Summary
Cytosolic recognition of viral RNA by the RNA helicases RIG-I and MDA5 is thought to be the major pathway for IFN-α/β induction by RNA viruses. However, other cytoplasmic RNA sensors have historically been implicated in IFN-α/β production. Here we have re-examined the role of the double-stranded RNA binding protein kinase R (PKR). Using cells expressing non-functional PKR or reduced levels of kinase, we show that PKR is required for production of IFN-α/β proteins in response to a subset of RNA viruses including encephalomyocarditis, Theiler’s murine encephalomyelitis and Semliki Forest virus but not influenza or Sendai virus. Surprisingly, induction of IFN-α/β mRNA is largely normal in PKR-deficient cells. However, much of that mRNA lacks the poly A tail, indicating that its integrity is compromised. Our results suggest that PKR plays a non-redundant role in IFN-α/β production in response to some but not all viruses, in part by regulating IFN-α/β mRNA stability.
Keywords: PKR, type I interferon, innate immunity, RNA viruses
Introduction
Immunity to viral infection is characterized by the production of anti-viral cytokines, in particular type I interferons (IFN-α/β), which induce innate antiviral resistance and contribute to adaptive immunity (Le Bon and Tough, 2002; Samuel, 2001; Stark et al., 1998). IFN-α/β is primarily induced following the recognition of viral nucleic acids by host pattern recognition receptors (PRRs) that survey the extra- and intra-cellular milieu for the presence of viral genomes or viral replication intermediates (Pichlmair and Reis e Sousa, 2007; Takeuchi and Akira, 2008). For instance, several members of the toll-like receptor (TLR) family including TLR3, TLR7 and TLR9 access the endosomal compartment, where they are triggered by viral double-stranded (ds) RNA, single-stranded RNA and DNA, respectively, entering the cell from the extracellular milieu (Pichlmair and Reis e Sousa, 2007; Uematsu and Akira, 2007). In addition, all cells can respond to the presence of viral nucleic acids that have accessed the cytoplasm. The cytosolic PRRs involved in this process have not been fully identified but recently, the RNA helicases retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated factor-5 (MDA5) were shown to be essential for recognition of RNA viruses (Gitlin et al., 2006; Kato et al., 2005; Kato et al., 2006; Yoneyama et al., 2004). After binding to agonistic RNAs, both RIG-I and MDA5 interact with the adaptor IPS-1/MAVS/VISA/Cardif and initiate a signalling cascade that leads to activation of the transcription factors IRF-3 and IRF-7, which control transcription of the IFN-α and IFN-β genes (Pichlmair and Reis e Sousa, 2007; Takeuchi and Akira, 2008). Although both RIG-I and MDA5 can mediate responses to the synthetic dsRNA analog poly I:C in vitro, analysis of mice deficient for MDA5 has shown that this helicase is critical for responses to picornaviruses such as encephalomyocarditis virus (EMCV) and Theiler’s murine encephalomyelitis virus (TMEV), whereas RIG-I mediates responses to other RNA viruses including Newcastle disease virus (NDV), vesicular stomatitis virus (VSV), Sendai virus (SeV) and influenza virus (Gitlin et al., 2006; Kato et al., 2005; Kato et al., 2006). This has led to the hypothesis that MDA5 and RIG-I have different specificities and discriminate between virus-specific forms of viral RNA. Consistent with that notion, we and others have recently shown that RIG-I but not MDA-5 triggering can ensue from recognition of 5′ triphosphate groups present in the genomes of many RNA viruses (Habjan et al., 2008; Hornung et al., 2006; Pichlmair et al., 2006; Rehwinkel et al., 2010). RNA agonists for MDA-5 have not been fully characterized but are thought to correspond to long molecules of double stranded (ds) RNA.
DsRNA can be found in the genome of some viruses, but can also be generated during the process of viral replication. Interestingly, immunodetectable dsRNA (i.e. dsRNA >30bp) is seen primarily in cells infected with DNA or positive strand RNA viruses (Pichlmair et al., 2006; Weber et al., 2006), many of which are recognized by MDA5. Such dsRNA also binds to and activates PKR, an enzyme that confers antiviral resistance but differs structurally and functionally from the RNA helicases (Williams, 1999, 2001). PKR belongs to the family of kinases that regulate cellular functions in response to stress and viral infection (Holcik and Sonenberg, 2005). The main function of PKR is to limit viral replication through phosphorylation of the α-subunit of the translation initiation factor eIF-2, which results in shut-down of cellular and viral protein synthesis (Williams, 1999, 2001). Long before the discovery of MDA5 and RIG-I, PKR was proposed to act as a virus PRR involved in type I IFN induction. Some studies, including our own, found that PKR-deficient cells can be defective in IFN-α/β production in response to poly I:C (Diebold et al., 2003; McAllister and Samuel, 2008; Smith et al., 2001; Yang et al., 1995). However, PKR deficiency did not prevent IFN-α/β responses to NDV and, in some cases, IFN-α/β production could be restored in PKR−/− cells by pre-treatment with type I IFN (Yang et al., 1995). The subsequent discovery of RIG-I and MDA5 led to the conclusion that PKR is largely unimportant for IFN-α/β induction (Pichlmair and Reis e Sousa, 2007; Takeuchi and Akira, 2008). However, some recent studies support a non-redundant role for the kinase in IFN-α/β responses to viral infection (Barry et al., 2009; Carpentier et al., 2007; Gilfoy and Mason, 2007) and raise the possibility that some but not all viruses induce IFN-α/β in a PKR-dependent manner.
Here we report that PKR is required for IFN-α/β production in response to a specific subset of RNA viruses including EMCV, TMEV and Semliki Forest Virus (SFV). Surprisingly, unlike RIG-I and MDA5, PKR does not act by inducing IFN-α/β gene transcription but rather by indirectly maintaining the integrity of newly synthesized IFN-α/β mRNA, thereby permitting its translation. Our data reveal a key and non-redundant role for PKR in sustaining the IFN response to some RNA viruses and help reconcile apparently conflicting observations in the literature on the role of PKR in type I IFN induction.
