Abstract
The oncogenic protein BCL-3 activates or represses gene transcription through binding with the NF-κB proteins p50 and p52 and is degraded through a phospho- and GSK3-dependent pathway. However, the mechanisms underlying its degradation remain poorly understood. Yeast two-hybrid analysis led to the identification of the proteasome subunit PSMB1 as a BCL-3-associated protein. The binding of BCL-3 to PSMB1 is required for its degradation through the proteasome. Indeed, PSMB1-depleted cells are defective in degrading polyubiquitinated BCL-3. The N-terminal part of BCL-3 includes lysines 13 and 26 required for the Lys48-linked polyubiquitination of BCL-3. Moreover, the E3 ligase FBW7, known to polyubiquitinate a variety of substrates phosphorylated by GSK3, is dispensable for BCL-3 degradation. Thus, our data defined a unique motif of BCL-3 that is needed for its recruitment to the proteasome and identified PSMB1 as a key protein required for the proteasome-mediated degradation of a nuclear and oncogenic IκB protein.
Keywords: Cancer Tumor Promoter, Gene Regulation, Gene Transcription, NF-Kappa B, Ubiquitination, BCL-3, Proteasome
Introduction
BCL-3 is an nuclear protein that belongs to the IκB family and regulates gene transcription when bound to the NF-κB proteins p50 or p52 (1–5). BCL-3 was originally identified through molecular cloning of the breakpoint of the t(14; 19) chromosomal translocation found in a subset of human B-cell chronic lymphotic leukemias (6). This translocation triggers BCL-3 overexpression and consequently the deregulation of many target genes that may be involved in survival and cell proliferation (7). Since then, it has been shown that overexpressed BCL-3 contributes to the constitutive NF-κB activation seen in most solid and hematological malignancies (8). Indeed, BCL-3 overexpression has been reported in multiple myelomas and subtypes of lymphomas, even in the absence of any t(14; 19) chromosomal translocation (9–12). Interestingly, deregulated BCL-3 expression also has been reported in solid tumors such as breast cancers, nasopharyngeal and hepatocarcinomas, although the underlying mechanisms remain unclear (13–15). Familial cylindromatosis, a genetic disease characterized by benign tumors of hair follicle keratinocytes (called cylindromas) results from loss-of-function mutations of CYLD,4 a deubiquitin ligase that specifically removes Lys63-linked polyubiquitin chains from various substrates, including BCL-3 (16, 17). As a result, the nuclear import of Lys63-linked polyubiquitinated BCL-3 is enhanced and cell proliferation occurs through cyclin D1 gene induction (17). Therefore, multiple mechanisms that include chromosomal rearrangements or mutations of BCL-3-associated proteins ultimately trigger BCL-3 overexpression in the nucleus, a key event in BCL-3-mediated oncogenesis. This IκB protein has been defined formally as an oncoprotein based on the ability of BCL-3-expressing NIH3T3 cells to form foci and induce tumor growth in nude mice and also because bcl-3 transgenic mice developed lymphoproliferative disorders (18, 19).
To gain more insight on BCL-3 functions, we conducted yeast two-hybrid analysis and defined the proteasome subunit PSMB1 as a BCL-3-interacting protein that binds an unique sequence within the N-terminal domain of this oncoprotein. This interaction is required for BCL-3 degradation, as evidenced by the stabilization of BCL-3 in PSMB1-depleted cells. Therefore, our data defined PSMB1 as a key protein required for the proteasome-dependent degradation of a nuclear IκB protein.
MATERIALS AND METHODS
Cell Culture, Biological Reagents, and Treatments
293, HeLa, and Karpas cells were cultured as described previously (18).
LiCl, TNFα, and MG132 were purchased from Sigma, Peprotech (Rocky Hill, NJ) and A&E Scientific (Marcq, Belgium), respectively. Polyclonal anti-HA, -Hsp90, -Myc, -BCL-3, and anti-IκBα and monoclonal anti-α-tubulin, -Myc, and -PML, and anti-phospho-IκBα antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-FLAG antibodies and beads were purchased from Sigma. Monoclonal anti-NBS1 antibody was from BD Biosciences. The polyclonal anti-PSMB1 antibody was from Biomol (Plymouth Meeting, PA). Polyclonal anti-p50 and monoclonal anti-p52 were from Millipore (Temecula, CA). GFP and PSMB1 siRNAs were purchased from Dharmacon (Lafayette, CO). pMT2T p50, pMT2T p52, pMT2T BCL-3, pMT2T BCL-3 K13–26R, pMT2T BCL-3 MTS, pMT2T BCL-3 ΔN (which lacks the first 112 amino acids), FLAG-BCL-3, -ΔN, -ΔC, and -ΔNΔC, and MTS were described previously, as were FLAG-IκBα and FLAG-IκBα NLS MT (1, 18, 20). FLAG-BCL-3 ΔN10, -ΔN30, -ΔN43, -ΔN67, -ΔN92, and -ΔN120 constructs were generated by PCR, whereas FLAG-BCL-3 ANK M1, -M12, and -M123 and BCL-3 5KR mutants were generated by mutagenesis. The BCL-3 Δ67–92 mutant that corresponds to the BCL-3 ΔPSMB1 mutant was generated by subcloning the cDNA sequence corresponding to amino acids 1 to 66 and subsequently the domain corresponding to amino acid 93 to the stop codon of BCL-3 into the pcDNA3-FLAG construct. FLAG-BCL-3 S394A/S398A/S419A was generated by mutagenesis using the FLAG-BCL-3 MTS mutant as template. Myc-PSMB1 was generated by PCR amplification and subcloning into the pCMV-Myc vector (Invitrogen), using the PSMB1 cDNA clone pulled out from yeast two-hybrid analysis as template. HA-Ub, Ub K63R Myc were described previously (21). Myc-FBW1 was a gift from Dr. Nakayama (Department of Molecular and Cellular Pathology, Medical Institute of Bioregulation, Kyushu University, Japan), whereas Myc-FBW7 and Myc-FBXW8 were generated by PCR using FLAG-FBW7 (a gift from Dr. Clurman, Department of Medicine University of Washington, Seattle, WA) and HA-FBXW8 (a gift from Dr. Jung, Institute für Pathologie, Bochum, Germany) as template, respectively.
