Abstract
A mouse parvovirus (designated MPV1f) was identified in a commercial laboratory mouse colony in Australia. The infection had not been detected by using an rNS1 parvovirus ELISA antigen even though the virus was genetically similar to other MPV1 variants reported previously. A recombinant biotinylated protein based on a truncated VP1 protein of the MPV1 strain was produced and used as antigen for ELISA and Western immunoblots to detect virus infection and determine the seroprevalence of infection in a colony of approximately 45,000 mice. Antibody-positive mice were detected in 8 of 11 rooms sampled, indicating that infection was widespread in the facility. Antibody was detected in 16.2% of 1161 sera obtained from 20 strains of mice. Seroprevalence varied among mouse strains, suggesting genetic variation in the susceptibility of mice to MPV1 or in their antibody response to infection, as has been reported previously in experimentally infected mice. Seroprevalence was high in some inbred strains, including DBA/2JArc and the random-bred strains Hsd:NIH and Arc:Arc(s). Antibody was not detected inC57BL/6J strains, and BALB/c strains showed low seroprevalence of MPV1f.
Abbreviation: MPV, mouse parvovirus; MVM, minute virus of mice; PBST, PBS + Tween 20; rNS1, recombinant NS1 nonstructural virus protein; rVP1, recombinant MPV1 viral capsid protein
Murine parvoviruses that have been identified and characterized include minute virus of mice (MVM)8 and mouse parvovirus 1 (MPV1).2 Several genotypic variants of MPV1 are recognized: MPV1a and MPV1b that originated from a murine cytotoxic T-cell line2,4,13 and MPV1c and MPV1e from naturally infected laboratory mice.3,4 Strains of MPV that have been designated MPV2,5 MPV3,5 MPV4 (GenBank accession nos., FJ440683 and FJ445512), and MPV5 (NC011618 and FJ440683) have also been described. MPV1 is the most common MPV type detected in mouse colonies,5 and the prevalence of infection in infected colonies is usually high.12 Although infection is typically asymptomatic, MPV1 is considered an important pathogen of laboratory mice, causing dysfunction of T lymphocytes and altering the pattern of graft and tumor rejection.13
The genome of MPV1 resembles MVM in size and organization, with conservation of open reading frames and similar promoter sequences, splice sites, packaging of the 5-kb minus (V) strand, replication intermediates, and nonstructural proteins (NS).2 However, there is little conservation in the structural virion proteins (VP) of MVM and MVP1, which accounts for the antigenic differences between the 2 viruses.2
Despite the availability of several methods, serologic detection of MPV1 infection remains problematic.3 MPV1 was detected by PCR in the laboratory mouse colony of our institution in 2003 even though routine serologic surveillance by using a then-available recombinant parvovirus NS1 protein antigen for ELISA consistently had provided negative results. Here we describe the genomic characteristics of the virus detected in Australia, the development of serologic tests using a recombinant truncated VP1 protein antigen, and determination of the prevalence of infection in various mouse strains in the colony prior to eradication of the virus.
Materials and Methods
Viral DNA.
Fecal samples and spleen tissue were obtained from mice in our institution's breeding facility. DNA was extracted from spleen tissue by using a QIAamp tissue kit (Qiagen, Hilden, Germany), and DNA was extracted from feces by using a QIAamp DNA stool minikit (Qiagen) according to the manufacturer's instructions. The extracted DNA was stored at −20 °C.
PCR for detection of MPV1 DNA.
Oligonucleotide primers (Proligo, Boulder, CO) for detection of virus were based on the previously reported sequence of other known strains of MPV1 and are shown in Table 1. These primers were designed to amplify a 653-nt fragment of the VP1 open reading frame and encompassed the putative allotropic domain.4
Table 1.
