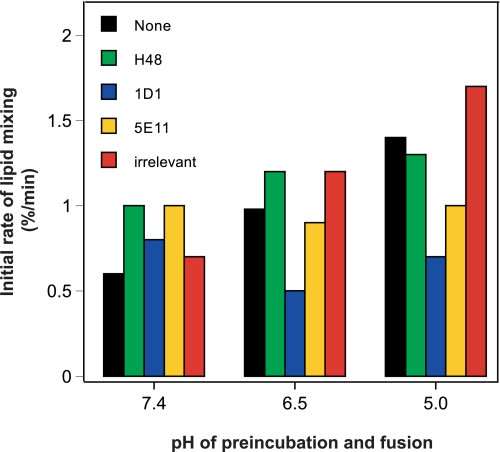
It is established that HCV-mediated fusion is a pH-dependent mechanism, and we have shown above that eLVP display a similar dependence of their membrane fusion toward acidic pH. Cell entry of HCV particles is, thus, achieved by low pH-dependent processes, most likely endocytosis. Therefore, we next sought to investigate whether the inhibiting effect of antibodies upon eLVP-mediated fusion varied with pH. This was assayed by preincubating eLVP at three different pH values (7.4, 6.5, and 5.0) in the presence or absence of the antibodies tested above. After this preincubation at low pH, we measured lipid mixing as described above at the same nominal pH value. As shown in Fig. 7 for one representative experiment (of three), the initial rate of fusion increased when pH decreased in the absence of antibody (black bars) or in the presence of the irrelevant antibody (anti-H) (red bars). Interestingly, the inhibition of fusion by anti-apoB antibodies became more prominent at lower pH values of preincubation (compare black with blue and yellow bars), with 1D1 displaying the highest inhibiting activity over 5E11. This suggests that eLVP preincubation at low pH could lead to the unmasking of apoB regions involved in fusion. The H48 antibody displays an intermediate phenotype; under the conditions of this assay, no inhibition of fusion could be measured till pH 5.0. At pH 4.5 however, a strong reduction in fusion was observed comparable with that of 5E11 anti-apoB antibody (data not shown). This reinforces our previous observation that blocking HCV E2 could lead to fusion inhibition but positions H48 as an antibody whose behavior and/or affinity is altered at low pH, as we already noticed (34).
FIGURE 7.
Inhibition of eLVP lipid mixing by monoclonal antibodies is dependent on pH. TRL particles produced in Caco-2 cells and positive for HCV E1-E2 were incubated for 20 min on ice at indicated pH without (black bars) or with 25 μg/ml of monoclonal antibodies against HCV E2 (H48, green bars), apoB 100 and apoB 48 (1D1, blue bars), apoB 100 (5E11, yellow bars), or with 60 μg/ml of the irrelevant antibody against measles virus H protein (irrelevant, red bars). The Ab/particles mixture was then added to a 37 °C-thermostated cuvette containing R18-labeled liposomes in a buffer at similar pH, and lipid mixing was recorded. Results shown for one representative set of data, obtained from one batch of eLVP (of three tested).
DISCUSSION
HCV particles circulating in the blood of infected patients were found associated to β-lipoproteins (7, 9, 14), and we recently introduced the hypothesis that highly infectious HCV represents in fact a LVP (or low density fractions containing HCV RNA) (11, 17). Our present study examines the structural details of eLVP and dissects for the first time one of their function, namely membrane fusion. eLVP observed by cryo-TEM appeared as very poorly contrasted spherical objects not delimited by a bilayer and devoid of internal density, closely resembling β-lipoproteins visualized by (cryo)-TEM (38–40) (see below). Our immunoelectron microscopy analyses highlighted the concomitant presence of HCV glycoproteins and apolipoprotein B on several particles. Because eLVPs do not contain the HCV capsid protein nor any HCV genetic material, they can be considered as defective viral particles. Interestingly, previous studies reported that purified LVP contained more apoB than HCV RNA molecules, suggesting that defective particles could circulate in the blood of chronically infected patients (11, 17) and as such constitute a “decoy” to the immune system. eLVPs also offer the unique opportunity to study a viral particle devoid of pathogenicity where the behavior of HCV envelope glycoproteins in the context of their association with lipoprotein components can be directly assessed.