Results
PKR requirement for IFN-α/β production is virus-specific
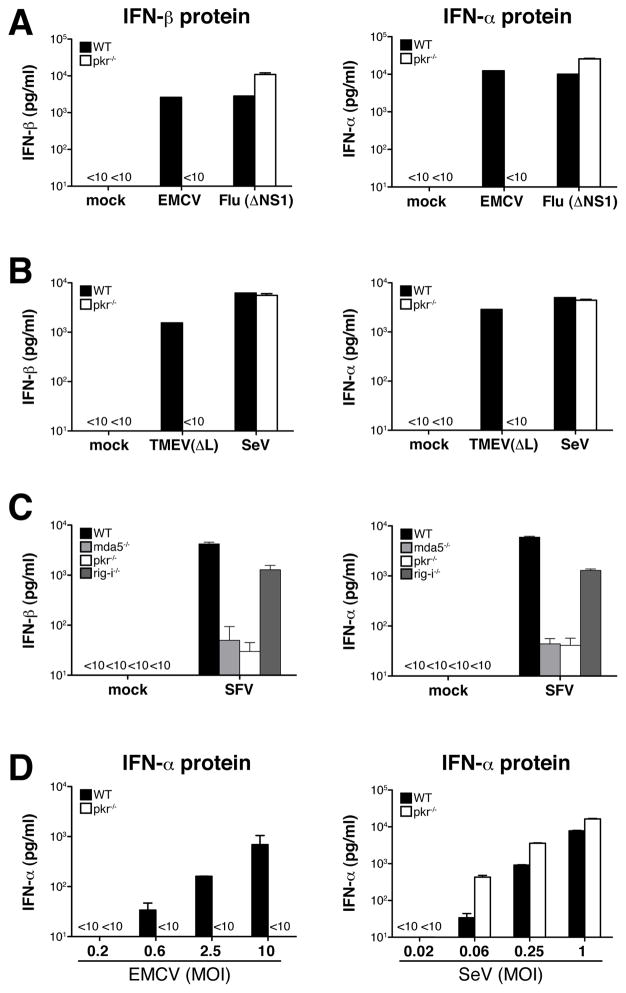
We re-examined the role of PKR in IFN production by infecting wild-type or PKR-deficient mouse bone-marrow derived dendritic cells (BM-DC) in vitro with a panel of RNA viruses that are sensed primarily by MDA5, including EMCV, TMEV, or viruses that are recognized by RIG-I such as influenza virus and SeV. Production of IFN-α/β by PKR-deficient BM-DC was normal in response to wild-type or ΔNS1 influenza virus (influenza mutant lacking the interferon inhibitor NS1; Fig. 1A and data not shown) or SeV (Fig. 1B), as measured by IFN-α and IFN-β protein accumulation in culture supernatants. In contrast, EMCV and TMEV(ΔL), a TMEV mutant lacking the interferon antagonistic leader (L) protein, failed to induce measurable levels of IFN-α or IFN-β protein in the absence of PKR (Fig. 1A, B). Like EMCV and TMEV, alphaviruses such as Semliki Forest Virus (SFV) generate considerable amounts of dsRNA in infected cells (Supplementary Fig. 1A). IFN-α/β production in response to SFV was barely detectable in the absence of MDA5 and was reduced by four-fold in RIG-I−/− BM-DCs (Fig. 1C), indicating that SFV is recognized by MDA5 with a modest contribution from RIG-I (see also Fig. 4). Importantly, IFN-α and IFN-β production after infection with SFV was also reduced by more than 100-fold in BM-DC lacking PKR (Fig. 1C). IFN-α/β induction by EMCV but not SeV was PKR-dependent at all MOIs tested (Fig. 1D), indicating that the virus-specific PKR dependence was not merely a quantitative effect.
Fig. 1. PKR is required for IFN-α/β production by BM-DCs in response to EMCV, TMEV and SFV.
BM-DCs (5×105/well) were infected with RNA viruses as indicated and IFN-α/β accumulation in the culture supernatants was measured by ELISA after overnight culture. Data are the mean ± SD of triplicate wells. (A) Production of IFN-β protein (left panel) and IFN-α protein (right panel) by BM-DCs infected with EMCV (MOI=10) or influenza virus (Flu (ΔNS1)) (MOI=1). Data are representative of at least three (influenza virus) and ten (EMCV) independent experiments. (B) Production of IFN-β protein (left panel) and IFN-α protein (right panel) by BM-DCs infected with TMEV(ΔL) (MOI=10) or SeV (MOI=1). Data are representative of at least three (TMEV) and ten (SeV) independent experiments. (C) IFN-α/β production in response to SFV is mainly dependent on MDA5. BM-DCs lacking either MDA5, RIG-I or PKR were infected with SFV (MOI=10). Data shown are one out of two experiments with similar results. (D) IFN-α production in relation to infectious dose for EMCV (left panel) and SeV (right panel). Data represent one of two experiments with similar results.
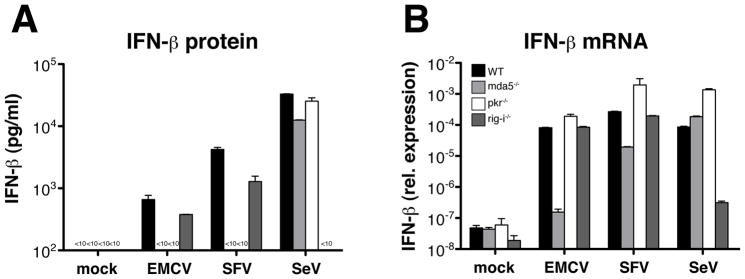
Fig. 4. PKR deficiency disproportionately reduces IFN-β protein versus mRNA.