Yeast Two-hybrid, Immunoprecipitations, and Immunofluorescence
The bait was generated by subcloning the cDNA sequence of BCL-3 coding for the first 112 N-terminal amino acids into the pGBKT7 vector (Clontech, Mountain View, CA) and yeast two-hybrid experiments were conducted as described previously (21), as were immunoprecipitations involving ectopically expressed proteins (18) as well as immunofluorescence (22).
RESULTS
N-terminal Domain of BCL-3 Harbors a Nuclear Localization Signal Dispensable for the Interaction with the NF-κB Proteins p50 and p52
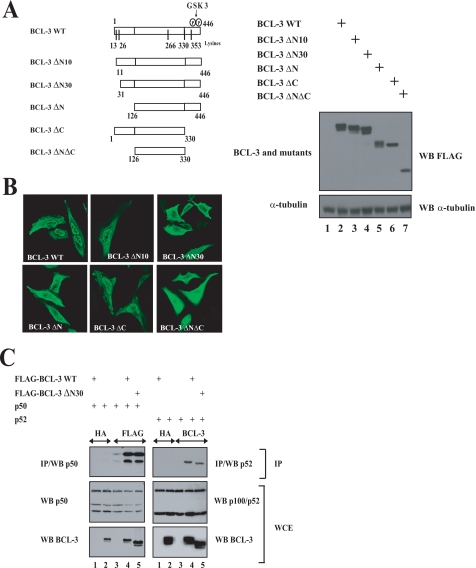
Both the N- and C-terminal domains of BCL-3 were defined previously as key regions for the regulation of the transactivation potential of this oncoprotein, yet the underlying mechanisms remain unclear (1). To learn more on the functions of these domains of BCL-3, we generated a variety of mutants lacking sequences either in the N- or C-terminal domain and first addressed their subcellular localization by immunofluorescence (Fig. 1, A and B). We identified a nuclear localization signal within the first 30 N-terminal amino acids. Indeed, whereas wild type (WT) BCL-3 was mainly but not exclusively located in the nucleus (Fig. 1B), removing the entire domain upstream of the ankyrin (BCL-3 ΔN) or the first 30 N-terminal amino acids of BCL-3 (BCL-3 ΔN30) localizes BCL-3 mainly in the cytoplasm (Fig. 1B). Although the N-terminal domain was required for the nuclear localization of BCL-3, its C-terminal domain was dispensable as a mutant that lacks the entire C-terminal domain downstream of the ankyrin repeats (BCL-3 ΔC) remained largely nuclear (Fig. 1B). Finally, removing both the N- and C-terminal domains of BCL-3 (BCL-3 ΔNΔC) led to an equal distribution of the resulting mutant in the cytoplasm and in the nucleus (Fig. 1B). Whereas the first 30 N-terminal amino acids were required for the nuclear localization of BCL-3, they were dispensable for the interaction with p50 or with p52 as the BCL-3 ΔN30 mutant still bound both NF-κB proteins, as judged by co-immunoprecipitation analysis (Fig. 1C, top panels, lane 5). Taken together, these data defined the N-terminal domain of BCL-3 as a key region for the nuclear localization of this oncoprotein.
FIGURE 1.
The N-terminal domain of BCL-3 harbors a nuclear localization signal. A, left, schematic representation of the BCL-3 constructs used in immunofluorescence analysis. The GSK3 phosphorylation sites (Ser394 and Ser398) as well as the lysine residues (13, 26, 266, 330, and 353) on BCL-3 are illustrated. On the right, anti-FLAG and α-tubulin (loading control) Western blots using extracts from 293 cells transfected with the indicated expression plasmids. B, the N-terminal domain of BCL-3 is critical for its nuclear localization. HeLa cells were transfected with the indicated FLAG-tagged expression plasmids, and immunofluorescence studies were carried out using the anti-FLAG antibody. C, the 30 N-terminal amino acids of BCL-3 are dispensable for the interaction with p50 and p52. 293 cells were transfected with the indicated expression plasmids, and cell extracts were subjected to anti-HA (negative control), anti-FLAG, or anti-BCL-3 immunoprecipitations (IP) followed by anti-p50 or anti-p100/p52 Western blots (WB; left and right top panels, respectively). Anti-p100/p52, anti-p50, and anti-BCL-3 Western blots were carried out with crude cell extracts (WCE) as well (bottom panels).
The Binding of BCL-3 with p50 and p52 Is Required for Its Constitutive Phosphorylation
Whereas the first 30 N-terminal domain of BCL-3, which harbors the nuclear localization signal, is dispensable for the interaction with the NF-κB proteins p50 and p52, the so-called ankyrin repeats are required for binding with those proteins (1).
The sequence of BCL-3 spanning from amino acids 161 to 174 is highly similar to the one of IκBα spanning from amino acids 108 to 121 (Fig. 2A). Interestingly, this second ankyrin repeat harbors the nuclear localization sequence (NLS) of IκBα and three point mutations within this domain indeed impaired IκBα nuclear localization as well as its interaction with p65 (23). To more precisely define the role of this highly conserved domain for the interaction with p50 and p52, we next generated BCL-3 mutants (referred to as BCL-3 ANK M1, BCL-3 ANK-M12, and BCL-3 ANK M123) that harbor one, two, or three point mutations identical to those generated on the corresponding residues of the IκBα sequence (Fig. 2A) (23). We first determined whether these point mutations interfered with BCL-3 binding to p50 or p52 by co-immunoprecipitations in 293 cells. Whereas WT BCL-3, and to a lesser extent BCL-3 ANK M1, associated with endogenous and ectopically expressed p50, both BCL-3 ANK M12 and BCL-3 ANK M123 failed to do so (Fig. 2B, top panel, compare lanes 4 and 5 with lanes 6 and 7). The same conclusion also applied for the ability of WT BCL-3 or BCL-3 mutants to interact with p52 (Fig. 2C, top panel, compare lane 5 with lanes 6 and 7). Of note, an IκBα mutant harboring three mutations (IκBα NLS MT) did not bind as efficiently as wild type IκBα to p50 (Fig. 2B, top panel, compare lanes 8 and 9). Importantly, both BCL-3 ANK M12 and -M123 mutants were no longer constitutively phosphorylated, as judged by their migrating profile on an SDS-PAGE gel (Fig. 2B, middle panel, compare lane 4 with lanes 6 and 7). Thus, our data suggest that binding of BCL-3 to p50 or p52 is required for constitutive BCL-3 phosphorylation. Point mutations within this ankyrin repeat also had consequences on BCL-3 subcellular localization, as the BCL-3 ANK M123 mutant was distributed equally in the cytoplasm and in the nucleus, whereas WT BCL-3 was located mainly in the nucleus (Fig. 2D). To further support the notion that the binding of BCL-3 to p50 or p52 is required for the GSK3-mediated phosphorylation of this oncogenic protein, we looked for these modified forms of BCL-3 in the cytoplasm and in the nucleus of Karpas cells left untreated or stimulated with LiCl, a GSK3 inhibitor. We noticed that BCL-3 was constitutively phosphorylated in a GSK3-dependent manner in both the nucleus and in the cytoplasm and also was bound to p52 in both cell compartments (Fig. 2E, top panels, lanes 2–4 and 6–8). Indeed, the phosphorylated forms of BCL-3 disappeared progressively upon LiCl treatment in the nucleus but also in the cytoplasm (Fig. 2E, third panel from the top, compare lanes 7 and 8 with lane 5 as well as lanes 3 and 4 with lane 1, respectively). Therefore, these data suggest that the binding of BCL-3 to p50 or p52 is the key issue for its constitutive GSK3-mediated phosphorylation rather than its nuclear localization.