The correlation between high infectivity and low density of plasma fractions containing HCV RNA was first reported in the early 90s (14, 41), and the notion of LVP gradually emerged that contained HCV components (core and RNA, E1 and E2) and apolipoproteins B, CII, CIII, and E but not the high density lipoprotein-associated apoA (9, 11, 17). Both apoB isoforms, apoB 100 and apoB 48, are present in LVPs with comparatively more apoB 48 in LVPs than in the plasma (17). Interestingly, these LVPs display the highest specific infectivity compared with viral particles with intermediate or high density (8). Because we showed that high infectivity could be related to high fusogenicity (20), here we sought to investigate the membrane fusion properties of eLVP produced by Caco-2 or by Huh-7.5 cells. Our data clearly show that eLVP from both cell types are able to fuse with liposomes in a pH- and E1/E2-dependent manner. Interestingly eLVP produced from Caco-2 cells displayed a high fusion capacity that increased with the amount of apoB present in the assay, whereas eLVP from Huh-7.5 cells displayed lower fusogenicity that did not depend on the apoB concentration. In addition, eLVP from Caco-2 cells exhibited an optimum for lipid mixing at pH 5.5, whereas Huh-7.5 cell eLVP-mediated lipid mixing peaked at pH 5.0. These discrepancies in fusion properties between both types of eLVP further emphasizes the defect in VLDL assembly reported for Huh-7 cells (19). This is also in line with studies on HCVcc, reporting association of apoE but not apoB to infectious particles (42). In fact this is still a matter of controversy, as other groups found HCVcc positive for apoE and apoB as well (21, 35). Interestingly, apoE was found essential to properly assembly and release of functional HCV particles through an interaction with NS5A (43). From our past and present data, it now becomes clear that particles obtained from cells fully functional for VLDL assembly (Caco-2) display a complete VLDL profile (19) and are fully mature to exhibit their fusion properties (this study). Conversely, particles produced from Huh-7.5 cells are somewhat defective in fusion, and the absence of correlation between apoB concentration and eLVP fusogenicity points to a defect in particle association with apoB. This defect might come from a different association of apoB with these Huh-7.5-derived eLVP compared with that of eLVP obtained from Caco-2 cells, as already observed. It is, thus, possible that Huh-7 cells might be deficient in key enzymes involved in triglyceride and TRL biosynthesis pathways (44, 45) and that, depending on the Huh-7 clone used, different enzymes might be lacking, leading to conflicting results (46).
Our cryo-TEM studies, allowing for analyses at high resolution, showed particles with an overall spherical shape and a homogeneous electron-scattering density. We could not visualize any bilayer delimiting these eLVP, suggesting that they are most likely delimited by a lipid monolayer, as are LDLs (47–49). This reinforces our previous observations that they are of a lipoprotein nature (19). Our present study is then to our knowledge the first report that a lipid monolayer could merge with a lipid bilayer. eLVP/liposome fusion is inhibited by the antiviral tripeptide Z-fFG in a dose-dependent manner. Fusion between bilayers has been proposed to be an ordered sequence of transition states (50). One intermediate in this process is the “fusion stalk,” in which the two outer leaflets have fused, resulting in a connection between the hydrophobic portions of the two bilayers or hemifusion (e.g. the viral membrane and the bilayer of the endosome, in pH-dependent viral fusion). Fusion between a monolayer and a bilayer could also proceed through the creation of a fusion stalk, which would allow contact between the hydrophobic core of the eLVP on one side and the inner leaflet of the liposome bilayer on the other side in a way reminiscent of that proposed by the Borén and co-workers (36, 37) for the fusion of lipid droplets. We show that this phenomenon leads to the mixing of internal contents of the liposomes engaged in fusion with eLVP; lipid and contents mixing depended upon the presence of HCV E1-E2. This illustrates that HCV glycoproteins, as exposed at the surface of eLVP, are fully functional for fusion completion.
