Abstract
According to a long-standing hypothesis, aging is mainly caused by accumulation of nuclear (n) DNA damage in differentiated cells such as neurons due to insufficient nDNA repair during lifetime. In line with this hypothesis it was until recently widely accepted that neuron loss is a general consequence of normal aging, explaining some degree of decline in brain function during aging. However, with the advent of more accurate procedures for counting neurons, it is currently widely accepted that there is widespread preservation of neuron numbers in the aging brain, and the changes that do occur are relatively specific to certain brain regions and types of neurons. Whether accumulation of nDNA damage and decline in nDNA repair is a general phenomenon in the aging brain or also shows cell-type specificity is, however, not known. It has not been possible to address this issue with the biochemical and molecular-biological methods available to study nDNA damage and nDNA repair. Rather, it was the introduction of autoradiographic methods to study quantitatively the relative amounts of nDNA damage (measured as nDNA single-strand breaks) and nDNA repair (measured as unscheduled DNA synthesis) on tissue sections that made it possible to address this question in a cell-type-specific manner under physiological conditions. The results of these studies revealed a formerly unknown inverse relationship between age-related accumulation of nDNA damage and age-related impairment in nDNA repair on the one hand, and the age-related, selective, loss of neurons on the other hand. This inverse relation may not only reflect a fundamental process of aging in the central nervous system but also provide the molecular basis for a new approach to understand the selective neuronal vulnerability in neurodegenerative diseases, particularly Alzheimer’s disease.
Keywords: Aging, Alzheimer’s disease, Brain, DNA damage, DNA repair
1. Introduction
Aging of the brain is an unavoidable process characterized by a large array of alterations in structure and function, eventually leading to brain dysfunction and cognitive decline. Importantly, many basic processes involved in aging of the central nervous system (CNS) contribute to prevalent age-related neurodegenerative diseases like Alzheimer’s disease (AD) and Parkinson’s disease (PD) [1]. Aggregation of modified proteins, disturbance in iron homeostasis, protein and DNA modifications and damage, oxidative stress and impairment of energy production are some of the key mechanisms linking aging to neurodegeneration [2–6]. It was thought for decades that neuron loss plays a key role in the aging process of the CNS [7]. However, more recent research applying advanced quantitative histologic techniques demonstrated that this is not the case (discussed in detail in Section 5). Accordingly, age-related cognitive decline cannot be simply explained as a result of age-related neuron loss.
Increasing evidence suggests in fact that accumulation of DNA damage (both nuclear (n) DNA and mitochondrial (mt) DNA) within specific types of neurons likely represents a critical contributor to the aging process [5,8–10] (discussed in detail in Section 4; the role of mtDNA damage in brain aging is, albeit interesting, beyond the scope of this paper). On the other hand, very little is yet known as to why age-related accumulation of nDNA damage causes age-related neurodegeneration. One intriguing possibility may be that this is due to an imbalance between increasing nDNA damage (mostly because of increased oxidative stress conditions during aging; discussed in detail in Section 2) and the cells’ limited capacity to repair this nDNA damage. Unrepaired or improperly impaired nDNA damage can then have deleterious consequences for neurons in the aging brain such as impaired transcription, genomic instability, dysregulation of cellular functions, cellular senescence and ultimately cell death [11].
However, if accumulation of nDNA damage would occur to the same amount in all types of neurons it would be difficult to assess its possible role in the selective neuronal vulnerability of aging and neurodegenerative diseases (discussed in detail in Section 6). In this regard it is important to note that nDNA damage and nDNA repair processes have been mostly investigated using biochemical or, more recently, molecular-biological techniques in tissue homogenates. This, however, prevents cell-type-specific analyses that are crucial in understanding the molecular mechanisms involved in brain aging. Astrocytes, oligodendrocytes, microglial cells and endothelial cells in blood vessels likely show substantial differences to neurons, and even within a given brain region different types of neurons may show differences in age-related accumulation of nDNA damage (such as granule and pyramidal cells in the hippocampus, or granule and Purkinje cells in the cerebellum). The introduction of autoradiographic methods by Korr and coworkers in the last 25 years to study quantitatively the relative amounts of nDNA damage and nDNA repair on tissue sections made it possible to get novel, important insights in this regard (details are discussed in Sections 2 and 3).
The present paper gives a short overview on the role of reactive oxidative species in generating nDNA damage, the techniques to study nDNA damage and nDNA repair, the age-related accumulation of nDNA damage and decline in nDNA repair in neurons, age-related neurodegeneration and potential roles of accumulation of nDNA damage and neuron loss in the selective neuronal vulnerability in neurodegenerative diseases, particularly AD. Therapeutic strategies aiming at preventing age-related nDNA damage, improving the efficacy of nDNA repair mechanisms and their capacity or, ultimately, eliminating those neurons that show the highest amount of nDNA damage may be of crucial importance in effective prevention of age-related neurodegeneration and age-related neurodegenerative diseases.
2. Reactive oxygen species and nuclear DNA damage
Reactive oxygen species (ROS) are continuously formed as a consequence of normal metabolism and in response to environmental factors such as UV light, ionizing radiation, heat and pollution [12–16]. ROS encompass a variety of chemical species including superoxide anions (O2•), hydroxyl radicals (•OH) and hydrogen peroxide (H2O2) [14]. Metabolic stress inevitably rises the levels of ROS in tissue. If the amount of ROS overwhelms the capacity of cells to counteract these harmful species, oxidative stress occurs and can induce various types of cell damage such as modifications of proteins, lipids and DNA leading to mitochondrial and ultimately cellular dysfunction [14,15,17].
Neurons seem to be particularly vulnerable to oxidative stress due to the substantial amount of oxygen consumption by the brain, the low glutathione content and a high proportion of polyunsaturated fatty acids in neurons [18,19]. Furthermore, neurons are postmitotic cells which implies that, in case they are irreversibly damaged or lost, they cannot be replaced.
Accumulation of nDNA damage in neurons has long been suggested to be one of the major forms of damage involved in brain aging and neurodegeneration [5,8,9,20–22]. More than 100 different types of nDNA lesions have been reported, including base modifications (for instance, 8-oxo-7,8-dihydro-2′-deoxyguanosine [8-oxo-dG], thymidine glycol, and 8-hydroxycytosine), single- and double-strand breaks, and interstrand cross-links [23–25]. Among these, nDNA single-strand breaks (SSBs) are the most common ones (thousands per cell per day) and are the most common lesion induced by ROS and exogenous genotoxins such as ionizing radiation and alkylating agents [25–29].
Many methods have been developed to detect nDNA damage (reviewed in, e.g., [25,30,31]). All these methods have advantages, yet all suffer from limitations and none of them is universally applicable. The major drawback of all these methods stems from the fact that identification of nDNA damage relies on a single parameter reflecting changes in biochemical or molecular attributes of the cells under study. Accordingly, these methods cannot be used to investigate nDNA damage of the brain in a cell-type-specific manner.
The Klenow method (Klenow fragment of DNA polymerase I-mediated biotin-dATP nick-end labeling [32,33] and the TUNEL method (terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick-end labeling [34,35]) detect nDNA damage on tissue sections. Thus, they make it possible to study nDNA damage in a cell-type-specific manner (note that the TUNEL method is widely applied to detect programmed cell death, i.e., apoptosis [34,36] even though this has been challenged in the literature [37,38]). However, due to the properties of the enzymes involved in the Klenow and TUNEL assays, multiple types of DNA breaks are labeled by these techniques [33,35].
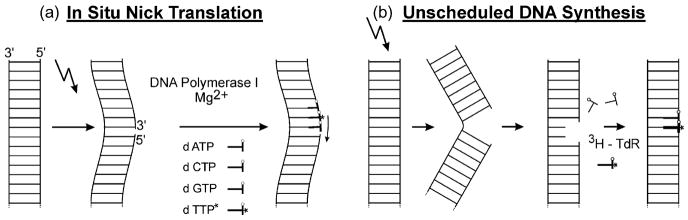
To overcome these limitations, in situ nick translation (ISNT) in combination with autoradiography was further developed by Korr et al. [39] to detect a certain type of nDNA damage (i.e., nDNA SSB) on tissue sections quantitatively, in a cell-type-specific manner. The basic concept of ISNT, originating from molecular biology [40], is to label nDNA SSBs in cell nuclei by insertion of tritiated deoxythymidine 5′-triphosphate ([3H]dTTP) into the nDNA mediated by E. coli DNA polymerase I [41,42] (Fig. 1a). The DNA polymerase I holoenzyme possesses three distinct elementary activities, namely a 5′ → 3′ exonuclease activity, a 3′ → 5′ exonuclease activity, and a 5′ → 3′ polymerase activity [41]. Accordingly, the ISNT method has the capacity to detect nDNA SSB by both “nick translation” and “gap filling”. This contrasts with the Klenow fragment of DNA polymerase I which contains the 5′ → 3′ polymerase activity [43] required for gap filling but does not contain the 5′ → 3′ exonuclease activity required for nick translation [44]. Thus, more nDNA strand breaks can be detected when using E. coli DNA polymerase rather than the Klenow fragment.
