Abstract
Ribosomal proteins play an important role in p53 activation in response to nucleolar stress. Multiple ribosomal proteins, including L5, L11, L23, and S7, have been shown to bind to and inhibit MDM2, leading to p53 activation. However, it is not clear whether ribosomal protein regulation of MDM2 is specific to some, but not all ribosomal proteins. Here we show that L29 and L30, two ribosomal proteins from the 60 S ribosomal subunit, do not bind to MDM2 and do not inhibit MDM2-mediated p53 suppression, indicating that the ribosomal protein regulation of the MDM2-p53 feedback loop is specific. Interestingly, direct perturbation of the 60 S ribosomal biogenesis by knocking down either L29 or L30 drastically induced the level and activity of p53, leading to p53-depedent cell cycle arrest. This p53 activation was drastically inhibited by knockdown of L11 or L5. Consistently, knockdown of L29 or L30 enhanced the interaction of MDM2 with L11 and L5 and markedly inhibited MDM2-mediated p53 ubiquitination, suggesting that direct perturbation of 60 S ribosomal biogenesis activates p53 via L11- and L5-mediated MDM2 suppression. Mechanistically, knockdown of L30 or L29 significantly increased the NEDDylation and nuclear retention of L11. Knocking down endogenous NEDD8 suppressed p53 activation induced by knockdown of L30. These results demonstrate that NEDDylation of L11 plays a critical role in mediating p53 activation in response to perturbation of ribosomal biogenesis.
Keywords: Cell Cycle, Nucleolus, p53, Ribosomes, Tumor Suppressor, L11, L5, MDM2, NEDD8, Ribosomal Biogenesis
Introduction
The tumor suppressor protein p53 plays a critical role in maintaining genomic integrity and preventing tumorigenesis. In response to diverse stressors, p53 is stabilized and activated to induce cell cycle arrest, apoptosis, or senescence (1, 2). The ubiquitin E32 ligase MDM2 plays a key role in inhibiting p53 under both physiological and stress conditions. MDM2 inhibits p53 by ubiquitinating p53 and targeting it for proteasomal degradation (3–5) as well as directly blocking its transactivation activity through binding to its N-terminal transactivation domain (6). MDM2 itself is a transcriptional target of p53, thus forming a feedback regulatory loop (7, 8). This loop is verified by studies showing that deletion of the p53 gene rescues the lethal phenotype of mdm2 knock-out mice (9, 10).
The importance of the MDM2-p53 feedback loop is also evident from the fact that diverse stressors activate p53 by interfering with this loop. For example, DNA damage, such as that induced by ionizing radiation and UV irradiation, triggers phosphorylation of both p53 and MDM2, blocking their physical and functional interaction and alleviating the inhibition of p53 by MDM2 (2). Aberrant proliferating signals induced by overexpression of oncogenes induce the expression of the ARF tumor suppressor (11). ARF binds to the central acidic domain of MDM2 and inhibits its ubiquitin E3 ligase activity toward p53, leading to p53 activation (11, 12). Recently, it has been shown that p53 is also activated by nucleolar stress (also called ribosomal stress) via inhibition of MDM2. This type of stress is induced by perturbation of ribosomal biogenesis, a multistep cellular process for making the ribosome, including ribosomal RNA synthesis, processing, and ribosomal assembly in the nucleolus as well as ribosome subunit export into the cytoplasm (13, 14). Ribosomal biogenesis is vital for cell growth and must be tightly coordinated with cell cycle progression. Deregulation of ribosomal biogenesis contributes to tumorigenesis (14, 15).
Accumulating evidence points to a key role for p53 in sensing ribosomal stress. Examples of such stress conditions include treatment of cells with a low dose of actinomycin D (Act D) (16), 5-fluorouracil (17, 18), or mycophenolic acid (MPA) (19), expression of dominant-negative mutant of the ribosomal RNA processing factor Bop1 (20), serum starvation or contact inhibition (21), genetic disruption of the polymerase I transcription initiation factor TIF-IA (22), or knockdown of either ribosomal protein S6 (23), or nucleostemin (24). Mechanistically, it has been shown that several ribosomal proteins, including L5, L11, L23, and S7, activate p53 by binding to MDM2 and inhibiting MDM2-mediated p53 ubiquitination and degradation in response to nucleolar stress (25–32). Reduction of these proteins by siRNA significantly attenuated the p53 activation induced by nucleolar stress. Interestingly, it has recently been shown that L11 and S7 are also required for p53 activation induced by DNA-damaging agents (32), suggesting that ribosomal proteins may play a crucial role in p53 activation in response to diverse stressors. Relevantly, mutations or deletions of ribosomal protein genes leading to haploinsufficiency of individual ribosomal proteins, including L5 and L11, contribute to Diamond-Blackfan anemia, a rare inherited anemia syndrome with increased incidence of tumors (15, 33, 34). Haploinsufficiency of several ribosomal proteins in zebrafish develop tumors as well (35), implying that these ribosomal proteins may possess intrinsic tumor suppressor function.
Currently, it is not known why multiple ribosomal proteins regulate the MDM2-p53 pathway. It is tempting to speculate that these proteins may act using different mechanisms or in concert with each other while controlling MDM2. Supporting the collaborative role of these ribosomal proteins is that L5 and L11 synergistically inhibit MDM2, leading to a robust activation of p53 compared with individual expression of L5 or L11 (36). Also, these ribosomal proteins appear to bind to different domains at the central region of MDM2 (27, 28, 37, 38), suggesting that they may form a multiprotein complex with MDM2. Another unanswered question is whether the ribosomal protein regulation of the MDM2-p53 pathway is specific to some, but not all, ribosomal proteins.
In this study, we show that two ribosomal proteins from the large ribosome subunit, L29 and L30, do not bind to MDM2 and do not inhibit MDM2-mediated p53 suppression, demonstrating that the ribosomal protein regulation of the MDM2-p53 pathway is specific. Interestingly, perturbation of 60 S ribosomal biogenesis by knocking down either L29 or L30 significantly induced p53 activity. This p53 activation requires L5 and L11, which are from the same 60 S ribosomal subunit, and the NEDDylation of L11. These results further demonstrate that L11 and L5 play a central role in p53 activation in response to nucleolar stress and that an increase in NEDDlyation and nucleolar retention of L11 may act as a signal for p53 activation.
