Abstract
Bacterial cultivation has been a mainstay of natural products discovery for the past 80 years. However, the majority of bacteria are recalcitrant to culture, providing an untapped source for new natural products. Metagenomic analysis provides an alternative method to directly access the uncultivated genome for natural products research and for the discovery of novel, bioactive substances. Applications of metagenomics to diverse habitats, such as soils and the interior of animals, are described.
Introduction
Extensive surveys of microbial 16S rRNA gene sequences derived directly from environmental samples suggest that only a small fraction of the bacteria present in nature is readily cultured in the laboratory.1-8 In fact, less than 1% of the bacteria present in most environmental samples is thought to be readily cultured using current fermentation technologies. More than 80 major bacterial divisions have now been identified in culture-independent analyses of environmental samples and fewer than half of these contain cultured isolates.9-11 This diverse collection of as yet uncultured environmental bacteria promises to be a rich, although challenging, source of novel natural products.
Like their free-living relatives, symbiotic bacteria living in close association with other organisms are largely uncultivated and have the genetic potential to synthesize diverse, novel natural products. In addition, symbiotic bacteria are known or suspected to synthesize numerous natural products previously isolated from whole animals and other organisms. A series of recent reviews in this journal and elsewhere documents the importance of “symbiotic” natural products, some of which are clinically used drugs or drug leads.12-14
In an attempt to circumvent the challenges associated with culturing environmental bacteria, whether free-living or symbiotic, culture-independent small molecule discovery strategies have been explored by a number of groups. In these studies, DNA is extracted directly from environmental samples and cloned into model cultured bacterial hosts (Figure 1). Environmental DNA (eDNA) or metagenomic DNA are collective terms used to describe DNA extracted directly from a given environment (e.g. soil, sediment, water, aquatic, plant, etc.) independent of host cultivation or fermentation. Clone libraries derived from eDNA are of potential interest to natural products chemists because all of the genes required for the biosynthesis of a natural product, including genes that code for biosynthetic, regulatory and resistance enzymes are typically found clustered on bacterial chromosomes. Large insert eDNA clones therefore have the potential to contain functionally intact biosynthetic gene clusters that might confer the production of metabolites to an easily cultured bacterial host.
Figure 1.
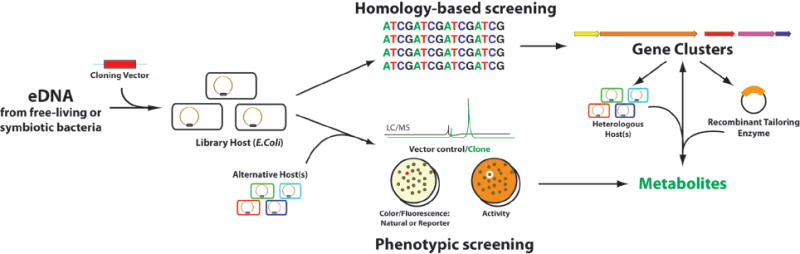
Overview of culture independent strategies used for the discovery of natural products and natural product gene clusters from eDNA libraries.
This review covers culture-independent studies that have focused on the discovery of natural products from either free-living bacterial communities or from bacteria closely associated with other organisms (symbionts).
1. Metagenomics of free-living bacterial communities
Two general strategies, phenotypic and homology (DNA) based screening have been employed to discover new natural products from large eDNA libraries. With phenotypic screening, eDNA libraries are initially examined in simple screens designed to identify clones that exhibit properties traditionally associated with the production of small molecules (i.e. color production, antibacterial activity or signaling phenomena). These clones are then examined in detail for the production of clone-specific small molecules. In homology-based screening, degenerate primers based on conserved natural product biosynthetic genes of interest are used to PCR amplify probe sequences from an eDNA library. The eDNA-specific probes are then used to recover individual clones from the library and the biosynthetic enzymes found on these clones are used generate novel small molecules. Results from both phenotype and homology-based screening of eDNA libraries are discussed here. This section of the review covers all reports of small molecules discovered from the heterologous expression of environmental DNA through January 2009.
2. Phenotypic screening of metagenomic libraries
2.1 The terragines, molecules from the functional expression eDNA clones in Streptomyces lividans
In 2000 a collaboration between the Anderson and Davies groups isolated terragines A-E (1-5) from soil eDNA clones hosted in Streptomyces lividans.15 Prior reports of metagenomic library construction and phenotypic screening had appeared in the literature but none of these studies identified small molecule producing clones.16, 17 Using DNA isolation techniques developed earlier by Davies and co-workers, environmental DNA was recovered directly from soil samples collected in British Colombia, Canada.18 This DNA was then used to construct a small collection of cosmid clones in Escherichia coli using a shuttle vector that would enable the facile intergenic transfer of environmental clones from the library host (E. coli) into S. lividans. Culture broth extracts from each of the resulting S. lividans recombinants were screened by HPLC-ESIMS and two clones were found to produce clone specific metabolites. The characterization of the metabolites produced by these clones resulted in the identification of terragines A-E (1-5) (Figure 2) as well as the re-isolation of the known microbial siderophore norcardamine (6).19
Figure 2.

The terragines (1-5) and norcardamine (6) were characterized from S. lividans transformed with soil eDNA cosmids.
Clones that produce terragine like metabolites were subsequently found at a very high frequency in this library. Eighteen of the approximately 1,000 S. lividans recombinants screened were found to produce members of the terragine/norcardamine families. None of these clones were sequenced and therefore the biosynthetic origin of the terragines is not known. Although terragines did not display any notable biological activity, this work represents the first example of small molecules being characterized from eDNA clones.
2.2 Molecules from the functional expression of eDNA clones in Escherichia coli
Metagenomic library construction methods have been extensively optimized for E. coli and the creation of genetically diverse cosmid and fosmid-based libraries is now easily attainable.16, 20, 21 Despite the potential limitations of using E. coli as an expression host since most secondary metabolites have been derived from phylogenetically distant bacteria, several studies have shown that novel metabolites can be heterologously expressed using this host.
N-acyl amino acids
The first novel bioactive small molecules derived from the heterologous expression of eDNA were a series of long-chain N-acyl amino acid antibiotics reported in 2000.22 In this study, an agar overlay assay against Bacillus subtilis was used to screen a cosmid library for antibacterially active clones. The antibacterial activity exhibited by one of the clones found in this screen was traced to the production of a family of N-acyltyrosine derivatives with saturated and monounsaturated fatty acid side chains eight to eighteen carbons in length (7). Sequencing and random transposon mutagenesis showed that a single open-reading frame (ORF) was necessary for the production of long-chain N-acyltyrosines (CSL12.1; Genbank No. AF324335) and subsequent subcloning of this ORF confirmed it was also sufficient for the production of these metabolites in E. coli. The closest relative identified in a BLAST search was a hypothetical N-acyl transferase (MJ1207) from the cultured bacteria Methanococcus janaschii.
Subsequent publications have reported eDNA clones that produce a variety of long chain N-acylated metabolites with antibacterial activity (Figure 3). These include clones that produce long N-acyltyrosines (7), N-acylphenylalanines (8), N-acyltryptophans (9), N-acylarginines (10) and N-acylputrescines (11).23-25 Transposon mutagenesis and sequence analysis indicated that in each case a single open reading frame was responsible for the biosynthesis of each family of metabolites.23-25 In many cases the N-acyl amino acid synthases (NAS) characterized from these clones show only limited sequence identity to each other.
Figure 3.

Families of N-acyl amino acid antibiotics characterized from eDNA clones. In each case a collection of metabolites with both saturated and monounsaturated side chains of various lengths are produced by the antibacterially active eDNA clone.
A detailed analysis of culture broth extracts from one N-acyltyrosine producing clone indicated that it produced two additional families of clone specific metabolties, long chain enamides (12) and long chain enol esters (13).26 Sequence analysis of the regions surrounding the N-acyl synthase (NAS) from this clone revealed a biosynthetic gene cluster containing 13 ORFs, designated feeA-M (fatty acid enol ester; GenBank No. AY128669). The characterization of intermediates that accumulate in key transposon mutants along with a detailed BLAST analysis revealed the presence of a fatty acid desaturase (feeF), an oxidative decarboxylase (feeG), an N, O-transferase (feeH), an acyl carrier protein (feel), a predicted fatty acid loading system (feeI-K) and the NAS (feeM).26 From these studies the authors proposed the biosynthetic scheme shown in Figure 4.
Figure 4.
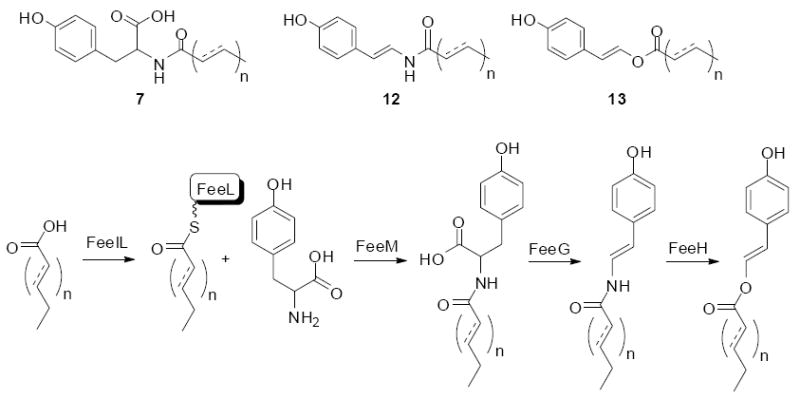
Long chain N-acyltyrosine antibiotics (7) have been characterized from a number of different antibacterially active eDNA clones. One of these clones was found to produce additional families of metabolites (8 and 9). In this clone the fee gene cluster is responsible for the biosynthesis of all three families of compounds.