Fig. 1.
Principles of in situ nick translation (ISNT) (a) and measuring UDS with autoradiography (b). For ISNT (a) tissue sections are incubated with a reaction solution containing buffer, MgCl2, 2-mercaptoethanol, endonuclease-free E. coli DNA polymerase I, dATP, dGTP, dCTP and [3H]dTTP. The reaction is terminated by washing the slides with buffer. Afterwards sections are Feulgen-stained (which also removes all [3H] activity not incorporated into the nDNA; [160–162]), dipped into liquid photoemulsion, exposed, and developed. Poststaining can be performed with hematoxyline among other standard histologic stains. For measuring UDS with autoradiography (b) animals are injected with [3H]TdR and sacrificed 1 or 2 h later. Tissue sections are then also Feulgen-stained, dipped into liquid photoemulsion, exposed, developed, and poststained. Further details can be found in [39].
Afterwards the process of inserting [3H]dTTP into the nDNA can be visualized by autoradiography [10,39]. By selecting a suitable exposure time of the autoradiographs and after employing several correction steps, single silver grains over the cell nuclei can be counted and results obtained on different cell types can be directly compared to each other (Fig. 2) [21,45–47]. (In a pilot experiment comparing the use of E. coli DNA polymerase with the use of the Klenow fragment we found a threefold higher autoradiographic signal with E. coli DNA polymerase than with the Klenow fragment; Korr et al., unpublished results). Of note, it should be recognized that ISNT can also be applied on archived sections for example from human postmortem brains. Visualization of ISNT with autoradiography is the only method to quantify ISNT on tissue sections, which cannot be achieved with immunohistochemistry as initially introduced by Iseki and Mori [41]. Furthermore, ISNT performed with autoradiography works linearly, an increased autoradiographic grain number being to the direct reflection of an increased number of nDNA SSB [48,49]. For technical details on the ISNT method and related autoradiographic techniques, see Refs. [39,45–47,50,51] by Korr and co-workers, and additional literature citations therein.
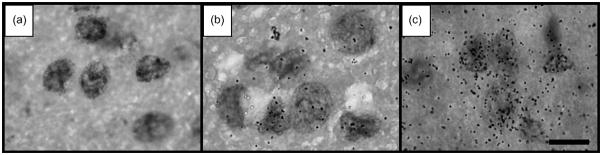
Fig. 2.
Autoradiographic detection of nDNA single-strand breaks in neurons in the thalamus of a 12-month-old mouse (a and b) and a 28-month-old mouse (c) with in situ nick translation (ISNT) (Brasnjevic et al., unpublished results; the corresponding experiments were described in detail in [10]). (a) Background signal in the autoradiographs after performing ISNT without use of E. coli DNA polymerase I. No silver grains were found after 5 days of exposure. (b and c) Autoradiographic signal after performing ISNT with E. coli DNA polymerase I. Note the clearly visible age-related increase in the number of silver grains over the nuclei in the investigated brain region, indicating an age-related increase in the relative amount of nDNA single-strand breaks. The scale bar represents 15 μm.
What is the neurobiological significance to measure the (relative) amount of nDNA SSB in neurons? As mentioned above, nDNA SSB are the most common endogenous lesions arising in cells and are also the most common lesions induced by exogenous genotoxins such as ionizing radiation and alkylating agents. Direct nDNA SSB are characterized by the disintegration of damaged sugars and arise primarily from attack by free radicals such as ROS. In contrast, indirect nDNA SSB are characterized by enzymatic cleavage of the phosphodiester backbone and are mainly normal intermediates of DNA base excision repair (BER) (addressed in Section 3). Nuclear DNA SSB are probably not permanent breaks but are repaired by a process called SSB repair [25,26,28]. Accordingly, a rise in the level of nDNA SSB can be the result of a shift in the balance between nDNA damage and nDNA repair, which has a very high fidelity. Thus, a higher level of nDNA SSB (and thus, a higher signal in ISNT autoradiographs) can be interpreted as a higher steady state level of damaged nDNA in the investigated tissue. Note that ISNT cannot be used to measure mtDNA SSBs in situ [10,39].
To summarize, ISNT is a powerful method to study the relative amount of nDNA SSB (i.e., the most common form of nDNA damage) on tissue sections quantitatively, cell type specifically and in situ. The impact of this technique to study age-related alterations in the amount of nDNA damage in the CNS and its possible impact on the development of neurodegenerative diseases is addressed in Sections 4 and 6.
3. Nuclear DNA repair
As mentioned in Section 2, the nDNA is continuously damaged by exogenous and endogenous agents and chemicals, which generate many types of lesions throughout the genome. To counteract this nDNA damage, cells harbor a complex system of enzymes collectively called nDNA repair systems (for review see [52–57]). The nature of the nDNA repair system that comes into operation when nDNA damage occurs depends upon the type of nDNA damage, the tissue under consideration and the replicative potential of the cells in the tissue. At least five partly overlapping nDNA repair pathways operate in mammals: (i) BER that repairs nDNA base alterations that have not distorted the nDNA helix, including oxidative base modifications and base alkylations (for review see [58,59]); (ii) nucleotide excision repair (NER) that repairs bulky nDNA helix-distorting lesions (see [60–62] for review); (iii) SSB repair (SSBR) that repairs nDNA SSBs (for review see [25,26,52]); (iv) double-strand break repair (DSBR) that repairs nDNA double-strand breaks (for review see [63]); and (v) mismatch repair (MMR) that removes mispaired bases and small insertion–deletion mispairs resulting from replication errors (for review see [64]). For sake of completeness it should be mentioned that DSBR consists of two subpathways, namely the homologous recombination and non-homologous end-joining [62,65].
There are two major differences between BER and NER. Firstly, during NER approximately 30 nucleotides around the nDNA lesion are excised and replaced but only one or two nucleotides around the nDNA lesion during BER [58–62]. Secondly, NER has been shown to occur preferentially coupled to transcription (reviewed in [66–68]). This is probably not the case for BER [69].
As in case of nDNA damage, a variety of methods have been developed to assess nDNA repair processes. Some of these methods are based on the detection of chromosome breakage (reviewed in [70]) including the Comet assay (see [71–73] for review). Other approaches measure the activity of nDNA repair enzymes [74,75] or the amount of damage localized to a plasmid (i.e., to exogenous DNA) that a cell can “reactivate” or repair (so-called host cell reactivation; reviewed in [76]). The common limitation of these methods is that they cannot be used to investigate nDNA repair processes ongoing in the brain in a cell-type-specific manner.
Importantly, the most common forms of nDNA damage caused by endogenous sources such as ROS are those repaired by BER and SSBR and these nDNA repair processes are (almost probably) not linked to transcription (as outlined above). It is therefore attractive to study nDNA repair in neurons by measuring so-called unscheduled DNA synthesis (UDS), i.e., nDNA synthesis outside the S-phase, particularly in non-proliferating cells [77,78]. Briefly, resynthesis of nDNA following the excision of nucleotides (which occurs in both BER and SSBR; [52,58]) is an essential step in UDS [77]. Therefore, UDS can be labeled by the incorporation of [methyl-3H]thymidine ([3H]TdR), for example, into the nDNA (Fig. 1b). Resynthesis of nDNA (i.e., filling the gap after the excision of nucleotides) is the primary responsibility of DNA polymerase β (Polβ) [29]. Usually Polβ inserts a single nucleotide. However, on occasion Polβ can extend the gap by 2–15 nucleotides. This has been most extensively studied during BER, which is consequently subclassified into two pathways, denoted single nucleotide repair or long patch repair, respectively [29]. A similar dichotomy is likely to exist for the repair of direct SSBs [29]. Accordingly, it is reasonable to assume that the majority of [3H]TdR incorporated into the nDNA during UDS represents long patch repair events during BER and SSBR rather than single nucleotide repair events during BER and SSBR. However, studies describing the relative amount of long patch repair events and single nucleotide repair events during BER and SSBR in vivo have not been published.