MATERIALS AND METHODS
Cell Lines, Plasmids, and Antibodies
Human p53-proficient osteosarcoma U2OS cells and human p53-null lung non-small cell carcinoma H1299 cells were cultured in DMEM supplemented with 10% FBS, 50 units/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in a 5% CO2 humidified atmosphere as previously described (24, 26). Human fibroblast WI38 cells (ATCC) were cultured in DMEM supplemented with 15% FBS and non-essential amino acids (Invitrogen). FLAG-tagged L11 (FLAG-L11), FLAG-L29, FLAG-L30, and HA-MDM2 encoding plasmids have been described (27, 39). His-NEDD8 plasmid has been described (40). The V5-NEDD8 was cloned by inserting full-length human NEDD8 cDNA into the pcDNA3-V5 vector at BamHI and EcoRI sites. Anti-p53 (DO-1, Santa Cruz Biotechnology), anti-p21 (NeoMarkers), anti-MDM2 (SMP14, Santa Cruz Biotechnology), and anti-L30 (G-12, Santa Cruz Biotechnology) antibodies were purchased. Anti-L5 (25), anti-L11 (39), and anti-NEDD8 (40) antibodies have been described.
Co-transfection, Immunoblot (IB), and Co-immunoprecipitation Analyses
Cells were transfected with plasmids as indicated in figure legends, using TransIT®-LT1 reagents following the manufacturer's protocol (Mirus Bio Corp.). Cells were harvested at 48 h post-transfection and lysed in lysis buffer consisting of 50 mm Tris/HCl (pH 8.0), 0.5% Nonidet P-40, 1 mm EDTA, 150 mm NaCl, 1 mm PMSF, 1 mm DTT, 1 μg/ml pepstatin A, and 1 mm leupeptin. Equal amounts of cleared cell lysates were used for IB analysis as described previously (24). Co-immunoprecipitation assays were conducted as described previously (27). Bound proteins were detected by IB using antibodies as indicated in figure legends.
RNAi
RNAi-mediated knockdown of endogenous L5, L11, L29, L30, and p53 was performed essentially as described (24, 25, 39). The target sequences for L5, L11, and the control scrambled II RNA were described previously (25, 27). The target sequences for other genes were 5′-AAGTTCCTGAGGAACATGCGC-3′ (L29), 5′-AACTGGTGTCCATCACTACAG-3′ (L30), and 5′-AAGACTCCAGTGGTAATCTAC-3′ (p53). The siRNA pool against NEDD8 has been described (40). All the siRNA duplexes with a 3′-dTdT overhang were synthesized by Dharmacon (Lafayette, CO). These siRNA duplexes were introduced into cells using siLentFect (Bio-Rad) following the manufacturer's protocol. Cells were harvested 48 h after transfection for IB, RT-qPCR, and cell cycle analyses.
RT and qPCR Analyses
Total RNA was isolated from cells using the RNeasy Mini Kits (Qiagen, Valencia, CA). Reverse transcriptions were performed as described (25). Quantitative (q) PCR was performed on an ABI StepOneTM real-time PCR system (Applied Biosystems) using SYBR Green Mix (Bio-Rad) as described previously (24, 39). All reactions were carried out in triplicate. Relative gene expression was calculated using the ΔCτ method following the manufacturer's instructions. The primers for bax were 5′-ACTCCCCCCGAGAGGTCTT-3′ and 5′-GCAAAGTAGAAAAGGGCGACAA-3′. The primers for p21, mdm2, and GAPDH were described (18, 39).
BrdU Incorporation Assays
BrdU incorporation assays were conducted as described (24). Cells were incubated in the presence of 10 μm of BrdU for 20 h. Cells were then fixed with 95% of ethanol and 5% of acetic acid and treated with 2 m HCl containing 1% Triton X-100. The cells were stained with the monoclonal anti-BrdU (Roche Applied Science) antibody followed by staining with Alexa Fluor 546 (red) goat anti-mouse antibodies (Molecular Probes, OR) and DAPI for DNA staining. Stained cells were analyzed under a Leica inverted fluorescence microscope.
Immunofluorescence Staining
Cells transfected with scrambled, L29, or L30 siRNA were fixed and stained with monoclonal anti-B23 antibody (Zymed Laboratories Inc.) followed by staining with Alexa Fluor 488 (green) goat anti-mouse antibody (Molecular Probes, OR) as well as DAPI for DNA staining. Stained cells were analyzed under a Leica inverted fluorescence microscope.
In Vivo Ubiquitination Assay
U2OS cells were transfected with V5-tagged ubiquitin (V5-Ub) with or without HA-MDM2 plasmid. Five hours later, the cells were transfected with scrambled, L29, or L30 siRNA using Lipofectamine 2000 (Invitrogen). The cells were treated with 40 μm MG132 for 6 h prior to harvest. The cells were harvested 48 h after siRNA transfection. Cleared cell lysates were immunoprecipitated with anti-p53 (DO-1) antibody followed by IB with anti-V5 antibody to detect p53 ubiquitination.
In Vivo NEDDylation Assay
U2OS cells were transfected with plasmids encoding His-NEDD8 and FLAG-L11 followed by scrambled, L29, or L30 siRNA. Cells were harvested 48 h after siRNA transfection. In vivo NEDDylation assay was conducted as previously described (40, 41). Briefly, 25% of the cells were used directly for IB. The rest of cells were lysed in buffer I (6 m guanidinium-HCl, 0.1 m Na2HPO4/NaH2PO4, 10 mm Tris-HCl, pH 8.0, 10 mm β-mercaptoethanol), and incubated with nickel-nitrilotriacetic acid beads (Qiagen) at room temperature for 4 h. After wash, NEDDylated proteins were eluted and analyzed by IB with anti-FLAG antibodies (40, 41). Alternatively, U2OS cells were transfected with FLAG-L11 and V5-NEDD8 followed by transfection with scrambled, L29, or L30 siRNA. The cell lysates were immunoprecipitated with anti-FLAG (M2) antibody followed by immunoblot with anti-V5 antibody.
Cell Cycle Analyses
Cells transfected with siRNAs as indicated in the figure legends were fixed and stained in 500 μl of propidium iodide (PI, Sigma) stain buffer (50 μg/ml PI, 200 μg/ml RNase A, 0.1% Triton X-100 in phosphate-buffered saline) at 37 °C for 30 min. The cells were then analyzed for DNA content using a BD Biosciences FACScan flow cytometer. Data were analyzed using the CellQuest and Modfit software programs.