Violacein and deoxyviolacein
The appearance of color in a bacterial colony is often an indication of small molecule biosynthesis. Simple visual inspection can thus be used as an initial screen to identify clones that might be producing clone-specific natural products. In 2001 the visual screening of a cosmid library constructed from soil eDNA inserts yielded several blue colored E. coli clones. Extensive HRMS and NMR analysis of the colored metabolites produced by this clone found that the color resulted from the production of violacein (14) and deoxyviolacein (15) (Figure 5). Violacein and deoxyviolacein are tryptophan dimers originally isolated from the cultured bacterium Chromobacterium violaceum. Sequencing of transposon insertions that knockout color production, increase color production or result in change in the color identified a cluster of four genes (VioA-D, GenBank No. AF367409) that resembled the violacein gene cluster sequenced from C. violaceum.27 While the gene organization of the violacein cluster from C. violaceum and eDNA is identical, the individual violacein biosynthetic enzymes (i.e. VioA-D) only show 48, 62, 71 and 69% amino acid identity, respectively.27, 28
Figure 5.
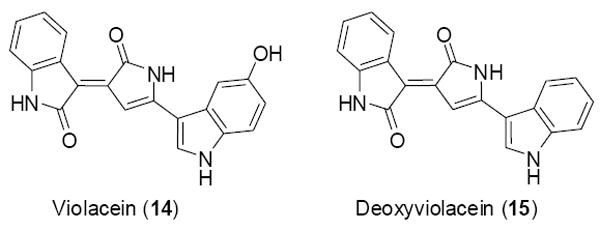
The previously reported metabolites violacein (14) and deoxyviolacein (15) are produced by a purple eDNA cosmid clone.
Turbomycins A and B
In a 2002 study, the visual screening of eDNA libraries prepared in E. coli yielded three pigmented clones. All three clones produced a brown pigment suggestive of melanin.29 One of the clones was examined in detail for the potential production of clone-specific small molecules. While no clone specific metabolites were identified in this study, acid precipitate from the melanin producing clones was shown to contain an increased concentration of two colored metabolites. High-resolution mass spectrometric (HRMS) and nuclear magnetic resonance (NMR) spectroscopic analysis of the colored material isolated from this acid precipitate indicated that the colored metabolites were a previously reported triaryl cation given the name turbomycin A (16) and a new analog given the name turbomycin B (17) (Figure 6). Turbomycin A (16) was originally described as a fungal metabolite from Saccharomyces cerevisiae in 1972.30
Figure 6.

Turbomycin A and B are tri-aryl cations that accumulate in the acid precipitate of melanin producing eDNA clones at a higher level than is observed in acid precipitates from vector control cultures.
Random transposon mutagenesis and subcloning showed that a single ORF (58% G+C) predicted to encode a 40kDa protein was necessary and sufficient for the increased production of the turbomycins. BLAST analysis indicated the putative protein was homologous to several known enzymes; 53% identity to legiolysin (L. pneumophila); 54% identity to hemolysin from Vibrio vulnificus; 49% identity to 4-hydroxyphenylpyruvate dioxygenase (4HPPD) from Pseudomonas sp. (P80064).29 4HPPD-related enzymes are known to catalyze the production of homogentisic acid (HGA), which under aerobic conditions undergoes spontaneous polymerization into HGA-melanin.31 The majority of the HGA undergoes spontaneous polymerization to melanin, which by an unknown mechanism appears to enhance the formation of the turbomycins under acidic conditions.29 The biosynthetic origin of 16 and 17 in this system is not clear.
Isocyanide metabolites
Antibacterial screening of an eDNA library constructed from soil collected in Massachusetts identified a clone that produces compound 18, a new isocyanide functionalized indole-based antibiotic.32 Transposon mutagenesis of the cosmid isolated from this clone suggested that two eDNA-derived open reading frames, isnA and isnB (GenBank No. DQ084328), were required for the production of this new metabolite in E. coli (Figure 7).32 While the isocyanide functional group had been seen previously in bacterial secondary metabolites, its biosynthesis and the source of its component atoms was not known.33, 34 The cloning and heterologous expression of a predicted eDNA derived isonitrile biosynthetic enzyme in E. coli made it possible for well-controlled feeding experiments to reveal the origin of these atoms in 18.32, 35 Extensive “inverse labeled” feeding studies, using E. coli strains with mutations in key primary metabolic steps were designed to systemically interrogate the E. coli metabolome for the source of the isocyanide carbon. These studies revealed that the isocyanide carbon was derived from the C2 carbon of five-carbon sugars from the pentose phosphate pathway. Additional feeding studies suggested that tryptophan is the source of the nitrogen atom in 18. In vitro reconstitution experiments using purified IsnA, IsnB, tryptophan and ribulose-5-phosphate confirmed the hypothetical origins of these isocyanide atoms (Figure 7).32, 35 In a subsequent DNA-based screening study, additional gene clusters harboring isnA from both cultured and uncultured bacteria were identified. This study led to the identification of 12 predicted isnA homologs from eDNA and sequenced bacterial genomes. Expression of these isonitrile synthase genes in various hosts ultimately yielded nine clone-specific small molecules.36
Figure 7.

The isonitrile functionalized indole derivative 18 (a) is produced in E. coli from tryptophan and a five-carbon sugar using the eDNA-derived enzymes IsnA and IsnB (b).
Indigo and indirubin producing clones
The most common classes of clones found when screening eDNA libraries for natural products have been antibacterially active clones that produce long chain N-acyl metabolites (discussed in Section 2.2a), red pigmented antibacterially active clones that express aminolevulinic acid syntheases (hemA), brown melanin producing clones (discussed in Section 2.2c), and blue pigmented clones that produce mixtures of indigo (19) and indirubin (20).23-25, 29, 37-39 Three reports of metagenomic clones that produce indigo and indirubin have appeared in the literature.37-39 A BAC clone derived from New England soil eDNA (GenBank No. DQ000460) was found to contain two predicted indole dioxygenases, each sufficient to produce the observed pigments.37 A fosmid clone constructed using DNA extracted from Korean soil (GenBank No. EF569599) was found to contain a single predicted monooxygenase that is responsible for observed pigments.38 A plasmid clone harboring eDNA from the gypsy moth mid gut (GenBank No. AR053980) contained a two-component flavin-dependent monooxygenase system (MoxZ/Y).39 While MoxY alone is sufficient for color production the color production is enhanced by MoxZ, a predicted NADH:flavin oxidoreductase.
The pigment clones from the New England and Korean soil libraries were identified in simple color screens while the clone from the gypsy moth library was found in an intracellular assay designed to identity small molecule quorum sensing mimics produced by clones in eDNA libraries (metabolite regulated expression, METREX).38 In the METREX assay activated LuxR (LuxR bound to an N-acylhomoserine lactone or an N-acylhomoserine lactone mimic) induces the expression of green fluorescent protein (GFP). Two METREX active eDNA clones that produce small molecules have been reported in the literature. One produces an N-acylhomoserine lactone that is reported to closely resemble N-(3-oxohexanoyl)-L-homoserine lactone and the other is the blue gypsy moth midgut clone described above. The blue clones produce indigo, indirubin and a METREX active metabolite that was not structurally characterized.39, 40 In the METREX assay heterologously produced metabolites do not have to exit the cell to be detected. Intracellular assays like the METREX are therefore potentially more sensitive and may in future help increase the frequency at which small molecule producing clones are found in large eDNA libraries.
2.3 Molecules from the functional expression eDNA clones in Ralstonia metallidurans
Historically, the heterologous expression of eDNA has been carried out in two model systems, E. coli and Streptomyces. The development of a more phylogenetically diverse collection of potential library hosts will likely be needed to fully access the genetic diversity available in any given metagenomic sample. Recent work with the β-proteobacterium Ralstonia metallidurans indicates that, as predicted, expanding the host repertoire is likely to increase the number of small molecule producing clones identified from eDNA library screening.41 In this study, phenotypic screening of an eDNA cosmid library hosted in R. metallidurans yielded two clones, one yellow pigmented clone (RM3) and one antibacterially active clone (RM57), that produced clone-specific small molecules (Figure 9). While the library used in this study was screened in R. metallidurans it was constructed in an new broad host range RK2-based cosmid (pJWC1) that should permit eDNA libraries to be screened in a phylogenetically diverse collection of bacterial hosts.42
Figure 9.
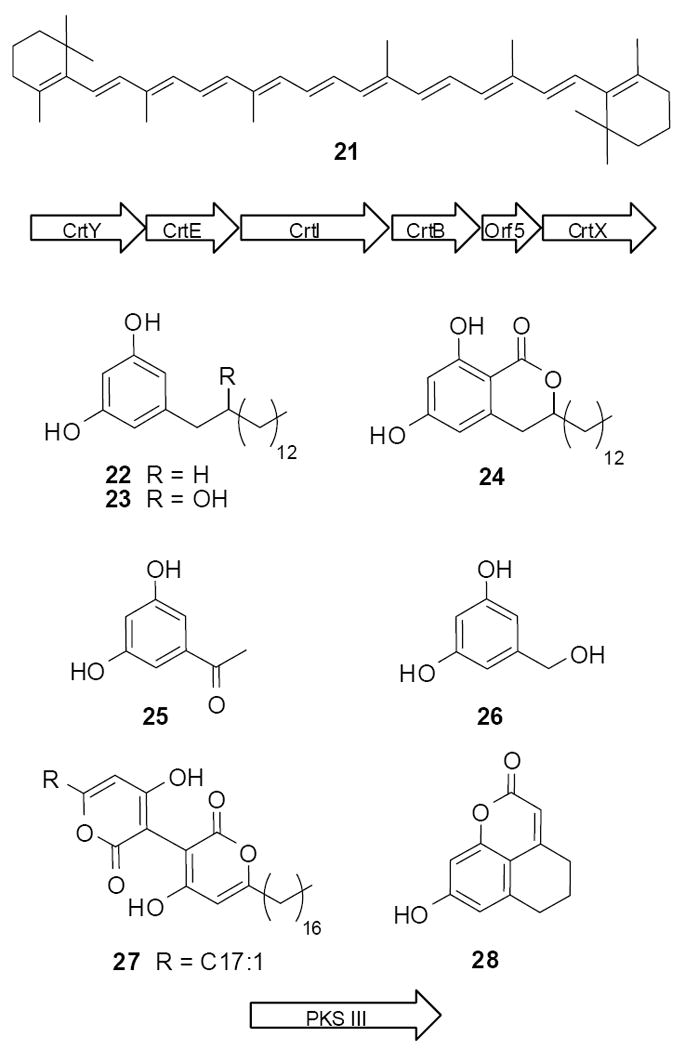
Both known (21-26) and novel (27 and 28) small molecules were characterized from clones found in phenotypic screens of R. metallidurans based libraries.