Incorporation of [3H]TdR into the nDNA can then be detected by liquid scintillation (see, e.g., [79]) and, as established by Korr et al. [20,21,39,80–82], with autoradiography on tissue sections after injection of [3H]TdR in animals. The latter provides a quantitative read-out of UDS in a cell-type-specific manner. Note that measuring UDS with autoradiography does also work linearly, with increased autoradiographic grain numbers being a direct reflection of increased amounts of UDS [81]. Unfortunately, [3H]TdR injected into animals is available in the tissue only for approximately 2 h. Accordingly, the relative amount of [3H]TdR incorporated into the nDNA during UDS is very small compared to the total amount of thymidine in the nDNA. However, due to the fact that the silver grains in the autoradiographs are generated as the product of 3H-activity multiplied by the autoradiographic exposure time, very small amounts of 3H-activity incorporated into the nDNA can also be demonstrated provided the exposure time is extended to several months (see [50] for details). As such, an exposure time of 250 days (approximately 9 months) is suitable to demonstrate UDS under physiological conditions in the brain of rodents [20,21,39,80–82]. Then single silver grains over the cell nuclei can be counted and, after correction for autoradiographic background, β-self-absorption, and nuclear size, the results obtained on different cell types can be directly compared to each other (for details see [21,50,80,81]). Several studies have addressed the validity of this method (see, e.g., [83–85]).
The neurobiological significance to measure the (relative) amount of UDS in neurons with autoradiography is that it is the only method available to get insights into the amount of nDNA repair processes ongoing in the brain under physiological or pathological conditions in a cell-type-specific manner. Differences in the signal in UDS autoradiographs obtained over cell nuclei can be interpreted as differences in the amount of nDNA repair ongoing in the tissue under study. Furthermore, this method can also be used to measure the extent of cytoplasmic mtDNA synthesis quantitatively in a cell-type-specific manner [20,21,82]. As there is no UDS-like pathway for repairing mtDNA damages that could be detected with autoradiography, mtDNA synthesis indicates the biogenesis of new mitochondria, occurring in all types of cells during the replacement of damaged, lysosomally digested mitochondria as well as for increasing the number of mitochondria per cell in order to improve cellular energy supply (details are provided in [82]). It should be mentioned that there are two major limitations related to the analysis of UDS with autoradiography: it does not allow to distinguish between the different nDNA repair pathways outlined above, and it does obviously not allow to study nDNA repair in a gene-specific manner. Nevertheless, the impact of this method on the study of age-related alterations in the amount of nDNA repair in the CNS and on the development of neurodegenerative diseases is addressed in Sections 5 and 6.
4. Age-related accumulation of nDNA damage and decline in nDNA repair in neurons
According to a hypothesis by Gensler and Bernstein [86] aging is mainly caused by accumulation of nDNA damage in differentiated cells such as neurons due to insufficient nDNA repair during lifetime. To explore further this hypothesis studies focused on tissue-specific changes in nDNA damage profiles during aging but yielded conflicting results [10,87] (for review see [20]). The use of different methods for detecting nDNA damage and nDNA repair as well as differences between experimentally induced nDNA damage and nDNA damage occurring in physiological conditions might have contributed to these discrepancies. It was the introduction of autoradiographic methods by Korr et al. [10,20,21,39,47,80–82] to study quantitatively the relative amounts of nDNA damage and nDNA repair on tissue sections that made it possible to test Gensler and Bernstein’s [86] hypothesis about the aging brain in a cell-type-specific manner under physiological conditions. Interestingly, it turned out that the situation in the aging brain is more complex than predicted by Gensler and Bernstein [86]. Specifically, the relative amount of spontaneous nDNA repair (measured as UDS) in the mouse brain decreased during aging in hippocampal pyramidal and granule cells as well as in cortical layer V pyramidal cells and neurons in the striatum and thalamus but not in Purkinje cells, mitral cells in the olfactory bulb and large neurons in the lateral vestibular nucleus [20,21]. A similar pattern was then found for age-related accumulation of nDNA SSBs in the mouse brain: hippocampal pyramidal and granule cells as well as cortical layer V pyramidal cells, and neurons in the striatum and thalamus showed an age-related increase in the relative amount of nDNA SSB whereas Purkinje cells did not [10,88, Brasnjevic et al., unpublished results]. These results agree with Gensler and Bernstein’s [86] hypothesis of age-related accumulation of nDNA damage in differentiated cells such as neurons due to insufficient nDNA repair during lifetime (although no causal relation between the age-related decline in the relative amount of nDNA repair and the age-related increase in the relative amount of nDNA SSB could be established in the mentioned studies). However, the results obtained on the Purkinje cells apparently disagreed with Gensler and Bernstein’s [86] hypothesis. Furthermore, the cerebellar Purkinje cells displayed the highest relative amount of nDNA SSB of all investigated cell types (in young animals approximately three times more than the hippocampal pyramidal cells; [10]) although the relative amount of nDNA repair was comparable among Purkinje cells and hippocampal pyramidal cells [20]. The high relative amount of nDNA SSB observed in Purkinje cells was most probably due to the fact that these cells are among the largest neurons in the CNS and have an exceptionally high metabolic demand [89–91]. On the other hand, this high metabolic demand has also been hypothesized to be involved in the high vulnerability of Purkinje cells under conditions such as acute hypoxia or seizures [89–91]. Based on these data we have hypothesized that the lack of age-related increase in the relative amount of nDNA SSB in Purkinje cells does not reflect an exception from Gensler and Bernstein’s [86] hypothesis but rather indicates a fundamental difference in the selective neuronal vulnerability of Purkinje cells during aging compared to the other types of neurons mentioned above. This will be explained in detail in Section 5.
In summary, the application of the autoradiographic methods introduced by Korr and co-workers (as mentioned above) has demonstrated that neurons do indeed accumulate nDNA damage during aging, and this could be due to insufficient nDNA repair during lifetime [86]. However, age-related accumulation of nDNA damage does not occur in all types of neurons but shows cell-type specificity. It is important to note that the causal relation between age-related accumulation of nDNA damage and age-related decline in the amount of nDNA repair has not been established yet. Further studies are needed in this regard, as well as further studies to understand the molecular mechanisms particularly underlying the age-related decline in the amount of nDNA repair and the molecular events that lead from accumulation of nDNA damage to loss of cellular function and, ultimately, neurodegeneration.
Furthermore, it has been proposed that accumulation of nDNA damage (or, in a broader sense, the inability of neurons to appropriately handle nDNA damage) serves as molecular trigger in the etiology of age-related neurodegenerative disorders [92–95]). This aspect will be discussed in Section 6.
5. Age-related neurodegeneration
Until recently it was widely accepted that neuron loss is a general consequence of normal aging, explaining the functional decline of the brain during aging (for review see [7,96,97]). However, with the advent of more accurate procedures for counting neurons, particularly the application of design-based stereologic techniques (for review see [98–100]), this view has been modified over the last years. The current view is that there is widespread preservation of neuron numbers in the aging brain, and the changes that do occur are relatively specific to certain brain regions and types of neurons [10,96,97,101–108]. For instance, data obtained in ours and other labs on postmortem brains from humans and non-human primates showed no evidence of age-related neuron loss in the entorhinal cortex or the hippocampal CA1 region, which are crucial for memory function [101,102,105]. Similarly, the numbers of pyramidal cells in the entorhinal cortex and hippocampus are conserved in aged rats with memory deficits [107]. Furthermore, we could recently show that in the aging mouse brain, total numbers of hippocampal pyramidal and granule cells as well as cortical layer V pyramidal cells and neurons in the striatum and thalamus are preserved, whereas the total number of Purkinje cells in the brains from these same mice decrease by approximately 25% during aging [10,88, Brasnjevic et al., unpublished results]. Importantly, these types of neurons for which no age-related alterations in total numbers were found were the same that showed age-related accumulation of nDNA SSB and age-related decline in the relative amount of nDNA repair (as outlined in Section 4). In contrast, the Purkinje cells were reduced in number during aging but showed neither age-related accumulation of nDNA SSB nor age-related decline in the relative amount of nDNA repair. From these data the question arises whether the described negative correlation between age-related accumulation of nDNA damage and decline in nDNA repair on the one hand, and age-related neuron loss on the other, reflect a causal link between these fundamental processes in the aging mammalian brain. However, this question cannot be currently answered.