Our novel finding that targeting the apolipoprotein B moiety of the eLVP with monoclonal antibodies leads to (partial) fusion inhibition opens new perspectives in our comprehension of the structure/function relationship of eLVP and, more generally, of the low density fractions of HCV. In a previous study we indeed showed that fusogenicity, in addition to infectivity, is highest in low density fractions of HCVcc (20), which further emphasizes the notion that virus composition has a pronounced impact on virus infectivity and suggests that this may in part be caused by differential fusion properties of the virions. In the context of the natural infection process, a physical association and/or an association of effect between HCV glycoproteins and the apolipoprotein part of LVP may play a role at the plasma membrane through modulation of interactions with the LDL or SR-BI receptors, for instance, thus facilitating HCV internalization (10, 11, 13). This might in addition influence trafficking of the virus·receptor complex on the cell surface and into the cells via endocytosis and, finally, the actual fusion step, through subtle HCV-lipid and apolipoprotein-lipid interactions with the endosomal membrane. Our results indeed suggest that the apolipoprotein part of the eLVP might play a role in HCV membrane fusion through a subtle interplay between lipids on one hand and HCV glycoproteins and apolipoproteins present at the surface of the same particle on the other hand. This might explain the relative independence upon apoB concentrations of the observed fusion of Huh-7.5-produced eLVP, assuming the two precursor models of lipoprotein synthesis in Huh-7.5 cells (19). At which stage of the process and whether it is a direct or indirect effect remains to be determined.
In conclusion, this study is the first molecular characterization of eLVP together with the investigation of their membrane fusion features, i.e. E1 and E2 carried by apoB-positive lipoproteins are capable of fusion with lipid bilayers. It is also the first description that the apolipoprotein portion of these particles could play a role in the fusion process, which would indicate that a subtle interplay between HCV glycoproteins, apolipoproteins, and lipids might govern the fusion properties of these particles. Whether this would explain the high specific infectivity of LVP from patient's serum remains to be clarified and is now under investigation in our laboratory.
Acknowledgments
We thank François Penin for helpful discussions and continuous support. Electron microscopy was performed at the Centre Technologique des Microstructures, with special thanks to Béatrice Burdin and Xavier Jaurand. We thank Martine Carreras for expert technical assistance.
This work was supported by the CNRS, INSERM, and the Agence Nationale de Recherche contre le SIDA et les hépatites virales (to E.-I. P. and P. A.).
- HCV
- hepatitis C virus
- HCVcc
- cell culture-grown HCV
- TRL
- triglyceride-rich lipoproteins
- LVP
- lipo-viral particle
- eLVP
- empty LVP
- DPA
- dipicolinic acid
- R18
- octadecylrhodamine B chloride
- TEM
- transmission electron microscopy
- Z-fFG
- carbobenzoxy-d-phenyl-l-phenylglycine
- Ab
- antibody.
REFERENCES
- 1.Alter M. J. (2007) World J. Gastroenterol. 13, 2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindenbach B. D., Rice C. M. (2001) in Fields Virology (Knipe D. M., Howley P. M. eds) Vol. 1, pp. 991–1041, Lippincott-Raven, Philadelphia [Google Scholar]
- 3.Moradpour D., Penin F., Rice C. M. (2007) Nat. Rev. Microbiol. 5, 453–463 [DOI] [PubMed] [Google Scholar]
