Abstract
Acetylcholinesterase (AChE) is a remarkably efficient serine hydrolase responsible for the termination of impulse signaling at cholinergic synapses. By employing Born-Oppenheimer molecular dynamics simulations with B3LYP/6-31G(d) QM/MM potential and the umbrella sampling method, we have characterized its complete catalytic reaction mechanism for hydrolyzing neurotransmitter acetylcholine (ACh) and determined its multi-step free energy reaction profiles for the first time. In both acylation and deacylation reaction stages, the first step involves the nucleophilic attack to the carbonyl carbon with the triad His447 serving as the general base, and leads to a tetrahedral covalent intermediate stabilized by the oxyanion hole. From the intermediate to the product, the orientation of His447 ring needs to be adjusted very slightly, and then the proton transfers from His447 to the product and the break of the scissile bond happen spontaneously. For the three-pronged oxyanion hole, it only makes two hydrogen bonds with the carbonyl oxygen at either the initial reactant or the final product state, but the third hydrogen bond is formed and stable at all transition and intermediate states during the catalytic process. At the intermediate state of the acylation reaction, a short and low-barrier hydrogen bond (LBHB) is found to be formed between two catalytic triad residues His447 and Glu334, and the spontaneous proton transfer between two residues has been observed. However, it is only about 1 ~ 2 kcal/mol stronger than the normal hydrogen bond. In comparison with previous theoretical investigations of the AChE catalytic mechanism, our current study clearly demonstrates the power and advantages of employing Born-Oppenheimer ab initio QM/MM MD simulations in characterizing enzyme reaction mechanisms.
1. Introduction
Enzymes are remarkable catalysts provided by nature and play essential roles in cells of all organisms. Most enzymes catalyze more than one chemical process involving multiple transition states and intermediates. To fully characterize their complicated reaction mechanisms is a fundamental goal and challenge in biological chemistry1,2, and has important implications in the rational design of mechanism-based inhibitors3–6 and discovery of novel biocatalysts7–12. One important enzyme that has attracted significant interest for mechanistic investigations is Acetylcholinesterase (AChE, EC 3.1.1.7) 13–17.
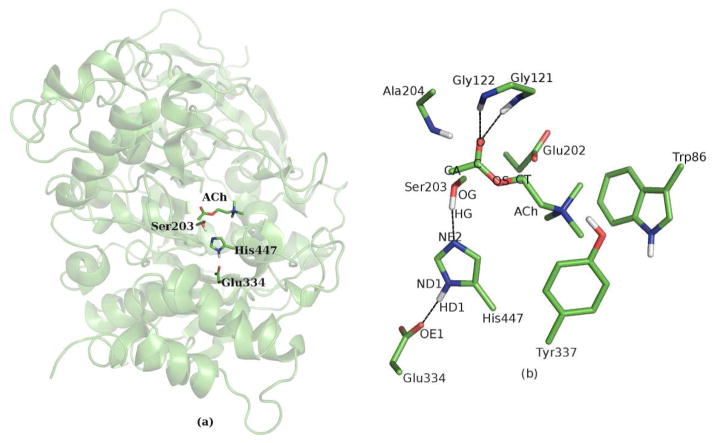
AChE is a serine hydrolase and the key enzyme in the central nervous system responsible for the termination of impulse signaling at cholinergic brain synapses13,14. It is one of the most efficient enzymes in nature, and catalyzes the hydrolysis of the neurotransmitter acetylcholine (ACh) with a reaction rate close to the diffusion controlled limit13. The active site of AChE is located at the base of a long gorge of 20 Å, and consists of two subsites18–20, as illustrated in Figure 1. An “esteratic” subsite includes the catalytic triad (side chains of Ser203, His447 and Glu334) and the oxyanion hole (peptidic NH groups of Gly121, Gly122, and Ala204), and is the essential catalytic functional unit of AChE13–20. The other “anionic” subsite is formed by side chains of Glu202, Trp86 and Tyr337, and is mainly responsible for binding the quaternary trimethylammonium tail group of ACh21,22. (Throughout this article, the sequence numbers of mouse AChE is employed). Like many other members of the serine hydrolase and serine protease families23–25, the catalytic process of AChE13 has been suggested to proceed in two successive stages, acylation and deacylation, as shown in Figure 2. Due to its high efficiency, transition states and intermediates formed in the catalytic process are very short-lived, and would be almost impossible for direct experimental characterization.13,26,27 On the other hand, most theoretical studies only investigated the first step of either acylation28–31 or deacylation reaction.32 In one recent ab initio QM/MM study,33 although it was claimed to characterize the full cycle of AChE catalytic process with minimizations and transition state searching, the determined tetrahedral intermediates were found to have a much lower energy than the reactants in both reaction stages, which are not consistent with experimental results13,26,27. Meanwhile, no transition state for the second step in each reaction stage has been located33. It is very likely that their determined reactants, transition states and intermediates may have totally different MM conformation sub-states, due to the weakness in their computational protocol33. Thus their characterized AChE catalytic process is likely to be erroneous. On the other hand, although the acylation mechanism was determined by QM studies of model complexes34,35, the heterogeneous enzyme environment of AChE has been totally neglected and its accuracy is quite limited. Thus, we can see that in spite of significant efforts28–35, it remains a significant challenge for theoretical studies to fully determine this important multi-step acetylcholine hydrolysis process catalyzed by AChE.
Figure 1.
Illustration of the AChE-ACh complex. (a) Cartoon representation of the enzyme and stick representation of the active site, (b) stick representation of important residues.
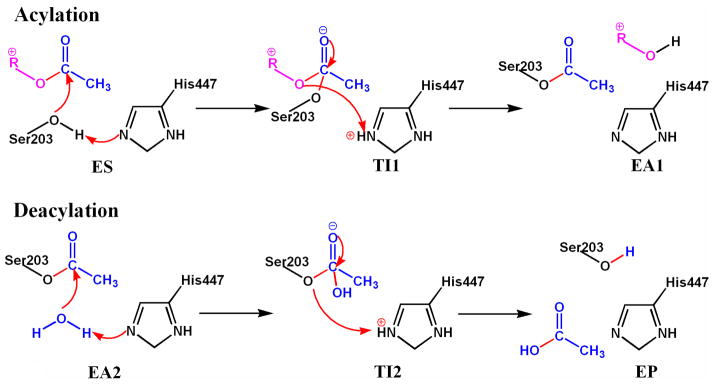
Figure 2.
Reaction mechanism of ACh hydrolysis catalyzed by AChE.
In the present work, our main aim is to characterize the complete catalytic reaction mechanism of AChE and determine the multi-step free energy reaction profile of ACh hydrolysis for the first time. Here we have employed Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling, a state-of-the-art approach to simulate reactions in complex systems. It allows for a first-principle description of chemical bond formation/breaking and dynamics of the enzyme active site, while properly including effects of the heterogeneous and fluctuating protein environment. Our simulation results are consistent with experimental studies, and have provided detailed novel insights into the inner workings of AChE.