BM-DCs (5×105/well) were infected with RNA viruses for 12 hours as indicated. IFN-β protein in the culture supernatants and IFN-β mRNA in total cDNA from lysed cells (synthesized using random hexamer primers) were measured by ELISA and quantitative PCR, respectively. Data are the mean ± SD of triplicate wells. (A) Production of IFN-β protein and (B) IFN-β mRNA from DCs lacking MDA5, RIG-I or PKR following infection with EMCV (MOI=10), SFV (MOI=10) or SeV (MOI=1). Data are representative of two independent experiments with similar results.
We measured dsRNA content as well as cell viability after viral infection. The percentage of cells expressing dsRNA following infection with EMCV or SFV was higher for PKR-deficient than for wild-type BM-DC (Supplementary Fig. 1A), in line with the established role of PKR in suppressing viral replication (Williams, 1999, 2001). We found a slight increase in the percentage of dead cells in cultures of BM-DC lacking PKR (Supplementary Fig. 1B), although this effect was too small to account for the lack of IFN-α/β secretion by these cells. Thus, lack of virus replication or cell death cannot explain the impaired IFN-α/β responses in PKR-deficient DC infected with picornaviruses or SFV.
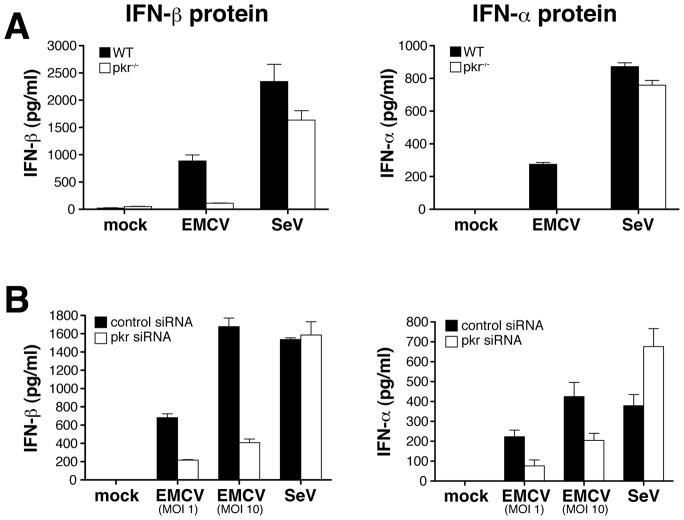
To confirm these observations in another cell type, we analyzed IFN-α/β expression in MEFs. To circumvent virus-induced cytotoxicity in those cells, which was much more pronounced than in DC (data not shown), MEFs were pre-treated with IFN-α/β. We used IFN A/D, a human IFN hybrid that does not cross-react with the ELISA antibodies used for measuring mouse IFN-α/β but acts like other IFNs to promote an antiviral state that protects the cells from cytopathic effects of viral infection (Rehberg et al., 1982). Like DCs, PKR−/− MEFs treated with IFN A/D mounted a normal IFN-α/β response after SeV infection, but showed a severe defect in the response to EMCV (Fig. 2A). We also used siRNA-mediated gene silencing to rule out that the effect was due to a secondary phenotype of the PKR-deficient cells. When infected with EMCV, MEFs treated with PKR-specific siRNA showed a 3-fold reduction in PKR levels (data not shown) and a 60% reduction in the level of IFN-α and IFN-β compared to MEFs treated with a control siRNA (Fig. 2B). Consistent with the results in PKR-deficient cells, the PKR-specific siRNA did not impair the response to SeV (Fig. 2B). Altogether, these observations suggest that PKR is required for IFN-α/β production in response to some RNA viruses and that the PKR-dependence of the IFN-α/β response to any given virus correlates with reliance on the MDA5 pathway.
Fig. 2. PKR mediates IFN-α/β production by MEFs in response to EMCV.
Immortalized MEFs (5×105/well) were stimulated with IFN A/D (1000 U/ml) for two hours prior to infection with EMCV or SeV. IFN-α/β accumulation in the culture supernatants was measured by ELISA after overnight culture. Data are the mean ± SD of triplicate wells. (A) IFN-β protein (left panel) and IFN-α protein (right panel) from PKR−/− and wild-type MEFs infected with EMCV (MOI=10) or SeV (MOI=1). (B) IFN-β protein (left panel) and IFN-α protein (right panel) from siRNA-treated MEFs after infection with EMCV or SeV (MOI=1). Data are representative of four independent experiments.
Phosphorylation of eIF-2α is not necessary for IFN-α/β production in response to PKR-dependent viruses
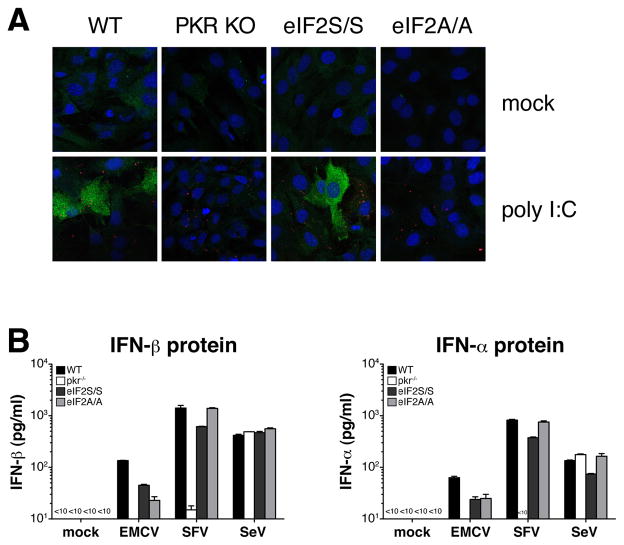
Most biological effects of PKR are mediated through phosphorylation of eIF-2α (Holcik and Sonenberg, 2005; Williams, 1999, 2001). To test whether eIF-2α phosphorylation is required for the PKR-dependent production of IFN-α/β in response to EMCV and SFV, we used cells with a homozygous Ser to Ala mutation at the eIF-2α phosphorylation site (Ser51) (Scheuner et al., 2001). In contrast to wild-type cells, eIF-2α mutant MEFs (eIF2A/A) did not phosphorylate eIF-2α in response to poly I:C (Fig. 3A). Nevertheless, eIF2A/A MEFs showed no defect in their ability to produce IFN-α or IFN-β proteins in response to stimulation with poly I:C (data not shown) or upon infection with EMCV or SFV (Fig. 3B). We conclude that phosphorylation of eIF-2α is not required for PKR-dependent IFN-α/β production.