Primer pairs used for detection of virus and analysis of the MPV1f genome
| Primer pair | Primer sequence (3′→5′) | Amplicon size (bp) | Purpose |
| MPV1_3505F | GGC TAA TGC CAG CGG GCT G | 653 | Viral detection and cloning |
| MPV1_4158R | CAG TTA GTG GTC CTT AGC | ||
| MPV1_147F | AGG CGC GAA AAG GAA GTG | 541 | Cloning |
| MPV1_688R | CTT GAG CTT GAC TAA AGT C | ||
| MPV1_1818F | ATT GCC TGG TGA CTT TGG T | 669 | Cloning |
| MPV_2487R | GCA GCG GCG TCA GAT G | ||
| MPV1_264F | CAT GGC TGG AAA TGC TTA C | 2019 | Cloning |
| MPV1_2283R | TTA GTC CAA GTT CAG CGG | ||
| MPV1_2271F | GAA CTT GGA CTA AGG TAC G | 2310 | Cloning |
| MPV1_4581R | GAA AGA AAG AAC ATG GTT GG | ||
| MPV1_4468F | ATA CAT GAG CAT TCA CAA ATG | 418 | Cloning |
| MPV1_4886R | AGA TTT TAT TGT TTT TTT GGT C |
All PCR reactions were performed in a 20-µL reaction volume in a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA). For detection of viral infection, reaction mixes consisted of 1× Platinum Taq reaction buffer (Invitrogen, Carlsbad, CA), 1.5 mM Mg2+, 0.15 mM dNTPs, 0.15 µM MPV1_3505F, 0.15 µM MPV1_4158R, and 0.046 U/µL Platinum Taq polymerase (Invitrogen). The PCR cycling consisted of 1 cycle of 94 °C for 3 min followed by 30 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s, with 1 final cycle of 72 °C for 7 min. The PCR reaction products were separated by electrophoresis on a 1% agarose gel, stained with ethidium bromide, and visualized by using a UV transilluminator. Visual images were produced by using Molecular Analyst software version 1.4 (Bio-Rad).
DNA sequencing and analysis.
DNA extracted from spleen tissue of an Hsd:NIH mouse identified as PCR-positive for MPV1 was used as a template for sequencing of the virus. This isolate provisionally was designated MPV1f. The entire genome but excluding the terminal palindromic regions was amplified for cloning by using 5 sets of primers (Table 1) to produce 5 overlapping amplicons. The reaction conditions were similar to those used for detection of viral infection. Each product was separated by using agarose gel electrophoresis and purified from the gel by using the Wizard SV Gel and PCR Clean-up System (Promega, Madison, WI). These products were cloned into the pGEM-T Easy vector (Promega) according to the manufacturer's instructions. Plasmid DNA was isolated by using the QIAprep Spin Miniprep Kit (Qiagen).
The cloned products were sequenced by using either vector-specific primers or primers specific to the cloned virus DNA, with the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PerkinElmer, Waltham, MA). Each region of viral DNA was sequenced from 4 different plasmid clones, each obtained from separate PCR reactions. The resulting sequences were modified to remove sequence arising from the vector and primers and were combined by using CAP3.10 Sequence alignments and similarity plots with other MPV types were performed by using ClustalW14 and sequences obtained from GenBank.
Recombinant truncated MPV1f VP1 capsid protein.
Oligonucleotide primer pair MPV1_3505F and MPV1_4158R (Table 1) were based on the sequence of MPV1a (GenBank accession no., MPU_12469) and were designed to amplify a region of the VP1 gene of MPV1f that encoded the amino acids considered the primary determinants of tissue tropism of MVM, referred to as the allotropic determinants.4 Amplification of this region by PCR was performed in 20-μL volumes in an automated Thermal Cycler (Bio-Rad) by using 200-μL flat-top PCR tubes (Sarstedt, Nümbrecht, Germany) and commercial reagents (Invitrogen). Each reaction contained 1 μL of the extracted DNA eluate as template, 0.916 U Platinum Taq DNA polymerase, 0.2 mM of each dNTP (dATP, dCTP, dGTP, dTTP), 10× PCR buffer, 1.5 mM MgCl2, 20 pmol/μL of each oligonucleotide primer, and ultrapure water. The thermal cycling conditions were an initial cycle of 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s, with a final period of 72 °C for 7 min.