2. Methods
Here our computational approaches center on Born–Oppenheimer MD simulations with ab initio QM/MM potential36–45 and the umbrella sampling method46–48. The QM sub-system is described by B3LYP functional with 6-31G* basis set, and the employed MM force fields are amber99SB49,50 for protein residues and TIP3P model51 for water molecules. The QM/MM interface is based on the pseudobond approach40,41,52–55. All QM/MM calculations were carried out with modified versions of Q-Chem56 with Tinker57 programs. For both acylation and deacylation reaction stages, almost the same simulation protocol has been employed. First, each prepared initial reactant system was equilibrated by extensive molecular dynamics simulations with periodic boundary condition. With an equilibrated MD snapshot, the QM/MM model was prepared by deleting the ions and waters beyond 25 Å from the reaction center, which was chosen as the hydroxyl oxygen atom of Ser203. Each resulted system contained about 10,000 atoms. For the acylation stage reaction, the QM sub-system includes the substrate ACh and the catalytic triad (side chains of Ser203, His447 and Glu334); while in the deacylation stage, the QM sub-system consists of the reactive water molecule and side chains of acetyl-Ser203, His447 and Glu334, as illustrated in Figure S1 in the Supporting Information. For all QM/MM calculations, the spherical boundary condition was applied, and only the atoms within 20 Å from the reaction center were allowed to move. The prepared system was first minimized and then employed to map out a reaction path with B3LYP/6-31G* QM/MM optimizations. For each determined structure along the path, a 500 ps MD simulation with MM force field was performed to equilibrate the MM subsystem, with the QM subsystem being frozen. Finally, the resulting snapshot was used as the starting structure for Born-Oppenheimer B3LYP/6-31G* QM/MM MD simulation with umbrella sampling. Each window was simulated for at least 30 ps. A total of more than 3000 ps ab initio QM/MM MD simulations have been carried out in this study. From these biased simulations, the free energy profile for each reaction was obtained with the weighted histogram analysis method (WHAM).58–60 This computational protocol has successfully applied to study several enzymes as well as chemical reactions in aqueous solution61–68. Full computational details are given in the Supporting Information.
3. Results
3.1 Acylation Reaction Stage
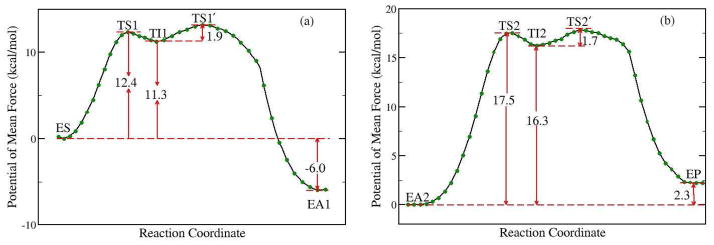
By employing Born-Oppenheimer molecular dynamics simulations with B3LYP/6-31G* QM/MM potential and the umbrella sampling method, first we characterized the acylation stage for ACh hydrolysis catalyzed by AChE and determined its complete free energy reaction profile, as shown in Figure 3a. We can see that the free energy curve is smooth and continuous, and the calculated activation barrier is 12.4 kcal/mol. In the initial step, the nucleophilic attack of the Ser203 O atom to the carbonyl C atom of acetylcholine is concerted with the proton transfer from Ser203 to His447, and forms a tetrahedral intermediate (TI1). Then the proton transfers to the leaving group of ACh and the scissile bond breaks down to form a covalent acyl-enzyme complex (EA1) with the release of the choline. The intermediate TI1 is located in a shallow well, and the free energy barrier for the second step is only 1.9 kcal/mol. Consistent with the determined free energy profile, the unrestrained QM/MM-MD simulations for the intermediate was only stable for a few picoseconds, and then led to form EA1. To further check whether there is another meta-stable state between the reactant and the intermediate TS1, we have carried out the unrestrained ab initio QM/MM molecular dynamics simulations with 50 random snapshots around the transition state region TS1. For each snapshot, two simulations have been run, one with the original velocities and the other with the reversed ones. Among all simulations, there is no other stable state observed except for the reactant ES and the intermediate TI1. For 46% of the snapshots (23 out of 50) one trajectory led to the reactant while the other went to the intermediate. These results further support that the nucleophilic attack and proton transfer is concerted, and confirm that our characterized transition state is meaningful.
Figure 3.
Free energy profile for (a) the acylation reaction stage, (b) the deacylation reaction stage determined by B3LYP(6-31G*) QM/MM molecular dynamics simulations and umbrella sampling.
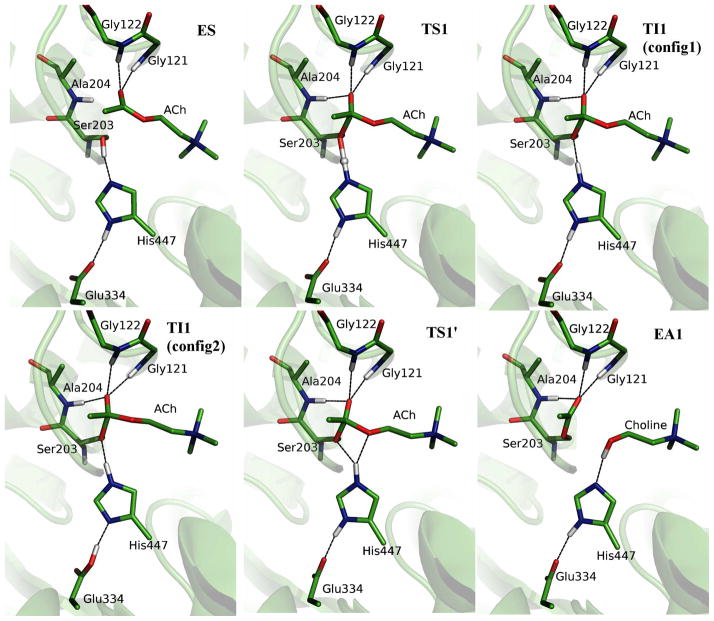
The structures of the reactant, transition states, intermediates and acyl-enzyme during the acylation stage are illustrated in Figure 4, and the key geometric parameters are listed in Table 1. At the AChE-ACh reactant complex, only two hydrogen bonds are formed between the carbonyl oxygen of ACh and the NH groups of Gly121 and Gly122, with N-O distances of 2.86 ± 0.12 and 2.82 ± 0.10 Å, respectively. As the reaction proceeds, the distance between the carbonyl oxygen of ACh and the NH group of Ala204 becomes smaller and forms a hydrogen bond in both the transition state and the tetrahedral intermediate, with the changing of O-N distance from 3.22 ± 0.22 Å in the enzyme-substrate complex (ES) region to 2.86 ± 0.10 Å at the tetrahedral intermediate (TI1). These results are consistent with the previous ab initio QM/MM studies of the initial step of the acylation reaction29,30.
Figure 4.