Nevertheless, this negative correlation between nDNA damage, nDNA repair, and neuron loss indicates that at least in the rodent brain, certain neurons with high amount of nDNA damage (such as Purkinje cells) will be removed during physiological aging, whereas other neurons with lower amount of nDNA damage will not. The molecular and cellular mechanisms underlying this phenomenon in the selective neuronal vulnerability during aging are currently not understood. There may be a certain threshold level of nDNA damage beyond which the cells are eliminated. This explanation is in line with our finding that already at 12 months of age, the relative amount of nDNA SSB in Purkinje cells was approximately threefold compared to the relative amount of nDNA SSB in hippocampal and cortical layer V pyramidal cells [10,88]. It is important to realize that even a substantial age-related amount of neuron loss may not per se result in clinically detectable functional decline. (At least ataxia, which occurs in ataxia-telangiectasia as a result of severe loss of Purkinje cells [109] is not a typical clinical feature of aging. On the other hand, a possible role of Purkinje cell loss in age-related cognitive decline according to the current understanding of cerebellocerebral interactions in cognition [110,111] cannot be ruled out and needs to be explored further.) Rather, age-related cognitive decline seems to be associated with those types of neurons that accumulate nDNA damage but are not reduced in number over time, i.e., those types of neurons that may not possess appropriate mechanisms to induce cell death in highly damaged cells [5,10,107]. With respect to the latter, preservation of brain function during aging might hypothetically some day be reached by specifically eliminating those neurons that show the highest amount of nDNA damage. It may be interesting to explore possible treatment strategies to prevent age-related nDNA damage and improve the efficacy of nDNA repair mechanisms. In this context, mimicking the molecular and cellular effects of caloric restriction by novel drugs could represent a promising approach (see [112–114]). However, a detailed discussion of this topic is beyond the scope of the current paper.
Given the fact that there is almost no neuron loss in the aging cerebral cortex, recent studies on human postmortem brains and brains from laboratory animals have suggested that age-related neuronal dysfunction, which must underlie the observed age-related decline in cognitive function, probably involves a host of other subtle changes within these structures, such as regression in the complexity of dendrite arborization and dendritic spines, as well as age-related disturbances in synaptic function and integrity [115–128]. A number of studies showed that the dendritic arbors and dendritic spines of cortical pyramidal cells undergo age-related regressive changes in specific regions and layers of the cerebral cortex of humans and non-human primates [115–117,120,122–125]. Furthermore, it has been demonstrated that decreased dendritic branching due to aging is accompanied by spine loss in the mouse visual cortex [118]. In the prefrontal cortex, such dendritic and spine alterations have been correlated with impairments of memory and cognitive function in aged monkeys [124,126,129]. Because the dendritic surface receives most of the synaptic input in a given neuron, alterations in dendritic morphology, and particularly in spine numbers, may lead to concomitant changes in synaptic density. Interestingly, such a parallel decrease in spine numbers and synaptic density during aging has been observed in quantitative electron microscopic studies [124,128].
Accordingly, structural changes of neurons in the aging brain have been well documented, but relatively little is known about the possible mechanisms underlying these changes. In this regard the effects of oxidative stress on the integrity of the nDNA in neurons appear as one of the most important mechanisms related to neurodegeneration. In line with this view, a recent study showed that in cultured neurons, the promoters of genes related to synaptic function, vesicular transport and mitochondrial function displayed specifically an increased susceptibility to accumulate oxidative stress damage, leading to downregulation of genes involved in synaptic integrity [6].
In summary, the combined application of autoradiographic methods to study quantitatively relative amounts of nDNA damage and nDNA repair on tissue sections, design-based stereology to assess adequately age-related neuron loss, and novel techniques for imaging and analysis of three-dimensional neural morphology (see also [130]) provide insights into the relations between molecular events associated with age-related nDNA damage (according to Gensler and Bernstein’s [86] hypothesis) and age-related neurodegeneration. However, the molecular mechanisms underlying these age-related changes remain to be established.
6. Accumulation of nDNA damage or neuron loss: implications for the selective neuronal vulnerability in neurodegenerative diseases
It is likely that the neurons selectively vulnerable in neurodegenerative diseases share key biochemical and cellular properties linked to their vulnerability. One such property is the aggregation of abnormal protein components leading to their selective loss in disorders such as AD (intra-and extracellular aggregation of amyloid β [Aβ], as well as intracellular aggregation of hyperphosphorylated tau protein, leading to the formation of neurofibrillary tangles (NFTs)), Parkinson’s disease (PD; aggregation of α-synuclein, leading to the formation of intracellular Lewy bodies) and Huntington’s disease (intracellular aggregates of huntingtin protein) [131–133].
The formation of NFT in AD shows preferential involvement of certain cortical and subcortical areas, suggesting systematic differences in regional neuronal vulnerability [134,135]. Furthermore, the spatiotemporal development of NFT in AD is correlated to the clinical severity of the disease and the cognitive decline of the patients [136]. Initial NFT formation is found in the transentorhinal region in the temporal lobe. From there, this process spreads to the entorhinal region, hippocampus and neocortex [106,135,137]. The heightened vulnerability of neurons in the entorhinal and association cortices has been related to their high expression level of somatodendritic dephosphorylated neurofilament protein [138,139] or to their close synaptic relationship with limbic areas [140]. Other factors discussed in the context of the selective neuronal vulnerability in AD comprise, among others, early region-specific alterations in the blood–brain barrier [141], high gene expression of mitochondrial DNA-encoded ND4 (a subunit of complex I of oxidative phosphorylation) [142], a low portion of L-amyloid β precursor protein (L-APP, the APP obtained from mRNAs lacking exon 15 of the APP gene by alternative splicing in the presence of a high APP content) [143], a decrease in some glutamate receptor subunits expression (particularly GluR2/3) with concomitant alterations of calcium conductance through AMPA-selective channels and destabilization of intracellular calcium homeostasis [144], a mutation early in development (blastocyst stage or before) in a mDNA molecule and/or in the nDNA of the precursor cells of neurons later involved in AD, that impairs oxidative phosphorylation and increases production of superoxide radical and H2O2 [145], or the particular vulnerability of brain regions that show accelerated progression during primate evolution [146,147]. However, none of these possible mechanisms provides a comprehensive elucidation of the molecular basis of the selective neuronal vulnerability in AD.
In this regard it is important to note that several studies have provided evidence that nDNA damage plays an important role in AD as well as in other neurodegenerative diseases [93,148–150]. For instance, many TUNEL-positive neurons are detected in postmortem brains from patients with AD [37,151] and a subset of these cells is known to accumulate truncated tau [152]. Interestingly, the neurons that show NFT formation in AD are the same that show age-related accumulation of nDNA damage and decreased amount of nDNA repair, but no age-related reduction in number, in the mouse brain (as outlined in Sections 4 and 5 above) [5,10,20,21]. It is therefore interesting to hypothesize that the selective neuronal vulnerability in AD is due at least in part to the fact that in the human brain certain types of neurons which accumulate nDNA damage but are not removed during aging become increasingly vulnerable to the NFT formation. Conversely, other types of neurons (such as cerebellar Purkinje cells) may be protected from NFT formation in AD by the cell loss during physiological aging (note that this cell loss during aging must not to be confused with the complex mechanism and pattern of pathological neuron loss of AD [108,138,153]).
PD shows a different selective neuronal vulnerability than AD [135,154] and it is unknown to which extend the cell-type-specific pattern of Levy body formation and neurodegeneration in PD matches the pattern of neurons that show accumulation of nDNA damage but no cell loss during aging. However, the most prominent cell loss in PD affects the dopaminergic neurons in the substantia nigra pars compacta (SNpc) [154,155]. A recent study found, using the Comet assay, an age-related increase in the amount of nDNA damage in this brain region in rats [156]. The high amount of iron found in the SNpc [157,158] may contribute (via the generation of ROS [157,159]) to age-related accumulation of nDNA damage and, eventually, loss of the dopaminergic neurons in the SNpc in PD. Further research is needed in this regard.
In summary, age-related accumulation of nDNA damage in specific populations of neurons might be among the most important molecular mechanisms in triggering the onset of neurodegenerative diseases such as AD. Consequently, possible therapeutic strategies to prevent age-related nDNA damage, to improve the efficacy of nDNA repair mechanisms or to eliminate those neurons that show the highest amount of nDNA damage may be of crucial importance in the effective prevention of age-related neurodegenerative diseases.
Acknowledgments
Work by the authors discussed in this paper was supported by the Dutch Brain Foundation (Hersenstichting, the Netherlands), the Alzheimer Forschung Initiative (AFI, Germany), the International Alzheimer’s Research Foundation (ISAO; the Netherlands), EC/NRPB Association Contract No. F14P-CT95-0008, and NIH grants AG02219 and AG05138.