Fig. 3. Phosphorylation of eIF-2α is not necessary for IFN-α/β production in response to PKR-dependent viruses.
(A) Confocal analysis of eIF-2α phosphorylation in Ser(51) mutant MEFs (eIF2A/A) and control MEFs. Cells were stimulated with poly I:C for 6 hours and stained with mAbs specific for phospho-eIF-2α and dsRNA (K1), as well as the nuclear dye DRAQ5. Phospho-eIF-2α and dsRNA staining are shown in green and red, respectively. (B) IFN-β protein (left panel) and IFN-α protein (right panel) from eIF-2A/A and control eIF-2S/S MEFs infected with EMCV (MOI=10), SFV (MOI=10) or SeV (MOI=1). Data shown are from one out of three experiments.
PKR regulates IFN-β mRNA integrity rather than transcriptional activation in cells infected with EMCV
We next asked whether PKR is necessary for EMCV- or SFV-induced transcriptional induction of the IFN-α/β genes. Because the IFN-α ELISA detects multiple IFN-α subtypes, we focused on IFN-β, which is encoded by a single gene, allowing us to directly correlate the protein and transcript levels. The latter were determined by quantitative PCR of cDNA generated by random hexamer oligonucleotide priming of RNA extracted from infected cells. As expected (Kato et al., 2005; Kato et al., 2006), SeV infection induced the appearance of IFN-β mRNA in a RIG-I dependent, MDA-5-independent manner, which correlated with IFN-β protein expression (Fig. 4A, B). Consistent with the role of MDA5 as a PRR for SFV and EMCV, MDA5−/− cells failed to accumulate IFN-β mRNA or produce IFN-β protein in response to either virus (Fig. 4A, B). In contrast, to our surprise, IFN-β mRNA was induced >1000 fold in PKR-deficient cells infected with EMCV (Fig. 4B) even though IFN-β protein was not detectable (Fig. 4A). Similarly, infection with SFV led to up-regulation of IFN-β mRNA in PKR−/− DCs at levels 100-fold and 10-fold higher than in DCs lacking either MDA5 or RIG-I, respectively (Fig. 4B), but did not result in IFN-β protein production (Fig. 4A). Taken together, our data suggest that, although MDA5 and PKR are both essential for driving IFN-α/β production in response to EMCV and SFV, the mechanism of their action is different. Unlike MDA5, PKR is largely dispensable for the induction of IFN-α/β mRNA but appears to act post-transcriptionally to regulate protein production.
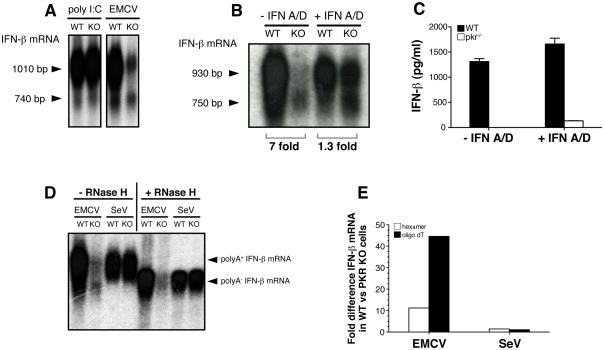
It was intriguing that PKR−/− cells produced little IFN-β protein despite seemingly normal induction of IFN-β mRNA following infection with EMCV. To gain further insight into this issue, we analyzed IFN-β mRNA from EMCV infected cells by Northern blot. This analysis indicated that IFN-β mRNA levels were in fact 6–8 fold lower in PKR−/− BM-DC compared to PKR+/+ cells (Fig. 5A, B). It was conceivable that the lower IFN-β mRNA levels in PKR-deficient cells arose from a failure to up-regulate MDA5, which is itself IFN-inducible (Kang et al., 2002). Consistent with this notion, PKR−/− cells showed decreased levels of MDA5 mRNA upon infection with EMCV but not SeV, which could be corrected by pre-treatment with IFN-A/D (Suppl. Fig. 2). Notably, IFN A/D pre-treatment also restored IFN-β mRNA levels in EMCV-infected PKR-deficient DC to within one-to three-fold of the levels in wild-type cells (Fig. 5B). However, IFN A/D pre-treatment failed to rescue IFN-β protein production by DC (Fig. 5C), as noted for MEFs (see above).
Fig. 5. Deadenylated IFN-β mRNA in EMCV-infected PKR−/− cells.