RESULTS
Ribosomal Proteins L29 and L30 Do Not Bind to MDM2 and Do Not Inhibit MDM2-mediated p53 Degradation
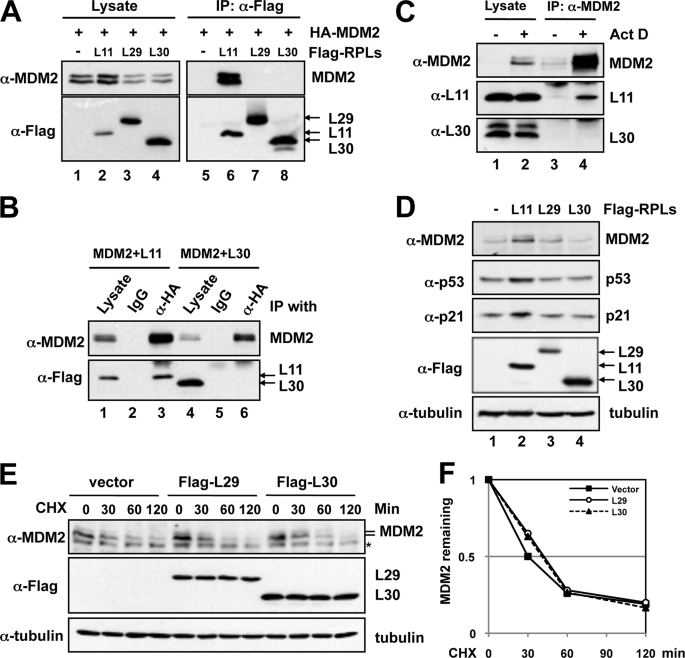
To determine whether ribosomal protein regulation of MDM2 is specific to some, but not all, ribosomal proteins, we sought to identify ribosomal proteins that do not bind to MDM2. In our previous study, we reported that, unlike L11, L29 and L30 do not bind to c-Myc (39). Thus, we tested whether L29 and L30 bind to MDM2. H1299 cells were transfected with MDM2 together with L11, L29, or L30, followed by co-immunoprecipitation-IB assays. As shown in Fig. 1A, MDM2 was co-immunoprecipitated with L11 (lane 6), but not L29 (lane 7) and L30 (lane 8), using anti-FLAG antibody. Reverse co-immunoprecipitation using anti-HA antibody also failed to co-immunoprecipitate MDM2 with L30, whereas L11 can be specifically co-immunoprecipitated with MDM2 using anti-HA, but not control, antibodies (Fig. 1B). The reverse co-immunoprecipitation between L29 and MDM2 was not determined as FLAG-L29 overlaps with the IgG light chain (data not shown). Furthermore, endogenous L11, but not endogenous L30, was co-immunoprecipitated with endogenous MDM2 using anti-MDM2 antibodies in U2OS cells treated with Act D, which induces the interaction between MDM2 and L11 (30) (Fig. 1C). Taken together, these results indicate that L29 and L30 do not bind to MDM2 in cells, supporting the previous report that MDM2 does not interact with intact ribosomes (27).
FIGURE 1.
L29 and L30 do not bind to MDM2 and do not inhibit MDM2-mediated p53 suppression. A, ectopic L29 and L30 do not bind to ectopic MDM2. H1299 cells were transfected with MDM2 in the absence or presence of FLAG-L11, FLAG-L29, or FLAG-L30 plasmid. Cell lysates were immunoprecipitated with anti-FLAG antibody followed by IB with anti-MDM2 and anti-FLAG antibodies. B, L11, but not L30, co-immunoprecipitates with MDM2 in cells. H1299 cells were transfected with HA-MDM2 together with FLAG-L11 or FLAG-L30 plasmid. The cell lysates were immunoprecipitated with anti-HA antibody followed by IB using anti-FLAG antibody. C, endogenous L30 does not interact with endogenous MDM2. U2OS cells were treated with DMSO or 5 nm Act D for 12 h. The cell lysates were immunoprecipitated with anti-MDM2 (SMP14) antibody followed by IB using anti-L11 and anti-L30 antibodies. D, L11, but not L29 and L30, induces p53 activity in cells. U2OS cells were transfected with FLAG-tagged L11, L29, or L30 plasmid. The cell lysates were assayed for the expression of p53, p21, and MDM2 by IB. E and F, overexpression of L29 or L30 does not affect MDM2 protein stability. U2OS cells transfected with the indicated plasmids were treated with 50 μg/ml cycloheximide (CHX) and harvested at different time points as indicated. The cell lysates were assayed for levels of MDM2 and tubulin using IB (E). The asterisk indicates a nonspecific antibody-reacting band. The relative levels of MDM2 were normalized against the expression of tubulin and plotted in F.
To test whether L29 and L30 could regulate the p53 pathway through MDM2-independent mechanisms, we transfected U2OS cells with FLAG-L29 or FLAG-L30 plasmid, and FLAG-L11 was used as a positive control. As shown in Fig. 1D, overexpression of L11 induced the level of endogenous p53 and its target genes p21 and MDM2, consistently with previous studies (26, 29, 30). However, overexpression of either L29 or L30 did not induce the level and activity of p53 as the levels of MDM2 and p21 were not significantly changed (lanes 3 and 4, Fig. 1D). Consistently, overexpression of either L29 or L30 did not affect the protein stability of MDM2 (Fig. 1, E and F) and p53 (data not shown). These results suggest that L29 and L30 do not directly regulate the MDM2-p53 pathway and reveal that ribosomal protein regulation of the MDM2-p53 feedback loop is specific to some, but not all, ribosomal proteins.
Knockdown of L29 or L30 Stabilizes and Activates p53
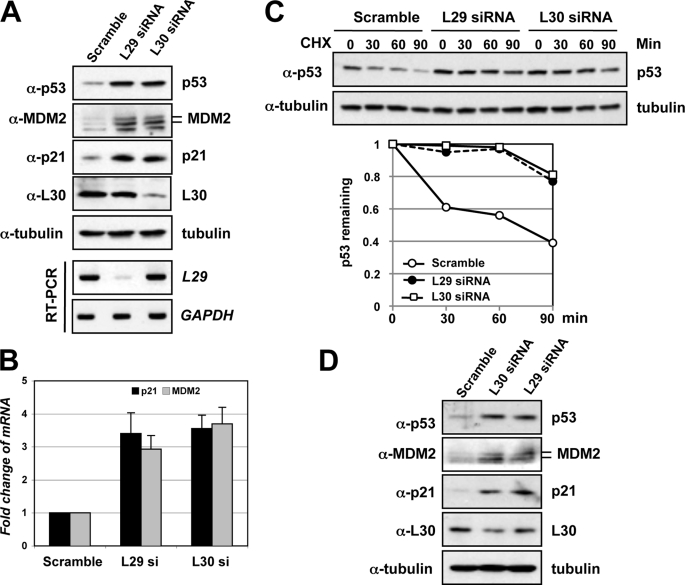
It has been shown that ribosomal proteins play a critical role in p53 activation in response to nucleolar stress, such as that induced by Act D, 5-fluorouracil, MPA, and serum starvation (18, 19, 21, 25, 27, 29, 30). To further explore the significance of ribosomal proteins in regulating p53 signaling in response to perturbation of ribosomal biogenesis, we next asked whether direct perturbation of ribosomal biogenesis by knocking down L29 or L30 would induce ribosomal stress-p53 response, because both proteins do not directly regulate the MDM2-p53 pathway (Fig. 1). It has previously been shown that knockdown of L29 in colon cancer cells induces differentiation and up-regulation of p21 and p53. However, the p53 signaling upon knockdown of L29 was not explored (42). As shown in Fig. 2A, knockdown of either L29 or L30 significantly induced the levels of p53 as well as its targets, p21 and MDM2. Consistently, knockdown of L29 or L30 induced the mRNA expression of p21 and MDM2, as determined by RT-qPCR assays (Fig. 2B). The induction of p53 by knockdown of L29 or L30 was due to stabilization of p53, because transfection of either L29 or L30 siRNA significantly prolonged the half-life of p53 compared with scrambled RNA transfected cells (Fig. 2C). Knocking down L29 or L30 in human normal fibroblast WI38 cells also markedly induced the levels of p53, MDM2, and p21 (Fig. 2D), indicating that induction of p53 by knocking down L29 or L30 is not cell-type-specific effect. Taken together, these results suggest that perturbation of 60 S ribosomal biogenesis by knocking down either L29 or L30 could induce nucleolar stress.