Footnotes
Dedicated to Dr. Hubert Korr, Professor of Anatomy and Cell Biology, RWTH Aachen University (Aachen, Germany), on the occasion of his 65th birthday, for his contribution to research on DNA damage and DNA repair in the central nervous system.
References
- 1.Maccioni RB, Munoz JP, Barbeito L. The molecular bases of Alzheimer’s disease and other neurodegenerative disorders. Arch Med Res. 2001;32:367–381. doi: 10.1016/s0188-4409(01)00316-2. [DOI] [PubMed] [Google Scholar]
- 2.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 3.Barnett YA, King CM. An investigation of antioxidant status, DNA repair capacity and mutation as a function of age in humans. Mutat Res. 1995;338:115–128. doi: 10.1016/0921-8734(95)00017-z. [DOI] [PubMed] [Google Scholar]
- 4.Floyd RA, Hensley K. Oxidative stress in brain aging. Implications for therapeutics of neurodegenerative diseases. Neurobiol Aging. 2002;23:795–807. doi: 10.1016/s0197-4580(02)00019-2. [DOI] [PubMed] [Google Scholar]
- 5.Rutten BPF, Korr H, Steinbusch HWM, Schmitz C. The aging brain: less neurons could be better. Mech Ageing Dev. 2003;124:349–355. doi: 10.1016/s0047-6374(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 6.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 7.Haug H. History of neuromorphometry. J Neurosci Methods. 1986;18:1–17. doi: 10.1016/0165-0270(86)90110-x. [DOI] [PubMed] [Google Scholar]
- 8.Bohr V, Anson RM, Mazur S, Dianov G. Oxidative DNA damage processing and changes with aging. Toxicol Lett. 1998;102/103:47–52. doi: 10.1016/s0378-4274(98)00280-x. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci USA. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutten BPF, Schmitz C, Gerlach OH, Oyen HM, de Mesquita EB, Steinbusch HWM, Korr H. The aging brain: accumulation of DNA damage or neuron loss? Neurobiol Aging. 2007;28:91–98. doi: 10.1016/j.neurobiolaging.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa MF, Alt W. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 12.Ames BN, Saul RL. Oxidative DNA damage as related to cancer and aging. Prog Clin Biol Res. 1986;209A:11–26. [PubMed] [Google Scholar]
- 13.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 3. Oxford University Press; Oxford: 1999. [Google Scholar]
- 15.Young IS, Woodside JV. Antioxidants in health and disease. J Clin Pathol. 2001;54:176–186. doi: 10.1136/jcp.54.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergamini CM, Gambetti S, Dondi A, Cervellati C. Oxygen, reactive oxygen species and tissue damage. Curr Pharm Des. 2004;10:1611–1626. doi: 10.2174/1381612043384664. [DOI] [PubMed] [Google Scholar]
- 17.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 18.Christen Y. Oxidative stress and Alzheimer’s disease. Am J Clin Nutr. 2000;71:621S–629S. doi: 10.1093/ajcn/71.2.621s. [DOI] [PubMed] [Google Scholar]
- 19.Rutten BPF, Steinbusch HWM, Korr H, Schmitz C. Antioxidants and Alzheimer’s disease: from bench to bedside (and back again) Curr Opin Clin Nutr Metab Care. 2002;5:645–651. doi: 10.1097/00075197-200211000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz C, Axmacher B, Zunker U, Korr H. Age-related changes of DNA repair and mitochondrial DNA synthesis in the mouse brain. Acta Neuropathol. 1999;97:71–81. doi: 10.1007/s004010050957. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz C, Materne S, Korr H. Cell-type-specific differences in age-related changes of DNA repair in the mouse brain—molecular basis for a new approach to understand the selective neuronal vulnerability in Alzheimer’s disease. J Alzheimers Dis. 1999;1:387–407. doi: 10.3233/jad-1999-1604. [DOI] [PubMed] [Google Scholar]
- 22.Karanjawala ZE, Lieber MR. DNA damage and aging. Mech Ageing Dev. 2004;25:405–416. doi: 10.1016/j.mad.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Vijg J, Uitterlinden AG. A search for DNA alterations in the aging mammalian genome: an experimental strategy. Mech Ageing Dev. 1987;41:47–63. doi: 10.1016/0047-6374(87)90053-4. [DOI] [PubMed] [Google Scholar]
- 24.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 25.Caldecott KW. Eukaryotic DNA Damage Surveillance and Repair. Kluwer Academic/Plenum Publishers; New York: 2004. [Google Scholar]
- 26.Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays. 2001;23:447–455. doi: 10.1002/bies.1063. [DOI] [PubMed] [Google Scholar]
- 27.Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic Biol Med. 2002;32:804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 28.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 29.Caldecott KW. DNA single-strand breaks and neurodegeneration. DNA Repair. 2004;3:875–882. doi: 10.1016/j.dnarep.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Guetens G, De Boeck G, Highley M, van Oosterom AT, de Bruijn EA. Oxidative DNA damage: biological significance and methods of analysis. Crit Rev Clin Lab Sci. 2002;39:331–457. doi: 10.1080/10408360290795547. [DOI] [PubMed] [Google Scholar]
- 31.Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 32.Lindahl T. Recognition and processing of damaged DNA. J Cell Sci Suppl. 1995;19:73–77. doi: 10.1242/jcs.1995.supplement_19.10. [DOI] [PubMed] [Google Scholar]
- 33.van Dierendonck JH. DNA damage detection using DNA polymerase I or its Klenow fragment. Applicability, specificity, limitations. Methods Mol Biol. 2002;203:81–108. doi: 10.1385/1-59259-179-5:81. [DOI] [PubMed] [Google Scholar]
- 34.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker PR, Carson C, Leblanc J, Sikorska M. Labeling DNA damage with terminal transferase. Applicability, specificity, and limitations. Methods Mol Biol. 2002;203:3–19. doi: 10.1007/978-1-59259-179-4_1. [DOI] [PubMed] [Google Scholar]
- 36.Allen RT, Hunter WJ, III, Agrawal DK. Morphological and biochemical characterization and analysis of apoptosis. J Pharmacol Toxicol Methods. 1997;37:215–228. doi: 10.1016/s1056-8719(97)00033-6. [DOI] [PubMed] [Google Scholar]
- 37.Jellinger KA, Stadelmann CH. The enigma of cell death in neurodegenerative disorders. J Neural Transm Suppl. 2000;60:21–36. doi: 10.1007/978-3-7091-6301-6_2. [DOI] [PubMed] [Google Scholar]
- 38.Jellinger KA, Stadelmann C. Problems of cell death in neurodegeneration and Alzheimer’s disease. J Alzheimers Dis. 2001;3:31–40. doi: 10.3233/jad-2001-3106. [DOI] [PubMed] [Google Scholar]
- 39.Korr H, Rohde HT, Benders J, Dafotakis M, Grolms N, Schmitz C. Neuron loss during early adulthood following prenatal low-dose X-irradiation in the mouse brain. Int J Radiat Biol. 2001;77:567–580. doi: 10.1080/09553000010028467. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch EF, Maniatis T. A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. Molecular Cloning. [Google Scholar]
- 41.Iseki S, Mori T. Histochemical detection of DNA strand scissions in mammalian cells by in situ nick translation. Cell Biol Int Rep. 1985;9:471–477. doi: 10.1016/0309-1651(85)90155-9. [DOI] [PubMed] [Google Scholar]
- 42.Iseki S. DNA strand breaks in rat tissues as detected by in situ nick translation. Exp Cell Res. 1986;167:311–326. doi: 10.1016/0014-4827(86)90172-2. [DOI] [PubMed] [Google Scholar]
- 43.Setlow P, Brutlag D, Kornberg A. Deoxyribonucleic acid polymerase: two distinct enzymes in one polypeptide. I. A proteolytic fragment containing the polymerase and 3′ leads to 5′ exonuclease functions. J Biol Chem. 1972;247:224–231. [PubMed] [Google Scholar]