Cosmids from RM3 and RM57 were isolated and sequenced. The sequencing of RM3 (GenBank No. FJ151553) revealed a six-gene operon with homology to carotenoid gene clusters and the yellow metabolite purified from RM3 cultures was found to be spectroscopically identical to β-carotene (21). Analysis of the RM57 sequence (GenBank No. FJ151552) did not suggest an obvious biosynthetic source of the antibacterial activity produced by this clone. Transposon mutagenesis of the cosmid from RM57 indicated that the antibiotic activity was dependent on a putative type III polyketide synthase. The closest relatives of this type III polyketide synthase are from Acidobacteria, a largely uncultured group of bacteria. Seven small molecules were isolated from cultures of clone RM57, 23-28 were elucidated as long- and short chain substituted resorcinol-like derivatives, while 27 was deduced as a pyrone heterodimer with both a saturated and a monounsaturated side chains. Compound 28 was determined to be a novel tricyclic isocoumarin-based carbon skeleton. The novel compounds 27 and 28 were reported to exhibit antibacterial activity against Staphylococcus aureus and against Bacullis subtilis.41
3. DNA-based Screening of Metagenomic Libraries
3.1 Type II ketosynthase genes from eDNA: PCR-based study
The first report of attempting to access small molecules from the genetic information present in the genomes of uncultured bacteria did not rely on the recovery of intact gene clusters to access small molecules but instead used a PCR based strategy to recover novel type II polyketide (PKS) ketoacyl synthase sequences.43 Bacterial type II PKS biosynthetic gene clusters encode a minimal-PKS that consists of three enzymes: two β-ketoacyl synthases, KSα, which catalyzes the Claisen-type condensation between the growing polyketide and incoming acyl-CoA subunits and KSβ, which controls polyketide chain length (i.e. chain length factor) and an acyl carrier protein (ACP), which anchors the growing polyketide chain during chain elongation. In type II PKS systems, the minimal PKS gene cassette is sufficient to generate the parent polyketide, while additional enzymes (e.g. ketoreductases (KR), cyclases, aromatases) are required to fold and cyclize the poly-β-ketoacyl intermediate into the final structure. Structural diversity in type II PKS natural products therefore originates from variation within the minimal-PKS domains and their combination with assorted modification enzymes. Following the principle that structural diversity can be generated by constructing new combinations of type II PKS biosynthetic elements, Hutchinson and co workers turned to eDNA as a source of novel genes.43 Knowing that all three constituents of the minimal-PKS (KSα-KSβ-ACP) are required for function, a system was designed to determine how heterologous KSαβ pairs affect overall polyketide chain length and structure.
Degenerate PCR primers based on conserved regions within KSα and ACP genes in type II PKSs known to produce 16- and 24-carbon chains were used to amplify KSβ sequences directly from soil DNA. The four KSβ genes found in this study were distinct from any previously described KSβ sequences. By swapping each new KSβ gene for the corresponding gene in the tetracenomycin (TCM) and actinorhodin (ACT) biosynthetic pathways from Streptomyces glaucescens and Streptomyces coelicolor, respectively, hybrid PKS cassettes were constructed (Figure 10). S. lividans and S. glaucescens were then employed as heterologous expression hosts. Ultimately, the known octaketide shunt products UWM1 (29), SEK4 (30), SEK4b (31) and the decaketide TCM F2 (32) (Figure 10) were characterized from these hybrid systems.43 Although this study did not result in the discovery of novel small molecules from eDNA, it was the first study to demonstrate the potential utility of the vast genetic diversity encoded within the genomes of uncultured bacterial communities for the discovery of small molecules.
Figure 10.
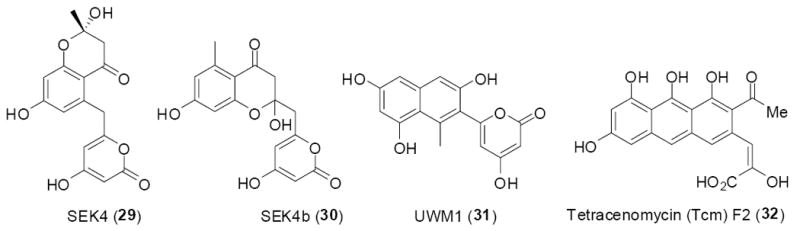
KSβ sequences were PCR amplified from eDNA and these eDNA derived KSβ sequences were used to construct hybrid minimal PKSs. The resulting hybrid minimal PKS cassettes were then shuttled into Streptomyces and found to produce compounds 29-32.
3.2 Type I PKS
The dienic alcohol isomers 33 and 34 were derived from a study that used both DNA and phenotype based strategies to screen a 5,000-member eDNA cosmid clone library (Figure 11). In this study the cosmid library was initially screened by PCR for the presence of Type I polyketide ketosynthase (KS) domains and then screened extracts from a subset of clones were screened by HPLC.44 Eleven novel PKS I like sequences were identified in a screen of the library using two different sets of ketosynthase (KS) specific degenerate primers. The eDNA-derived KS sequences showed similarity to known type I KS sequences from a variety of bacteria including myxobacteria (Stigmatella aurantiaca), cyanobacteria (Microcystis and Nostoc) and various Mycobacteria. One KS containing clone was fully sequenced and found to contain six predicted NRPS and PKS megasynthases. These included a truncated NRPS gene, a mixed NRPS-PKS gene, three full-length PKS genes and a truncated PKS gene. Individual clones from this library (both KS containing clones as well additional randomly selected clones) were individually mobilized into Streptomyces lividans TK24 for heterologous expression studies. Each S. lividans clone was initially screened for antibacterial activity however no antibacterially active clones were detected. Extracts from cultures of 40 S. lividans recombinants grown under a variety of cultures conditions were subsequently screened by HPLC for the presence of clone specific metabolites. Two clones that yielded identical chromatographic profiles were found to produce six related clone specific metabolites. Liquid chromatography-mass spectrometry analysis of the clone metaboiltes found in organic extracts from these S. lividans exoconjugants led to the identification of the novel aliphatic diene alcohol isomers 33 and 34.44
Figure 11.
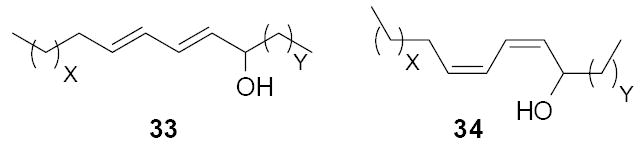
Diene alcohols (33, 34) found during the analysis of culture broth extracts from cultures of S. lividans transformed with a soil eDNA cosmid clone (X+Y=12).
3.3 Targeting OxyC genes
In addition to screening eDNA libraries for completely novel small molecules it also possible to image screening large eDNA libraries for new derivatives of many pharmacologically important natural products. With this in mind, a recent study set out to identify eDNA-derived gene clusters that might encode the biosynthesis of new glycopeptide congeners related to the clinically relevant antibiotics vancomycin and teicoplanin.45 Vancomycin and teicoplanin are nonribosomal peptide synthetase (NRPS) derived, oxidatively cross-linked, heptapeptide antibiotics. OxyC is a conserved cytochrome P450 that catalyzes an oxidative C-C coupling between the hydroxyphenylglycine at position 5 and the dihydroxyphenylgylcine at position 7 in both vancomycin and teicoplanin.46 PCR screening of a 10,000,000 membered eDNA mega-library using degenerate primers based on known OxyC sequences yielded two OxyC-like amplicons. These sequences were then used as probes to recover OxyC containing cosmid clones from the mega-library. Successive rounds of library screening were then carried out to recover overlapping cosmid clones representing two new eDNA-derived glycopeptide gene clusters.
Bioinformatic analysis suggested that the VEG (vancomycin-like eDNA gene cluster, Genbank No. EU874252) and TEG (teicoplanin-like eDNA gene cluster, Genbank No. EU874253) pathways both encode the biosynthesis of new glycopeptide congeners (Figure 12). The VEG pathway contains the complete complement of genes required for the biosynthesis of a cross-linked, glycosylated heptapeptide (hydroxyphenylglycine (HPG),betahydroxytyrosine (BHT), HPG, HPG, HPG, BHT-diydroxyphenylglycine (DPG)).45 The TEG cluster encodes a predicted heptapeptide that only differs from the VEG product by the substitution of DPG for HPG at position three. While the VEG pathway is rich in methyl- and glycosyltranferase finishing enzymes, the TEG pathway contains three predicted sulfotranferase genes (Teg12, 13 and 14). The presence of these sulfotransferases suggests TEG might encode the biosynthesis of an unprecedented polysulfated glycopeptide congener (Figure 12). The TEG pathway is predicted to produce a heptapeptide that only differs from the teicoplanin aglycone (35) by the substitution of BHT for the tyrosine at residue 2. Based on this predicted structural homology the authors elected to use the three TEG sulfotransferases in an attempt to modify the teicoplanin substrate with novel sulfation patterns in vitro. In total, seven sulfated glycopeptide congeners, the sulfo-teicoplanins A-G (36-42), were generated in this study (Figure 12).45 Sulfo-teicoplanins A-G showed activity against MRSA strain USA 300 and VRSA strain JH2. A general trend of increased MIC was observed with the addition of more sulfates to the teicoplanin aglycone (Figure 12).45
Figure 12.