FIGURE 2.
Knocking down L29 or L30 stabilizes p53 and induces p53 activation. A, p53 induction and activation by knockdown of L29 or L30. U2OS cells were transfected with scrambled, L29, or L30 siRNA. Cell lysates were assayed for expression of p53, p21, MDM2, and L30 by IB using antibodies as indicated. The knockdown of endogenous L29 was determined by RT-PCR detection of the L29 mRNA (bottom panels). B, knockdown of endogenous L29 or L30 increases the mRNA expression of p53 targets mdm2 and p21. Total RNAs were extracted from U2OS cells transfected with siRNAs as in A and subjected to RT-qPCR assays. Relative expression of p21 and mdm2 genes was normalized against the expression of GAPDH. C, knockdown of endogenous L29 or L30 stabilizes p53. U2OS cells transfected with siRNAs as in A were treated with 50 μg/ml CHX and harvested at different time points as indicated. The cell lysates were assayed for levels of p53 and tubulin by IB. The bands were quantified and normalized with loading controls determined by tubulin expression and plotted in the bottom panel. D, knockdown of L29 or L30 induces p53 in WI38 cells. WI38 cells were transfected with scrambled, L29, or L30 siRNA. Cell lysates were assayed for expression of p53, p21, MDM2, and L30 by IB using antibodies as indicated.
Knockdown of L29 or L30 Induces p53-dependent Cell Cycle Arrest
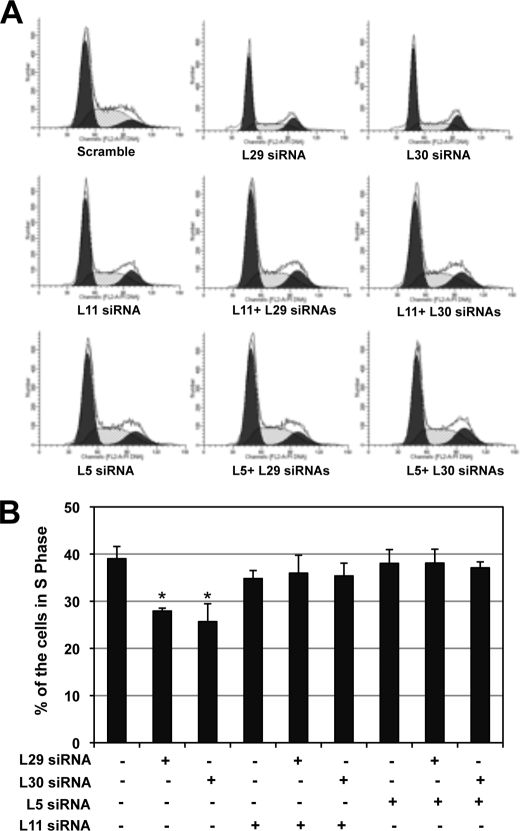
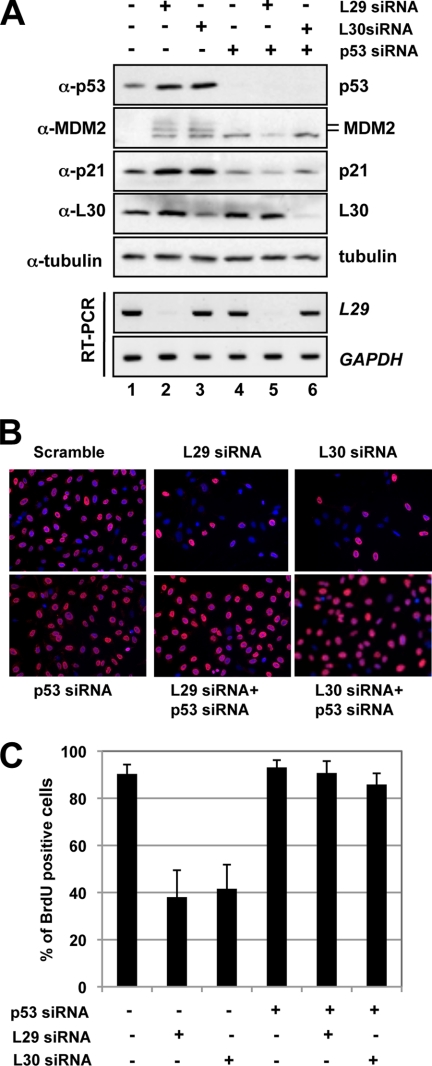
Next, we tested whether p53 activation induced by knockdown of L29 or L30 would result in cell cycle arrest. Initially, we observed that knockdown of either L29 or L30 drastically induced cell cycle arrest (see Fig. 6 and data not shown). To determine if the cell cycle arrest was dependent on p53, we transfected cells with p53 siRNA to ablate endogenous p53. As shown in Fig. 3A, transfection of p53 siRNA efficiently knocked down the endogenous p53 (last three lanes of the top panel). Again, knockdown of either L29 or L30 induced the levels of p53, p21, and MDM2, while further knockdown of p53 abolished the induction of p21 and MDM2. The knockdown of L30 and L29 was confirmed by IB and RT-PCR, respectively (bottom panels of Fig. 3A). Then we used this system to observe whether knockdown of L29 or L30 affects cell proliferation using BrdU incorporation assays. As shown in Fig. 3B, knockdown of either L29 or L30 resulted in dramatic decrease of BrdU-labeled cells, suggesting that knockdown of either L29 or L30 significantly inhibited cell proliferation. Interestingly, further knockdown of p53 abolished this inhibition of cell proliferation. These results are summarized in the Fig. 3C and suggest that knockdown of L29 or L30 induces p53-dependent cell cycle arrest in cells.
FIGURE 6.
Knockdown of L5 or L11 abolishes the cell cycle arrest induced by knockdown of L29 or L30. U2OS cells were transfected with scrambled, L29, L30, L5, or L11 siRNA as indicated. The cells were harvested 48 h post-transfection and stained with PI for cell cycle analysis. The histograms of PI staining from one representative experiment are shown in panel A, The mean percentages of cells in S phase from three independent experiments are shown in panel B; *, p < 0.01, compared with scrambled RNA control (first bar).