The PCR products were electrophoretically separated in 1.2% (w/v) electrophoresis-grade agarose (Progen, Toowong, Australia) containing ethidium bromide (Invitrogen) at 80 V for 1 h by using TAE buffer (1 mM EDTA, 40 mM Tris-acetate, pH 8.0). The amplified PCR products was excised and purified by using the Wizard PCR Purification System (Promega) and ligated into the PinPoint Xa1 vector (Promega) as specified by the manufacturer. The recombinant vector was transformed into high-efficiency E. coli JM109 cells (Promega) by using a heat-shock method. The transformed cells were grown overnight on LB agar plates containing ampicillin at a final concentration of 100 μg/mL. Colonies were picked by using sterile toothpicks into individual 1.5-mL sterile microcentrifuge tubes containing 50 μL PCR-grade water. The resuspended cells were heat-treated at 90 °C for 5 min with occasional vortexing to lyse the cells and then centrifuged (14,000 × g for 5 min). The plasmid DNA was purified with a QIAprep Spin Miniprep Kit (Promega) by using the protocol described by the manufacturer. The orientation of the insert was determined by restriction endonuclease mapping and was based on the known sequence of the PinPoint Xa1 Vector and VP1 sequence. The endonuclease PvuII, with the recognition sequence CAGCTG, was chosen because there was a single PvuII restriction site within the multiple cloning region of the PinPoint Xa1 Vector and a single PvuII restriction site within the putative allotropic determinant.
Small-scale cultures of E. coli JM109 carrying the PinPoint recombinant plasmid were inoculated into 5 mL LB medium containing 100 μg/mL ampicillin and 2 μM biotin and incubated overnight at 37 °C with shaking. The cultures were diluted 1:100 by adding 50 μL to 5 mL LB containing 100 μg/mL ampicillin and 2 μM biotin, incubated for 1 h at 37 °C with shaking until cultures yielded an A600 of approximately 0.4 (midlog phase). Expression was induced by adding isopropyl-beta-D-thiogalactopyranoside (Sigma-Aldrich, St Louis, MO) to a final concentration of 100 μM and further incubation for 5 h at 37 °C with shaking. Bacterial cells were pelleted by centrifugation (8000 × g for 10 min at 4 °C), resuspended in PBS, and washed by centrifugation (8000 × g for 10 min at 4 °C). The cells were then suspended in 10 volumes of PBS and 0.1 volume of lysozyme (Sigma-Aldrich) solution (10 mg in 25 mM Tris-HCl, pH 8) and frozen at –20 °C until needed.
To recover the recombinant protein, determined to be insoluble and present predominantly within inclusion bodies within the bacterial cells, the bacterial suspension was thawed on ice and then sonicated. After sonication, the crude lysate was centrifuged (10,000 × g for 15 min at 4 °C) to remove contaminant proteins, the pellet was washed twice with 0.1% Triton X100 in PBS, and twice with 0.1% deoxycholate in PBS. After a final centrifugation with 0.1% deoxycholate in PBS, only the top layer was collected, which contained the required protein, and the bottom layer was discarded because, when analyzed by electrophoresis and Western immunoblotting, it contained mainly host cell proteins; this manipulation reduced protein yields but improved the efficiency of the purification process. The washed inclusion bodies then were solubilized by suspending them in extraction buffer (50 mM Tris-HCl [pH 7.5], 8 M urea, 1 mM dithiothreitol [Promega], 1 mM phenylmethyl sulfonyl fluoride [Promega]) for 60 min at room temperature.
The solubilized proteins were diluted approximately 10-fold with extraction buffer so that the final protein concentration was approximately 0.1 mg/mL and dialyzed overnight against a 100-fold volume of wash buffer (50 mM Tris-HCl, pH 7.5; 200 mM NaCl). The final dialysate was clarified by centrifugation (14,000 × g for 30 min at 4 °C). The concentration of the solubilized protein was determined by using protein acidic dye binding assay (Bio-Rad) as recommended by the manufacturer.