FIGURE 2.
BCL-3 binding to p50 or p52 is required for constitutive BCL-3 phosphorylation. A, mutation of residues within the second ankyrin repeats of BCL-3. Both IκBα and BCL-3 are illustrated schematically. The NLS sequence within the second ankyrin repeat of IκBα is shown. The FLAG-IκBα NLS MT construct encodes a FLAG-tagged IκBα protein harboring three mutations within the NLS sequence (23). Single, double, or triple mutants where the corresponding leucine and isoleucine residues within the second ankyrin repeats of BCL-3 were mutated into alanines, as indicated (BCL-3 ANK M1, -M12, or M123, respectively). B and C, the integrity of the second ankyrin repeat of BCL-3 is required for binding to p50 (B) or p52 (C). 293 cells were transfected with the indicated expression plasmids and anti-HA (negative control) -FLAG, or -BCL-3 immunoprecipitations (IP) followed by anti-p50 or p52 Western blot analysis were carried out (top panels). Cell extracts also were subjected to anti-FLAG, -p50, and -p52 Western blot analysis (bottom panels). D, mutations within the second ankyrin repeat alter the nuclear localization of BCL-3. HeLa cells were transfected with the indicated expression plasmids and the resulting cells were subjected to immunofluorescence analysis using the anti-FLAG antibody. E, constitutive GSK3-mediated BCL-3 phosphorylation in the cytoplasm and in the nucleus of Karpas cells. Lymphoma-derived Karpas cells were left untreated (lanes 1, 2, 5, and 6) or stimulated with LiCl for the indicated periods of times (lanes 3, 4, 7, and 8), and cell extracts were subjected to anti-HA (negative control, lanes 1 and 5) or anti-BCL-3 immunoprecipitations (lanes 2–4 and 6–8) followed by anti-p52/p100 Western blot (WB) analysis (top panel). Crude cell extracts were subjected to anti-p52/p100, anti-BCL-3, α-tubulin and anti-NBS1 Western blots as well (bottom panels). WCE, whole cell extract.
The Degradative Lys48-linked Polyubiquitination of BCL-3 Requires N-terminal Lysine 13 and 26 Residues
The critical role of the N-terminal domain of BCL-3 does not only result from the presence of this nuclear localization signal. Indeed, this domain also is targeted by post-translational modifications such as polyubiquitination (18). In fact, BCL-3 is subjected to both degradative (Lys48-linked) and nondegradative (Lys63-linked) polyubiquitination, but the signaling pathways involved in those modifications are characterized poorly (17, 18). To prove that BCL-3 is indeed polyubiquitinated in vivo, we performed immunoprecipitation experiments in denaturing conditions to exclusively detect the polyubiquitinated forms of BCL-3 but not of the associated molecules. Polyubiquitinated adducts of BCL-3 were still detectable even in denaturing conditions that prevented its binding to NF-κB protein p50 (Fig. 3A, compare lanes 2 and 6). As expected, MG132 treatment facilitated the detection of polyubiquitinated adducts of BCL-3, even in denaturing conditions, further supporting the notion that the proteasome is required for BCL-3 degradation (Fig. 3, A and B, respectively, compare lanes 2 and 4). To learn more about the residues required for BCL-3 degradation, we next tested the Lys48-linked polyubiquitination of WT BCL-3 or the BCL-3 ΔN mutant in the presence of MG132 and with a ubiquitin mutant that only makes Lys48 ubiquitin chains (Ub K63R). Removing the N-terminal domain of BCL-3 severely compromised the formation of Lys48-linked polyubiquitin chains on this protein (Fig. 3C, top panel, compare lanes 2 and 3). As this domain harbors two lysines (residues 13 and 26), we mutated both (BCL-3 K13–26R) and addressed the consequences on the Lys48-linked polyubiquitination of BCL-3 in presence of MG132. The detection of polyubiquitinated adducts on BCL-3 was impaired severely upon mutation of both lysine 13 and 26 residues (Fig. 3C, top panel, compare lanes 2 and 4). As expected, the mutations of all five lysine residues of BCL-3 (BCL-3 5KR) also impaired its Lys48-linked polyubiquitination (Fig. 3C, top panel, compare lanes 2 and 5). Interestingly, whereas MG132 did not modify BCL-3 localization, the mutant that lacks all lysine residues or the GSK3 phosphorylation sites (BCL-3 MTS) (18) was mostly nuclear with bigger dots, as judged by immunofluorescence analysis, presumably because of an impaired degradation (Fig. 3D). Taken together, our data defined lysines 13 and 26 as key residues for the degradative Lys48 polyubiquitination of BCL-3.
FIGURE 3.