FIGURE 3.
Knockdown of L29 or L30 induces p53-dependent cell cycle arrest. A, knockdown of either L29 or L30 induces p53-dependent induction of p21 and MDM2. U2OS cells were transfected with siRNAs as indicated. The cell lysates were assayed for the expression of p53, MDM2, p21, and L30 proteins using IB as well as L29 and GAPDH mRNA using RT-PCR. B and C, knockdown of either L29 or L30 results in p53-dependent inhibition of cell proliferation. U2OS cells were transfected with siRNAs as in A. At 48 h post-transfection, the cells were incubated with BrdU for another 20 h. The cells were fixed and stained with anti-BrdU antibodies (red) and DAPI (blue). The average of BrdU-positive cells is shown in C.
Knocking Down L29 or L30 Inhibits MDM2-mediated p53 Ubiquitination
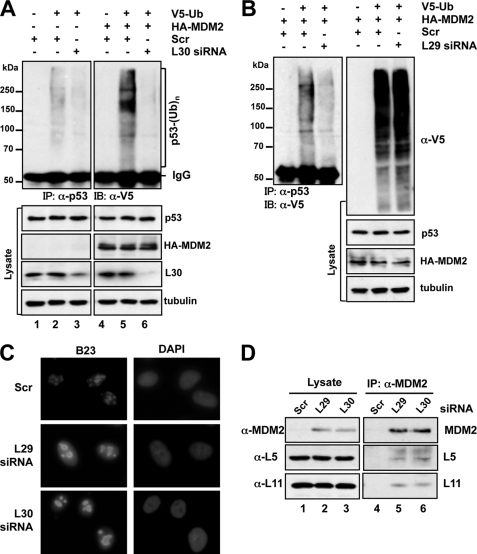
To test whether the stabilization of p53 is due to inhibition of MDM2-mediated p53 ubiquitination and degradation, U2OS cells were transfected with V5-Ub with or without MDM2, as well as scrambled, L29, or L30 siRNA, followed by co-immunoprecipitation assays. As shown in Fig. 4, overexpression of MDM2 enhanced p53 ubiquitination (compare lane 5 to 2 of the top panels, Fig. 4A). Knockdown of L30 drastically inhibited MDM2-mediated p53 ubiquitination. Similarly, knockdown of L29 also significantly reduced the ubiquitinated species of p53, whereas the global ubiquitination was not significantly affected (Fig. 4B). These results clearly suggest that inhibition of p53 ubiquitination contributes to the stabilization of p53 by knockdown of L29 or L30.
FIGURE 4.
Knockdown of L29 or L30 results in nucleolar disruption, enhances the interaction of MDM2 with L5 and L11, and inhibits MDM2-mediated p53 ubiquitination. A, U2OS cells were transfected with V5-Ub with or without MDM2, together with scrambled or L30 siRNA as indicated. Cell lysates were immunoprecipitated with anti-p53 (DO-1) antibody followed by IB with anti-V5 antibody (top panels). The cell lysates were also directly immunoblotted with antibodies as indicated in the bottom panels. B, U2OS cells were transfected with V5-Ub and MDM2, together with scrambled or L29 siRNA. The cell lysates were immunoprecipitated with anti-p53 (DO-1) antibody followed by IB with anti-V5. The cell lysates were also directly immunoblotted with antibodies as indicated in the right panels. C, knockdown of L29 or L30 results in nucleolar disruption. U2OS cells transfected with scrambled, L29, or L30 siRNA were immunostained with anti-B23 antibodies (green) and DAPI (blue). D, knockdown of L29 or L30 enhances the interaction of MDM2 with L5 and L11. U2OS cells transfected with scrambled, L29, or L30 siRNA were subjected to immunoprecipitation with anti-MDM2 (SMP14) antibodies, followed by IB using the indicated antibodies.
Knocking Down L29 or L30 Disrupts the Nucleolus and Enhances the Interaction of MDM2 with L5 and L11
To examine how knockdown of L29 or L30 inhibits MDM2-mediated p53 ubiquitination and degradation, we reasoned that perturbation of ribosomal biogenesis by ablation of individual ribosomal proteins could trigger nucleolar stress and activate p53, unless the ribosomal protein is essential for p53 activation (e.g. L5 and L11). Nucleolar stress is often accompanied by the disruption of the nucleolus (43), although nucleolar disruption is not absolutely required for nucleolar stress-induced p53 activation as in the case of knockdown of S6 (23). To test whether knockdown of L29 or L30 could affect the integrity of the nucleolus, we examined the cellular localization of the nucleolar marker nucleophosmin (B23) in cells transfected with scrambled, L29, or L30 siRNA using immunofluorescence staining. Compared with scrambled RNA-transfected cells, L29 siRNA- or L30 siRNA-transfected cells displayed aberrantly enlarged, distorted, and merged nucleoli with increased distribution of B23 in the nucleoplasm (Fig. 4C), similar to that in L23 siRNA-transfected cells (28), suggesting that ablation of L29 or L30 disrupts the nucleolus.
Because L5 and L11 are required for p53 activation in response to nucleolar stress (18, 19, 21, 24, 30), we asked if knockdown of L29 or L30 could induce the interaction of MDM2 with L5 and L11. Co-immunoprecipitation assays using anti-MDM2 antibodies showed that knockdown of L29 or L30 drastically enhanced the interaction of MDM2 with L5 and L11 (Fig. 4D). Of note, knockdown of L29 or L30 does not significantly affect the total levels of L5 and L11 in cells (Figs. 3A, 5A, and 5B). Altogether, these results suggest that knockdown of L29 or L30 activates p53 through enhancing the binding of L5 and L11 to MDM2.
FIGURE 5.
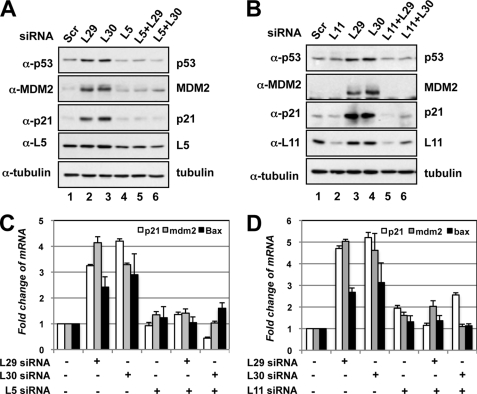
p53 activation induced by knockdown of L29 or L30 requires L5 and L11. A, knockdown of L5 abolished the induction of p53 by knockdown of L29 or L30. U2OS cells were transfected with siRNAs as indicated. Cell lysates were subjected to IB to detect the expression of p53, MDM2, p21, or L5, as indicated. B, knockdown of L11 abolished the induction of p53 by knockdown of L29 or L30. U2OS cells transfected with the indicated siRNAs were subjected to IB to detect the expression of p53, MDM2, p21, or L11. C and D, knockdown of L5 or L11 abolished the induced expression of p21, mdm2, and bax mRNAs by knockdown of L29 or L30. U2OS cells were transfected with scrambled, L29 siRNA, L30 siRNA, L5 siRNA (C), or L11 (D) siRNA as indicated. Total RNAs were extracted and subjected to RT-qPCR assays. Relative expression of p21 and mdm2 genes was normalized against the expression of GAPDH.