ELISA for detection of MPV1 antibody.
ELISA plates (Maxisorb; Nunc, Roskilde, Denmark) were coated with 50 μL per well of 5 μg/mL streptavidin (Promega) in ultrapure PCR grade water (Fisher Biotec, Wembley, Australia) overnight at 37 °C. Fifty microliters of blocking buffer (1.5% dry skim milk in PBST [0.05% Tween 20 in PBS]) was added to each well and incubated for 1 h at room temperature. The buffer was discarded, and the wells were washed 3 times with PBST. The solubilized antigen was diluted to 3.538 μg/μL in PBST and 50 μL added to each well. The plates were covered and left for 1 h at room temperature to allow the protein to bind to the plates. The wells then were washed 3 times with PBST.
Each test and appropriate control sera (1:20 dilutions; 50 μL each) was added to duplicate wells and incubated at room temperature for 90 min. The plates were then washed 3 times in PBST and 50 μL of horseradish peroxidase-conjugated goat antimouse IgG secondary antibodies (ICN Biochemicals, Irvine, CA) diluted at 1:1000 in PBST was added to each well, and the plates were incubated at room temperature for 1 h. The plates were washed 3 times in PBST, 50 μL diluted horseradish peroxidase substrate (Bio-Rad) was added per well, plates incubated at room temperature for 10 min, and the reaction was stopped by the addition of 50 μL 2% oxalic acid to each well. The absorbance at a wavelength of 405 nm for each well was determined by using a microplate reader (Bio-Rad). The mean A405 was determined from the duplicate wells.
A standard curve was established by testing the A405 values of known MPV1-positive sera. A pool of MPV1-positive mouse sera was stored in small volumes and kept at −80 °C until use. Serial 2-fold dilutions of an initial 1:20 dilution were prepared, and each dilution was added in duplicate to the wells of the first 2 columns of an ELISA microtiter plate. The cut-off value was derived for each microtiter plate based on 2 standard deviations above the mean of 30 MPV1-negative serum samples divided by the A405 value at 1:160 dilution of the positive curve. To obtain a cutoff point specific for each individual plate, the obtained value was multiplied by the A405 value at a 1:160 dilution.
The source of the standard negative sera was from Western-immunoblot-negative and PCR-negative Hsd:NIH mice, whereas the initial source of the reference MPV1-positive sera was from naturally MPV1f-infected Hsd:NIH mice, which were positive according to the described methods; subsequently, a pool of sera from experimentally infected mice was used. All sera were tested in duplicate, and the mean of the 2 values was used.
Western immunoblotting.
Recombinant VP1 protein underwent SDS-PAGE by using a 12.5% separating gel and electroblotted from the gel to nitrocellulose membranes. The membranes were clamped into a Mini-PROTEAN II multiscreen apparatus (Bio-Rad), and 600 µL of a 1:1000 dilution of streptavidin A (Promega) or standard positive MPV1 control and 1:50 dilutions of the test sera and negative sera in PBST were applied into different channels of the multiscreen apparatus and incubated for 1 h at 37 °C. The streptavidin A was removed by vacuum aspiration, and this channel was washed 3 times with PBST. Substrate (500 μL) stabilized in Western Blue (Bio-Rad) was applied to the same channel and left for 2 to 3 min for color development to occur. The sera were removed by vacuum aspiration, the multiscreen apparatus disassembled, and the membrane washed 3 times with PBST. Horseradish-peroxidase-conjugated antibody (Sigma-Aldrich) was added to the membrane at a 1:1000 dilution and incubated with gentle agitation for 1 h at room temperature. The conjugates were decanted, the blot was washed 2 times with PBST and once with PBS, and chromagen reagent (3 mg chloronaphthalene [Bio-Rad] solubilized in 1 mL 100% methanol, 5 mL PBS, 2 μL H2O2) was applied to the blot. The membrane was left in the dark for about 3 min or until the color developed sufficiently; the reaction stopped by rinsing the membrane in water.