Illustration of the structures for the characterized acylation reaction stage. ES, enzyme-substrate complex; TS1, the first transition state; TI1, tetrahedral intermediate; TS1′, the second transition state in acylation; EA1, acylated enzyme. Two representative configurations of TI1 are shown.
Table 1.
List of key geometric parameters for the reactant, transition states, intermediate and acylenzyme for acylation reaction from B3LYP/6-31G* QM/MM MD simulations. Atom names are the same as labeled on Figure 1.
| Distance (Å) | ES | TS1 | TI1 | TS1′ | EA1 |
|---|---|---|---|---|---|
| OG (S203) – C (ACh) | 2.59 ± 0.04 | 1.82 ± 0.07 | 1.58 ± 0.05 | 1.47 ± 0.05 | 1.34 ± 0.03 |
| HG (S203) – OG(S203) | 1.00 ± 0.02 | 1.37 ± 0.07 | 1.63 ± 0.05 | 2.00 ± 0.09 | 2.78 ± 0.09 |
| HG (S203) – NE2(H447) | 1.74 ± 0.09 | 1.17 ± 0.05 | 1.07 ± 0.03 | 1.04 ± 0.03 | 1.74 ± 0.11 |
| HG (S203) – OS(ACh) | 3.01 ± 0.18 | 2.84 ± 0.18 | 2.77 ± 0.20 | 1.99 ± 0.10 | 1.01 ± 0.03 |
| C (ACh) – OS (ACh) | 1.35 ± 0.03 | 1.44 ± 0.04 | 1.51 ± 0.05 | 1.70 ± 0.10 | 2.83 ± 0.09 |
| ND1 (H447) – OE1(E334) | 2.65 ± 0.08 | 2.61 ± 0.08 | 2.57 ± 0.07 | 2.63 ± 0.08 | 2.74 ± 0.10 |
| N (G121) – O(ACh) | 2.86 ± 0.12 | 2.83 ± 0.09 | 2.83 ± 0.09 | 2.87 ± 0.10 | 2.90 ± 0.11 |
| N (G122) – O(ACh) | 2.82 ± 0.10 | 2.90 ± 0.11 | 2.91 ± 1.00 | 2.89 ± 0.11 | 2.91 ± 0.10 |
| N (A204) – O(ACh) | 3.24 ± 0.22 | 2.93 ± 0.11 | 2.85 ± 0.09 | 2.90 ± 0.10 | 2.89 ± 0.10 |
| ∠ NE2 (H447) – HG (S203) – OS (ACh) degree | 105.4 ± 6.1 | 126.8 ± 5.4 | 132.6 ± 5.8 | 151.6 ± 7.9 | 167.2 ± 6.2 |
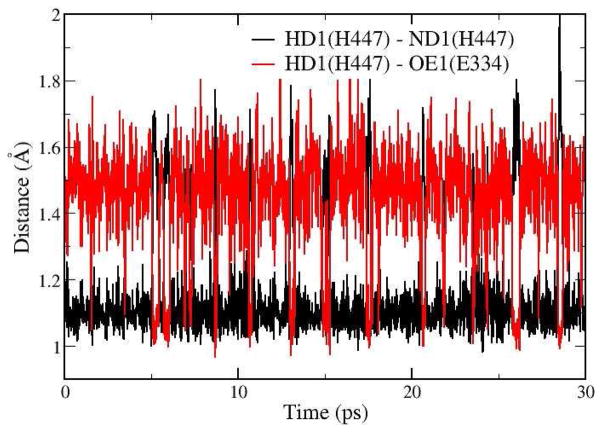
At the intermediate state TI1, the bond between OG of Ser203 and C atom of ACh is formed and the HG atom of Ser203 has transferred to NE2 atom of His447. (Atom names are labeled on Figure 1). There is a short hydrogen bond formed between His447 and Glu334, in which the distance between ND1 (H447) and OE1 (D334) is only 2.57 ± 0.07 Å. In addition, it is found that although HD1 atom stays with ND1 atom of His447 most of the time, spontaneous transfer between His447 and Glu334 has occurred transiently at the TI1 state, as shown in Figure 5. Thus two representative configurations of TI1 have been illustrated in Figure 4 as TI1 (config1) and TI1 (config2). It should be noted that such spontaneous proton transfer does not happen at other states during the acylation stage. For the rest of the acylation reaction, our simulation results indicate that the His447 ring first adjusts its orientation a little bit without breaking the HG-NE2 bond. At the TS1′ state, the distance between HG and OS of ACh is decreased to 1.99 ± 0.10 Å while the angle of donor-hydrogen-acceptor (NE2-HG-OS) is 151.6 ± 7.9°, which shows that the proton is in the better direction to attack the OS atom of ACh than at the intermediate state. Meanwhile, the distance between HG atom and OG of Ser203 is going up to 2.00 ± 0.09 Å, and the scissile bond is a little elongated to 1.70 ± 0.10 Å. After adjusting the orientation of the His447 ring, the proton transfer from His447 to OS atom and the breaking of the scissile bond is a spontaneous downhill process, which leads to a stable product EA1. We can see that three hydrogen bonds between the carbonyl oxygen and the oxyanion hole are always formed and stable in this second step of the acylation reaction.
Figure 5.
Calculated distances RN-H (the distance between HD1 and ND1 of His447) and RO-H (distance between HD1 and carboxyl oxygen OE1 of Glu334) in QM/MM-MD simulation at the intermediate TI1.