Reduction of Endogenous L5 or L11 by siRNA Alleviates p53 Activation and Cell Cycle Arrest Induced by Knockdown of L29 or L30
Next, we examined whether p53 activation induced by knockdown of L29 or L30 requires L5 and L11. To this end, we performed siRNA-mediated ablation experiments. Indeed, reduction of either L5 (Fig. 5A) or L11 (Fig. 5B) levels by siRNA markedly inhibited the level of p53 induced by knockdown of L29 or L30, compared with that in scrambled RNA-transfected cells. Consistently, knocking down either L5 or L11 abrogated the induction of p21 and MDM2 proteins by knockdown of L29 or L30 (Fig. 5, A and B) as well as the mRNA levels of p21, mdm2, and bax as measured by RT-qPCR assays (Fig. 5, C and D). To test whether L5 and L11 are required for cell cycle arrest induced by knockdown of either L29 or L30, we performed cell cycle analysis. As shown in Fig. 6A, knocking down either L5 or L11 significantly inhibited cell cycle arrest induced by knockdown of L29 or L30. These results were summarized in Fig. 6B. Taken together, these results demonstrate that L5 and L11 are required for the p53 activation and cell cycle arrest induced by knockdown of either L29 or L30.
Knocking Down L29 or L30 Results in Nuclear and Nucleolar Retention of L11
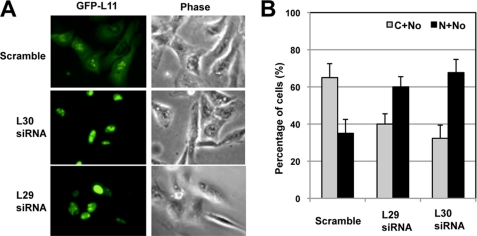
Because knockdown of L29 or L30 does not affect the total levels of L5 and L11, the enhanced binding of MDM2 to L11 and L5 upon knockdown of L29 or L30 could be due to the change in cellular localization of the proteins. It has been shown that L11 could be released from the nucleolus into the nucleoplasm where it binds to MDM2 (21). Thus, we examined the cellular localization of L11 upon knockdown of L29 or L30. Ectopically expressed GFP-L11 is typically expressed in the cytoplasm and the nucleolus, with mild expression in the nucleoplasm (top panels, Fig. 7A). To our surprise, upon knockdown of L29 or L30, the majority of GFP-L11 was localized in the nucleus with dramatic enrichment in the nucleolus (Fig. 7A), although the structure of the nucleolus is disrupted (Fig. 4C). This observation was summarized in Fig. 7B and suggests that knockdown of L30 or L29 results in the nuclear and nucleolar retention of L11 where it might associate with MDM2 and suppress MDM2-mediated p53 ubiquitination, leading to p53 activation.
FIGURE 7.
Knockdown of L29 or L30 results in the nuclear retention of L11. U2OS cells were transfected with GFP-L11 followed by scrambled, L29, or L30 siRNA. The green fluorescence (GFP-L11) and phase contrast images were shown in A. The quantification of GFP-L11 localization is shown in B. C, cytoplasm; N, nucleoplasm; No, nucleolus.
Knocking Down L29 or L30 Enhances the NEDDylation of L11
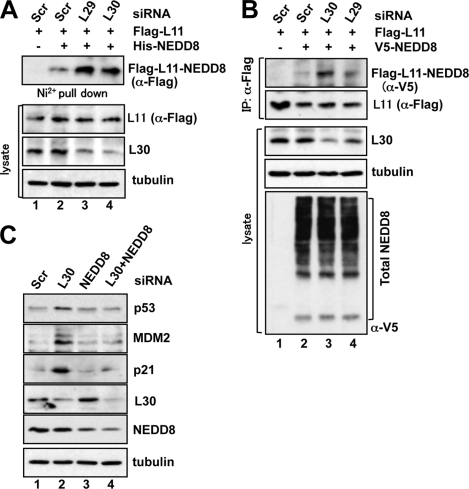
To examine how L11 is accumulated in the nucleus and the nucleolus, we examined whether L11 could be post-translationally modified upon knockdown of L29 or L30. It has been shown that ribosomal proteins are targets for NEDDylation (41), and NEDDylation of L11 promotes nucleolar localization of L11 (40). Thus, we examined whether knockdown of L29 or L30 could affect the NEDDylation of L11. U2OS cells were transfected with FLAG-L11 and His-NEDD8 followed by scrambled, L29, or L30 siRNA. The NEDDylated proteins were purified using nickel-nitrilotriacetic acid pulldown and assayed for L11 NEDDylation by IB with anti-FLAG antibody. As shown in Fig. 8A, knockdown of L29 or L30 significantly increased the NEDDylation of L11 (compare lanes 3 and 4 with lane 2). Similar results were also observed using co-immunoprecipitation assays in cells transfected with FLAG-L11 and V5-NEDD8 (compare lanes 3 and 4 to lane 2), whereas the total NEDDylation in cells were not significantly changed (bottom panel of Fig. 8B). These results indicate that knockdown of L30 or L29 enhances the NEDDylation of L11.
FIGURE 8.
Knockdown of L29 or L30 enhances L11 NEDDylation. A, U2OS cells were transfected with FLAG-L11 and His-NEDD8 plasmids, together with scrambled, L29, or L30 siRNA as indicated. In vivo NEDDylation assay was conducted using nickel-nitrilotriacetic acid beads pulldown, followed by IB with anti-FLAG antibodies. The expression of FLAG-L11, L30, and tubulin is shown in the bottom panels. B, U2OS cells were transfected with FLAG-L11 and V5-NEDD8 plasmids, together with scrambled, L29, or L30 siRNA as indicated. The cell lysates were immunoprecipitated with anti-FLAG antibodies followed by IB with the anti-V5 and anti-FLAG antibodies. The expression of L30, tubulin, and total NEDDylated proteins is shown in the bottom panels. C, knockdown of NEDD8 attenuates p53 induction by knockdown of L30. U2OS cells were transfected with siRNAs as indicated. Cell lysates were subjected to IB analysis to detect the expression of the indicated proteins.