Serologic sampling of mice.
The breeding facility contained approximately 45,000 mice, mostly of 23 different strains and housed in 11 rooms that opened to a common central corridor. The facility received HEPA-filtered air at 18 °C to 26 °C. Fresh (100%) conditioned air flowed from the rooms and into the corridor. Mice were housed in open-topped cages with autoclaved pine shavings as bedding and were fed autoclaved proprietary mouse chow (Specialty Feeds, Glen Forrest, Australia) prepared to withstand autoclaving and acidified (pH 2.6) water. Bedding was changed weekly. Staff showered into the facility; dressed in autoclaved clothing, hoods, masks, and gloves; and had access to all rooms. The mice were free of mouse hepatitis virus, MVM, murine rotavirus, pneumonia virus of mice, Theiler encephalomyelitis virus, murine cytomegalovirus, ectromelia virus, Hantaan virus, lymphocytic choriomeningitis virus, Sendai virus, reovirus 3, mouse adenovirus, polyoma virus, Mycoplasma pulmonis, other known pathogenic bacteria, endoparasites, and ectoparasites.
Approximately 100 mice per room were sampled on the basis that this frequency would provide a 99% probability of detecting at least 1 seropositive mouse in the population if virus was present at an expected prevalence of 5%.7 For collection of blood samples, mice were euthanized in 20% oxygen in carbon dioxide gas. Blood samples were collected by means of cardiocentesis from 1161 retired breeders and stock mice of 8 wk of age or older from the following mouse strains: outbred Swiss Arc:Arc(s) (originally CD1), Quackenbush Swiss (Arc:Q(s)), Hsd:NIH; inbred A/JArc, C3H/HeJArc, C57BL/6JArc, C57BL/10ScSnArc, DBA/2JArc, FVB/NJ, SJL/JArc, Vm/Dk; hybrid B6D2F1, CBB6F1; congenic B6.SJL-PtprcaPep3b/BoyJArc, B10.D2/N2Sn; mutant C57BL/10ScSn-Dmdmdx/Arc and induced mutant C57BL/6J-Tg(A2Kb HLA)6HsdArc, B6.129P2-Apoetm1Unc/Arc, CBA-TgIl5 1Glx, B6;129S-Gt(ROSA)26Sor, FVB-TgK14mCD806Irw and BALB/cArc-Foxn1nu/+. Sera were separated and kept at −20 °C until analyzed.
Sera were tested first for MPV1-induced antibody by ELISA, and samples that were ELISA-positive were further tested by Western immunoblotting. A positive antibody result was interpreted as one with a positive result with both assays.
All animal work undertaken conformed to the requirements of the Murdoch University Animal Ethics Committee (license R1110/04).
Results
Identification of MPV1 in spleen tissue.
The PCR reaction used for detection of virus produced a 653-bp from MPV1 DNA extracted from spleen tissue.
DNA and amino acid sequence analysis.
The sequence of the Australian MPV1f strain corresponding to nucleotides 148 to 4864 of MPV1a was determined from DNA prepared from the spleen tissue of a PCR-positive Hsd:NIH mouse. The sequence included all the expected protein coding regions as well as part of the 5′ and 3′ regions of MPV1a but excluded the terminal palindromic regions. The sequence was deposited in GenBank (accession no., 1157220).