3.2 Deacylation Reaction Stage
With the computationally determined EA1 product of the acylation stage, the initial structure for simulating the deacylation reaction was prepared by removing choline from the binding pocket since experimental studies have indicated that the release of choline is faster than the deacylation reaction.69,70 With the departure of the quaternary trimethylammonium tail group from the active site, one open question is regarding the protonation state of Glu202 in the deacylation reaction28,32–34. In a previous study, it has been suggested that Glu202 is likely to be protonated in the deacylation stage based on both pKa calculations and EVB simulations.32 Here in order to examine both possibilities, we have prepared and simulated two reactant complex models for the deacylation stage, which only differ in the protonation state of Glu202, and determined their free energy profiles for the first step of the deacylation. In the extensive MD simulations of the model with deprotonated Glu202, it is found that one sodium ion outside of the active site would move into the binding pocket, while this is not the case for the system with protonated Glu202. For the model of protonated Glu202, even if a sodium ion is manually put in the binding pocket, it would be released to the bulk solvent during the MD simulation. These results suggest that the total charge of the binding pocket would be same regardless of the protonation state of Glu202, and it needs a positive charge in the binding pocket with the departure of choline. With the determined free energy profiles for both models, it is found that the reaction barrier with Glu202 protonated would 5 kcal/mol lower than the one with Glu202 deprotonated. (see Figure S3 in the Supporting Information.) These results further support the previous suggestion that Glu202 should be protonated in the deacylation stage, and are consistent with the experimental result that the mutation of Glu202 to Gln has only a minimal effect on the kcat of ACh hydrolysis.71–73
The determined complete free energy profile for the deacylation stage with B3LYP(6-31G*) QM/MM MD simulations is shown in Figure 3b. Like in the acylation stage, it also involves a high energy tetrahedral intermediate (TI2). The calculated activation energy barrier is 17.5 kcal/mol, which is higher than that for the acylation stage. Thus our results indicate that the first step of deacylation reaction is the rate-limiting step for the ACh hydrolysis catalyzed by AChE, which is consistent with the experimental results.26 It should be noted that our determined free energy profiles for both reaction stages are totally different from the results of a recent ab initio QM/MM study33 by Nemukhin and his co-workers. One concern is that this significant discrepancy between our results and Nemukhin’s33 may be due to the choice of density functional and basis set since we have employed B3LYP(6-31G*) instead of PBE0/6-31+G* in Nemukhin’s work33. To address this concern, we have also performed both PBE0/6-31+G* and B3LYP/6-31+G* QM/MM MD simulations to determine the free energy profile of the first step of the deacylation reaction, as shown in Fig. S4. We can see that our results from different functionals and basis sets are very consistent. With the same PBE0/6-31+G* QM method and basis set, the free energy difference between the intermediate of the deacylation step and its reactant is 11.4 kcal/mol from our QM/MM MD simulations. This is totally different from the Nemukhin’s result of −9.7 kcal/mol for the energy difference between the intermediate of the deacylation step, in which the intermediate is more stable than its deacylation reactant, contradictory to experimental results13,26,27. Thus, these results further confirm that the significant difference between our results and Nemukhin’s is not due to the choice of density functional and basis set, but more likely comes from the weakness in their computational protocol33 which leads to different conformational sub-states for reactants, transition state and intermediates, resulting an erroneous potential energy reaction profile.
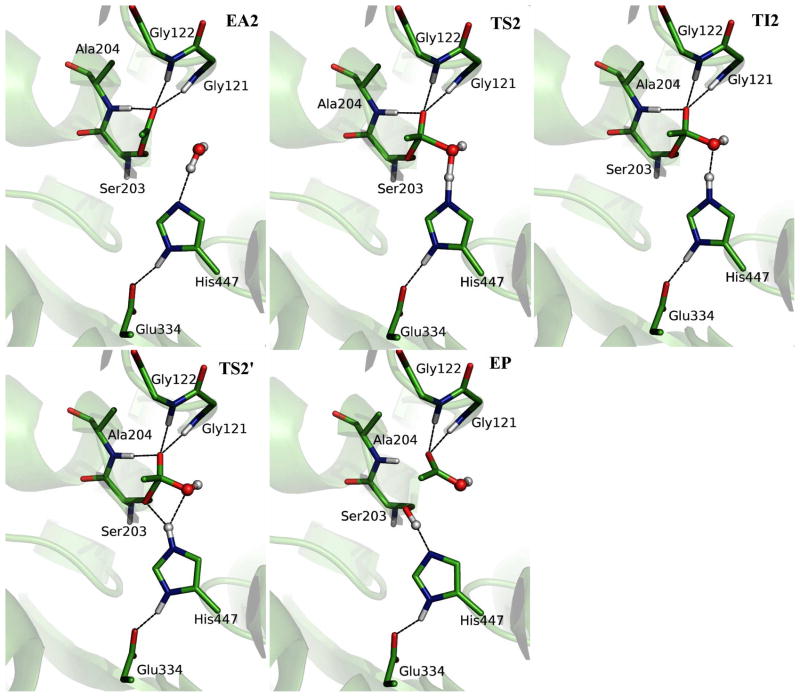
Figure 6 illustrates the structures of the acylenzyme (EA2), transition states, intermediate and product (EP) from our ab initio QM/MM MD simulations, and the key geometric parameters are listed in Table 2. We can see that the deacylation has a similar mechanism as the acylation. First the water molecule attacks the carbonyl carbon of the acetyl group, which is facilitated by the proton transfer to His447, leading to a high energy tetrahedral intermediate (TI2). In the following step, the proton transfers from His447 to the OG atom of acetyl-serine and the scissile bond breaks, yielding an acetic acid and the wild-type AChE. At the EA2 state, the acetyl head group is held in place by forming three hydrogen bonds with the NH group of oxyanion hole. The water molecule in the active site forms the hydrogen bond with His447, and is close to the carbonyl-carbon of the acetyl serine. At TS2, the proton is located in the middle of water and His447. At the intermediate state TI2, the bond between O atom of water and carbonyl C atom of acetyl-serine is almost formed and the proton has transferred to NE2 atom of His447. From TI2 to TS2′, it does not involve covalent bond forming and breaking, and the barrier is mainly due to the slight adjustment of the orientation of His447 ring. We can see that in the region of the second transition state TS2′, the bond between proton and NE2 of His447 is almost the same as that in the intermediate, and the scissile bond has been slightly elongated to 1.64 ± 0.09 Å. However, the distance between H atom and O atom of water is going up to 2.06 ± 0.12 Å with the distance between proton and OG of acetyl-serine decreasing to 1.89 ± 0.10 Å. Meanwhile, the angle of donor-hydrogen-acceptor (NE2-HW-OG) becomes 145.2 ± 9.1o. Thus the proton is in a better direction to be transferred to the OG atom of acyl-serine. In the product region, the C-O scissile bond was broken with an average distance of 2.68 ± 0.11 Å while the Ser203 re-forms the hydrogen bond with His447. The distance between H of Ser203 and N of His447 is 1.78 ± 0.11 Å, which is close to the value (1.74 ± 0.09 Å) at ES state. Throughout the whole deacylation stage, the third catalytic residue Glu334 forms a normal H-bond with His447 and almost no spontaneous proton transfer between Glu334 and His447 has been observed, including at the intermediate state TI2.
Figure 6.
Illustration of the structures for the characterized deacylation reaction stage. EA2, acylated enzyme in deacylaton; TS2, first transition state; TI2, tetrahedral intermediate; TS2′, the second transition state; EP, enzyme-product complex.
Table 2.