Knocking Down NEED8 Attenuates the p53 Activation by Knockdown of L30
To test whether NEDDylation of L11 plays a role in p53 activation in response to direct perturbation of ribosomal biogenesis, we conducted NEDD8 knockdown assays. As shown in Fig. 8C, knockdown of endogenous NEDD8 drastically alleviated the induction of p53 as well as p21 and MDM2 by knockdown of L30. These results suggest that L11 NEDDylation plays a critical role in mediating p53 signaling induced by knockdown of L30.
DISCUSSION
Recent studies on ribosomal protein regulation of MDM2 have revealed a critical signaling pathway leading to p53 activation in cells in response to nucleolar stress (18, 19, 21, 25–32, 44). A number of ribosomal proteins, including L5, L11, L23, L26, S7, and S3, have been shown to bind to MDM2 and activate p53 by either blocking p53 ubiquitination and degradation by MDM2 (25–32), enhancing p53 translation (45, 46), or modulating DNA repair (47). The current working model is that, in response to the nucleolar stress, these ribosomal proteins may be released from the nucleolus to the nucleoplasm where they can target MDM2 (44). However, it is not clear whether ribosomal protein regulation of MDM2 is specific only to some ribosomal proteins. Our current data reveal for the first time that this ribosomal protein regulation of the MDM2-p53 loop is specific, as two tested ribosomal proteins, L29 and L30, do not bind to MDM2 and do not regulate p53 level and activity in cells (Fig. 1). This finding supports the previous observation that MDM2 does not associate with intact ribosomes (27). Instead, MDM2 associates with a group of ribosomal proteins, forming an MDM2-multiribosomal protein complex (27, 48).
Another finding of this study is that direct perturbation of 60 S ribosomal biogenesis by knocking down either L29 or L30 results in the nucleolar stress-p53 activation pathway, which requires L5 and L11. Knocking down either L5 or L11 by siRNA drastically suppressed L29 or L30 siRNA-mediated p53 activation and cell cycle arrest (Figs. 5 and 6). Also, the knockdown of L29 or L30 markedly enhanced the interaction of MDM2 with L5 and L11 (Fig. 4D) and inhibited MDM2-mediated ubiquitination of p53 (Fig. 4, A and B). Therefore, these data provide firm evidence supporting the critical role for L5 and L11 in mediating the p53 checkpoint in response to nucleolar stress, ensuring a fine coordination between cell cycle progression and ribosomal biogenesis.
Recently, it has been shown that perturbation of 40 S ribosomal biogenesis by the knockdown of S6 leads to p53-dependent cell cycle arrest that also requires L11 (23). In this study, knockdown of S6 enhanced the translation of L11, leading to increased binding of L11 to MDM2 and subsequent p53 activation. In contrast, we did not observe a change in the levels of both L5 and L11 upon knockdown of L29 or L30. However, the interaction of MDM2 with L5 and L11 was clearly increased when either L29 or L30 was knocked down. Rather, this observation might be explained by the change in cellular localization of these proteins, as knockdown of L29 or L30, unlike the knockdown of S6 (23), disrupts the integrity of the nucleolus (Fig. 4C). Indeed, knockdown of L29 or L30 by siRNA caused significant nuclear and nucleolar retention of L11 (Fig. 7), suggesting that L11 might inhibit MDM2 in these compartments. Further analysis showed that knockdown of L29 or L30 enhanced the NEDDylation of L11. Ablation of endogenous NEDD8 significantly attenuated the p53 activation induced by knockdown of L30 (Fig. 8). Thus, L11 NEDDylation plays a crucial role in mediating p53 activation in response to perturbation of 60 S ribosomal biogenesis. Supporting this notion, NEDDylation of L11 is essential for its nucleolar localization as well as its role in mediating p53 signaling in response to treatment with a low dose of Act D (40). Of note, both knockdown of L30 or L29 and Act D treatment require L11 and NEDD8 to activate p53 but the mechanism is different. Act D treatment results in a decrease of L11 NEDDylation and its nucleoplasmic localization (40). It has been shown that the dynamics of ubiquitin ligases, such as MDM2, are regulated by the nucleolus such that ligases in a mobile state are more efficient at degrading substrate (49). Thus, altering the dynamic nature of cellular localization and mobility of L11 and L5, by either increasing or decreasing, may change MDM2 from a mobile to a static state, leading to its inactivation and p53 activation. Future studies would focus on deciphering how perturbation of 60 S ribosomal biogenesis results in the increase of L11 NEDDylation.
Of note, it is not true that individual knockdown of every ribosomal protein induces p53. Apparently, knockdown of L5, L11, or S7 does not do so. This observation can be simply explained by the fact that these three ribosomal proteins are required for p53 activation induced by nucleolar stress (18, 19, 21, 24, 30, 32), even though their knockdown triggers nucleolar stress. Thus, examination of the effect of knockdown of individual ribosomal proteins on p53 induction could serve as a useful strategy to identify ribosomal proteins that possess a non-redundant role in inhibiting MDM2. If a ribosomal protein is essential for p53 activation, its knockdown will block such p53 activation in cells in response to stress. Likewise, if a ribosomal protein is not essential, its knockdown would trigger nucleolar stress and activate p53.
Thus, we have learned that L5, L11, L23, L26, S7, and S3 (21, 25–32, 45, 47), but not L29 and L30 (this study), interact with MDM2. To understand the complete picture about ribosomal protein regulation of MDM2, it is important to identify all ribosomal proteins that regulate the MDM2-p53 pathway. Thus future studies would include defining ribosomal proteins into the following categories: 1) Non-redundant MDM2 regulators. These proteins bind to and inhibit MDM2, leading to p53 activation, and are essential for p53 activation in response to nucleolar stress. Therefore, individual knockdown of these ribosomal proteins would not induce and activate p53 (e.g. L5, L11, and S7). 2) Redundant MDM2 regulators. These proteins bind to MDM2 and inhibit MDM2, leading to p53 activation, but are not absolutely required for p53 activation in response to stress, as knockdown of these ribosomal proteins also induces and activates p53 (e.g. L23). 3) Non-MDM2 regulators. These ribosomal proteins do not bind to MDM2 and do not inhibit MDM2, but their knockdown induces nucleolar stress and activates p53 (e.g. L29 and L30). 4) Other ribosomal proteins that may bind to MDM2, albeit with different regulatory functions on MDM2 or that do not bind to MDM2 but induce p53 activation when overexpressed via pathways different from the inhibition of MDM2.
Acknowledgments
We thank Dr. Hua Lu for generously providing reagents and Dr. Jayme R. Gallegos and Tiffany DeVine for carefully reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R00 CA127134 from the NCI. This work was also supported by a grant from Medical Research Foundation of Oregon and a startup fund from Oregon Health & Science University (to M.-S. D.).