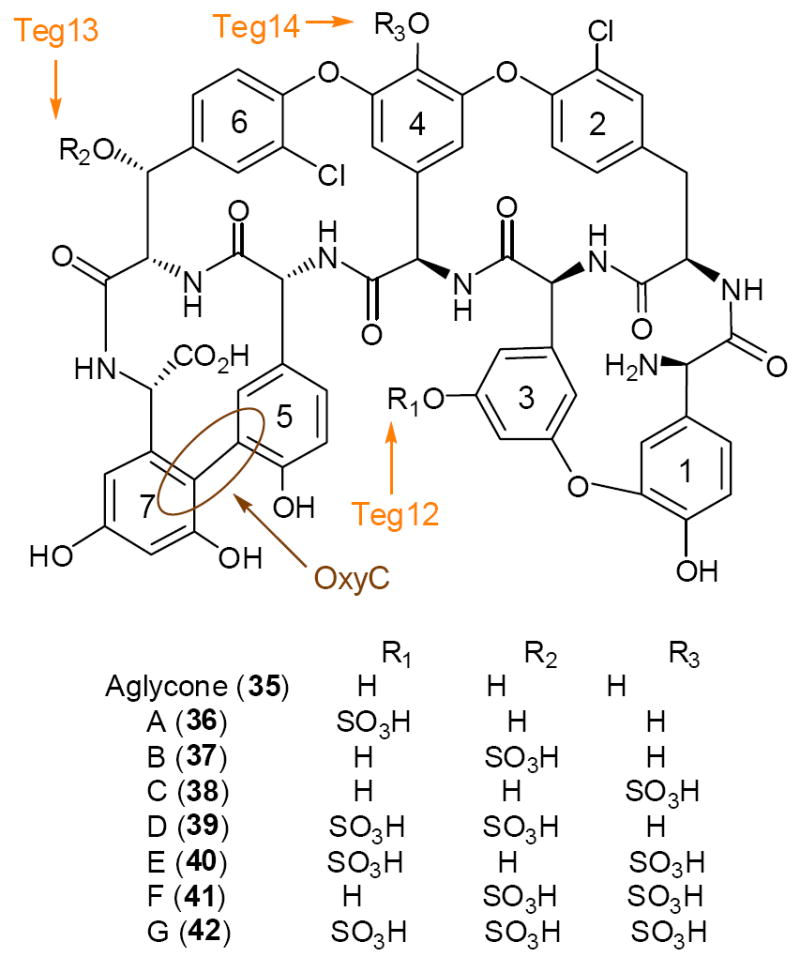
The VEG and TEG glycopeptide gene clusters were cloned from a soil eDNA mega library. Sulfo-teicoplanins A-G (36-42) were produced from the teicoplanin aglycone (35) using three sulfotransferases (Teg 13, 14, and 15) found in the eDNA-derived TEG gene cluster. Gene cluster color coding: nonribosomal peptide synthetase (green), amino acid biosynthesis (green hashed arrows), glycosyl transferase (red), sugar biosynthesis (red hashed arrows), oxidative coupling (brown), methyl transferase (blue), sulfotransferase (orange), halogenase (yellow).
4. Metagenomics of Symbiotic Bacteria
In the previous section, we focused on pathway cloning from environments such as soils, waters, and so on. Here, the focus is on metagenomic approaches that examine organisms as a habitat — in short, metagenomic approaches to symbiosis. There are also many other ways to examine natural products from symbioses, the most important being cultivation-based approaches to obtain pure strains of bacteria. These will not be discussed, as they are covered by an excellent recent series of reviews in this journal.12, 13
Symbiosis is often broadly defined as the intimate association of two or more different organisms. For example, mutualistic symbioses in which each partner benefits are obvious examples of the classical definition of symbiosis, but casual and pathogenic associations are also sometimes considered to be symbioses. The definition of symbiosis is subject to some debate, but for practical reasons, any situation in which the source of genes is a whole (mixed) organism will be considered a symbiosis in this review. Bacteria are arguably the most important “symbionts” from the natural products perspective, although symbiotic fungi, dinoflagellates, and other organisms can also produce important natural products. Bacteria live in virtually every animal and plant, as well as in many other types of organisms such as other bacteria, fungi, and other eukaryotic microbes.
There are two primary ways of looking at organisms as a habitat in the pursuit of natural products. The first, a gene-directed method, is essentially identical philosophically and methodologically to homology-based and phenotypic approaches described in previous sections of this review. For example, natural product biosynthetic genes can be cloned and expressed from uncultivated bacteria inhabiting a whole animal, just as they can be cloned and expressed from soils. A good example of this close relationship can be found in the gypsy moth story earlier in this review.39 The second approach involves looking at natural products first, then engaging in directed gene cloning. For this reason, it is chemistry-directed (also called “structure-directed”) rather than gene directed. For example, there are many potently bioactive marine natural products isolated from animals, and often these compounds are thought to have a microbial origin. Biogenetic hypotheses based upon these chemical structures can be used to clone or identify biosynthetic genes. In terms of technology, chemistry-directed approaches are being used to solve the so-called “supply problem” of marine natural products. Instead of harvesting animals from reef environments to obtain compounds, genetic techniques are used to synthesize the compounds in transgenic bacteria. Chemistry-directed methods also have certain biotechnological advances, described in section 7.
5. Chemistry-directed approaches to symbiosis
Often, a known chemical structure from a whole organism, such as an animal or plant, is suspected to be of microbial origin. In these cases, metagenomics can be used in the discovery and expression of the biosynthetic pathway to the known compound. This bears an exact analogy to pathway cloning from cultured bacteria, and the underlying methods are similar. A major difference is that metagenomes in animals are enormously complicated, with hundreds or thousands of bacterial strains sometimes inhabiting a single animal. This increased complexity has required the development of new techniques and ideas, the basics of which will be reviewed in this section.
Primarily, the complexity of metagenomes in whole organisms has necessitated approaches that simplify the problem. As the cost of sequencing decreases and bioinformatics becomes more powerful, it is increasingly possible to avoid these simplifying schemes, using shotgun sequencing and informatics to identify candidate clones, but this remains challenging. Two basic simplifying approaches have been described: 1) Identification of candidate producing microbial strains prior to cloning; 2) Identification of genes leading to extremely highly homologous compounds or biochemical modifications in simpler systems. Application of these methods has led to the major successes in chemistry-directed metagenomics thus far.
5.1 Candidate strain approaches
Bryostatins
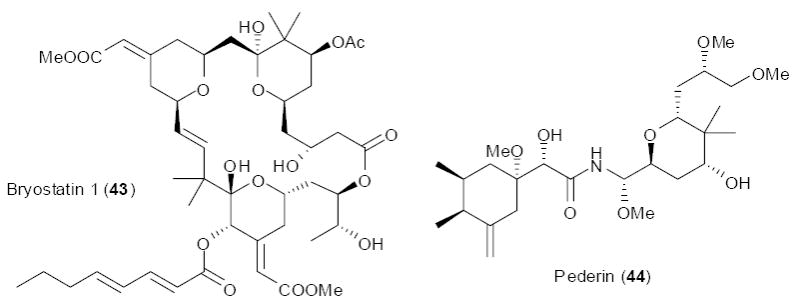
The first genes to be identified using simplifying approaches were those to the highly potent anticancer compounds, bryostatins (e.g., 43). In this example, candidate producing bacteria were identified prior to gene cloning. Bryostatins were isolated in low quantities from the marine bryozoan, Bugula neritina, where they serve to chemically defend the animal and its larvae from predation.47 It was known that B. neritina maintained a single bacterial strain using vertical transmission.48 That is, the larvae of B. neritina contained this strain, ensuring passage from one generation to the next. The bacteria have yet to be cultivated, but they were identified by methods including 16S gene sequence analysis and in situ hybridization to be Candidatus ‘Endobugula sertula’.49 Because it was clear that these were the major (and essentially only) bacteria transmitted in the larvae, larvae were obtained and used to greatly enrich the uncultivated ‘E. sertula’. Subsequently, bacterial type I PKS degenerate primers were used to clone candidate ketosynthase (KS) genes.50 Later, these genes enabled identification and sequencing of the whole gene cluster.51 Fragments of this gene cluster were functionally expressed in E. coli, adding to the probability that the correct cluster had been identified.52 The identification of a single candidate strain, and subsequent enrichment of that strain prior to gene cloning, greatly enabled whole pathway identification.
Pederin and trans-AT genes
Candidate uncultivated bacteria were also identified in Paederus fuscipes beetles, greatly enabling pathway cloning. These beetles secrete a noxious substance containing the highly potent cytotoxin, pederin (44). Microbiological data implicated uncultivated bacteria, Pseudomonas sp., in the synthesis of these compounds, and the beetles themselves were relatively enriched in this uncultivated strain.53-55 This identification enabled cloning of candidate biosynthetic genes using a PCR-based approach. KS-specific degenerate primers were used to identify a candidate cluster from whole beetle metagenomic DNA, leading to the first publication of a “symbiotic” biosynthetic gene cluster in a groundbreaking study.56 Because the genes matched the expected domain architecture for pederin, as type I modular PKS genes, it seemed highly likely that the correct genes were identified. More recently, direct biochemical evidence for this pathway function has been obtained.57 A difficulty in pathway analysis was that the pederin PKS genes lacked acyltransferase (AT) domains, indicating that they belong to a growing family of “trans-AT” PKS genes. The first group of bacterial type I modular PKSs to be cloned were all “cis-AT”, having the AT domain in cis with the remainder of the PKS polypeptide. By contrast, the trans-AT group has a distinct set of AT polypeptides that dock with the rest of the PKS polypeptide at specific docking sites. At the time of the initial pederin pathway discovery, no such other genes were in the literature, complicating analysis. However, the discovery proved to be a boon to chemistry-directed cloning approaches, as described below.