List of key geometric parameters for the reactant, transition states, intermediate and product of deacylation reaction from B3LYP/6-31G* QM/MM MD simulations. Acyl is short for acetyl, and ASer is short for acylated Ser203. Atom names are the same as labeled on Figure 1. OW is the oxygen atom of reactive water, and HW is the leaving hydrogen of water.
| Distance (Å) | EA2 | TS2 | TI2 | TS2′ | EP |
|---|---|---|---|---|---|
| OW (WAT) – C (acyl) | 2.84 ± 0.05 | 1.70 ± 0.06 | 1.54 ± 0.04 | 1.47 ± 0.05 | 1.34 ± 0.03 |
| HW (WAT) – OW(WAT) | 0.99 ± 0.03 | 1.35 ± 0.06 | 1.59 ± 0.05 | 2.06 ± 0.12 | 2.98 ± 0.12 |
| HW (WAT) – NE2(H447) | 1.90 ± 0.13 | 1.18 ± 0.06 | 1.08 ± 0.03 | 1.05 ± 0.03 | 1.78 ± 0.11 |
| HW (WAT) – OG(ASer) | 2.93 ± 0.18 | 2.48 ± 0.13 | 2.40 ± 0.15 | 1.89 ± 0.10 | 1.00 ± 0.03 |
| C (acyl) – OG (ASer) | 1.35 ± 0.03 | 1.45 ± 0.04 | 1.52 ± 0.05 | 1.64 ± 0.09 | 2.68 ± 0.11 |
| ND1 (H447) – OE1(E334) | 2.77 ± 0.11 | 2.68 ± 0.09 | 2.67 ± 0.09 | 2.62 ± 0.08 | 2.66 ± 0.09 |
| N (G121) – O(acyl) | 3.02 ± 0.18 | 2.87 ± 0.11 | 2.84 ± 0.10 | 2.83 ± 0.09 | 2.87 ± 0.12 |
| N (G122) – O(acyl) | 2.85 ± 0.09 | 2.86 ± 0.10 | 2.85 ± 0.09 | 2.86 ± 0.10 | 2.82 ± 0.09 |
| N (A204) – O(acyl) | 2.85 ± 0.10 | 2.88 ± 0.10 | 2.86 ± 0.09 | 2.89 ± 0.11 | 3.23 ± 0.22 |
| ∠ NE2 (H447) – HW (WAT) – OG (ASer) degree | 85.6 ± 6.5 | 113.7 ± 5.3 | 119.3 ± 5.5 | 145.2 ± 9.1 | 168.3 ± 6.3 |
4. Discussion
Like many other members of the serine hydrolase and serine protease families, AChE employs the catalytic triad (Ser203, His447 and Glu334) as the essential catalytic functional unit. Ser203 serves as a nucleophilic attacking group to initiate the whole ACh hydrolysis process, and His447 is generally assumed to act a general acid-base element in both acylation and deacylation stages. However, for the third residue of the catalytic triad, Glu334, its detailed catalytic role has been quite controversial and at least three different hypotheses have been put forward. The first is the “charge-relay” mechanism74,75, in which the carboxylate of Glu334 functions as a general base to accept a proton from His447 and would be protonated at the intermediate. Early work of other serine proteases also suggested this double proton transfer,76–78 but subsequent experimental studies have provided evidences against it.75,79–85 The second is the “low-barrier hydrogen bond (LBHB) or short-strong hydrogen bond (SSHB)” mechanism, 86–92 which suggests that the formation of LBHB or SSHB at the transition state or intermediate can supply 10~20 kcal/mol extra stabilization energy. The main experimental evidence comes from the proton NMR experiments86,89,91 which indicate that a short hydrogen bond (2.62–2.63 A) is formed between Glu and His of the catalytic triad in an AChE-transition state analogue inhibitor complex. It should be noted that there is no direct evidence for the strength of such hydrogen bonds. The third suggestion is that the electrostatic interaction between the carboxylate group of Glu334 and the incipient imidazolium cation is mainly responsible for stabilizing the transition state and intermediate28–30,32,93.
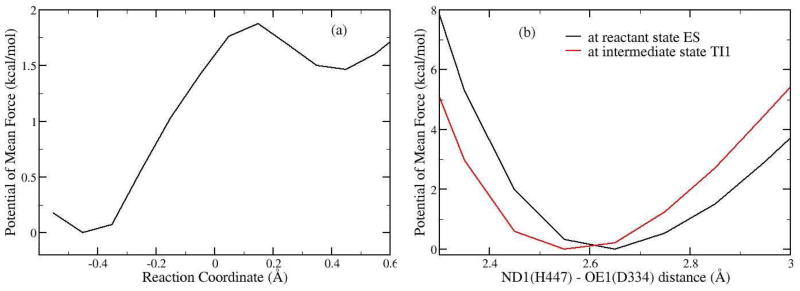
From our current ab initio QM/MM MD simulations, one very interesting finding is that at the intermediate state of the acylation stage, there is a short hydrogen bond formed between His447 and Glu334, in which the distance between ND1 (His447) and OE1 (Glu334) is only 2.57 ± 0.07 Å. Meanwhile, although HD1 atom stays with ND1 atom of His447 most of the time, spontaneous transfer from His447 to Glu334 has occurred transiently at the TI1 state, as shown in Fig. 5. In order to provide further insights into this short hydrogen bond, we have determined the free energy profile for the proton transfer between His447 to Glu334 at the TI1 state with B3LYP(6-31G*) QM/MM MD simulations and umbrella sampling, as shown in Figure 7a. We can see that the free energy barrier for the proton transfer is less than 2.0 kcal/mol, which is quite low. Thus the short hydrogen bond formed at the TI1 state of the acylation can be considered as a LBHB. Meanwhile, the proton transfer between His447 and Glu334 would destabilize the tetrahedral intermediate, which does not support the charge-relay mechanism.
Figure 7.
(a) Free energy profile for the proton transfer between Glu334 and His447 at the tetrahedral intermediate state (TI1) of the acylation stage. The reaction coordinate is RND1-HD1 – ROE1-HD1. From left to right, the proton transfer from His447 to the carboxyl oxygen of Glu334. (b) Free energy profile for the elongation of H-bond formed between Glu334 and His447 at ES state and TI1 state. Reaction coordinate is the distance between the carboxyl oxygen atom OE1 of Glu334 and the nitrogen atom ND1 of His447.
With the discovery of a short and low barrier hydrogen bond at the intermediate state of the acylation stage, the next essential question is regarding its strength and its catalytic role in stabilizing the intermediate. In an enzyme system, it would not be possible to calculate the absolute strength of a hydrogen bond by pulling two fragments too far apart. Typically, it would be reasonable to assume that a normal N-H…O hydrogen bond would have a N-O distance of 2.8 Å, while the distance of 3.0 Å would be considered a weak hydrogen bond. Thus here we employed the N-O distance between His447 and Glu334 as the reaction coordinate, and elongate it to 3.0 Å at both the reactant (EA) and intermediate (TI1) states with B3LYP/6-31G* QM/MM-MD simulations and umbrella sampling. The calculated free energy profile is shown in Figure 7b. We can see that although the hydrogen bond between His447 and Glu334 at the TI1 is a short and low-barrier hydrogen bond, it is only 1 ~ 2 kcal/mol stronger than a normal hydrogen bond at 2.8 Å. Meanwhile, by comparing the free energy curve at the reactant state and that at the intermediate state, the free energy difference at 3.0 Å is only 1.8 kcal/mol between the two states, which indicates that the extra stabilization coming from the formation of this LBHB at the intermediate is limited. Meanwhile, it should be noted that the deacylation stage is rate-limiting and no LBHB is formed at the tetrahedral intermediate TI2. Thus our simulations support that the main catalytic role of Glu334 is to stabilize the positive charge on the imidazole ring of His447 at transition states and intermediates through electrostatic interactions. 28–30,32,93
For the triad histidine residue His447, our characterized mechanism indicates that it serves the same role in two successive stages, acylation and deacylation. Initially, it serves as a general base to abstract a proton from the nucleophilic attacking oxygen atom, and is doubly protonated at the tetrahedral intermediate. In the second step of the reaction, the His447 ring adjusts its orientation a little bit so that the proton on His447 is in a better position to transfer to the oxygen of the scissile bond. Then the proton transfer and the breaking of the scissile bond become a spontaneous downhill process to yield the product. It should be noted for serine protease, it has been quite controversial regarding the motion of the triad histidine residue in the reaction process. One recent hypothesis is that the ring of triad histidine residue needs to be flipped at the tetrahedral intermediate to facilitate the second step,94,95 while others suggest that only minimal motion of His ring is needed.23–25,96 Here for the AChE, our simulations indicate that the orientation His447 ring only needs to be adjusted very slightly during the reaction process.