(A, B, C) BM-DCs (5×105/well) were transfected with poly I:C (10μg/well) for six hours or infected with EMCV (MOI=10) for 9 hours in the absence or presence of IFN A/D (1000U/ml) as indicated. (A, B) IFN-β mRNA was analyzed by Northern blot and the position of two distinct species of IFN-β mRNA is indicated by arrow heads. The size of the different mRNA species was calculated by comparing their electrophoretic mobility to a molecular size marker in the form of a ssRNA ladder. Numbers below picture represent fold difference between samples from PKR+/+ and PKR−/− cells as determined by phosphoimager analysis. (C) IFN-β protein in the supernatant from the same cultures as in (B) was measured by ELISA. Data are the mean ± SD of triplicate wells and are representative of three independent experiments with similar results. (D) Detection of polyA+ and polyA− forms of IFN-β mRNA by Northern blot. Total RNA was isolated from BM-DCs infected with EMCV (MOI=10) or SeV (MOI=1) for 12 and 6 hours respectively. Where indicated, RNA samples were treated with oligo-dT ± RNase H. Arrow heads indicate position of polyA+ and polyA− IFN-β mRNA. (E) RNA from cells treated as in (D) was analyzed by quantitative PCR using random hexamer (open bars) or oligo dT oligonucleotides (filled bars) to generate cDNA. Relative expression of IFN-β mRNA in PKR+/+ versus PKR−/− cells was calculated by analysing the Ct values for each amplification curve and converting them into fold difference using the formula 2(−ΔCt). Data in (D) and (E) are representative of two experiments.
Unexpectedly, Northern blot analysis of RNA from cells infected with EMCV revealed the presence of two bands for IFN-β with different apparent molecular sizes (Fig. 5A, B). The lower molecular size band was especially prominent in PKR-deficient cells: it accounted for 10–20% of the total IFN-β mRNA in PKR+/+ DC but up to 60% in PKR−/− DC (Fig. 5A, B, D). The shorter form of IFN-β mRNA was not seen upon infection with SeV and was not especially prominent in response to transfection with poly I:C (Fig. 5A, B, D). The size difference between the short and long forms was a few hundred bases, which could correspond to the poly A tract of the mRNA. Consistent with that notion, the shorter form had an apparent molecular size of around 750 bases (Fig. 5A, B), similar to that reported for the IFN-β cDNA lacking the poly A tail (Higashi et al., 1983). To test the possibility that the lower molecular size band corresponded to IFN-β mRNA lacking a poly A tail, we hybridized dT oligonucleotides to RNA from infected cells and digested with RNase H. After enzymatic removal of the poly A tail, all IFN-β mRNA showed an electrophoretic mobility similar to that of the lower band seen in RNA from PKR−/− cells not treated with RNase H (Fig. 5D). Importantly, the mRNA from EMCV-infected PKR−/− cells did not show a further shift in electrophoretic mobility, indicating that it was already deadenylated (Fig. 5D). Furthermore, when dT oligonucleotides were used instead of random hexamer oligonucleotides to generate complementary DNA, quantitative PCR revealed a profound difference in the levels of IFN-β mRNA between PKR+/+ and PKR−/− cells (Fig. 5E). This effect was EMCV-specific because we found no difference between hexamer and oligo dT-primed IFN-β cDNA in cells infected with SeV (Fig. 5E), consistent with the Northern blot analysis (Fig. 5B, D). We also compared hexamer and oligo dT-primed cDNA for several other inducible or constitutively-expressed genes, including GAPDH, IL-6, TNF-α and RPS-16, and observed that the ratio of cDNA between wild-type and PKR−/− cells following infection with EMCV or SeV remained unchanged (Suppl. Fig. 3). These results indicate that the lack of a poly A tail in EMCV-infected cells is specific to IFN-β mRNA and not reflective of a generic deadenylation of all cellular mRNAs. We conclude that, in the absence of PKR, most of the IFN-β mRNA generated in response to EMCV infection lacks the poly A-tail. This could impair its translation and explain the absence of IFN-β protein synthesis by EMCV-infected PKR−/− cells.
Serum levels of IFN-β are reduced in PKR-deficient mice after infection with EMCV
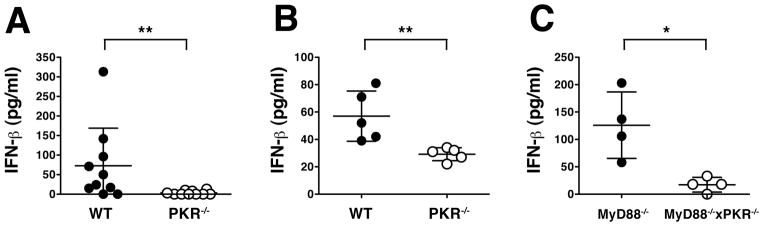
To extend these observations to an in vivo setting, we infected wild-type and PKR-deficient mice with EMCV and measured IFN-β in the serum at 20 hours post-infection. Serum IFN-β levels in wild-type mice were generally low regardless of genetic background (Fig. 6A–C) and mRNA for IFN-β in tissues was below the detection limit, precluding analysis of its adenylation status (not shown). Nevertheless, we consistently observed even lower levels of serum IFN-β protein in mice lacking PKR (Fig. 6A–C). This was true in experiments when wild type 129/Sv mice were compared to PKR−/− mouse on a mixed 129 Sv/C57BL/6 background (Fig. 6A), but also when PKR−/− mice that had been backcrossed more than ten times to the C57BL/6 strain were compared to wild type mice of that strain (Fig. 6B). Finally, PKR deficiency led to a reduction in serum IFN-β levels in MyD88−/− mice infected with EMCV (Fig. 6C), excluding the possibility that the effect of PKR was downstream of the TLR7 pathway, which can contribute to IFN responses to ssRNA viruses in vivo. In conclusion, our results and those of others (Barry et al., 2009) confirm a role for PKR in generating an optimal IFN response to EMCV and SFV in vivo.
Fig. 6. PKR promotes IFN-β protein production in response to infection with EMCV in vivo.
IFN-β protein was measured in the serum of mice that had been infected with EMCV 20 hours earlier. (A) Wild-type (129 Sv) and PKR−/− mice (mixed 129 Sv/C57BL/6 background), p<0.006 (n=10). (B) Wild-type (C57BL/6) and PKR−/− mice (C57BL/6 background), p<0.008 (n=5). (C) MyD88−/− and MyD88−/− × PKR−/− mice, p<0.03 (n=4). (A–C) Dots represent individual mice, error bars are mean ± SD. Data in each panel are representative of two independent experiments with similar results. Paired data were analysed using a two-tailed Mann-Whitney test.