Cyanobactins
The importance of obtaining a simplified system, in which candidate producing bacteria can be enriched, is clearly demonstrated by the above studies. A more recent series of studies relying on enriched uncultivated symbionts involved the relationship between didemnid family ascidians and Prochloron spp. cyanobacteria.58 These ascidians inhabit shallow tropical waters, where they often contain high concentrations of cytotoxic cyclic peptides, such as patellamide A (45), ulithiacyclamide (46), and trunkamide (47) (this family of compounds has recently been named “cyanobactins”).59 Didemnids were also well known for harboring Prochloron spp., which have been extensively studied for their biological properties. For example, they contribute nutritionally to the survival of their host animals.58 Because of this relationship, the first structure paper on cyanobactins identified Prochloron as possible cyanobactin producers.60 Later cell-separation evidence further implicated Prochloron in cyanobactin biosynthesis, although conflicting evidence was obtained.61-63 Prochloron is easily enriched by simply squeezing the host. In our hands, enriched samples containing 50-90% Prochloron DNA are obtained by this method within a 10-min period.
Using this advantage, two different approaches were employed to identify cyanobactin producing genes. A large-insert library of Prochloron-enriched DNA was transferred to E. coli and screened for synthesis of cyanobactins, leading to identification of clones that were capable of cyanobactin synthesis.64 A chemistry-directed cloning approach was also used. In principle, the cyanobactins could be synthesized either ribosomally or nonribosomally. PCR and fosmid analysis of Prochloron metagenomes from 17 samples made the NRPS-based pathway extremely unlikely.65 Therefore, the Prochloron metagenome was directly shotgun sequenced, leading to identification of ribosomal precursor peptides encoding cyanobactins. Fosmids containing these sequences were expressed in E. coli, leading to production of the predicted cyanobactins.66 These studies marked the first pathway expression of biosynthetic genes from uncultivated symbionts. With the sequence context available from metagenome sequencing, the producing organism was confirmed to be Prochloron. This is a simplified system, since the ribosomal peptide sequences are directly encoded and thus readily verified, and the DNA is greatly enriched for Prochloron, but it shows the power of direct sequencing in metagenomics applications. The availability of a gene cluster has also enabled biosynthetic pathway engineering, as discussed later in this review.
Fungal-bacterial symbiosis
Symbioses involving fungi have been examined using the chemistry-targeting approach. Rhizoxin (48) and rhizonin (49) are polyketide-peptide and peptide toxins, respectively, found in the fungus, Rhizopus microsporus. When attempting to clone biosynthetic genes for rhizoxin using a priming strategy, only bacteria-like PKS genes were identified.67-70 By contrast, fungal-type PKS genes were not found. In fact, these PKSs were trans-AT genes that were localized to endosymbiotic bacteria, ‘Burkholderia rhizoxina’, living within the fungal mycelia. Later, rhizonins were also linked to endosymbiotic Burkholderia.68 This symbiosis has since been found to be of global occurrence.70
Leaf-cutting ants
A final example of the chemistry-directed approach involves PCR of whole environments after discovery of pathways in cultivated bacteria. An example of this approach concerns symbiotic Streptomyces sp. living with leaf-cutting ants. These ants cultivate fungal gardens, in which leaves are digested first by fungi, then consumed by ants. Because these beneficial fungal monocultures are sometimes infected by other fungi, the ants have developed a strategy to control fungal infection. The ants have very specific symbiotic relationships with actinomycete bacteria that synthesize antifungal compounds, enabling the ants to maintain their fungal gardens.71 From Streptomyces sp. isolated from various leaf-cutting ant species, the known antifungal agent candicidin A (50) was identified.72 This amphotericin-like polyene significantly inhibited pathogenic fungi, but not beneficial fungi. A metagenomics approach was then applied in which candicidin biosynthetic genes were amplified both from cultivated bacteria and from whole ant DNA, clearly indicating the presence of these genes in a single worker ant.
Discodermolide – the need for simplicity
Clearly, identification of putative producing strains greatly accelerates metagenomic approaches in animals. An example of the difficulty of pathway discovery without prior candidate strain identification can be found in attempts to clone the discodermolide (51) gene cluster from the sponge, Discodermia dissoluta.73 Discodermolide is an important lead anticancer compound of complex, presumed polyketide, biosynthetic origin. Work at the Kosan company employed a unique hybridization array strategy to discover hundreds of different PKS and nonribosomal peptide synthetase (NRPS) gene clusters from the sponge, yet none could be definitively tied to discodermolide production. This study was very important because it revealed for the first time the true complexity of natural product genes in these metagenomes.
Enabling technologies for identification of producing strains
Clearly, as the above examples illustrate relatively precise gene-targeting methods have been required to obtain the desired genes from symbiotic metagenomes. Below, some of the most important non-genetic advances in biosynthetic pathway targeting will be described. These targeting technologies have enormous potential to speed the rate of chemistry-directed cloning in symbiosis and metagenomic research. However, it should be kept in mind that as core genetic technologies continue to decrease in cost, simpler direct sequencing, DNA synthesis, and expression methods will likely compete favorably with these targeting strategies.
In the past 30 years, cell separation has been used to determine which cells contain the natural product of interest within complex symbiotic mixtures.74 These studies impacted later metagenomic approaches, such as those used with Prochloron-ascidians and Oscillatoria-sponges. In this type of study, cells are physically separated by various methods, and then extracted with solvents and analyzed. These cell separation studies could be supplemented with genetic approaches that helped to narrow in on the target organisms. An early example of this type of research connecting cell separation to genetics involved symbiosis between Theonella swinhoei (and related sponges) and filamentous heterotrophic bacteria. Cell separation studies implicated these filamentous bacteria in production of complex cyclic — and presumably nonribosomal — peptides such as theopalauamide (52).75 Using 16S methodology, the bacteria were identified as Candidatus ‘Entotheonella palauensis’, a deeply branching group distantly related to known delta-proteobacteria such as myxobacteria and sulfate reducers.76 ‘E. palauensis’ has since been found as a symbiont of various sponge species, where it harbors a considerably diverse set of natural product biosynthetic genes.73, 77
A substantial improvement to the cell separation procedure was recently reported, in which individual cells are directly examined by MALDI MS.78 Since single cells can be readily obtained and sequenced or used for PCR, this technology promises to greatly accelerate metagenomics research. It could also greatly reduce errors associated with cell separation techniques, most of which derive from the ability of bacteria to secrete natural products and the difficulty of localizing extremely hydrophobic, membrane-associated compounds. These problems make misidentification of producing cells a very real possibility if cell separation is employed. By contrast, a lack of specificity is very readily ruled out by the MALDI method.
Another series of advances derive from immunohistochemical approaches to compound localization. The first such experiment localized latrunculin-family compounds (e.g., 53) to sponge cells.79 Later, the excitatory amino acid derivative, dysiherbaine (54) was localized to Synechocystis sp. cyanobacteria within the sponge Ledenfeldia chondrodes (formerly D. herbacea).80 As has been found in other cyanobacterial symbioses in sponges and ascidians, natural product synthesis was only associated with a subset of Synechocystis cells, despite essentially identical 16S genes. A clear caveat to all types of localization studies — both histochemistry and mass spectrometry based — was found in immunohistochemical examination of bryostatin location in B. neritina bryozoans.81 Although the compounds are almost certainly produced by certain bacteria, the compounds did not co-localize with the bacteria and instead were distributed in host tissues. This study clearly demonstrates the complexity inherent in symbiotic natural products research.
5.2 Chemical and biochemical homology approaches
Onnamide and trans-AT
An alternative chemistry-directed approach involves identification of very precise biosynthetic gene relatives, usually to highly homologous compounds. For example, PKS probes sometimes identify hundreds or thousands of candidate genes in metagenomes without enrichment; this is simply too much to handle using current technology. Identification of much more precise chemical motifs allows the number of candidate genes to be cut down greatly, potentially even to single hits. This approach has been successfully applied to several symbiotic metagenome problems. The first example of such an approach was the cloning of the onnamide gene cluster. Onnamide A (55), found in marine sponges of the genus Theonella, is very similar in structure to pederin (44) from Paederus beetles. In fact, much of the polyketide portion of this molecule is essentially identical to that of pederin. A major difference is the addition of a terminal arginine, potentially added by NRPS domains, in the sponge compounds. In the initial identification of pederin biosynthetic genes, NRPS domains potentially activating arginine were identified.56 However, the gene cluster was “interrupted” by an oxidase domain, making it very likely that the NRPS domains were in fact not functional in the pederin pathway. The very clear homology between pederin and onnamide genes were used to identify the candidate onnamide gene cluster.82 Briefly, the trans-AT feature of this gene cluster was used in the cloning strategy. Specific primers for KS domains were applied. Subsequent phylogenetic analysis allowed identification of trans-AT PKSs, which were just a small subset of the total number of identified PKS gens in the metagenome. Fosmid clones were then screened to provide a partial candidate onnamide cluster. The fosmids also contained sequences for other bacterial gene types, indicating that these “sponge” compounds were more probably of bacterial origin.
The onnamide study also led to the development of yet another enabling technology. The very complex, whole-sponge metagenome of Theonella was analyzed using a new pooling method, in which pools of clones were partitioned into semi-liquid media.83 The pools were then screened by PCR, leading to identification of pools containing the target gene of interest. Pools were diluted, then re-screened by PCR allowing identification of pure fosmids containing the target gene cluster in just a few steps. This widely applicable method allows the rapid, PCR-based identification of fosmids from highly complex mixtures. By contrast, other methods examining large libraries previously relied on hybridization, a much more expensive and potentially even problematic approach in some cases.
Palmerolide
A recent study of the palmerolide (56)-containing Antarctic ascidian, Synoicum adareanum, followed a similar strategy in the first report of ascidian metagenomic PKS genes.84, 85 Palmerolide is a potent antitumor natural product of presumed hybrid PKS-NRPS origin. KS primers were used to identify PKS gene fragments clustering most strongly with the trans-AT group. cis-AT genes were not cloned in this study, and the identified KS genes shared similar features such as GC percentage, indicating that they could derive from similar or identical bacteria or gene clusters. These results are very promising for the ultimate identification of palmerolide biosynthetic genes, although much remains to be done to determine whether the correct genes have been found.