The degradation of BCL-3 through the proteasome requires lysines 13 and 26. A and B, the proteasome is required for BCL-3 degradation. 293 cells were transfected with the indicated expression plasmids and subsequently left untreated or stimulated for 2 h with MG132 (20 μm) the next day. Cell extracts (non-SDS lysis buffer (lanes 1–4 (A)) or SDS lysis buffer (A, lanes 5–6, and B)) were subjected to anti-FLAG immunoprecipitations (IP) followed by anti-HA (A and B) and anti-p50 (A) Western blot (WB) analysis (top panels). Cell extracts also were subjected to anti-FLAG (A and B), anti-p50 (A), and anti-HA (A and B) Western blot analysis (bottom panels). C, critical roles of lysines 13 and 26 for the Lys48-linked polyubiquitination (poly-Ub) of BCL-3. 293 cells were transfected with the indicated expression plasmids and subsequently treated with MG132 (20 μm) for 4 h. Cell extracts were subjected to anti-BCL-3 immunoprecipitations followed by anti-Myc Western blot analysis (top panel). Anti-BCL-3 and anti-Myc Western blots also were carried out on the crude cell extracts (bottom panels). D, subcellular localization of wild type and BCL-3 mutants. HeLa cells were transfected with the indicated FLAG-tagged expression plasmid, and immunofluorescence studies were carried out using the anti-FLAG antibody. WCE, whole cell extract.
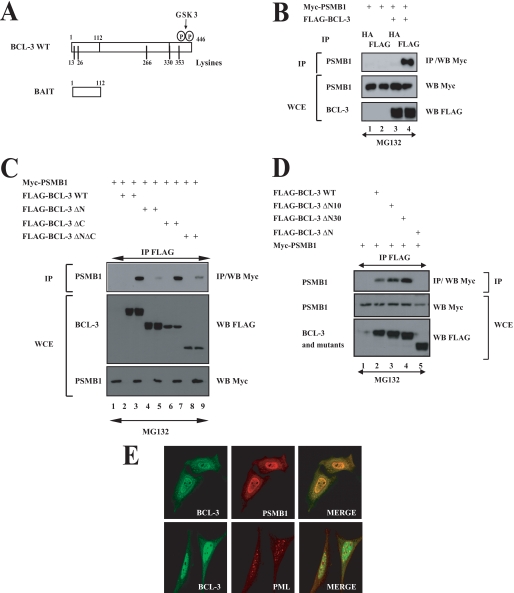
N-terminal Domain Is Required for Association of BCL-3 with Proteasome Subunit PSMB1
To further identify BCL-3-interacting partners involved in its Lys48-degradative polyubiquitination, yeast two-hybrid experiments were conducted using the N-terminal domain of BCL-3 (amino acids 1–112) as bait (Fig. 4A). Positive clones included the co-repressor N-CoR, SUMO 1 (data not shown), as well as the entire coding sequence of PSMB1, a subunit of the 20 S proteasome. This latter interaction also occurred in mammalian cells as overexpressed FLAG-BCL-3 associated with Myc-PSMB1, as judged by co-immunoprecipitation experiments performed in 293 cells (Fig. 4B, top panel, lane 4). The N-terminal domain of BCL-3 was confirmed to be required for binding to PSMB1 as the BCL-3 ΔN mutant failed to bind this proteasome subunit (Fig. 4C, top panel, lane 5). Interestingly, the BCL-3 products lacking the first 10 or 30 N-terminal amino acids (BCL-3 ΔN10 and BCL-3 ΔN30) still interacted with PSMB1 (Fig. 4D, top panel, lanes 3 and 4, respectively). Therefore, these results suggest that BCL-3 interacts with the proteasome via the N-terminal domain between amino acids 31 and 112. We next performed immunofluorescence analysis in HeLa cells and observed that ectopically expressed BCL-3 colocalized with PSMB1 mainly but not exclusively in the nucleus (Fig. 4E, right top panel). Importantly, BCL-3 did not colocalize with the PML bodies in the nucleus (Fig. 4E, right bottom panel). Thus, the association of BCL-3 with the proteasome subunit requires its N-terminal domain, mainly occurs in the nucleus, and does not involve any shuttling into the PML bodies.
FIGURE 4.
PSMB1 is a BCL-3-interacting protein. A, a schematic representation of both BCL-3 and the bait used for yeast two-hybrid analyses. The GSK3 phosphorylation sites (Ser394 and Ser398) as well as the lysine residues (13, 26, 266, 330, and 353) on BCL-3 are illustrated. B–D, ectopically expressed BCL-3 and PSMB1 interact in mammalian cells through the N-terminal domain of BCL-3. 293 cells were transfected with the indicated expression plasmids. Cells were treated with MG132 (20 μm) for 4 h, and anti-HA (B) (negative control) or anti-FLAG (B–D) immunoprecipitations (IP) followed by an anti-Myc Western blot (WB) performed on the immunoprecipitates were carried out (top panels). Crude cell extracts were subjected to anti-Myc and anti-FLAG Western blots as well. E, PMSB1 and BCL-3 mainly co-localize in the nucleus. HeLa cells were transfected with FLAG-BCL-3 and with Myc-PMSB1, and their localizations were revealed through anti-FLAG and anti-Myc immunofluorescence, respectively. PML bodies also were visualized using the corresponding anti-PML antibody. WCE, whole cell extract.