- E3
- ubiquitin-protein isopeptide ligase
- Act D
- actinomycin D
- ARF
- alternative reading frame
- IB
- immunoblot
- NEDD8
- neural precursor cell expressed, developmentally down-regulated 8
- PI
- propidium iodide
- RP
- ribosomal protein
- rRNA
- ribosomal RNA
- qPCR
- quantitative PCR
- MDM2
- murine double minute 2
- MPA
- mycophenolic acid.
REFERENCES
- 1.Oren M. (2003) Cell Death Differ. 10, 431–442 [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B., Lane D., Levine A. J. (2000) Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 3.Fuchs S. Y., Adler V., Buschmann T., Wu X., Ronai Z. (1998) Oncogene 17, 2543–2547 [DOI] [PubMed] [Google Scholar]
- 4.Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000) J. Biol. Chem. 275, 8945–8951 [DOI] [PubMed] [Google Scholar]
- 5.Honda R., Tanaka H., Yasuda H. (1997) FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 6.Oliner J. D., Pietenpol J. A., Thiagalingam S., Gyuris J., Kinzler K. W., Vogelstein B. (1993) Nature 362, 857–860 [DOI] [PubMed] [Google Scholar]
- 7.Barak Y., Juven T., Haffner R., Oren M. (1993) EMBO J. 12, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X., Bayle J. H., Olson D., Levine A. J. (1993) Genes Dev. 7, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 9.Montes de Oca Luna R., Wagner D. S., Lozano G. (1995) Nature 378, 203–206 [DOI] [PubMed] [Google Scholar]
- 10.Jones S. N., Roe A. E., Donehower L. A., Bradley A. (1995) Nature 378, 206–208 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Xiong Y. (2001) Cell Growth & Differ. 12, 175–186 [PubMed] [Google Scholar]
- 12.Sherr C. J., Weber J. D. (2000) Curr. Opin. Genet. Dev. 10, 94–99 [DOI] [PubMed] [Google Scholar]
- 13.Rudra D., Warner J. R. (2004) Genes Dev. 18, 2431–2436 [DOI] [PubMed] [Google Scholar]
- 14.Ruggero D., Pandolfi P. P. (2003) Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 15.Dai M. S., Lu H. (2008) J. Cell Biochem. 105, 670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashcroft M., Taya Y., Vousden K. H. (2000) Mol. Cell Biol. 20, 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilkes D. M., Chen L., Chen J. (2006) EMBO J. 25, 5614–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun X. X., Dai M. S., Lu H. (2007) J. Biol. Chem. 282, 8052–8059 [DOI] [PubMed] [Google Scholar]
- 19.Sun X. X., Dai M. S., Lu H. (2008) J. Biol. Chem. 283, 12387–12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strezoska Z., Pestov D. G., Lau L. F. (2000) Mol. Cell Biol. 20, 5516–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhat K. P., Itahana K., Jin A., Zhang Y. (2004) EMBO J. 23, 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan X., Zhou Y., Casanova E., Chai M., Kiss E., Gröne H. J., Schütz G., Grummt I. (2005) Mol. Cell 19, 77–87 [DOI] [PubMed] [Google Scholar]
- 23.Fumagalli S., Di Cara A., Neb-Gulati A., Natt F., Schwemberger S., Hall J., Babcock G. F., Bernardi R., Pandolfi P. P., Thomas G. (2009) Nat. Cell Biol. 11, 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai M. S., Sun X. X., Lu H. (2008) Mol. Cell Biol. 28, 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai M. S., Lu H. (2004) J. Biol. Chem. 279, 44475–44482 [DOI] [PubMed] [Google Scholar]
- 26.Dai M. S., Shi D., Jin Y., Sun X. X., Zhang Y., Grossman S. R., Lu H. (2006) J. Biol. Chem. 281, 24304–24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Mol. Cell Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin A., Itahana K., O'Keefe K., Zhang Y. (2004) Mol. Cell Biol. 24, 7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Mol. Cell Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D., Zhang Z., Li M., Wang W., Li Y., Rayburn E. R., Hill D. L., Wang H., Zhang R. (2007) Oncogene 26, 5029–5037 [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y., Poyurovsky M. V., Li Y., Biderman L., Stahl J., Jacq X., Prives C. (2009) Mol. Cell 35, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cmejla R., Cmejlova J., Handrkova H., Petrak J., Petrtylova K., Mihal V., Stary J., Cerna Z., Jabali Y., Pospisilova D. (2009) Hum. Mutat. 30, 321–327 [DOI] [PubMed] [Google Scholar]
- 34.Gazda H. T., Sheen M. R., Vlachos A., Choesmel V., O'Donohue M. F., Schneider H., Darras N., Hasman C., Sieff C. A., Newburger P. E., Ball S. E., Niewiadomska E., Matysiak M., Zaucha J. M., Glader B., Niemeyer C., Meerpohl J. J., Atsidaftos E., Lipton J. M., Gleizes P. E., Beggs A. H. (2008) Am. J. Hum. Genet. 83, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai K., Amsterdam A., Farrington S., Bronson R. T., Hopkins N., Lees J. A. (2009) Dev. Dyn. 238, 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horn H. F., Vousden K. H. (2008) Oncogene 27, 5774–5784 [DOI] [PubMed] [Google Scholar]
- 37.Itahana K., Bhat K. P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. (2003) Mol. Cell 12, 1151–1164 [DOI] [PubMed] [Google Scholar]
- 38.Elenbaas B., Dobbelstein M., Roth J., Shenk T., Levine A. J. (1996) Mol. Med. 2, 439–451 [PMC free article] [PubMed] [Google Scholar]
- 39.Dai M. S., Arnold H., Sun X. X., Sears R., Lu H. (2007) EMBO J. 26, 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundqvist A., Liu G., Mirsaliotis A., Xirodimas D. P. (2009) EMBO Rep. 10, 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xirodimas D. P., Sundqvist A., Nakamura A., Shen L., Botting C., Hay R. T. (2008) EMBO Rep. 9, 280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J. J., Huang B. H., Zhang J., Carson D. D., Hooi S. C. (2006) J. Cell Physiol. 207, 287–292 [DOI] [PubMed] [Google Scholar]
- 43.Rubbi C. P., Milner J. (2003) EMBO J. 22, 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Lu H. (2009) Cancer Cell 16, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M. B., Oren M. (2008) Mol. Cell 32, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Cell 123, 49–63 [DOI] [PubMed] [Google Scholar]
- 47.Yadavilli S., Mayo L. D., Higgins M., Lain S., Hegde V., Deutsch W. A. (2009) DNA Repair 8, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindström M. S., Deisenroth C., Zhang Y. (2007) Cell Cycle 6, 434–437 [DOI] [PubMed] [Google Scholar]
- 49.Mekhail K., Khacho M., Carrigan A., Hache R. R., Gunaratnam L., Lee S. (2005) J. Cell Biol. 170, 733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]