Halogenated peptides from sponge symbionts
An early example of genetically targeting a precise chemical motif can be found in the barbamide (57) / dysidenin (58) group of nonribosomal peptide natural products. The dysidenin family was isolated from marine sponges of the genera Dysidea and Lamellodysidea, which harbor photosynthetic symbiotic cyanobacteria of the genus Oscillatoria. Cell separation studies implicated O. spogeliae in the synthesis of these nonribosomal peptides,86 but cell separation methods can at times be misleading. Barbamide (57) was isolated from Lyngbya majuscula, a tropical marine cyanobacterium that forms mats containing diverse bacteria. The identification of the barbamide biosynthetic gene cluster from L. majuscula facilitated identification of candidate dysidenin biosynthetic genes within marine sponges.87 This family of compounds contains a very rare chemical motif, a multiply chlorinated leucine residue. By sequence alignment and analysis, it proved possible to design specific primers aimed at this chemical motif, targeting the halogenase gene. In two separate studies, this feature was used to isolate candidate halogenases from marine sponges containing the dysidenin family of compounds. In one study, phylogenetic analysis and comparison was used to identify halogenases that were present only in tandem with the dysidinen-group compounds.88, 89 In another study, the identified halogenase fragments were hybridized with mRNA expressed in Oscillatoria sp. within whole sponge tissue, using a method known as catalyzed reporter deposition-fluorescence in situ hybridization.90 This study closed the loop between pathway expression and cell separation studies for the first time. Presumably, these genetic features could possibly be used to obtain intact dysidenin family biosynthetic gene clusters, although this has not been reported. Through these studies, it was clear that the sponges contain multiple strains of Oscillatoria, some of which contained halogenase genes and some of which did not despite some otherwise identical genetic features.
trans-AT genes in sponges
Very recently, a twist on the trans-AT approach was described. A major difference between trans-AT and cis-AT pathways is that in the trans-AT group, phylogenetic analysis of KS domains can be used to predict chemistry.91 Conversely, certain chemical features imply conserved sequence features within KS domains. When analyzing a group of sponges, Fisch et al. found that general KS primers amplified too many diverse PKS gene sequences, making it very difficult to identify the correct gene cluster.92 A nested PCR strategy was then applied. First, the degenerate KS primers were used. Subsequently, a trans-AT specific PCR primer pair was applied. The primers were designed such that one primer was specific to 84% of all trans-AT KSs, while the second primer was specific for certain chemical motifs in the resulting polyketide product. Using this strategy, probable gene clusters were identified for pederin relatives, mycalamide (59) and psymberin (60), both from marine sponge metagenomes. In the case of psymberin, over 400,000 clones were screened using the pooling strategy described above, leading to rapid identification of 5 fosmids containing the putative psymberin cluster.
6. Gene-directed approaches to symbiosis
In the above examples, chemical structures were used to guide metagenomic approaches to gene cloning. Relatively less information is available concerning gene-directed approaches in whole organisms. Recently, it has become extremely common to perform metagenomic analysis on whole animals, plants, and other organisms. However, application of this approach to natural products has relatively rarely been discussed.
Perhaps the first example of a metagenomic approach applied to symbiosis involved lichens, which are symbiotic mixtures of fungi, photosynthetic algae or cyanobacteria, and other bacteria, where primarily fungal PKSs are targeted.93, 94 More recently, the metagenomic approach has been applied to marine sponges. Sponges are considered to be good targets of this approach because they contain abundant and highly diverse bacterial symbionts; in some cases up to 60% of their mass is bacteria.74, 94, 95 The diversity of bacteria in sponges has been recently reviewed.96 In several cases, sponge symbiont PKS and NRPS genes have been cloned by PCR without using chemical guidance. This approach gives a broad picture of the metabolic diversity and potential of marine sponges.
Chemistry-based screening of sponges using KS probes, as described in the preceding section, provided much insight into the diversity of PKS genes in sponges. Much has also been learned by gene-based approaches. An early example examined Pseudoceratina clavata, a sponge found on the Great Barrier Reef.97 Using KS primers, 5 different KS-containing clones were identified. In addition, 5 fosmids containing additional KS sequences were identified by multiplex PCR screening. Bacteria were cultivated from the same sponge, providing a comparison set. While the cultivated bacteria contained both cis- and trans-AT PKSs related to other bacterial genes, the 10 sequenced sponge metagenome clones branched in a new, sponge-specific bacterial clade. This new clade appeared to be associated with lipid biosynthesis.
A concurrent study reported that 11 out of 20 examined sponge species contain this sponge-specific clade, dubbed sup.98, 99 KS domains were amplified by PCR, and 150 clones were identified with <97% sequence identity and thus considered unique. Of these, 127 were part of the sup clade, while 6 were in another apparently sponge-specific group. The remaining sequences fell into the cis-AT, trans-AT, or PKS-NRPS groups that are associated with natural products. Based upon sequencing of the sup cluster, the sponge-specific group was predicted to synthesize methyl-branched fatty acids. The existence of such a large, unanticipated metabolic group that has not been found elsewhere underscores the value of examining animal metagenomes directly. The fact that only a small percentage of sponge PKS genes were predicted to lead to natural products was considered very useful to chemistry-directed approaches, since the sup genes could be discarded as candidates.
A recent study examined the metabolic potential of the sponge, Haliclona simulans.100 Seven PKS genes and no NRPS genes were identified by PCR. All of the PKS genes appeared natural product-associated and did not fall in the sponge-specific clades.
7. Scientific and technological advances enabled by symbiosis metagenomics
It could be asked, given the number of natural products biosynthetic genes found in soil and other metagenomes, why even bother studying pathways from whole animals and other organisms? There are many scientific reasons to pursue such basic studies, but there is also a biomedical and technological rationale. For example, a widely pursued biomedical angle is solving the supply problem of compounds from marine and other animals.74 By specifically accessing genes to important marine invertebrate compounds, it is possible to synthesize the genes in culture instead of collecting animals from coral reefs. There are many other technical advances enabled by examination of symbiotic metagenomes. As one example, examination of the cyanobactin biosynthetic pathways in marine ascidians and their symbionts has led to facile genome mining and rational engineering of new compounds for drug discovery.101-103 Finally, certain types of natural product genes may be strongly associated with or essentially only found in certain animal habitats, as exemplified by the sup group.99
More than 60 cyanobactins have been isolated from ascidians harboring Prochloron bacteria.59 These compounds differ by their amino acid sequences and lengths, as well as by the presence or absence of certain chemical features such as prenylation or oxidation. By studying pathways from nearly 100 different Prochloron-ascidian symbioses, single mutations were identified that led to different chemical features.101, 103 By contrast, in most cultivated bacteria, there are numerous mutations even in pathways to nearly identical compounds. By examining the single mutations in cyanobactin pathways, the wholly unnatural natural product, eptidemnamide, was readily engineered for synthesis in E. coli. Potentially, large numbers of derivatives could be synthesized genetically using this method. Indeed, the natural variation in tandem with the eptidemnamide structure suggests an extremely relaxed substrate selectivity of nearly identical modifying enzymes.59 Another example of the application of this method is in drug supply. The rare cyanobactin derivative trunkamide has only been found in a small number of ascidians, potentially hindering its development as an anticancer drug lead. Using yeast recombination, this compound was introduced into E. coli, where the mature product was synthesized.103 These rapid genetic modifications take minimal work but introduce a great degree of structural variety into “unnatural natural product” libraries.59, 101 As a final example, the enzymes underlying cyanobactin synthesis are of demonstrated broad substrate selectivity and have potential for the synthesis of fine chemicals in vitro or in vivo.104
Symbiosis in cultivated organisms has also yielded engineering insights. For example, the human pathogen Mycobacterium ulcerans (and other mycobacterial strains) produce mycolactone and derivatives, which cause debilitating skin ulcers in humans. By genomic comparison (albeit using cultivated bacteria), sequence features were identified that could enable rational, recombination-based synthesis of new polyketides.105 Thus, symbiotic metagenomes are a promising area of study for biotechnological applications.
8. Conclusions and Future Directions
In recent years, proof of concept has been obtained for metagenomic approaches to natural products from diverse habitats. Many of the limiting technical challenges have been overcome, leaving cost as the major potential barrier to progress. However, as DNA sequencing and synthesis costs rapidly decrease, eDNA methods are becoming mainstays of natural products discovery and research. These methods thus promise a bright future, both for the discovery of novel pharmaceuticals from nature and for the better understanding of the roles these compounds play in nature.
Supplementary Material
Figure 8.

Three different metagenomic studies have identified eDNA clones that produce indigo and indirubin.
Figure 13.
Genes for the biosynthesis of two polyketides were more readily obtained because of guiding microbiology that simplified the problem.
Figure 14.

Cyanobactin biosynthetic genes were cloned from enriched samples of Prochloron symbiotic bacteria living in ascidian animals.
Figure 15.
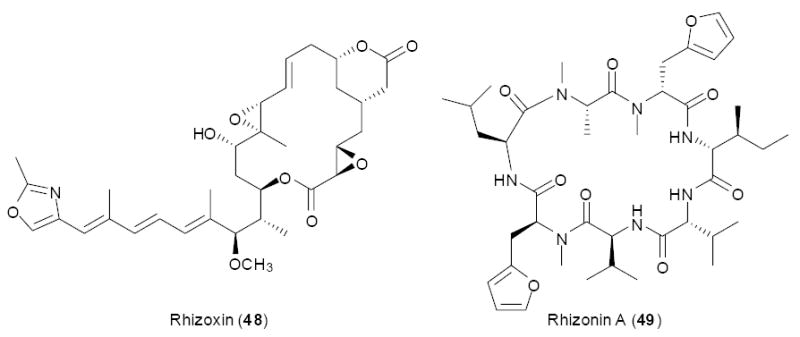
Two metabolites originally isolated from fungi were later shown to originate in endosymbiotic bacteria.