- 44.Setlow P, Kornberg A. Deoxyribonucleic acid polymerase: two distinct enzymes in one polypeptide. II. A proteolytic fragment containing the 5′ leads to 3′ exonuclease function. Restoration of intact enzyme functions from the two proteolytic fragments. J Biol Chem. 1972;247:232–240. [PubMed] [Google Scholar]
- 45.Stillström J. Grain count corrections in autoradiography. Int J Appl Radiat Isot. 1965;16:357–363. doi: 10.1016/0020-708x(65)90019-0. [DOI] [PubMed] [Google Scholar]
- 46.Korr H, Schmidt H. An improved procedure for background correction in autoradiography. Histochemistry. 1988;88:407–470. doi: 10.1007/BF00570302. [DOI] [PubMed] [Google Scholar]
- 47.Korr H, Bauer K, Bunzeck AS, Nacken M, Karbach FT. Correction factors of 3H β-self-absorption for quantitative autoradiography of different cell types in the brain of pre-and postnatal mice. Histochem Cell Biol. 1997;108:537–541. doi: 10.1007/s004180050194. [DOI] [PubMed] [Google Scholar]
- 48.Maehara Y, Anai H, Kusumoto T, Sakaguchi Y, Sugimachi K. Nick translation detection in situ of cellular DNA strand break induced by radiation. Am J Pathol. 1989;134:7–10. [PMC free article] [PubMed] [Google Scholar]
- 49.Wang RY, Hsu TC, Brock WA, Liang J. DNA fragility and repair capability are separate genetic phenotypes: studies on in situ nick translation and chromosome breakage. Int J Oncol. 1995;6:51–54. doi: 10.3892/ijo.6.1.51. [DOI] [PubMed] [Google Scholar]
- 50.Korr H. Light microscopical autoradiography of nervous tissue. In: Heym C, Forssmann WG, editors. Techniques in Neuroanatomical Research. Springer; Berlin: 1981. pp. 218–244. [Google Scholar]
- 51.Maurer W, Primbsch E. Größe der β-Selbstabsorption bei der 3H-Autoradiographie (magnitude of beta-autoabsorption with H-3 autoradiography) Exp Cell Res. 1964;33:8–18. doi: 10.1016/s0014-4827(64)81005-3. [DOI] [PubMed] [Google Scholar]
- 52.Caldecott KW. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair. 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 53.Caldecott KW. DNA single-strand break repair and spinocerebellar ataxia. Cell. 2003;112:7–10. doi: 10.1016/s0092-8674(02)01247-3. [DOI] [PubMed] [Google Scholar]
- 54.Wood RD. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 55.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 56.Mellon I. Transcription-coupled repair: a complex affair. Mutat Res. 2005;577:155–161. doi: 10.1016/j.mrfmmm.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 57.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD. DNA repair: from molecular mechanism to human disease. DNA Repair. 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 58.Mitra S, Hazra TK, Roy R, Ikeda S, Biswas T, Lock J, Boldogh I, Izumi T. Complexities of DNA base excision repair in mammalian cells. Mol Cells. 1997;7:305–312. [PubMed] [Google Scholar]
- 59.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 61.Batty DP, Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 62.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 63.Haber JE. Partners and pathways repairing a double-strand break. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 64.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 65.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 66.Lainé JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 67.Sarasin A, Stary A. New insights for understanding the transcription-coupled repair pathway. DNA Repair. 2007;6:265–269. doi: 10.1016/j.dnarep.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 68.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 69.Check E. Retracted papers damage work on DNA repair. Nature. 2005;435:1015. doi: 10.1038/4351015a. [DOI] [PubMed] [Google Scholar]
- 70.Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst. 2000;92:874–897. doi: 10.1093/jnci/92.11.874. [DOI] [PubMed] [Google Scholar]
- 71.Fairbairn DW, Olive PL, O’Neill KL. The comet assay: a comprehensive review. Mutat Res. 1995;399:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
- 72.Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 73.Frenzilli G, Scarcelli V, Fornai F, Paparelli A, Nigro M. The comet assay as a method of assessment of neurotoxicity: usefulness for drugs of abuse. Ann NY Acad Sci. 2006;1074:478–481. doi: 10.1196/annals.1369.048. [DOI] [PubMed] [Google Scholar]
- 74.Paterson MC, Lohman PHM, Sluyter ML. Use of UV endonuclease from Micrococcus luteus to monitor the progress of DNA repair in UV-irradiated human cells. Mutat Res. 1973;19:245–256. doi: 10.1016/0027-5107(73)90083-3. [DOI] [PubMed] [Google Scholar]
- 75.Ganesan AK. A method for detecting pyrimidine dimers in the DNA of bacteria irradiated with low doses of ultraviolet light. Proc Natl Acad Sci USA. 1973;70:2753–2756. doi: 10.1073/pnas.70.10.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson JM, Latimer JJ. Analysis of DNA repair using transfection-based host cell reactivation. Methods Mol Biol. 2005;291:321–335. doi: 10.1385/1-59259-840-4:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Djordjevic B, Tolmach LJ. Responses of synchronous populations of HeLa cells to ultraviolet irradiation at selected stages of the generation cycle. Radiat Res. 1967;32:327–346. [PubMed] [Google Scholar]
- 78.Setlow RB. Repair different basic mechanisms in DNA repair. Arch Toxicol Suppl. 1980;3:217–228. doi: 10.1007/978-3-642-67389-4_16. [DOI] [PubMed] [Google Scholar]
- 79.Mitchell AD, Casciano DA, Meltz ML, Robinson DE, San RH, Williams GM, Von Halle ES. Unscheduled DNA synthesis tests. A report of the U.S. Environmental Protection Agency Gene-Tox Program. Mutat Res. 1983;123:363–410. doi: 10.1016/0165-1110(83)90029-5. [DOI] [PubMed] [Google Scholar]
- 80.Korr H, Schultze B. Unscheduled DNA synthesis in various types of cells of the mouse brain in vivo. Exp Brain Res. 1989;74:573–578. doi: 10.1007/BF00247359. [DOI] [PubMed] [Google Scholar]
- 81.Korr H, Koeser K, Oldenkott S, Schmidt H, Schultze B. X-ray dose–effect relationship on unscheduled DNA synthesis and spontaneous unscheduled DNA synthesis in mouse brain cells studied in vivo. Radiat Environ Biophys. 1989;28:13–26. doi: 10.1007/BF01209719. [DOI] [PubMed] [Google Scholar]
- 82.Korr H, Philippi V, Helg C, Schiefer J, Graeber MB, Kreutzberg GW. Unscheduled DNA synthesis and mitochondrial DNA synthetic rate following injuring of the facial nerve. Acta Neuropathol. 1997;94:557–566. doi: 10.1007/s004010050750. [DOI] [PubMed] [Google Scholar]
- 83.Korr H, Jansen A, Roels C. Further characterization of DNA repair in vivo studied with autoradiography in the brain of the adult mouse. In: Elsner N, Singer W, editors. Dynamics and Plasticity in Neuronal Systems. Thieme; Stuttgart: 1989. p. 167. [Google Scholar]
- 84.Amano M, Messier B, Leblond CP. Specificity of labelled thymidine as a deoxyribonucleic acid precursor in radioautography. J Histochem Cytochem. 1959;7:153–155. doi: 10.1177/7.3.153. [DOI] [PubMed] [Google Scholar]
- 85.Hill M. Non-S-phase incorporation of 3H-thymidine into DNA of X-irradiated mammalian cells. Int J Radiat Biol. 1967;13:199–203. doi: 10.1080/09553006814550121. [DOI] [PubMed] [Google Scholar]
- 86.Gensler HL, Bernstein H. DNA damage as the primary cause of aging. Q Rev Biol. 1981;56:279–303. doi: 10.1086/412317. [DOI] [PubMed] [Google Scholar]
- 87.Mandavilli BS, Rao KS. Neurons in the cerebral cortex are most susceptible to DNA-damage in aging rat brain. Biochem Mol Biol Int. 1996;40:507–514. doi: 10.1080/15216549600201073. [DOI] [PubMed] [Google Scholar]