Discussion
The notion that PKR is involved in the induction of IFN-α/β has been largely dismissed by the discovery of the RNA helicases RIG-I and MDA5 and associated signaling pathways (Pichlmair and Reis e Sousa, 2007; Takeuchi and Akira, 2008). RNA viruses are increasingly classified by their ability to stimulate RIG-I, MDA5 or a combination of both (Gitlin et al., 2006; Kato et al., 2005; Kato et al., 2006). Here, we compared viruses representing different ends of this spectrum for their ability to induce IFN-α/β in the absence or presence of PKR. In agreement with previous studies using NDV (Smith et al., 2001; Yang et al., 1995), PKR was not required for IFN-α/β production in cells infected with RIG-I dependent viruses such as influenza and SeV. In contrast, IFN-α/β production in response to the picornaviruses EMCV and TMEV, both capable of triggering MDA5, was greatly reduced in cells lacking PKR. Furthermore, SFV, which we find to be primarily recognized via MDA5, also required PKR for IFN protein production as reported recently (Barry et al., 2009). Two previous reports have also implicated PKR in cytokine production after infection with TMEV and West Nile virus (Carpentier et al., 2007; Gilfoy and Mason, 2007), two viruses that are recognized, at least in part, via MDA5 (Fredericksen et al., 2008). Thus, it is intriguing to speculate that PKR may promote IFN-α/β production to MDA5- but not to RIG-I-dependent viruses. Interestingly, a requirement for PKR in IFN-α/β responses to the putative MDA5 agonist, poly I:C, had been noted in some studies (Diebold et al., 2003; McAllister and Samuel, 2008; Smith et al., 2001; Yang et al., 1995). We find that such PKR dependence is variable, perhaps because many preparations of poly I:C additionally stimulate RIG-I and TLR3 pathways, and is therefore best observed using viruses that show a strong dependence on MDA5 for IFN-α/β induction (OS, AP and CRS, unpublished observations). The correlation between MDA5- and PKR dependence of antiviral IFN-α/β responses may arise from the fact that PKR and MDA5 can be activated by long dsRNA (Lemaire et al., 2008), which is not produced to any great extent by many of the RIG-I dependent viruses (Pichlmair et al., 2006; Weber et al., 2006). However, it should be noted that PKR can also be activated upon infection with RIG-I dependent viruses, possibly through 5′-triphosphate RNA containing short stem-loops (Nallagatla et al., 2007). Therefore, one would need to envisage that different RNA agonists elicit different PKR responses and that PKR has a hitherto unappreciated role in controlling IFN-α/β production when it has been activated by infection with viruses that generate specific forms of dsRNA.
EMCV has emerged as the prototypic MDA5-dependent virus (Gitlin et al., 2006; Kato et al., 2006) and a previous study showed a reduction in IFN-α/β expression following EMCV infection of cells expressing a dominant-negative form of PKR (Der and Lau, 1995). This is consistent with our findings although, in contrast to that earlier report, we could find no evidence that PKR contributes significantly to the induction of IFN-α/β genes. Although PKR can promote the activation of nuclear factor kappa B (Kumar et al., 1994), its ability to activate IRF-3 and 7, the critical transcription factors in induction of IFN-α/β genes (Honda and Taniguchi, 2006; Sato et al., 2000), has not been formally demonstrated. Our data are consistent with the current concept that signaling via RIG-I-like helicases, but not through PKR, leads to transcription of the IFN-α and IFN-β genes (Yoneyama et al., 2005). Nevertheless, our results support a role for PKR in regulating the quality of IFN-α/β mRNA in the context of viral infection. How PKR regulates this process or what are the molecular targets downstream of PKR is currently not understood. We have been able to rule out a role for the translation initiation factor eIF-2α, the best characterised PKR substrate, by showing that phosphorylation of eIF-2α is not required for IFN production in response to EMCV infection. Given that eIF-2α phosphorylation is crucial for the virus-induced translation stop (Garcia et al., 2006) and apoptosis (Kaufman, 1999; Scheuner et al., 2006), these results indicate that PKR promotes IFN-α/β protein expression by a different mechanism.
This mechanism appears to involve regulation of IFN-α/β mRNA integrity as we found that much of the IFN-β mRNA from PKR−/− cells after infection with EMCV appears truncated. Any role for PKR in preventing this process is not only specific to infection with some viruses but also mRNA specific as we did not observe loss of integrity in other mRNAs, be they from inducible or constitutively expressed genes. Further analysis of IFN-β mRNA from EMCV-infected cells suggests that the truncated form of IFN-β mRNA might correspond to mRNA that has lost its poly A tail. The expression of many cytokines is controlled by post-transcriptional mechanisms that regulate various aspects of RNA biology, especially mRNA stability and decay (Anderson, 2008). Indeed, IFN-β mRNA contains several destabilizing elements, including a class II AU-rich element that promotes asynchronous deadenylation and leads to the accumulation of poly A-negative intermediates (Paste et al., 2003). The molecules that bind to the destabilizing elements in IFN-β mRNA or that regulate the degradation of the poly A tail remain poorly characterized (Anderson, 2008; Raj and Pitha, 1993) and, therefore, leave open many possibilities as to how PKR regulates IFN-β mRNA integrity in the context of EMCV infection. For example, PKR impacts on eIF4E phosphorylation via the B56α regulatory subunit of protein phosphatase 2A thereby potentially affecting translation and deadenylation of mRNA in a manner independent of eIF2α (Xu and Williams, 2000). Alternatively, PKR may act by antagonizing virus-specific factors that promote destabilization of IFN-β mRNA rather than having a direct role in regulating IFN-α/β mRNAs. Although we do not understand the mechanism at present, our data nevertheless provide a possible explanation as to why IFN-β mRNA, despite being present in PKR−/− cells, is not expressed at the protein level.