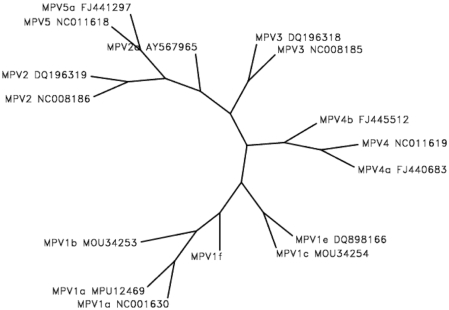
The genomic sequence of MPV1f had 98% to 99% nucleotide identity with that of MPV1a, 1b, 1c, and 1e. The NS1 open reading frame had 97.9% to 99.7% nucleotide identity with that of MPV1a, 1b, 1c, and 1e. The VP1 open reading frame had 99% to 99.9% nucleotide identity with that of MPV1a, 1b, 1c, and 1e.The phylogenetic relationship between the entire genome of MPV1f and other genotypes of MPV1 and MPV2, MPV3, MPV4, and MPV5 (Figure 1) indicates that MPV1f is phylogenetically distinct from previously reported strains.
Figure 1.
Unrooted phylogenetic tree demonstrating the relationship among the nucleotide sequence of the Australian MPV1f strain and MPV genotypes reported previously in GenBank (genotype and GenBank accession numbers are shown). The alignment was generated by using the ClustalW program14 and demonstrated that the Australian MPV1f strain was phylogenetically distinct.
Serologic assays.
Antibody was detected by both ELISA and Western immunoblotting in 188 (16.2%) of the 1161 sera examined; these results were regarded as true positives. An additional 7 sera were ELISA-positive but Western-negative and were considered false positives. The seroprevalence varied among strains of mice: the highest seroprevalence occurred in Hsd:NIH mice, followed by B6D2F1 hybrids and Arc:Arc(s). Antibody was not detected in mice of C57BL lineage other than in hybrids with DBA/2J (B6D2F1), which had a high seroprevalence, and only a low seroprevalence was detected in heterozygous mutant mice on a BALB/c background (Table 2). Antibody-positive animals were present in 8 of the 11 mouse breeding rooms sampled (Table 2). The Arc:Arc(s) strain had a high seroprevalence in both of the 2 rooms from which samples were obtained. In addition, C57BL/6JArc animals were negative in both of the 2 rooms of mice sampled; one of these rooms (Room 64) also contained Arc:Arc(s) mice, which showed a 97.9% seroprevalence. Two of the rooms (nos. 46 and 62) in which antibody was not detected contained mice of C57BL lineage only, and the other (no. 54) housed B10.D2/N2Sn, B6.SJL-Ptprca Pep3b /BoyJArc, and A/JArc mice only.
Table 2.
MPV1 antibody detection in mouse strains in the rodent breeding facility
| Mouse strain | Room | No. of sera | Positive by ELISA and Western blotting |
| B6D2F1 | 40 | 92 | 39 (42.4%) |
| DBA/2JArc | 40 | 8 | 1 (12.5%) |
| Hsd:NIH | 41 | 118 | 63 (53.4%) |
| BALB/cArc-Foxn1nu/+ | 43 | 99 | 7 (7.1%) |
| B6.129P2-Apoetm1Unc/Arc | 44 | 26 | 0 |
| C57BL/10ScSn-DmdmdxArc | 44 | 6 | 0 |
| CBB6F1 | 44 | 23 | 0 |
| SJL/JArc | 44 | 42 | 1 (2.3%) |
| C57BL/10ScSnArcArc | 46 | 14 | 0 |
| C57BL/6J-Tg(A2Kb HLA)6HsdArc | 46 | 32 | 0 |
| B6;129S-Gt (ROSA) 26Sor | 47 | 21 | 0 |
| FVB-TgK14mCD806lrw | 47 | 13 | 0 |
| CBA-TgIl51Glx | 47 | 66 | 9 (13.6%) |
| Vm/Dk | 47 | 19 | 3 (15.7%) |
| B10.D2/N2Sn, B6.SJL-PtprcaPep3b/BoyJArc, A/JArc | 54 | 117 | 0 |
| C3H/HeJ | 60 | 25 | 5 (20%) |
| FVB/NJ | 60 | 77 | 27 (35.1%) |
| Arc:Arc(s) | 61 | 87 | 54 (62%) |
| C57BL/6JArc | 62 | 104 | 0 |
| Arc:Arc(s) | 64 | 48 | 47 (97.9%) |
| Arc:Q(s) | 64 | 77 | 1 (1.3%) |
| C57BL/6JArc | 64 | 41 | 0 |
| Total | 1161 | 188 (16.2%) |
Discussion
The presence of MPV1 antibody in a commercial laboratory mouse colony in Australia that was not detected by appropriate use of a recombinant parvovirus NS1 MVM antigen suggested the presence of antigenic differences between the Australian strain (MPV1f) and those described previously. However, the high nucleotide homology between MPV1f and previously described variants suggested only minimal antigenic differences among MPV strains. Sequence analysis of the NS1 open reading frame of MPV1f indicated a close relationship those of other MPV1 genotypes. Why an antibody response to MPV1f was not detected by using an ELISA based on a recombinant NS1 protein of MVM is unknown.