Besides the triad, another important component of catalytic functional unit is the oxyanion role which is to stabilize the oxyanion in transition states and intermediates. In contrast to two-pronged oxyanion hole in most serine proteases and hydrolases,24,25,97,98 it is a three-pronged oxyanion hole in AChE, formed by peptidic NH groups from Gly121, Gly122 and Ala204. In the X-ray structure of an AChE-inhibitor complex, the respective distances from N of the NH groups to the carbonyl oxygen of the inhibitor are 2.9, 2.9 and 3.2 Å.18
In our current ab initio QM/MM MD simulations of the AChE-ACh reactant complex, the respective NO distances are 2.86 ± 0.12, 2.82 ± 0.10 Å and 3.22 ± 0.22 Å, which is quite consistent with the experimental results.18 We can see that only two hydrogen bonds are formed between the carbonyl oxygen of ACh and the NH groups of Gly121 and Gly122 in the reactant complex. As the reaction proceeds, the distance between the carbonyl oxygen of ACh and the NH group of Ala204 becomes smaller and forms a hydrogen bond in both the transition state and the tetrahedral intermediate, with the changing of O-N distance from 3.22 ± 0.22 Å in the enzyme-substrate complex (ES) region to 2.86 ± 0.10 Å at the tetrahedral intermediate (TI1). These results are consistent with the previous ab initio QM/MM studies of the initial step of acylation reaction29. For the rest of the acylation reaction stage, three hydrogen bonds between the carbonyl oxygen and the oxyanion hole are always formed and stable.
For the deacylation stage, we found that the formation of hydrogen bonds between the carbonyl oxygen and the oxyanion hole is a little different from the acylation stage. At the reactant of the deacylation reaction, three hydrogen bonds are formed, with N-O distances at 3.02 ± 0.17, 2.86 ± 0.10 Å and 2.89 ± 0.11 Å respectively, and the hydrogen bond formed by Gly121 is a little weaker than the other two. When the reaction processes, the hydrogen bond formed by Gly121 becomes stronger and there are always three hydrogen bonds formed before the scissile bond breaks. From the TS2′ to the final product, the hydrogen bond formed by NH group of Ala204 becomes weaker, and the N-O distance is elongated to 3.23 ± 0.22 Å at the EP state. Thus interestingly, there are only two hydrogen bonds formed between the oxyanion hole and the carbonyl oxygen atom of acetic acid in the final product state, which is almost the same as in the reactant state of the acylation reaction.
In the active site of AChE, besides the catalytic triad and oxanion hole which serve as the essential catalytic machinery, there is an “anionic” sub-site responsible for recognition of the quaternary ammonium moiety of substrates and inhibitors. This “anionic” sub-site consists of side chains of Trp86, Glu202 and Tyr337. Both Trp86 and Tyr337 are aromatic residues, and their binding to the cationic moiety have been attributed to cation-pi and dispersion/hydrophobic interactions21,22. In our characterized mechanism, Glu202 is only negatively charged in the acylation stage, and plays an important role in binding the positively charged substrate acetylcholine21,22; with the release of the positively charged choline from the active site after the acylation, Glu202 would be protonated during the deacylation stage, which is also the rate-determining step of this whole catalytic reaction process. This would be consistent with the finding from the experimental mutation results that the negative charge of Glu202 is important for substrate binding,71 but has a minimal effect on the catalytic reaction rate.71–73
Conclusion
By employing Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling, a state-of-the-art approach to simulate enzyme reactions, we have characterized the complete catalytic reaction mechanism of AChE and determined the multi-step free energy reaction profile of ACh hydrolysis for the first time. The calculated activation barriers with B3LYP(6-31 G*) QM/MM MD simulations are 12.3 kcal/mol and 17.5 kcal/mol for acylation and deacylation reactions respectively, and the initial step of deacylation is the rate-limiting step. In both reaction stages, the first step involves the nucleophilic attack to the carbonyl carbon facilitated by the triad histidine residue His447 serving as the general base, and leads to a high-energy tetrahedral covalent intermediate stabilized by the oxyanion hole; from the intermediate to the product, the orientation of His447 ring needs to be adjusted slightly, and then the proton transfer from His447 to the product and the break of the scissile bond happen spontaneously without any activation barriers. One very interesting and significant finding is that although a short and low-barrier hydrogen bond (LBHB) is formed at the intermediate state of the acylation stage, it is only about 1 ~ 2 kcal/mol stronger than the normal hydrogen bond and the catalytic effect of LBHB is quite limited in the AChE hydrolysis. Our current study clearly demonstrates the significant progress made in computational enzymology, and illustrates the power and advantages of employing ab initio QM/MM MD simulations in characterizing enzyme mechanisms.
Supplementary Material
Acknowledgments
We are grateful for the support of National Institute of Health (R01-GM079223), and National Science Foundation (CHE-CAREER-0448156, TeraGrid resources provided by NCSA and LONI). We also thank NYU-ITS for providing computational resources and support.