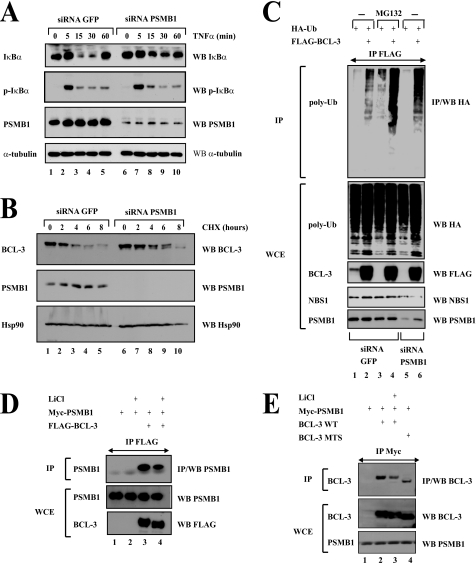
PSMB1 Is Required for BCL-3 Degradation
We next assessed the role of this proteasome subunit in the degradative polyubiquitination of BCL-3 through PSMB1-depleted 293 cells. Because IκBα also is degraded through the proteasome pathway upon TNFα stimulation (24), we first determined whether PSMB1 depletion impaired TNFα-mediated IκBα degradation. Whereas PSMB1 depletion did not interfere with TNFα-mediated IκBα phosphorylation (Fig. 5A, second panel from the top, compare lanes 2 to 5 with lanes 7 to 10), IκBα degradation triggered by this proinflammatory cytokine was attenuated in PSMB1-depleted 293 cells (Fig. 5A, top panel, compare lanes 3 and 4 with lanes 8 and 9). Thus, PSMB1 is required for signal-induced IκBα degradation. The half-life of BCL-3 also is extended in PSMB1-depleted Karpas cells (Fig. 5B, top panel, compare lanes 1 to 5 with lanes 6 to 10). In conclusion, PSMB1 also is required for BCL-3 degradation. A role for PSMB1 in BCL-3 degradation was further supported by the accumulation of BCL-3 polyubiquitinated adducts upon PSMB1 depletion. Indeed, we transfected FLAG-BCL-3 and HA-Ub in siRNA GFP (negative control) or PSMB1 293 cells, isolated the nuclear proteins, and looked for BCL-3 polyubiquitinated forms in anti-FLAG immunoprecipitates. As a positive control, we pretreated the siRNA GFP cells with the proteasome inhibitor MG132. As expected, MG132 treatment dramatically increased the amount of BCL-3 polyubiquitinated forms (Fig. 5C, top panel, compare lanes 2 and 4). Interestingly, an accumulation of BCL-3 polyubiquitinated forms also was detected upon PSMB1 depletion (Fig. 5C, top panel, compare lanes 2 and 6). Of note, this accumulation was not as dramatic as the one seen upon MG132 treatment (Fig. 5C, top panel, compare lanes 4 and 6). This may most likely be due to the partial PSMB1 loss of function as residual amounts of this proteasome subunit were still detectable in the siRNA PSMB1 cells (Fig. 5C, bottom panel, compare lanes 5 and 6 with lanes 1–4). In conclusion, our data provide experimental evidence for a role of PSMB1 in BCL-3 proteasomal degradation.
FIGURE 5.
PSMB1-deficient cells show impaired BCL-3 degradation. A, PSMB1 depletion interferes with TNFα-mediated IκBα degradation but not its phosphorylation. 293 cells were transfected with a siRNA targeting either GFP (negative control, lanes 1–5) or PSMB1 (lanes 6–10). Cells subsequently were left untreated (lanes 1 and 6) or stimulated with TNFα (100 units/ml) for the indicated periods of time. Cell extracts were subjected to anti-IκBα, -phospho-IκBα, -PSMB1, and α-tubulin, as indicated. B, BCL-3 half-life is increased upon PSMB1 depletion. Karpas cells were transfected with the indicated siRNA and subsequently left untreated (lanes 1 and 6) or stimulated with cycloheximide (CHX; 50 μg/ml) (lanes 2–5 and 7–10) for the indicated periods of time. Anti-BCL-3, -PSMB1, and -Hsp90 (used for normalization purposes) Western blots (WB) were carried out on the crude cell extracts, as indicated. C, Lys48-linked polyubiquitinated (poly-Ub) forms of BCL-3 are accumulating upon MG132 treatment or PSMB1 depletion. siRNA GFP or PSMB1 293 cells were transfected with the indicated expression plasmids and subsequently left untreated (lanes 1, 2, 5, and 6) or stimulated with MG132 (20 μm) (lanes 3 and 4) for 4 h. Cell extracts were subjected to anti-FLAG immunoprecipitates (IP) followed by anti-HA Western blot analysis (top panel). Anti-HA, -FLAG, -NBS1, and -PSMB1 Western blots also were carried out with the crude cell extracts (bottom panels). D and E, GSK3 enhances the association of BCL-3 to the proteasome. 293 cells were transfected with the indicated expression plasmids and subsequently left untreated (lanes 1 and 3 (D) and lanes 1, 2, and 4 (E)) or stimulated with LiCl (50 mm) for 2 h (lanes 2 and 4 (D) and lane 3 (E)). Anti-FLAG (D) or -Myc (E) immunoprecipitates were subjected to anti-PSMB1 (D) or BCL-3 (E) Western blot analysis (top panels). Crude cell extracts also were subjected to anti-PSMB1, -FLAG (D), and -BCL-3 (E) Western blots (bottom panels). WCE, whole cell extract.
As the degradation of BCL-3 through the proteasome is regulated by GSK3-mediated phosphorylation (18), we next explored whether GSK3 inhibition by pharmacological means impaired the association of BCL-3 with PSMB1. To address this issue, 293 cells were transfected with FLAG-BCL-3 and Myc-PSMB1 and subsequently treated with LiCl, a GSK3 inhibitor. This inhibitor indeed impaired constitutive BCL-3 phosphorylation (Fig. 5D, bottom panel, compare lanes 3 and 4) and also interfered with the binding of Myc-PSMB1 to FLAG-BCL-3 (Fig. 5D, top panel, compare lanes 3 and 4). This conclusion also was true when Myc-PSMB1 instead of FLAG-BCL-3 was immunoprecipitated in unstimulated or LiCl-treated cells (Fig. 5E, top panel, compare lanes 2 and 3, respectively). Thus, our results suggest that GSK3 phosphorylation triggers the recruitment of BCL-3 to the proteasome via binding to PSMB1. It is, however, important to note that the BCL-3 MTS mutant, which lacks both GSK3 phosphorylation sites, still bound PSMB1, although less efficiently than the wild type protein (Fig. 5E, top panel, compare lanes 4 and 2, respectively). Thus, BCL-3 also may be recruited to the proteasome through GSK3-independent pathways.