Figure 16.

Polyketides from ant symbionts (50) and, putatively, a sponge symbiont (51).
Figure 17.

Compounds that helped with enabling technologies.
Figure 18.
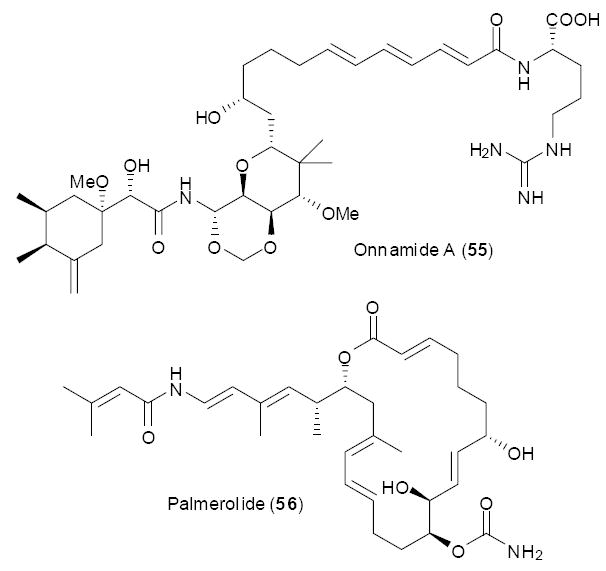
Some polyketides from sponges and ascidians.
Figure 19.
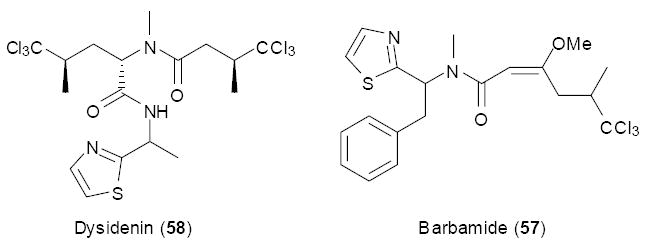
Halogenated peptides, putatively from sponge symbionts.
Figure 20.

Further probable trans-AT products from sponges.
Figure 21.
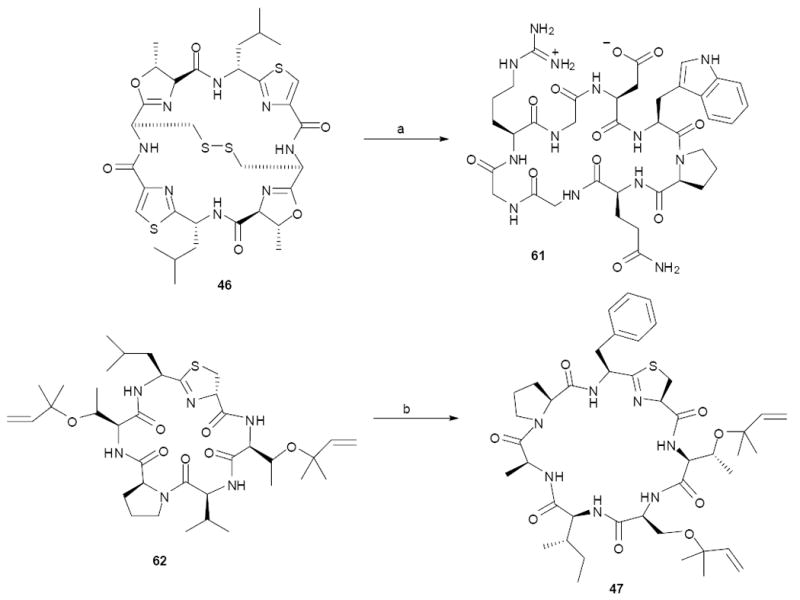
Use of metagenomics in engineering. Top: The natural product ulithiacyclamide (46) was “converted” to the wholly unnatural compound eptidemnamide (61) by (a) one-step PCR mutagenesis and synthesized in E. coli. Bottom: The natural product patellin 2 was converted to the rare natural product trunkamide (47) by (b) one-step recombination in yeast, followed by production in E. coli. These results and the natural selectivity of the enzymes indicate that large libraries of products can be synthesized by identical enzymes.
Contributor Information
Sean F. Brady, Email: sbrady@rockefeller.edu.
Eric W. Schmidt, Email: ews1@utah.edu.
References
- 1.Gans J, Wolinsky M, Dunbar J. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 2.Torsvik V, Goksoyr J, Daae FL. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torsvik V, Ovreas L, Thingstad TF. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 4.Jannasch HW, Jones GE. Limnol Oceanography. 1959;4:128–139. [Google Scholar]
- 5.Kaeberlein T, Lewis K, Epstein SS. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 6.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 7.Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M. Proc Natl Acad Sci U S A. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappe MS, Giovannoni SJ. Ann Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 9.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. Nucleic Acids Res. 2007;35:D169–172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webster G, Yarram L, Freese E, Koster J, Sass H, Parkes RJ, Weightman AJ. FEMS Microbiol Ecol. 2007;62:78–89. doi: 10.1111/j.1574-6941.2007.00372.x. [DOI] [PubMed] [Google Scholar]
- 12.Piel J. Nat Prod Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- 13.Piel J. Nat Prod Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt EW. Nat Chem Biol. 2008;4:466–473. doi: 10.1038/nchembio.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang GY, Graziani E, Waters B, Pan W, Li X, McDermott J, Meurer G, Saxena G, Anderson RJ, Davies J. Org Lett. 2000;2:2401–2404. doi: 10.1021/ol005860z. [DOI] [PubMed] [Google Scholar]
- 16.Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henne A, Daniel R, Schmitz RA, Gottschalk G. Appl Environ Microbiol. 1999;65:3901–3907. doi: 10.1128/aem.65.9.3901-3907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yap WH, Li X, Soong TW, Davies JE. J Ind Microbiol Biotechnol. 1996;17:179–184. [Google Scholar]
- 19.DeBoer C, Dietz A. J Antibiot. 1976;29:1182–1188. doi: 10.7164/antibiotics.29.1182. [DOI] [PubMed] [Google Scholar]
- 20.Lorenz P, Eck J. Nature Rev. 2005;3:510–516. doi: 10.1038/nrmicro1161. [DOI] [PubMed] [Google Scholar]
- 21.Brady SF. Nature Protocols. 2007;2:1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- 22.Brady SF, Clardy J. J Am Chem Soc. 2000;122:12903–12904. [Google Scholar]
- 23.Brady SF, Chao CJ, Clardy J. Appl Environ Microbiol. 2004;70:6865–6870. doi: 10.1128/AEM.70.11.6865-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady SF, Clardy J. J Nat Prod. 2004;67:1283–1286. doi: 10.1021/np0499766. [DOI] [PubMed] [Google Scholar]
- 25.Brady SF, Clardy J. Org Lett. 2005;7:3613–3616. doi: 10.1021/ol0509585. [DOI] [PubMed] [Google Scholar]
- 26.Brady SF, Chao CJ, Clardy J. J Am Chem Soc. 2002;124:9968–9969. doi: 10.1021/ja0268985. [DOI] [PubMed] [Google Scholar]
- 27.August PR, Grossman TH, Minor C, Draper MP, MacNeil IA, Pemberton JM, Call KM, Holt D, Osburne MS. J Mol Microbiol Biotechnol. 2000;2:513–519. [PubMed] [Google Scholar]
- 28.Brady SF, Chao CJ, Handelsman J, Clardy J. Org Lett. 2001;3:1981–1984. doi: 10.1021/ol015949k. [DOI] [PubMed] [Google Scholar]
- 29.Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J. Appl Environ Microbiol. 2002;68:4301–4306. doi: 10.1128/AEM.68.9.4301-4306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budzikiewicz H, Eckau H, Ehrenberg M. Tetrahedron Lett. 1972;13:3807–3810. [Google Scholar]
- 31.Hegedus ZL. Toxicology. 2000;145:85–101. doi: 10.1016/s0300-483x(00)00157-8. [DOI] [PubMed] [Google Scholar]
- 32.Brady SF, Clardy J. Angewandte Chem. 44:7063–7065. doi: 10.1002/anie.200501941. [DOI] [PubMed] [Google Scholar]
- 33.Garson MJ, Simpson JS. Nat Prod Rep. 2004;21:164–179. doi: 10.1039/b302359c. [DOI] [PubMed] [Google Scholar]
- 34.Edenborough MS, Herbert RB. Nat Prod Rep. 1988;5:229–245. doi: 10.1039/np9880500229. [DOI] [PubMed] [Google Scholar]
- 35.Brady SF, Clardy J. Angewandte Chem. 2005;44:7045–7048. doi: 10.1002/anie.200501942. [DOI] [PubMed] [Google Scholar]
- 36.Brady SF, Bauer JD, Clarke-Pearson MF, Daniels R. J Am Chem Soc. 2007;129:12102–12103. doi: 10.1021/ja075492v. [DOI] [PubMed] [Google Scholar]
- 37.MacNeil IA, Tiong CL, Minor C, August PR, Grossman TH, Loiacono KA, Lynch BA, Phillips T, Narula S, Sundaramoorthi R, Tyler A, Aldredge T, Long H, Gilman M, Holt D, Osburne MS. J Mol Microbiol Biotechnol. 2001;3:301–308. [PubMed] [Google Scholar]
- 38.Lim HK, Chung EJ, Kim JC, Choi GJ, Jang KS, Chung YR, Cho KY, Lee SW. Appl Environ Microbiol. 2005;71:7768–7777. doi: 10.1128/AEM.71.12.7768-7777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan C, Ju J, Borlee BR, Williamson LL, Shen B, Raffa KF, Handelsman J. Appl Environ Microbiol. 2007;73:3669–3676. doi: 10.1128/AEM.02617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson LL, Borlee BR, Schloss PD, Guan C, Allen HK, Handelsman J. Appl Environ Microbiol. 2005;71:6335–6344. doi: 10.1128/AEM.71.10.6335-6344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craig JW, Chang FY, Brady SF. ACS Chem Biol. 2009;4:23–28. doi: 10.1021/cb8002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinstein M, Roberts RC, Helinski DR. J Bacteriol. 1992;174:7486–7489. doi: 10.1128/jb.174.22.7486-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seow KT, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson CR, Davies J. J Bacteriol. 1997;179:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Courtois S, Cappellano CM, Ball M, Francou FX, Normand P, Helynck G, Martinez A, Kolvek SJ, Hopke J, Osburne MS, August PR, Nalin R, Guerineau M, Jeannin P, Simonet P, Pernodet JL. Appl Environ Microbiol. 2003;69:49–55. doi: 10.1128/AEM.69.1.49-55.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banik JJ, Brady SF. Proc Natl Acad Sci U S A. 2008;105:17273–17277. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pylypenko O, Vitali F, Zerbe K, Robinson JA, Schlichting I. J Biol Chem. 2003;278:46727–46733. doi: 10.1074/jbc.M306486200. [DOI] [PubMed] [Google Scholar]
- 47.Lopanik N, Lindquist N, Targett N. Oecologia. 2004;139:131–139. doi: 10.1007/s00442-004-1487-5. [DOI] [PubMed] [Google Scholar]
- 48.Woollacott RM. Mar Biol. 1981;65:155–158. [Google Scholar]
- 49.Davidson SK, Haygood MG. Biol Bull. 1999;196:273–280. doi: 10.2307/1542952. [DOI] [PubMed] [Google Scholar]