Footnotes
Supporting Information Available: Full computational details, Figure S1, S2, S3 and S4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Frey PA, Hegeman AD. Enzymatic Reaction Mechanisms. Oxford University Press; Oxford: 2007. [Google Scholar]
- 2.Kraut DA, Carroll KS, Herschlag D. Annu Rev Biochem. 2003;72:517–571. doi: 10.1146/annurev.biochem.72.121801.161617. [DOI] [PubMed] [Google Scholar]
- 3.Potashman MH, Duggan ME. J Med Chem. 2009;52:1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 4.Robertson JG. Biochemistry. 2005;44:5561–5571. doi: 10.1021/bi050247e. [DOI] [PubMed] [Google Scholar]
- 5.Schramm VL. Accounts Chem Res. 2003;36:588–596. doi: 10.1021/ar0200495. [DOI] [PubMed] [Google Scholar]
- 6.Schramm VL. J Biol Chem. 2007;282:28297–28300. doi: 10.1074/jbc.R700018200. [DOI] [PubMed] [Google Scholar]
- 7.Nanda V, Koder RL. Nat Chem. 2010;2:15–24. doi: 10.1038/nchem.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng F, Yang WC, Ko MC, Liu JJ, Cho H, Gao DQ, Tong M, Tai HH, Woods JH, Zhan CG. J Am Chem Soc. 2008;130:12148–12155. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190–U4. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 11.Yeung N, Lin YW, Gao YG, Zhao X, Russell BS, Lei LY, Miner KD, Robinson H, Lu Y. Nature. 2009;462:1079–U144. doi: 10.1038/nature08620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roca M, Vardi-Kilshtain A, Warshel A. Biochemistry. 2009;48:3046–3056. doi: 10.1021/bi802191b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinn DM. Chemical Reviews. 1987;87:955–979. [Google Scholar]
- 14.Rosenberry TL. Adv Enzymol Relat Areas Mol Biol. 1975;43:103–218. doi: 10.1002/9780470122884.ch3. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberry TL. J Mol Neurosci. 2010;40:32–39. doi: 10.1007/s12031-009-9250-3. [DOI] [PubMed] [Google Scholar]
- 16.Cheng YH, Cheng XL, Radic Z, McCammon JA. J Am Chem Soc. 2007;129:6562–6570. doi: 10.1021/ja070601r. [DOI] [PubMed] [Google Scholar]
- 17.Silman I, Sussman JL. Chem-Biol Interact. 2008;175:3–10. doi: 10.1016/j.cbi.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 18.Harel M, Quinn DM, Nair HK, Silman I, Sussman JL. J Am Chem Soc. 1996;118:2340–2346. [Google Scholar]
- 19.Bourne Y, Taylor P, Marchot P. Cell. 1995;83:503–512. doi: 10.1016/0092-8674(95)90128-0. [DOI] [PubMed] [Google Scholar]
- 20.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 21.Kua J, Zhang YK, Eslami AC, Butler JR, McCammon JA. Protein Sci. 2003;12:2675–2684. doi: 10.1110/ps.03318603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinn DM, Feaster SR, Nair HK, Baker NA, Radic Z, Taylor P. J Am Chem Soc. 2000;122:2975–2980. [Google Scholar]
- 23.Ishida T, Kato S. J Am Chem Soc. 2003;125:12035–12048. doi: 10.1021/ja021369m. [DOI] [PubMed] [Google Scholar]
- 24.Hedstrom L. Chemical Reviews. 2002;102:4501–4523. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 25.Topf M, Richards WG. J Am Chem Soc. 2004;126:14631–14641. doi: 10.1021/ja047010a. [DOI] [PubMed] [Google Scholar]
- 26.Froede HC, Wilson IB. J Biol Chem. 1984;259:11010–11013. [PubMed] [Google Scholar]
- 27.Malany S, Sawai M, Sikorski RS, Seravalli J, Quinn DM, Radic Z, Taylor P, Kronman C, Velan B, Shafferman A. J Am Chem Soc. 2000;122:2981–2987. [Google Scholar]
- 28.Fuxreiter M, Warshel A. J Am Chem Soc. 1998;120:183–194. [Google Scholar]
- 29.Zhang YK, Kua J, McCammon JA. J Am Chem Soc. 2002;124:10572–10577. doi: 10.1021/ja020243m. [DOI] [PubMed] [Google Scholar]
- 30.Zhang YK, Kua J, McCammon JA. J Phys Chem B. 2003;107:4459–4463. [Google Scholar]
- 31.Gao DQ, Zhan CG. J Phys Chem B. 2005;109:23070–23076. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 32.Vagedes P, Rabenstein B, Aqvist J, Marelius J, Knapp EW. J Am Chem Soc. 2000;122:12254–12262. [Google Scholar]
- 33.Nemukhin AV, Lushchekina SV, Bochenkova AV, Golubeva AA, Varfolomeev SD. Journal of Molecular Modeling. 2008;14:409–416. doi: 10.1007/s00894-008-0287-y. [DOI] [PubMed] [Google Scholar]
- 34.Wlodek ST, Antosiewicz J, Briggs JM. J Am Chem Soc. 1997;119:8159–8165. [Google Scholar]
- 35.Manojkumar TK, Cui CZ, Kim KS. J Comput Chem. 2005;26:606–611. doi: 10.1002/jcc.20199. [DOI] [PubMed] [Google Scholar]
- 36.Warshel A, Levitt M. J Mol Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 37.Singh UC, Kollman PA. J Comput Chem. 1986;7:718–730. [Google Scholar]
- 38.Field MJ, Bash PA, Karplus M. J Comput Chem. 1990;11:700–733. [Google Scholar]
- 39.Mulholland AJ. Drug Discovery Today. 2005;10:1393–1402. doi: 10.1016/S1359-6446(05)03611-1. [DOI] [PubMed] [Google Scholar]
- 40.Zhang YK. Theor Chem Acc. 2006;116:43–50. [Google Scholar]
- 41.Hu H, Yang WT. Annu Rev Phys Chem. 2008;59:573–601. doi: 10.1146/annurev.physchem.59.032607.093618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friesner RA, Guallar V. Annu Rev Phys Chem. 2005;56:389–427. doi: 10.1146/annurev.physchem.55.091602.094410. [DOI] [PubMed] [Google Scholar]
- 43.Gao JL, Truhlar DG. Annu Rev Phys Chem. 2002;53:467–505. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- 44.Senn HM, Thiel W. Angew Chem-Int Edit. 2009;48:1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 45.Shurki A, Warshel A. Protein Simulations. 2003;66:249–313. doi: 10.1016/s0065-3233(03)66007-9. [DOI] [PubMed] [Google Scholar]
- 46.Patey GN, Valleau JP. J Chem Phys. 1975;63:2334–2339. [Google Scholar]
- 47.Roux B. Comput Phys Commun. 1995;91:275–282. [Google Scholar]
- 48.Boczko EM, Brooks CL. J Phys Chem. 1993;97:4509–4513. [Google Scholar]
- 49.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 50.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins-Structure Function and Bioinformatics. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 52.Zhang YK, Lee TS, Yang WT. J Chem Phys. 1999;110:46–54. [Google Scholar]
- 53.Zhang YK, Liu HY, Yang WT. J Chem Phys. 2000;112:3483–3492. [Google Scholar]
- 54.Zhang YK. J Chem Phys. 2005;122:024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 55.Liu HY, Zhang YK, Yang WT. J Am Chem Soc. 2000;122:6560–6570. [Google Scholar]