- 88.Brasnjevic I, Klocke B, Hoppenburg I, Bulic B, Bode GH, Hof PR, Steinbusch HWM, Schmitz C. Evidence for transcription-coupled nuclear DNA repair in brain aging. submitted for publication. [Google Scholar]
- 89.Dam M, Bolwig T, Hertz M, Bajorec J, Lomax P, Dam AM. Does seizure activity produce Purkinje cell loss? Epilepsia. 1984;25:747–751. doi: 10.1111/j.1528-1157.1984.tb03486.x. [DOI] [PubMed] [Google Scholar]
- 90.Cervós-Navarro J, Diemer NH. Selective vulnerability in brain hypoxia. Crit Rev Neurobiol. 1991;6:149–182. [PubMed] [Google Scholar]
- 91.Welsh JP, Yuen G, Placantonakis DG, Vu TQ, Haiss F, O’Hearn E, Molliver ME, Aicher SA. Why do Purkinje cells die so easily after global brain ischemia? Aldolase C, EAAT4, and the cerebellar contribution to posthypoxic myoclonus. Adv Neurol. 2002;89:331–359. [PubMed] [Google Scholar]
- 92.Bohr VA, Sander M, Kraemer KH. Rare diseases provide rare insights into DNA repair pathways, TFIIH, aging and cancer center. DNA Repair. 2005;4:293–302. doi: 10.1016/j.dnarep.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 93.Rao KS. DNA repair in aging rat neurons. Neuroscience. 2007;145:1330–1340. doi: 10.1016/j.neuroscience.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 94.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 95.Markesbery WR, Lovell MA. Damage to lipids, proteins, DNA, and RNA in mild cognitive impairment. Arch Neurol. 2007;64:954–956. doi: 10.1001/archneur.64.7.954. [DOI] [PubMed] [Google Scholar]
- 96.Wickelgren I. For the cortex, neuron loss may be less than thought. Science. 1996;273:48–50. [PubMed] [Google Scholar]
- 97.Wickelgren I. Is hippocampal cell death a myth? Science. 1996;271:1229–1230. doi: 10.1126/science.271.5253.1229. [DOI] [PubMed] [Google Scholar]
- 98.West MJ. New stereological methods for counting neurons. Neurobiol Aging. 1993;14:275–285. doi: 10.1016/0197-4580(93)90112-o. [DOI] [PubMed] [Google Scholar]
- 99.Schmitz C, Hof PR. Design-based stereology in neuroscience. Neuroscience. 2005;130:813–831. doi: 10.1016/j.neuroscience.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 100.Glaser J, Greene G, Hendricks S. Stereology for Biological Research with a Focus on Neuroscience. MBF Press; Williston: 2007. [Google Scholar]
- 101.West MJ, Gundersen HJ. Unbiased stereological estimation of the number of neurons in the human hippocampus. J Comp Neurol. 1990;296:1–22. doi: 10.1002/cne.902960102. [DOI] [PubMed] [Google Scholar]
- 102.West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer’s disease. Lancet. 1994;344:769–772. doi: 10.1016/s0140-6736(94)92338-8. [DOI] [PubMed] [Google Scholar]
- 103.Giannakopoulos P, Hof PR, Kövari E, Vallet PG, Herrmann FR, Bouras C. Distinct patterns of neuronal loss and Alzheimer’s disease lesion distribution in elderly individuals older than 90 years. J Neuropathol Exp Neurol. 1996;55:1210–1220. doi: 10.1097/00005072-199612000-00004. [DOI] [PubMed] [Google Scholar]
- 104.Rapp PR, Gallagher M. Preserved neuron number in the hippocampus of aged rats with spatial learning deficits. Proc Natl Acad Sci USA. 1996;93:9926–9930. doi: 10.1073/pnas.93.18.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gazzaley AH, Thakker MM, Hof PR, Morrison JH. Preserved number of entorhinal cortex layer II neurons in aged macaque monkeys. Neurobiol Aging. 1997;18:549–553. doi: 10.1016/s0197-4580(97)00112-7. [DOI] [PubMed] [Google Scholar]
- 106.Pakkenberg B, Gundersen HJ. Neocortical neuron number in humans: effect of sex and age. J Comp Neurol. 1997;384:312–320. [PubMed] [Google Scholar]
- 107.Merrill DA, Chiba AA, Tuszynski MH. Conservation of neuronal number and size in the entorhinal cortex of behaviorally characterized aged rats. J Comp Neurol. 2001;438:445–456. doi: 10.1002/cne.1327. [DOI] [PubMed] [Google Scholar]
- 108.Morrison JH, Hof PR. Selective vulnerability of corticocortical and hippocampal circuits in aging and Alzheimer’s disease. Prog Brain Res. 2002;136:467–486. doi: 10.1016/s0079-6123(02)36039-4. [DOI] [PubMed] [Google Scholar]
- 109.Gatti RA. Speculations on the ataxia-telangiectasia defect. Clin Immunol Immunopathol. 1991;61:S10–S15. doi: 10.1016/s0090-1229(05)80032-7. [DOI] [PubMed] [Google Scholar]
- 110.Manto MU. On the cerebello–cerebral interactions. Cerebellum. 2006;5:286–288. doi: 10.1080/14734220601003955. [DOI] [PubMed] [Google Scholar]
- 111.Timmann D, Daum I. Cerebellar contributions to cognitive functions: a progress report after two decades of research. Cerebellum. 2007;6:159–162. doi: 10.1080/14734220701496448. [DOI] [PubMed] [Google Scholar]
- 112.Pallàs M, Verdaguer E, Tajes M, Gutierrez-Cuesta J, Camins A. Modulation of sirtuins: new targets for antiageing. Recent Patents CNS Drug Discov. 2008;3:61–69. doi: 10.2174/157488908783421492. [DOI] [PubMed] [Google Scholar]
- 113.Wolf FI, Fasanella S, Tedesco B, Cavallini G, Donati A, Bergamini E, Cittadini A. Peripheral lymphocyte 8-OHdG levels correlate with age-associated increase of tissue oxidative DNA damage in Sprague–Dawley rats. Protective effects of caloric restriction. Exp Gerontol. 2005;40:181–188. doi: 10.1016/j.exger.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 114.Prapurna DR, Rao KS. Long-term effects of caloric restriction initiated at different ages on DNA polymerases in rat brain. Mech Ageing Dev. 1996;92:133–142. doi: 10.1016/s0047-6374(96)01815-5. [DOI] [PubMed] [Google Scholar]
- 115.Scheibel ME, Lindsay RD, Tomiyasu U, Scheibel AB. Progressive dendritic changes in aging human cortex. Exp Neurol. 1975;47:392–403. doi: 10.1016/0014-4886(75)90072-2. [DOI] [PubMed] [Google Scholar]
- 116.Cupp CJ, Uemura E. Age-related changes in prefrontal cortex of Macaca mulatta: quantitative analysis of dendritic branching patterns. Exp Neurol. 1980;69:143–163. doi: 10.1016/0014-4886(80)90150-8. [DOI] [PubMed] [Google Scholar]
- 117.Uemura E. Age-related changes in prefrontal cortex of Macaca mulatta: synaptic density. Exp Neurol. 1980;69:164–172. doi: 10.1016/0014-4886(80)90151-x. [DOI] [PubMed] [Google Scholar]
- 118.Leuba G. Aging of dendrites in the cerebral cortex of the mouse. Neuropathol Appl Neurobiol. 1983;9:467–475. doi: 10.1111/j.1365-2990.1983.tb00130.x. [DOI] [PubMed] [Google Scholar]
- 119.Nakamura S, Akiguchi I, Kameyama M, Mizuno N. Age-related changes of pyramidal cell basal dendrites in layers III and V of human motor cortex: a quantitative Golgi study. Acta Neuropathol. 1985;65:281–284. doi: 10.1007/BF00687009. [DOI] [PubMed] [Google Scholar]
- 120.Jacobs B, Scheibel AB. A quantitative dendritic analysis of Wernicke’s area in humans. I. Lifespan changes. J Comp Neurol. 1993;327:83–96. doi: 10.1002/cne.903270107. [DOI] [PubMed] [Google Scholar]
- 121.Barnes CA. Normal aging: regionally specific changes in hippocampal synaptic transmission. Trends Neurosci. 1994;17:13–18. doi: 10.1016/0166-2236(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 122.Jacobs B, Driscoll L, Schall M. Life-span dendritic and spine changes in areas 10 and 18 of human cortex: a quantitative Golgi study. J Comp Neurol. 1997;386:661–680. [PubMed] [Google Scholar]
- 123.de Brabander JM, Kramers RJ, Uylings HB. Layer-specific dendritic regression of pyramidal cells with ageing in the human prefrontal cortex. Eur J Neurosci. 1998;10:1261–1269. doi: 10.1046/j.1460-9568.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- 124.Peters A, Sethares C, Moss MB. The effects of aging on layer 1 in area 46 of prefrontal cortex in the rhesus monkey. Cereb Cortex. 1998;8:671–684. doi: 10.1093/cercor/8.8.671. [DOI] [PubMed] [Google Scholar]