The recombinant truncated VP1 protein was developed and used as an antigen because a commercial recombinant VP2 antigen was not available when the survey was conducted in 2003. The recombinant VP1 protein provided an antigen that detected virus-associated antibody in some strains of MPV1-infected mice by both ELISA and Western immunoblotting. The VP1 ELISA subsequently was determined to be of approximately equal sensitivity to the commercially available VP2 ELISA (Research Animal Diagnostic Laboratory, Columbia, MO) when tested on sera collected for routine MPV surveillance (Animal Resources Centre), and neither VP-based ELISA detected significant antibody in mice with a BALB/c background that were experimentally infected with MPV1f.9 Both VP1 and VP2 exhibit type-specific reactivity, in contrast to the group-specific reactivity of the nonstructural proteins.2 In addition, ELISA that used recombinant VP2 expressed with a baculovirus system12 or in bacteria1 were reported to be much more specific than recombinant NS1 MVM antigens and to outperform them significantly.12 The selection of the truncated region of VP1 used as the antigen was based on a combination of yield and antigenicity relative to several other truncated VP1 proteins that were produced (results not reported). Although the recombinant protein was expressed intracellularly in an insoluble form, presumably in inclusion bodies, these characteristics enabled the use of centrifugation to separate the VP1-laden structures from the remainder of the bacterial cells. For solubilization of rVP1 protein, 8 M urea and prolonged dialysis were used to maximize correct refolding of the proteins.15
The overall serologic prevalence of 16.2% in the 1161 sera tested from the Animal Resources Centre was not an accurate indication of the prevalence of infection in the facility at the time of sampling. The number of samples (100 or greater) obtained per room was based on a 99% probability of detecting at least 1 seropositive mouse in the population if present at an expected prevalence of 5%,7 however this figure assumes that all animals tested were uniformly susceptible to the virus, that all infected animals would respond serologically, and that all strains in the same room were uniformly susceptible.
Seroprevalence varied from 0% to 53.4% among mouse strains. High seroprevalence rates were detected in inbred DBA/2JArc and its hybrid B6D2F1 as well as outbred Hsd:NIH and Arc:Arc(s) strains, no antibody was detected in C57BL strains (other than hybrids with DBA/2), and only a low seroprevalence was detected in BALB/c strains. Although location within the facility may have had some influence on seroprevalence, this factor does not seem to have had a great effect. Infection was detected in mice from 8 of the 11 rooms sampled, and similar results were obtained from mice of the same strain located in different rooms. The strain variations in seroprevalence reflected those seen in the antibody response to MPV1 reported previously in experimentally infected mice. Specifically, these reports describe an absence of seroconversion in C57BL/6 mice; seroconversion in all C3H mice but only some BALB/c, ICR, and DBA/2 mice;6 and a poor antibody response to MPV1 infection in mice with a BALB/c background.11 However, caution needs to be exercised in the interpretation of the serologic data from naturally infected mice. Experimental infection with MPV1f demonstrated that mice of C57BL background can develop antibody detectable by using the same serologic test as that we used here.9 Experimental infection of mice with the MPV1f strain also demonstrated that the inability to detect antibody in mice with a BALB/c background is associated, at least in part, with the serologic assay used.9 Mice with a BALB/c background developed a prolonged infection and antibody response; the antibody response, however, was not detected or detected in only a few mice when ELISA with recombinant virus protein antigens were used but was detected in a majority of infected mice when an MVM–MPV immunofluorescent assay was used.