- 56.Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O’Neill DP, DiStasio RA, Lochan RC, Wang T, Beran GJO, Besley NA, Herbert JM, Lin CY, Van Voorhis T, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen PE, Korambath PP, Adamson RD, Austin B, Baker J, Byrd EFC, Dachsel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu CP, Kedziora G, Khalliulin RZ, Klunzinger P, Lee AM, Lee MS, Liang W, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta E, Sherrill CD, Simmonett AC, Subotnik JE, Woodcock HL, Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Hehre WJ, Schaefer HF, Kong J, Krylov AI, Gill PMW, Head-Gordon M. Phys Chem Chem Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 57.Ponder JW. 2004 [Google Scholar]
- 58.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 59.Souaille M, Roux B. Comput Phys Commun. 2001;135:40–57. [Google Scholar]
- 60.Ferrenberg AM, Swendsen RH. Phys Rev Lett. 1988;61:2635–2638. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- 61.Wang SL, Hu P, Zhang YK. J Phys Chem B. 2007;111:3758–3764. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu P, Wang S, Zhang Y. J Am Chem Soc. 2008;130:3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu P, Wang SL, Zhang YK. J Am Chem Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu ZY, Zhang YK. J Chem Theory Comput. 2008;4:1237–1248. doi: 10.1021/ct800116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ke ZH, Wang SL, Xie DQ, Zhang YK. J Phys Chem B. 2009;113:16705–16710. doi: 10.1021/jp9080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ke ZH, Zhou YZ, Hu P, Wang SL, Xie DQ, Zhang YK. J Phys Chem B. 2009;113:12750–12758. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng H, Wang SL, Zhang YK. J Comput Chem. 2009;30:2706–2711. doi: 10.1002/jcc.21296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu RB, Hu P, Wang SL, Cao ZX, Zhang YK. J Chem Theory Comput. 2010;6:337–343. doi: 10.1021/ct9005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Froede HC, Wilson IB, Kaufman H. Arch Biochem Biophys. 1986;247:420–423. doi: 10.1016/0003-9861(86)90601-6. [DOI] [PubMed] [Google Scholar]
- 70.Szegletes T, Mallender WD, Thomas PJ, Rosenberry TL. Biochemistry. 1999;38:122–133. doi: 10.1021/bi9813577. [DOI] [PubMed] [Google Scholar]
- 71.Radic Z, Gibney G, Kawamoto S, Macpheequigley K, Bongiorno C, Taylor P. Biochemistry. 1992;31:9760–9767. doi: 10.1021/bi00155a032. [DOI] [PubMed] [Google Scholar]
- 72.Ordentlich A, Barak D, Kronman C, Ariel N, Segall Y, Velan B, Shafferman A. J Biol Chem. 1995;270:2082–2091. doi: 10.1074/jbc.270.5.2082. [DOI] [PubMed] [Google Scholar]
- 73.Shafferman A, Velan B, Ordentlich A, Kronman C, Grosfeld H, Leitner M, Flashner Y, Cohen S, Barak D, Ariel N. EMBO J. 1992;11:3561–3568. doi: 10.1002/j.1460-2075.1992.tb05439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blow DM, Birktoft JJ, Hartley BS. Nature. 1969;221:337. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- 75.Bachovchin WW, Roberts JD. J Am Chem Soc. 1978;100:8041–8047. [Google Scholar]
- 76.Dewar MJS, Storch DM. Proc Natl Acad Sci U S A. 1985;82:2225–2229. doi: 10.1073/pnas.82.8.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scheiner S, Kleier DA, Lipscomb WN. Proc Natl Acad Sci U S A. 1975;72:2606–2610. doi: 10.1073/pnas.72.7.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Umeyama H, Imamura A, Nagata C. J Theor Biol. 1973;41:485–502. doi: 10.1016/0022-5193(73)90057-x. [DOI] [PubMed] [Google Scholar]
- 79.Bachovchin WW. Biochemistry. 1986;25:7751–7759. doi: 10.1021/bi00371a070. [DOI] [PubMed] [Google Scholar]
- 80.Kossiakoff AA, Spencer SA. Biochemistry. 1981;20:6462–6474. doi: 10.1021/bi00525a027. [DOI] [PubMed] [Google Scholar]
- 81.Steitz TA, Shulman RG. Annual Review of Biophysics and Bioengineering. 1982;11:419–444. doi: 10.1146/annurev.bb.11.060182.002223. [DOI] [PubMed] [Google Scholar]
- 82.Robillar G, Shulman RG. J Mol Biol. 1974;86:519–540. doi: 10.1016/0022-2836(74)90178-8. [DOI] [PubMed] [Google Scholar]
- 83.Robillar G, Shulman RG. J Mol Biol. 1974;86:541–558. doi: 10.1016/0022-2836(74)90179-x. [DOI] [PubMed] [Google Scholar]
- 84.Bachovchin WW. Proc Natl Acad Sci U S A. 1985;82:7948–7951. doi: 10.1073/pnas.82.23.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kossiakoff AA, Spencer SA. Nature. 1980;288:414–416. doi: 10.1038/288414a0. [DOI] [PubMed] [Google Scholar]
- 86.Massiah MA, Viragh C, Reddy PM, Kovach IM, Johnson J, Rosenberry TL, Mildvan AS. Biochemistry. 2001;40:5682–5690. doi: 10.1021/bi010243j. [DOI] [PubMed] [Google Scholar]
- 87.Gerlt JA, Gassman PG. Biochemistry. 1993;32:11943–11952. doi: 10.1021/bi00096a001. [DOI] [PubMed] [Google Scholar]
- 88.Cleland WW, Kreevoy MM. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 89.Guthrie JP. Chem Biol. 1996;3:163–170. doi: 10.1016/s1074-5521(96)90258-6. [DOI] [PubMed] [Google Scholar]
- 90.Ash EL, Sudmeier JL, DeFabo EC, Bachovchin WW. Science. 1997;278:1128–1132. doi: 10.1126/science.278.5340.1128. [DOI] [PubMed] [Google Scholar]
- 91.Perrin CL, Nielson JB. Annu Rev Phys Chem. 1997;48:511–544. doi: 10.1146/annurev.physchem.48.1.511. [DOI] [PubMed] [Google Scholar]
- 92.Warshel A, Papazyan A, Kollman PA. Science. 1995;269:102–104. doi: 10.1126/science.7661987. [DOI] [PubMed] [Google Scholar]
- 93.Warshel A, Narayszabo G, Sussman F, Hwang JK. Biochemistry. 1989;28:3629–3637. doi: 10.1021/bi00435a001. [DOI] [PubMed] [Google Scholar]
- 94.Ash EL, Sudmeier JL, Day RM, Vincent M, Torchilin EV, Haddad KC, Bradshaw EM, Sanford DG, Bachovchin WW. Proc Natl Acad Sci U S A. 2000;97:10371–10376. doi: 10.1073/pnas.97.19.10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scheiner S. J Phys Chem B. 2008;112:6837–6846. doi: 10.1021/jp710617w. [DOI] [PubMed] [Google Scholar]
- 96.Topf M, Varnai P, Schofield CJ, Richards WG. Proteins-Structure Function and Genetics. 2002;47:357–369. doi: 10.1002/prot.10097. [DOI] [PubMed] [Google Scholar]
- 97.Henderson R. J Mol Biol. 1970;54:341–354. doi: 10.1016/0022-2836(70)90434-1. [DOI] [PubMed] [Google Scholar]
- 98.Waugh SM, Harris JL, Fletterick R, Craik CS. Nature Structural Biology. 2000;7:762–765. doi: 10.1038/78992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.