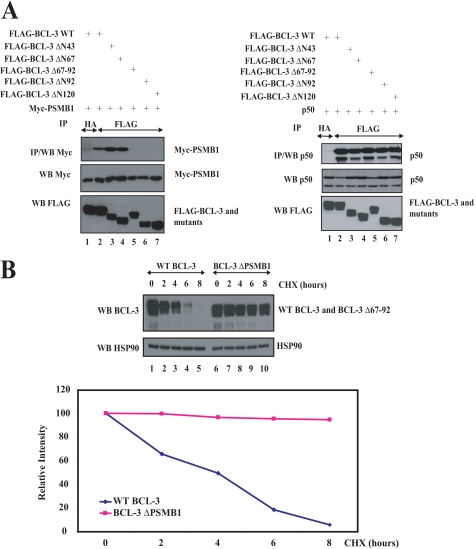
To further confirm that PSMB1 is required for BCL-3 degradation, we next generated a BCL-3 mutant that specifically lacks the PSMB1-interacting domain (“BCL-3 ΔPSMB1”) and subsequently addressed its half-life. We first deleted the first 43, 67, 92, or 120 N-terminal amino acids of BCL-3 and noticed that the BCL-3 ΔN67 but not the ΔN92 mutant bound PSMB1 in co-immunoprecipitation experiments in 293 cells (Fig. 6A, left top panel, compare lanes 4 and 6, respectively). Of note, all of these BCL-3 mutants still bound the NF-κB protein p50 (Fig. 6A, right top panel, lanes 4 and 6). The sequence of BCL-3 between amino acids 68 and 92 harbors the PSMB1-interacting domain as the BCL-3 Δ67-92 mutant (also named “BCL-3 ΔPSMB1”) failed to bind this proteasome subunit but still bound p50 (Fig. 6A, top panels, lane 5). Having generated the BCL-3 ΔPSMB1 mutant, we next addressed its half-life when expressed in 293 cells treated or not with cycloheximide. Whereas WT BCL-3 almost totally disappeared after 6 h of cycloheximide treatment, the BCL-3 Δ67–92 remained easily detectable at that time, and its level of expression was decreased very moderately after 8 h of cycloheximide treatment, when compared with the levels of WT BCL-3 (Fig. 6B, compare lanes 6–10 with lanes 1–5, respectively). A quantification of WT BCL-3 or BCL-3 Δ67–92 levels confirmed this conclusion (Fig. 6B, bottom). Therefore, our data strongly suggest that this BCL-3 Δ67–92 mutant has a prolonged half-life due to its inability to bind PSMB1. This last result thus defines PSMB1 as a key protein required for BCL-3 degradation.
FIGURE 6.
PSMB1 is required for BCL-3 degradation. A, identification of the PSMB1-binding domain on BCL-3. 293 cells were transfected with the indicated expression plasmids an anti-HA (negative control) or -FLAG immunoprecipitations (IP), followed by anti-Myc or -p50 Western blots (WB), were carried out (left top panels and right top panels, respectively). Crude cell extracts were subjected to anti-Myc, -FLAG, and -p50 Western blots, as indicated (bottom panels). B, increased half-life of the BCL-3 mutant that fails to bind PSMB1 (BCL-3 Δ67–92 also named BCL-3 ΔPSMB1). WT or the BCL-3 ΔPSMB1 mutant were transfected in 293 cells, and the resulting cells were left untreated or stimulated with cycloheximide (CHX) for the indicated periods of time. Anti-BCL-3 and -Hsp90 (loading control) Western blots were performed on the crude cell extracts. Bottom panel, a quantification of WT BCL-3 or BCL-3 ΔPSMB1 levels is shown in transfected 293 cells. The signal intensity in unstimulated 293 cells is set to 100% for both experimental conditions.
E3 Ligases FBW1, FBW7, and FBXW8 Are Dispensable for GSK3-mediated BCL-3 Degradation
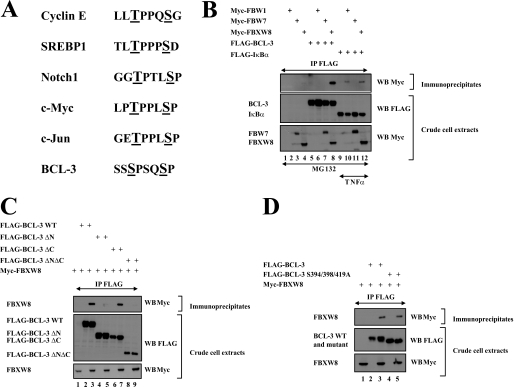
BCL-3 is degraded once phosphorylated by GSK3 on serines 394 and 398 located on its C-terminal domain (18). Importantly, we identified a phosphodegron within this C-terminal domain of BCL-3 that is similar to the ones seen in other proteins known to be phosphorylated by GSK3 and polyubiquitinated by the E3 ligase FBW7 (Fig. 7A) (25–28). Therefore, we investigated whether BCL-3 binds to this E3 ligase by co-immunoprecipitation analysis. As expected, FBW1, an E3 ligase required for TNFα-mediated IκBα degradation (29), efficiently bound this IκB protein in MG132- and TNFα-treated cells but failed to associate with BCL-3 (Fig. 7B, top panel, lanes 12 and 6, respectively). Moreover, FBW7 also failed to bind BCL-3, which indicates that this E3 ligase is dispensable for the GSK3-dependent pathway that leads to BCL-3 degradation. Of note, FBXW8, another E3 ligase, efficiently bound BCL-3 (Fig. 7B, top panel, lane 8). This binding required the N-terminal domain of BCL-3 as the BCL-3 ΔN mutant failed to bind this E3 ligase, whereas the BCL-3 ΔC did (Fig. 7C, top panel, lanes 5 and 7, respectively). However, FBXW8 also is dispensable for BCL-3 degradation through the GSK3-dependent pathway, as a BCL-3 mutant that lacks both GSK3 phosphorylation sites, as well as serine 419 still bound this E3 ligase similarly to WT BCL-3 (Fig. 7D, top panel, compare lanes 3 and 5). Taken together, our data indicate that BCL-3 binds to FBXW8, yet this binding is not required for the GSK3 phospho-dependent degradation of this nuclear oncoprotein.
FIGURE 7.
FBW1, FBW7, and FBXW8 are dispensable for BCL-3 degradation through the GSK3-dependent pathway. A, listing of the phosphodegrons seen in multiple FBW7 substrates and in BCL-3. The residues phosphorylated by GSK3 are underlined. B–D, FBXW8 but not FBW1 and FBW7 bind BCL-3. 293 cells were transfected with the indicated expression constructs and subsequently left untreated (C and D) or stimulated with MG132 and/or TNFα as indicated (B). Anti-FLAG immunoprecipitations followed by anti-Myc Western blot analysis were performed (top panels). Crude cell extracts were subjected to anti-FLAG and -Myc Western blots as well (bottom panels).