- 50.Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG. Appl Environ Microbiol. 2001;67:4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, Sherman DH, Haygood MG. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 52.Lopanik NB, Shields JA, Buchholz TJ, Rath CM, Hothersall J, Haygood MG, Hakansson K, Thomas CM, Sherman DH. Chem Biol. 2008;15:1175–1186. doi: 10.1016/j.chembiol.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kellner RL. Insect Biochem Mol Biol. 2002;32:389–395. doi: 10.1016/s0965-1748(01)00115-1. [DOI] [PubMed] [Google Scholar]
- 54.Kellner RL. J Insect Physiol. 2001;47:475–483. doi: 10.1016/s0022-1910(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 55.Kellner RL. J Hered. 2000;91:158–162. doi: 10.1093/jhered/91.2.158. [DOI] [PubMed] [Google Scholar]
- 56.Piel J. Proc Natl Acad Sci U S A. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmermann K, Engeser M, Blunt JW, Munro MH, Piel J. J Am Chem Soc. 2009 doi: 10.1021/ja808889k. [DOI] [PubMed] [Google Scholar]
- 58.Lewin RA, Cheng L. Prochloron: A Microbial Enigma. Chapman & Hall; 1989. [Google Scholar]
- 59.Schmidt EW, Donia MS. Methods Enzymol. 2009;458:575–596. doi: 10.1016/S0076-6879(09)04823-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ireland CM, Scheuer PJ. J Am Chem Soc. 1980;102:5688–5691. [Google Scholar]
- 61.Salomon CE, Faulkner DJ. J Nat Prod. 2002;65:689–692. doi: 10.1021/np010556f. [DOI] [PubMed] [Google Scholar]
- 62.Degnan BM, Hawkins CJ, Lavin MF, McCaffrey EJ, Parry DL, van den Brenk AL, Watters DJ. J Med Chem. 1989;32:1349–1354. doi: 10.1021/jm00126a034. [DOI] [PubMed] [Google Scholar]
- 63.Degnan BM, Hawkins CJ, Lavin MF, McCaffrey EJ, Parry DL, Watters DJ. J Med Chem. 1989;32:1354–1359. doi: 10.1021/jm00126a035. [DOI] [PubMed] [Google Scholar]
- 64.Long PF, Dunlap WC, Battershill CN, Jaspars M. Chembiochem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt EW, Sudek S, Haygood MG. J Nat Prod. 2004;67:1341–1345. doi: 10.1021/np049948n. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc Natl Acad Sci U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Partida-Martinez LP, Hertweck C. Nature. 2005;437:884–888. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 68.Partida-Martinez LP, de Looss CF, Ishida K, Ishida M, Roth M, Buder K, Hertweck C. Appl Environ Microbiol. 2007;73:793–797. doi: 10.1128/AEM.01784-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Partida-Martinez LP, Monajembashi S, Greulich KO, Hertweck C. Curr Biol. 2007;17:773–777. doi: 10.1016/j.cub.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 70.Lackner G, Mobius N, Scherlach K, Partida-Martinez LP, Winkler R, Schmitt I, Hertweck C. Appl Environ Microbiol. 2009;75:2982–2986. doi: 10.1128/AEM.01765-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Science. 2006;311:81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 72.Haeder S, Wirth R, Herz H, Spiteller D. Proc Natl Acad Sci U S A. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schirmer A, Gadkari R, Reeves CD, Ibrahim F, DeLong EF, Hutchinson CR. Appl Environ Microbiol. 2005;71:4840–4849. doi: 10.1128/AEM.71.8.4840-4849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haygood MG, Schmidt EW, Davidson SK, Faulkner DJ. J Mol Microbiol Biotechnol. 1999;1:33–43. [PubMed] [Google Scholar]
- 75.Bewley CA, Holland ND, Faulkner DJ. Experientia. 1996;52:716–722. doi: 10.1007/BF01925581. [DOI] [PubMed] [Google Scholar]
- 76.Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG. Mar Biol. 2000;136:969–977. [Google Scholar]
- 77.Bruck WM, Sennett SH, Pomponi SA, Willenz P, McCarthy PJ. ISME J. 2008;2:335–339. doi: 10.1038/ismej.2007.91. [DOI] [PubMed] [Google Scholar]
- 78.Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH, Dorrestein PC. Mol Biosyst. 2008;4:562–570. doi: 10.1039/b720018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gillor O, Carmeli S, Rahamim Y, Fishelson Z, Ilan M. Mar Biotechnol. 2000;2:213–223. doi: 10.1007/s101260000026. [DOI] [PubMed] [Google Scholar]
- 80.Sakai R, Yoshida K, Kimura A, Koike K, Jimbo M, Kobiyama A, Kamiya H. Chembiochem. 2008;9:543–551. doi: 10.1002/cbic.200700498. [DOI] [PubMed] [Google Scholar]
- 81.Sharp KH, Davidson SK, Haygood MG. ISME J. 2007;1:693–702. doi: 10.1038/ismej.2007.78. [DOI] [PubMed] [Google Scholar]
- 82.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Proc Natl Acad Sci U S A. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hrvatin S, Piel J. J Microbiol Methods. 2007;68:434–436. doi: 10.1016/j.mimet.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 84.Diyabalanage T, Amsler CD, McClintock JB, Baker BJ. J Am Chem Soc. 2006;128:5630–5631. doi: 10.1021/ja0588508. [DOI] [PubMed] [Google Scholar]
- 85.Riesenfeld CS, Murray AE, Baker BJ. J Nat Prod. 2008 doi: 10.1021/np800287n. [DOI] [PubMed] [Google Scholar]
- 86.Unson MD, Faulkner DJ. Experientia. 1993;49:349–353. [Google Scholar]
- 87.Chang Z, Flatt P, Gerwick WH, Nguyen VA, Willis CL, Sherman DH. Gene. 2002;296:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 88.Ridley CP, Bergquist PR, Harper MK, Faulkner DJ, Hooper JNA, Haygood MG. Chem Biol. 2005;12:397–406. doi: 10.1016/j.chembiol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 89.Ridley CP, Faulkner DJ, Haygood MG. Appl Environ Microbiol. 2005;71:7366–7375. doi: 10.1128/AEM.71.11.7366-7375.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flatt P, Gautschi J, Thacker R, Musafija-Girt M, Crews P, Gerwick W. Mar Biol. 2005;147:761–774. [Google Scholar]
- 91.Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 92.Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, Crews P, Piel J. Nat Chem Biol. 2009 doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- 93.Miao V, Coeffet-LeGal MF, Brown D, Sinnemann S, Donaldson G, Davies J. Trends Biotechnol. 2001;19:349–355. doi: 10.1016/s0167-7799(01)01700-0. [DOI] [PubMed] [Google Scholar]
- 94.Piel J, Butzke D, Fusetani N, Hui D, Platzer M, Wen G, Matsunaga S. J Nat Prod. 2005;68:472–479. doi: 10.1021/np049612d. [DOI] [PubMed] [Google Scholar]
- 95.Salomon CE, Magarvey NA, Sherman DH. Nat Prod Rep. 2004;21:105–121. doi: 10.1039/b301384g. [DOI] [PubMed] [Google Scholar]
- 96.Taylor MW, Radax R, Steger D, Wagner M. Microbiol Mol Biol Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim TK, Fuerst JA. Environ Microbiol. 2006;8:1460–1470. doi: 10.1111/j.1462-2920.2006.01040.x. [DOI] [PubMed] [Google Scholar]
- 98.Hochmuth T, Piel J. Phytochemistry. 2009 doi: 10.1016/j.phytochem.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 99.Fieseler L, Hentschel U, Grozdanov L, Schirmer A, Wen G, Platzer M, Hrvatin S, Butzke D, Zimmermann K, Piel J. Appl Environ Microbiol. 2007;73:2144–2155. doi: 10.1128/AEM.02260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kennedy J, Codling CE, Jones BV, Dobson AD, Marchesi JR. Environ Microbiol. 2008;10:1888–1902. doi: 10.1111/j.1462-2920.2008.01614.x. [DOI] [PubMed] [Google Scholar]
- 101.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 102.Sudek S, Haygood MG, Youssef DT, Schmidt EW. Appl Environ Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Donia MS, Ravel J, Schmidt EW. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. J Am Chem Soc. 2009;131:2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pidot SJ, Hong H, Seemann T, Porter JL, Yip MJ, Men A, Johnson M, Wilson P, Davies JK, Leadlay PF, Stinear TP. BMC Genomics. 2008;9:462. doi: 10.1186/1471-2164-9-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