In sum, our data support a role for PKR in regulating the expression of IFN-α/β proteins and demonstrate for the first time that PKR is required coordinately with a member of the RNA helicase family, MDA5, to induce an IFN response to certain RNA viruses. Our study further reveals that PKR may act primarily in establishing and/or maintaining the polyadenylation status of IFN-α/β mRNA, adding to our understanding of IFN-α/β induction after virus infection and revealing a hitherto unappreciated role for mRNA stability in the process.
Experimental Procedures
Reagents
IFN A/D was a gift from I. Kerr (Cancer Research UK). Poly I:C was from Amersham Biosciences. Recombinant murine GM-CSF was made at Cancer Research UK. SiRNA specific for mouse PKR (Prkr-3) and human PRMT (protein arginine methyltransferase; control siRNA) were from Qiagen. The target sequences for murine PKR and human PRMT are CAGCTCGTCTATGACAAGTAA and AAAGATTACTACTTTGACTCC, respectively. Anti-dsRNA antibody (clone K1) was from English & Scientific Consulting Bt. Polyclonal anti-phospho-eIF-2α (Ser51) was from New England Biolabs.
The plasmid pGEM-IFN-β was generated by PCR amplification of full-length IFN-β from BM-DCs stimulated with SeV and subsequent cloning of the PCR product into pGEM-T vector (Promega). The sequences for murine IFN-β forward and reverse primers were 5′-ATGAACAACAGGTGGATCC and 5′-GGCATCAACTGACAGGTCTT, respectively.
The plasmid pBABE-puro-LargeT was a gift from G. Peters (Cancer Research UK).
Animals and cells
129 SvEv mice were obtained from Taconic. PKR−/− mice (Yang et al., 1995) on a mixed 129 SvEv x C57Bl6 background were originally obtained from H. Unger (University of Veterinary Medicine, Vienna, Austria) and were maintained in the animal facility of CRUK (Clare Hall, South Mimms, UK) under specific pathogen-free conditions. For in vivo experiments, mice were fully backcrossed (10 generations) to C57BL/6 or to mice deficient in MyD88. BM-DC were generated in RPMI 1640 medium containing 10% FCS, 2 mM glutamine, 50 μM 2-mercaptoethanol, 100 units/ml penicillin, 100 μg/ml streptomycin and GM-CSF (~10-ng/ml). Wild-type (129-SvEv Tac) and PKR−/− MEFs were prepared from 12.5 day embryos by standard protocols. MEFs were used as primary cells or after immortalization with retrovirus expressing large T antigen prepared from supernatants of Phoenix cells transfected with pBABE-puro-LargeT. Immortalized MEFs were selected on puromycin (final concentration 2-μg/ml) for 2 weeks and were grown in DMEM medium containing 10% FCS, 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin. Primary MEFs were grown in DMEM containing 10% filtered, non heat-inactivated FCS, 1 × MEM amino acids, 1 × non-essential amino acids, 50 μM 2-mercaptoethanol, 100 units/ml penicillin, 100 μg/ml streptomycin.
Viruses and cytokine induction assays
Influenza A/PR/8/34 and ΔNS1 were a gift from T. Muster (Vienna). SFV and EMCV were a gift from I. Kerr. TMEV(ΔL) was a gift from T. Michiels (Brussels). SeV was obtained from LGC Promochem/ATCC.
BM-DCs or MEFs were seeded in 24 plates at 5×105 cells per ml and cultured with different viruses for 12–15 hours unless indicated otherwise. This time point was chosen because for some viruses (i.e. EMCV), IFN-α/β protein became detectable only at around 10 hours after infection (data not shown). IFN A/D (1000 U/ml) was added to MEF cultures two hours prior to infection. Mice were infected intravenously with EMCV (106 pfu/mouse). Supernatants or mouse sera were assayed for cytokine content by sandwich ELISA. IFN-α (multiple subtypes) was measured by ELISA as described previously (Diebold et al., 2003) and IFN-β was measured using an ELISA Kit (R+D Systems).
Flow cytometry
BM-DCs were seeded in 24 plates at 5×105 cells per ml and infected with EMCV or SFV for overnight. Cells were fixed in 4% paraformaldehyde, permeabilised using 0.1% saponin and stained with anti-CD11c mAb (Becton Dickinson) and anti-dsRNA mAb followed by appropriate secondary antibodies. For assessment of cell viability BM-DCs were fixed and stained with LIVE/DEAD fixable dead cell dye (Invitrogen) as recommended by the manufacturer. Samples were run on a FACS Calibur (Becton Dickinson) and data were analyzed using FlowJo software (Treestar).
Confocal microscopy
MEFs were grown on coverslips overnight and stimulated for 6 hours with dsRNA by transfecting poly I:C (1 μg/well) complexed with lipofectamine 2000 (Invitrogen). Cells were fixed in 4% paraformaldehyde, permeabilised in 0.1% Triton X-100 and stained with anti-dsRNA and anti-phospho-eIF-2α antibodies followed by secondary antibodies including Alexa488-conjugated anti-rabbit, Alexa546-conjugated anti-mouse and DRAQ5 (Invitrogen). Coverslips were mounted on a slide and images were taken with a laser scanning confocal microscope (LSM 510; Zeiss).
PKR knock-down
MEFs were seeded at 2.5×104 cells/well in 24-well plates in DMEM medium without antibiotics. Cells were transfected twice (at 0 and +24 hrs) with PKR-specific siRNA or control siRNA (40 pmol/well) using the transfection reagent lipofectamine 2000 (Invitrogen). At the end of 48hrs cells were either stimulated with IFN A/D (1000 U/ml) for 1-2 hrs prior to infection with viruses as indicated or stimulated with IFN A/D (1000 U/ml) alone overnight for analysis of PKR protein by Western Blot.