DISCUSSION
We report here the identification of the proteasome subunit PSMB1 as a BCL-3-associated protein that is critical for its degradation. This interaction requires the N-terminal domain of BCL-3, which also harbors a NLS sequence as well as the lysine residues required for the Lys48-linked polyubiquitination of this nuclear IκB protein. Our data therefore defined the N-terminal domain of BCL-3 as a key element for its activity.
The mechanisms underlying BCL-3 degradation remained poorly characterized. We identified previously a GSK3-dependent pathway that involves the phosphorylation of two C-terminal residues (18). We now show that BCL-3 is associated to the proteasome and interacts with the proteasome subunit PSMB1 once phosphorylated by GSK3. Interestingly, the GSK3-dependent pathway is negatively regulated by Akt (18), RAS, as well as by phosphatidylinositol 3-kinase (data not shown). Functional links, which appear to be cell-specific, were established previously between NF-κB and RAS. Although the NF-κB proteins p65 and c-Rel are dispensable for RAS-induced cellular transformation in NIH3T3 cells, they nevertheless potentiate RAS oncogenic potential (30). Indeed, some NF-κB-dependent genes such as Serpin B2 and TSLP require p65 and/or c-Rel for their induction of expression by RAS (30). Of note, these genes are not induced by cytokines known to activate NF-κB (TNFα, IL-1β, and etc.), which supports the idea that RAS induces the expression of some NF-κB-dependent genes while suppressing the ability of proinflammatory cytokines to activate NF-κB in fibroblasts (30, 31). Another study reported that oncogenic RAS induces IKK activation and therefore IκBα degradation in liver epithelial cells (32). These studies, combined with our report, therefore suggest that multiple cross-talks exist between oncoprotein RAS and NF-κB. The ability of RAS to stabilize BCL-3 defines a mechanism by which BCL-3 expression may be enhanced even in cases where the t(14; 19) chromosomal translocation does not occur. Elevated levels of BCL-3 in the nucleus can also be the result of loss-of-function mutations targeting the deubiquitine ligase CYLD (17). Similarly, human papillomavirus-positive cancers such as cervical as well as head and neck malignancies also have elevated levels of nuclear BCL-3 because the human papillomavirus-encoded E6 protein inactivates CYLD under hypoxic conditions (33). It also is tempting to speculate that loss-of-mutations of E3 ligases that target BCL-3 may ultimately cause the accumulation of this protein. These results thus considerably extent the pathological contexts that ultimately lead to BCL-3 overexpression in solid and hematological malignancies.
Multiple proteins whose GSK3-dependent phosphorylation triggers their subsequent degradation can be stabilized by RAS. Among those candidates is cyclin E, whose degradation requires the E3 ligase FBW7 (34). The FBW7 substrates such as c-Myc, c-Jun, Notch1, cyclin D1, and cyclin E share a so-called Cdc4 phosphodegron (35) that we actually found on BCL-3 (Fig. 7A). Yet, FBW7 does not bind phosphorylated BCL-3, and the half-life of this IκB oncogenic protein is not enhanced in FBW7-depleted cells (data not shown). Importantly, BCL-3 binds to FBXW8, another E3 ligase, through its N-terminal domain, yet this binding is not modulated by GSK3 phosphorylation. Thus, the E3 ligase that polyubiquitinates BCL-3 upon phosphorylation by GSK3 remains unknown. Additional interactomic studies are currently in process to isolate the E3 ligase that specifically polyubiquitinates BCL-3 through the GSK3-dependent pathway.
BCL-3 may actually be degraded through more than one pathway. Indeed, we show here that a mutant that lacks the GSK3 phosphorylation sites is still, although more weakly, associated to the proteasome. Although this recruitment may not be exclusively required for BCL-3 degradation and also may be involved for the modulation of the transactivation potential of BCL-3, this indicates that other pathways, potentially phospho-independent, also ultimately may trigger BCL-3 degradation. It also is tempting to speculate that some loss-of-function mutations targeting the E3 ligases required for BCL-3 degradation also may explain why the expression level of this oncoprotein is increased even in cases of malignancies where the translocation involving the bcl-3 gene is not observed. Future experimental strategies, mainly relying on interactomic studies, should shed more light on those issues.
Acknowledgments
We are grateful to Drs. Nakayama, Clurman, and Jung for the gift of the expression constructs for FBW1, FBW7, and FBXW8, respectively.
This work was supported by grants from the Fonds de la Recherche Scientifique (FNRS), TELEVIE, the Belgian Federation Against Cancer, the Concerted Research Action Program (04/09-323, University of Liège), the Inter-University Attraction Pole 6/12 (Federal Ministry of Science), the Centre Anti-Cancéreux, the Leon Frédéricq Foundation (ULg), and the King Baudouin Foundation (Brussels, Belgium).
- CYLD
- cylindromatosis
- TNF
- tumor necrosis factor
- GFP
- green fluorescent protein
- siRNA
- small interfering RNA
- Ub
- ubiquitin
- HA
- hemagglutinin
- WT
- wild type
- NLS
- nuclear localization sequence
- SUMO
- small ubiquitin-like modifier
- PMC
- promyelocytic leukemia gene product
- MTS
- mutant serine
- ANK
- ankyrin.