- 4.Gottwein J. M., Scheel T. K., Jensen T. B., Lademann J. B., Prentoe J. C., Knudsen M. L., Hoegh A. M., Bukh J. (2009) Hepatology 49, 364–377 [DOI] [PubMed] [Google Scholar]
- 5.Murphy D., Chamberland J., Dandavino R., Sablon E. (2007) Hepatology 46, Suppl. 1, 623 [Google Scholar]
- 6.Simmonds P., Bukh J., Combet C., Deléage G., Enomoto N., Feinstone S., Halfon P., Inchauspé G., Kuiken C., Maertens G., Mizokami M., Murphy D. G., Okamoto H., Pawlotsky J. M., Penin F., Sablon E., Shin-I T., Stuyver L. J., Thiel H. J., Viazov S., Weiner A. J., Widell A. (2005) Hepatology 42, 962–973 [DOI] [PubMed] [Google Scholar]
- 7.Thomssen R., Bonk S., Propfe C., Heermann K. H., Köchel H. G., Uy A. (1992) Med. Microbiol. Immunol. 181, 293–300 [DOI] [PubMed] [Google Scholar]
- 8.André P., Perlemuter G., Budkowska A., Bréchot C., Lotteau V. (2005) Semin. Liver Dis. 25, 93–104 [DOI] [PubMed] [Google Scholar]
- 9.Nielsen S. U., Bassendine M. F., Burt A. D., Martin C., Pumeechockchai W., Toms G. L. (2006) J. Virol. 80, 2418–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agnello V., Abel G., Elfahal M., Knight G. B., Zhang Q. X. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12766–12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.André P., Komurian-Pradel F., Deforges S., Perret M., Berland J. L., Sodoyer M., Pol S., Bréchot C., Paranhos-Baccalà G., Lotteau V. (2002) J. Virol. 76, 6919–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wünschmann S., Medh J. D., Klinzmann D., Schmidt W. N., Stapleton J. T. (2000) J. Virol. 74, 10055–10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molina S., Castet V., Fournier-Wirth C., Pichard-Garcia L., Avner R., Harats D., Roitelman J., Barbaras R., Graber P., Ghersa P., Smolarsky M., Funaro A., Malavasi F., Larrey D., Coste J., Fabre J. M., Sa-Cunha A., Maurel P. (2007) J. Hepatol 46, 411–419 [DOI] [PubMed] [Google Scholar]
- 14.Bradley D., McCaustland K., Krawczynski K., Spelbring J., Humphrey C., Cook E. H. (1991) J. Med. Virol. 34, 206–208 [DOI] [PubMed] [Google Scholar]
- 15.Diaz O., Cubero M., Trabaud M. A., Quer J., Icard V., Esteban J. I., Lotteau V., André P. (2008) J. Med. Virol. 80, 242–246 [DOI] [PubMed] [Google Scholar]
- 16.Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., Rice C. M. (2005) Science 309, 623–626 [DOI] [PubMed] [Google Scholar]
- 17.Diaz O., Delers F., Maynard M., Demignot S., Zoulim F., Chambaz J., Trépo C., Lotteau V., André P. (2006) J. Gen. Virol. 87, 2983–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pumeechockchai W., Bevitt D., Agarwal K., Petropoulou T., Langer B. C., Belohradsky B., Bassendine M. F., Toms G. L. (2002) J. Med. Virol. 68, 335–342 [DOI] [PubMed] [Google Scholar]
- 19.Icard V., Diaz O., Scholtes C., Perrin-Cocon L., Ramière C., Bartenschlager R., Penin F., Lotteau V., André P. (2009) PLoS ONE 4, e4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haid S., Pietschmann T., Pécheur E. I. (2009) J. Biol. Chem. 284, 17657–17667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang K. S., Jiang J., Cai Z., Luo G. (2007) J. Virol. 81, 13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavillette D., Bartosch B., Nourrisson D., Verney G., Cosset F. L., Penin F., Pécheur E. I. (2006) J. Biol. Chem. 281, 3909–3917 [DOI] [PubMed] [Google Scholar]
- 23.Dupuy F. P., Mouly E., Mesel-Lemoine M., Morel C., Abriol J., Cherai M., Baillou C., Nègre D., Cosset F. L., Klatzmann D., Lemoine F. M. (2005) J. Gene Med. 7, 1158–1171 [DOI] [PubMed] [Google Scholar]
- 24.Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. (1997) Nat. Biotechnol. 15, 871–875 [DOI] [PubMed] [Google Scholar]
- 25.Wilschut J., Papahadjopoulos D. (1979) Nature 281, 690–692 [DOI] [PubMed] [Google Scholar]