PCR and Northern blot
Total RNA was isolated from infected or uninfected cells using the RNeasy kit (Qiagen) combined with a DNA digestion step (DNase set, Qiagen). Single stranded cDNA was synthesized using SuperScript II (Invitrogen) and random hexamer primers. Quantitative PCR amplification was carried out using TaqMan universal master mix (Applied Biosystems) and pre-developed TaqMan assay reagents (containing primers and fluorescent probe) for murine IFN-β, MDA5, 18s rRNA, IL-6, TNF-α, RPS-16 and GAPDH (Applied Biosystems) on an ABI 7900HT thermal cycler (Applied Biosystems).
For Northern blotting, total RNA was separated by gel electrophoresis on an 1.5% agarose gel. RNA was transfered by capillary action to a nylon membrane (Hybond-N+; Amersham Biosciences), crosslinked to the membrane with UV light and hybridised with [a32P]-dCTP and [a32P]-dATP (Perkin Elmer) labelled probes. Anti-sense deoxyribo-probes specific for murine IFN-β were generated by linear PCR amplification of a full-length template (generated through conventional PCR amplification of pGEM-IFN-β) using the reverse primer for IFN-β. For poly A tail digestion, total RNA was hybridised to dT(15) oligonucleotides and treated with RNase H (USB Europe GMBH) for 2 h at 37°C.
Supplementary Material
Acknowledgments
This work was funded by Cancer Research UK and in part by NIH Grants DK042394, HL052173 and HL057346 to R.J.K. C.R.S. acknowledges the financial support of the Fondation Bettencourt-Schueller. R.J.K. is an investigator of the Howard Hughes Medical Institute. J.R. is a recipient of FEBS and HFSP long-term fellowships. We thank Ian Kerr for EMCV and SFV, Thomas Muster for Flu(ΔNS1), Thomas Michiels for TMEV(ΔL) and Hermann Unger for PKR−/− mice. We are grateful to Sandra S. Diebold and members of the Immunobiology lab for advice and critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- Barry G, Breakwell L, Fragkoudis R, Attarzadeh-Yazdi G, Rodriguez-Andres J, Kohl A, Fazakerley JK. PKR acts early in infection to suppress Semliki Forest virus production and strongly enhances the type I interferon response. J Gen Virol. 2009;90:1382–1391. doi: 10.1099/vir.0.007336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier PA, Williams BR, Miller SD. Distinct roles of protein kinase R and toll-like receptor 3 in the activation of astrocytes by viral stimuli. Glia. 2007;55:239–252. doi: 10.1002/glia.20450. [DOI] [PubMed] [Google Scholar]
- Der SD, Lau AS. Involvement of the double-stranded-RNA-dependent kinase PKR in interferon expression and interferon-mediated antiviral activity. Proc Natl Acad Sci U S A. 1995;92:8841–8845. doi: 10.1073/pnas.92.19.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Montoya M, Unger H, Alexopoulou L, Roy P, Haswell LE, Al-Shamkhani A, Flavell R, Borrow P, Reis e Sousa C. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfoy FD, Mason PW. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:11148–11158. doi: 10.1128/JVI.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Andersson I, Klingström J, Schümann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Mühlberger E, et al. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Sokawa Y, Watanabe Y, Kawade Y, Ohno S, Takaoka C, Taniguchi T. Structure and expression of a cloned cDNA for mouse interferon-beta. J Biol Chem. 1983;258:9522–9529. [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Double-stranded RNA-activated protein kinase mediates virus-induced apoptosis: a new role for an old actor. Proc Natl Acad Sci U S A. 1999;96:11693–11695. doi: 10.1073/pnas.96.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Haque J, Lacoste J, Hiscott J, Williams BR. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci U S A. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bon A, Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14:432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CS, Samuel CE. Protein kinase PKR enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J Biol Chem. 2008 doi: 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- Paste M, Huez G, Kruys V. Deadenylation of interferon-beta mRNA is mediated by both the AU-rich element in the 3′-untranslated region and an instability sequence in the coding region. Eur J Biochem. 2003;270:1590–1597. doi: 10.1046/j.1432-1033.2003.03530.x. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Raj NB, Pitha PM. 65-kDa protein binds to destabilizing sequences in the IFN-beta mRNA coding and 3′ UTR. Faseb J. 1993;7:702–710. doi: 10.1096/fasebj.7.8.8500695. [DOI] [PubMed] [Google Scholar]
- Rehberg E, Kelder B, Hoal EG, Pestka S. Specific molecular activities of recombinant and hybrid leukocyte interferons. J Biol Chem. 1982;257:11497–11502. [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. RIG-I Detects Viral Genomic RNA during Negative-Strand RNA Virus Infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Samuel C. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Scheuner D, Patel R, Wang F, Lee K, Kumar K, Wu J, Nilsson A, Karin M, Kaufman RJ. Double-stranded RNA-dependent protein kinase phosphorylation of the alpha-subunit of eukaryotic translation initiation factor 2 mediates apoptosis. J Biol Chem. 2006;281:21458–21468. doi: 10.1074/jbc.M603784200. [DOI] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Smith EJ, Marie I, Prakash A, Garcia-Sastre A, Levy DE. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by Vaccinia virus E3L protein. J Biol Chem. 2001;276:8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- Stark G, Kerr I, Williams B, Silverman R, Schreiber R. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- Williams BR. Signal integration via PKR. Sci STKE. 2001;2001:RE2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- Xu Z, Williams BR. The B56alpha regulatory subunit of protein phosphatase 2A is a target for regulation by double-stranded RNA-dependent protein kinase PKR. Mol Cell Biol. 2000;20:5285–5299. doi: 10.1128/mcb.20.14.5285-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. Embo J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.