REFERENCES
- 1.Bours V., Franzoso G., Azarenko V., Park S., Kanno T., Brown K., Siebenlist U. (1993) Cell 72, 729–739 [DOI] [PubMed] [Google Scholar]
- 2.Palmer S., Chen Y. H. (2008) Immunol. Res. 42, 210–218 [DOI] [PubMed] [Google Scholar]
- 3.Franzoso G., Bours V., Azarenko V., Park S., Tomita-Yamaguchi M., Kanno T., Brown K., Siebenlist U. (1993) EMBO J. 12, 3893–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr L. D., Duckett C. S., Wamsley P., Zhang Q., Chiao P., Nabel G., McKeithan T. W., Baeuerle P. A., Verma I. M. (1992) Genes Dev. 6(12A), 2352–2363 [DOI] [PubMed] [Google Scholar]
- 5.Nolan G. P., Fujita T., Bhatia K., Huppi C., Liou H. C., Scott M. L., Baltimore D. (1993) Mol. Cell. Biol. 13, 3557–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKeithan T. W., Rowley J. D., Shows T. B., Diaz M. O. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 9257–9260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohno H., Takimoto G., McKeithan T. W. (1990) Cell 60, 991–997 [DOI] [PubMed] [Google Scholar]
- 8.Karin M., Lin A. (2002) Nat. Immunol. 3, 221–227 [DOI] [PubMed] [Google Scholar]
- 9.Mathas S., Jöhrens K., Joos S., Lietz A., Hummel F., Janz M., Jundt F., Anagnostopoulos I., Bommert K., Lichter P., Stein H., Scheidereit C., Dörken B. (2005) Blood 106, 4287–4293 [DOI] [PubMed] [Google Scholar]
- 10.Brenne A. T., Fagerli U. M., Shaughnessy J. D., Jr., Våtsveen T. K., Rø T. B., Hella H., Zhan F., Barlogie B., Sundan A., Børset M., Waage A. (2009) Eur. J. Haematol. 82, 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canoz O., Rassidakis G. Z., Admirand J. H., Medeiros L. J. (2004) Mod. Pathol. 17, 911–917 [DOI] [PubMed] [Google Scholar]
- 12.Nishikori M., Maesako Y., Ueda C., Kurata M., Uchiyama T., Ohno H. (2003) Blood 101, 2789–2796 [DOI] [PubMed] [Google Scholar]
- 13.Cogswell P. C., Guttridge D. C., Funkhouser W. K., Baldwin A. S., Jr. (2000) Oncogene 19, 1123–1131 [DOI] [PubMed] [Google Scholar]
- 14.O'Neil B. H., Bůzková P., Farrah H., Kashatus D., Sanoff H., Goldberg R. M., Baldwin A. S., Funkhouser W. K. (2007) Oncology 72, 97–104 [DOI] [PubMed] [Google Scholar]
- 15.Thornburg N. J., Pathmanathan R., Raab-Traub N. (2003) Cancer Res. 63, 8293–8301 [PubMed] [Google Scholar]
- 16.Bignell G. R., Warren W., Seal S., Takahashi M., Rapley E., Barfoot R., Green H., Brown C., Biggs P. J., Lakhani S. R., Jones C., Hansen J., Blair E., Hofmann B., Siebert R., Turner G., Evans D. G., Schrander-Stumpel C., Beemer F. A., van Den Ouweland A., Halley D., Delpech B., Cleveland M. G., Leigh I., Leisti J., Rasmussen S. (2000) Nat. Genet. 25, 160–165 [DOI] [PubMed] [Google Scholar]
- 17.Massoumi R., Chmielarska K., Hennecke K., Pfeifer A., Fässler R. (2006) Cell 125, 665–677 [DOI] [PubMed] [Google Scholar]
- 18.Viatour P., Dejardin E., Warnier M., Lair F., Claudio E., Bureau F., Marine J. C., Merville M. P., Maurer U., Green D., Piette J., Siebenlist U., Bours V., Chariot A. (2004) Mol. Cell 16, 35–45 [DOI] [PubMed] [Google Scholar]
- 19.Ong S. T., Hackbarth M. L., Degenstein L. C., Baunoch D. A., Anastasi J., McKeithan T. W. (1998) Oncogene 16, 2333–2343 [DOI] [PubMed] [Google Scholar]
- 20.Viatour P., Legrand-Poels S., van Lint C., Warnier M., Merville M. P., Gielen J., Piette J., Bours V., Chariot A. (2003) J. Biol. Chem. 278, 46541–46548 [DOI] [PubMed] [Google Scholar]
- 21.Gatot J. S., Gioia R., Chau T. L., Patrascu F., Warnier M., Close P., Chapelle J. P., Muraille E., Brown K., Siebenlist U., Piette J., Dejardin E., Chariot A. (2007) J. Biol. Chem. 282, 31131–31146 [DOI] [PubMed] [Google Scholar]
- 22.Robert I., Aussems M., Keutgens A., Zhang X., Hennuy B., Viatour P., Vanstraelen G., Merville M. P., Chapelle J. P., de Leval L., Lambert F., Dejardin E., Gothot A., Chariot A. (2009) Oncogene 28, 1626–1638 [DOI] [PubMed] [Google Scholar]
- 23.Sachdev S., Hoffmann A., Hannink M. (1998) Mol. Cell. Biol. 18, 2524–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown K., Gerstberger S., Carlson L., Franzoso G., Siebenlist U. (1995) Science 267, 1485–1488 [DOI] [PubMed] [Google Scholar]
- 25.Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G., Jr. (2005) Cancer Cell 8, 25–33 [DOI] [PubMed] [Google Scholar]
- 26.Welcker M., Singer J., Loeb K. R., Grim J., Bloecher A., Gurien-West M., Clurman B. E., Roberts J. M. (2003) Mol. Cell 12, 381–392 [DOI] [PubMed] [Google Scholar]
- 27.Tetzlaff M. T., Yu W., Li M., Zhang P., Finegold M., Mahon K., Harper J. W., Schwartz R. J., Elledge S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3338–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grim J. E., Gustafson M. P., Hirata R. K., Hagar A. C., Swanger J., Welcker M., Hwang H. C., Ericsson J., Russell D. W., Clurman B. E. (2008) J. Cell Biol. 181, 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaron A., Hatzubai A., Davis M., Lavon I., Amit S., Manning A. M., Andersen J. S., Mann M., Mercurio F., Ben-Neriah Y. (1998) Nature 396, 590–594 [DOI] [PubMed] [Google Scholar]
- 30.Hanson J. L., Hawke N. A., Kashatus D., Baldwin A. S. (2004) Cancer Res. 64, 7248–7255 [DOI] [PubMed] [Google Scholar]
- 31.Hanson J. L., Anest V., Reuther-Madrid J., Baldwin A. S. (2003) J. Biol. Chem. 278, 34910–34917 [DOI] [PubMed] [Google Scholar]
- 32.Arsura M., Mercurio F., Oliver A. L., Thorgeirsson S. S., Sonenshein G. E. (2000) Mol. Cell. Biol. 20, 5381–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An J., Mo D., Liu H., Veena M. S., Srivatsan E. S., Massoumi R., Rettig M. B. (2008) Cancer Cell 14, 394–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minella A. C., Welcker M., Clurman B. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9649–9654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welcker M., Clurman B. E. (2008) Nat. Rev. Cancer 8, 83–93 [DOI] [PubMed] [Google Scholar]