- 26.Wilschut J., Düzgüneş N., Fraley R., Papahadjopoulos D. (1980) Biochemistry 19, 6011–6021 [DOI] [PubMed] [Google Scholar]
- 27.Op De Beeck A., Voisset C., Bartosch B., Ciczora Y., Cocquerel L., Keck Z., Foung S., Cosset F. L., Dubuisson J. (2004) J. Virol. 78, 2994–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoekstra D., de Boer T., Klappe K., Wilschut J. (1984) Biochemistry 23, 5675–5681 [DOI] [PubMed] [Google Scholar]
- 29.Segrest J. P., Jones M. K., De Loof H., Dashti N. (2001) J. Lipid Res. 42, 1346–1367 [PubMed] [Google Scholar]
- 30.Kelsey D. R., Flanagan T. D., Young J., Yeagle P. L. (1990) J. Biol. Chem. 265, 12178–12183 [PubMed] [Google Scholar]
- 31.Dentino A. R., Westerman P. W., Yeagle P. L. (1995) Biochim. Biophys. Acta 1235, 213–220 [DOI] [PubMed] [Google Scholar]
- 32.Theolis R., Jr., Weech P. K., Marcel Y. L., Milne R. W. (1984) Arteriosclerosis 4, 498–509 [DOI] [PubMed] [Google Scholar]
- 33.Milne R. W., Theolis R., Jr., Verdery R. B., Marcel Y. L. (1983) Arteriosclerosis 3, 23–30 [DOI] [PubMed] [Google Scholar]
- 34.Bonnafous P., Perrault M., Le Bihan O., Bartosch B., Lavillette D., Penin F., Lambert O., Pécheur E. I. (2010) J. Gen. Virol., in press [DOI] [PubMed] [Google Scholar]
- 35.Jiang J., Luo G. (2009) J. Virol. 83, 12680–12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.51 Olofsson S. O., Boström P., Andersson L., Rutberg M., Perman J., Borén J. (2009) Biochim. Biophys. Acta 1791, 448–458 [DOI] [PubMed] [Google Scholar]
- 37.Gastaminza P., Cheng G., Wieland S., Zhong J., Liao W., Chisari F. V. (2008) J. Virol. 82, 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Antwerpen R., Gilkey J. C. (1994) J. Lipid Res. 35, 2223–2231 [PubMed] [Google Scholar]
- 39.van Antwerpen R., Chen G. C., Pullinger C. R., Kane J. P., LaBelle M., Krauss R. M., Luna-Chavez C., Forte T. M., Gilkey J. C. (1997) J. Lipid Res. 38, 659–669 [PubMed] [Google Scholar]
- 40.Forte T., Norum K. R., Glomset J. A., Nichols A. V. (1971) J. Clin. Invest. 50, 1141–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hijikata M., Shimizu Y. K., Kato H., Iwamoto A., Shih J. W., Alter H. J., Purcell R. H., Yoshikura H. (1993) J. Virol. 67, 1953–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owen D. M., Huang H., Ye J., Gale M., Jr. (2009) Virology 394, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benga W. J., Krieger S. E., Dimitrova M., Zeisel M. B., Parnot M., Lupberger J., Hildt E., Luo G., McLauchlan J., Baumert T. F., Schuster C. (2010) Hepatology 51, 43–53 [DOI] [PubMed] [Google Scholar]
- 44.Smith S. J., Cases S., Jensen D. R., Chen H. C., Sande E., Tow B., Sanan D. A., Raber J., Eckel R. H., Farese R. V., Jr. (2000) Nat. Genet. 25, 87–90 [DOI] [PubMed] [Google Scholar]
- 45.Buhman K. K., Accad M., Novak S., Choi R. S., Wong J. S., Hamilton R. L., Turley S., Farese R. V., Jr. (2000) Nat. Med. 6, 1341–1347 [DOI] [PubMed] [Google Scholar]
- 46.Pawlotsky J. M., Cocquerel L., Durantel D., Lavillette D., Lerat H., Pécheur E. I., Polyak S. J., Tautz N., Thimme R. (2010) Gastroenterology 138, 6–12 e1–2 [DOI] [PubMed] [Google Scholar]
- 47.Smith L. C., Pownall H. J., Gotto A. M., Jr. (1978) Annu. Rev. Biochem. 47, 751–757 [DOI] [PubMed] [Google Scholar]
- 48.Hevonoja T., Pentikäinen M. O., Hyvönen M. T., Kovanen P. T., Ala-Korpela M. (2000) Biochim. Biophys. Acta 1488, 189–210 [DOI] [PubMed] [Google Scholar]
- 49.Ren G., Rudenko G., Ludtke S. J., Deisenhofer J., Chiu W., Pownall H. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1059–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chernomordik L., Kozlov M. M., Zimmerberg J. (1995) J. Membr. Biol. 146, 1–14 [DOI] [PubMed] [Google Scholar]