Acknowledgment
This project was funded by the Australian Research Council (Project LP0347146).
References
- 1.Ball-Goodrich LJ, Hansen G, Dhawan R, Paturzo FX, Vivas-Gonzalez BE. 2002. Validation of an enzyme-linked immunosorbent assay for detection of mouse parvovirus infection in laboratory mice. Comp Med 52:160–166 [PubMed] [Google Scholar]
- 2.Ball-Goodrich LJ, Johnson E. 1994. Molecular characterization of a newly recognized mouse parvovirus. J Virol 68:6476–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besselsen DG, Becker MD, Henderson KS, Wagner AM, Banu LA, Shek WR. 2007. Temporal transmission studies of mouse parvovirus 1 in BALB/c and C.B-17/Icr-Prkdc(scid) mice. Comp Med 57:66–73 [PubMed] [Google Scholar]
- 4.Besselsen DG, Pintel DJ, Purdy GA, Besch-Williford CL, Franklin CL, Hook RR, Jr, Riley LK. 1996. Molecular characterization of newly recognized rodent parvoviruses. J Gen Virol 77:899–911 [DOI] [PubMed] [Google Scholar]
- 5.Besselsen DG, Romero MJ, Wagner AM, Henderson KS, Livingston RS. 2006. Identification of novel murine parvovirus strains by epidemiological analysis of naturally infected mice. J Gen Virol 87:1543–1556 [DOI] [PubMed] [Google Scholar]
- 6.Besselsen DG, Wagner AM, Loganbill JK. 2000. Effect of mouse strain and age on detection of mouse parvovirus 1 by use of serologic testing and polymerase chain reaction analysis. Comp Med 50:498–502 [PubMed] [Google Scholar]
- 7.Cannon RM, Roe RT. 1982. Livestock disease surveys: a field manual for veterinarians. Canberra (Australia): Australian Government Publishing Service [Google Scholar]
- 8.Crawford LV. 1966. A minute virus of mice. Virology 29:605–612 [DOI] [PubMed] [Google Scholar]
- 9.Filipovska-Naumovska E, Thompson MJ, Hopwood D, Pass DA, Wilcox GE. 2010. Strain- and age-associated variation in viral persistence and antibody response to mouse parvovirus 1 in experimentally infected mice. J Am Assoc Lab Anim Sci 49:443–447 [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Madan A. 1999. CAP3: A DNA sequence assembly program. Genome Res 9:868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan IH, Kendall LV, Ziman M, Wong S, Mendoza S, Fahey J, Griffey SM, Barthold SW, Luciw PA. 2005. Simultaneous serodetection of 10 highly prevalent mouse infectious pathogens in a single reaction by multiplex analysis. Clin Diagn Lab Immunol 12:513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livingston RS, Besselsen DG, Steffen EK, Besch-Williford CL, Franklin CL, Riley LK. 2002. Serodiagnosis of mice minute virus and mouse parvovirus infections in mice by enzyme-linked immunosorbent assay with baculovirus-expressed recombinant VP2 proteins. Clin Diagn Lab Immunol 9:1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKisic MD, Paturzo FX, Smith AL. 1996. Mouse parvovirus infection potentiates rejection of tumor allografts and modulates T cell effector functions. Transplantation 61:292–299 [DOI] [PubMed] [Google Scholar]
- 14.Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsumoto K, Ejima D, Kumagai I, Arakawa T. 2003. Practical considerations in refolding proteins from inclusion bodies. Protein Expr Purif 28:1–8 [DOI] [PubMed] [Google Scholar]