- 125.Peters A, Moss MB, Sethares C. The effects of aging on layer 1 of primary visual cortex in the rhesus monkey. Cereb Cortex. 2001;11:93–103. doi: 10.1093/cercor/11.2.93. [DOI] [PubMed] [Google Scholar]
- 126.Duan H, Wearne SL, Morrison JH, Hof PR. Quantitative analysis of the dendritic morphology of corticocortical projection neurons in the macaque monkey association cortex. Neuroscience. 2002;114:349–359. doi: 10.1016/s0306-4522(02)00305-6. [DOI] [PubMed] [Google Scholar]
- 127.Duan H, Wearne SL, Rocher AB, Macedo A, Morrison JH, Hof PR. Age-related dendritic and spine changes in corticocortically projecting neurons in macaque monkeys. Cereb Cortex. 2003;13:950–961. doi: 10.1093/cercor/13.9.950. [DOI] [PubMed] [Google Scholar]
- 128.Hof PR, Morrison JH. The aging brain: morphomolecular senescence of cortical circuits. Trends Neurosci. 2004;27:607–613. doi: 10.1016/j.tins.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 129.Page TL, Einstein M, Duan H, He Y, Flores T, Rolshud D, Erwin JM, Wearne SL, Morrison JH, Hof PR. Morphological alterations in neurons forming corticocortical projections in the neocortex of aged Patas monkeys. Neurosci Lett. 2002;317:37–41. doi: 10.1016/s0304-3940(01)02428-4. [DOI] [PubMed] [Google Scholar]
- 130.Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience. 2005;136:661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
- 131.Butterfield DA, Kanski J. Brain protein oxidation in age-related neurodegenerative disorders that are associated with aggregated proteins. Mech Ageing Dev. 2001;122:945–962. doi: 10.1016/s0047-6374(01)00249-4. [DOI] [PubMed] [Google Scholar]
- 132.Shastry BS. Neurodegenerative disorders of protein aggregation. Neurochem Int. 2003;43:1–7. doi: 10.1016/s0197-0186(02)00196-1. [DOI] [PubMed] [Google Scholar]
- 133.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 134.Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16:271–278. doi: 10.1016/0197-4580(95)00021-6. [DOI] [PubMed] [Google Scholar]
- 135.Braak H, Rüb U, Schultz C, Del Tredici K. Vulnerability of cortical neurons to Alzheimer’s and Parkinson’s diseases. J Alzheimers Dis. 2006;9(3 Suppl):35–44. doi: 10.3233/jad-2006-9s305. [DOI] [PubMed] [Google Scholar]
- 136.Riley KP, Snowdon DA, Markesbery WR. Alzheimer’s neurofibrillary pathology and the spectrum of cognitive function: findings from the Nun study. Ann Neurol. 2002;51:567–577. doi: 10.1002/ana.10161. [DOI] [PubMed] [Google Scholar]
- 137.Cummings JL, Cole G. Alzheimer’s disease. J Am Med Assoc. 2002;287:2335–2338. doi: 10.1001/jama.287.18.2335. [DOI] [PubMed] [Google Scholar]
- 138.Morrison JH, Hof PR. Life and death of neurons in the aging brain. Science. 1997;278:412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- 139.Morrison BM, Hof PR, Morrison JH. Determinants of neuronal vulnerability in neurodegenerative diseases. Ann Neurol. 1998;44(Suppl 1):S32–S44. doi: 10.1002/ana.410440706. [DOI] [PubMed] [Google Scholar]
- 140.Stein DJ, Buchsbaum MS, Hof PJ, Siegel BV, Jr, Shihabuddin L. Greater metabolic rate decreases in hippocampal formation and proisocortex than in neocortex in Alzheimer’s disease. Neuropsychobiology. 1998;37:10–19. doi: 10.1159/000026471. [DOI] [PubMed] [Google Scholar]
- 141.Gibson GE, Calingasan NY, Baker H, Gandy S, Sheu KF. Importance of vascular changes in selective neurodegeneration with thiamine deficiency. Ann NY Acad Sci. 1997;826:516–519. doi: 10.1111/j.1749-6632.1997.tb48517.x. [DOI] [PubMed] [Google Scholar]
- 142.Fukuyama R, Hatanpaa K, Rapoport SI, Chandrasekaran K. Gene expression of ND4, a subunit of complex I of oxidative phosphorylation in mitochondria, is decreased in temporal cortex of brains of Alzheimer’s disease patients. Brain Res. 1996;713:290–293. doi: 10.1016/0006-8993(95)01517-5. [DOI] [PubMed] [Google Scholar]
- 143.Sandbrink R, Masters CL, Beyreuther K. APP gene family. Alternative splicing generates functionally related isoforms. Ann NY Acad Sci. 1996;777:281–287. doi: 10.1111/j.1749-6632.1996.tb34433.x. [DOI] [PubMed] [Google Scholar]
- 144.Armstrong DM, Ikonomovic MD, Sheffield R, Wenthold RJ. AMPA-selective glutamate receptor subtype immunoreactivity in the entorhinal cortex of non-demented elderly and patients with Alzheimer’s disease. Brain Res. 1994;639:207–216. doi: 10.1016/0006-8993(94)91732-9. [DOI] [PubMed] [Google Scholar]
- 145.Harman D. Free radical theory of aging: Alzheimer’s disease pathogenesis. Age. 1995;18:97–119. [Google Scholar]
- 146.Nimchinsky EA, Gilissen E, Allman JM, Perl DP, Erwin JM, Hof PR. A neuronal morphologic type unique to humans and great apes. Proc Natl Acad Sci USA. 1999;96:5268–5273. doi: 10.1073/pnas.96.9.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rapoport SI. Integrated phylogeny of the primate brain, with special reference to humans and their diseases. Brain Res Rev. 1990;15:267–294. doi: 10.1016/0165-0173(90)90004-8. [DOI] [PubMed] [Google Scholar]
- 148.Robison SH, Bradley WG. DNA damage and chronic neuronal degenerations. J Neurol Sci. 1984;64:11–20. doi: 10.1016/0022-510x(84)90051-0. [DOI] [PubMed] [Google Scholar]
- 149.Itzhaki RF. Possible factors in the etiology of Alzheimer’s disease. Mol Neurobiol. 1994;9:1–13. doi: 10.1007/BF02816099. [DOI] [PubMed] [Google Scholar]
- 150.Cotman CW, Su JH. Mechanisms of neuronal death in Alzheimer’s disease. Brain Pathol. 1996;6:493–506. doi: 10.1111/j.1750-3639.1996.tb00878.x. [DOI] [PubMed] [Google Scholar]
- 151.Anderson AJ, Su JH, Cotman CW. DNA damage and apoptosis in Alzheimer’s disease: colocalization with c-Jun immunoreactivity, relationship to brain area, and effect of postmortem delay. J Neurosci. 1996;16:1710–1719. doi: 10.1523/JNEUROSCI.16-05-01710.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ugolini G, Cattaneo A, Novak M. Co-localization of truncated tau and DNA fragmentation in Alzheimer’s disease neurons. Neuroreport. 1997;8:3709–3712. doi: 10.1097/00001756-199712010-00010. [DOI] [PubMed] [Google Scholar]
- 153.Thal DR, Del Tredici K, Braak H. Neurodegeneration in normal brain aging and disease. Sci Aging Knowledge Environ. 2004;23:pe26. doi: 10.1126/sageke.2004.23.pe26. [DOI] [PubMed] [Google Scholar]
- 154.Braak H, Braak E, Yilmazer D, Schultz C, de Vos RA, Jansen EN. Nigral and extranigral pathology in Parkinson’s disease. J Neural Transm Suppl. 1995;46:15–31. [PubMed] [Google Scholar]
- 155.Bernheimer H, Birkmayer W, Hornykiewicz O, Jellinger K, Seitelberger F. Brain dopamine and the syndromes of Parkinson and Huntington. Clinical, morphological and neurochemical correlations. J Neurol Sci. 1973;20:415–455. doi: 10.1016/0022-510x(73)90175-5. [DOI] [PubMed] [Google Scholar]
- 156.Giovannelli L, Decorosi F, Dolara P, Pulvirenti L. Vulnerability to DNA damage in the aging rat substantia nigra: a study with the comet assay. Brain Res. 2003;969:244–247. doi: 10.1016/s0006-8993(03)02275-3. [DOI] [PubMed] [Google Scholar]
- 157.Götz ME, Double K, Gerlach M, Youdim MB, Riederer P. The relevance of iron in the pathogenesis of Parkinson’s disease. Ann NY Acad Sci. 2004;1012:193–208. doi: 10.1196/annals.1306.017. [DOI] [PubMed] [Google Scholar]
- 158.Berg D, Roggendorf W, Schröder U, Klein R, Tatschner T, Benz P, Tucha O, Preier M, Lange KW, Reiners K, Gerlach M, Becker G. Echogenicity of the substantia nigra: association with increased iron content and marker for susceptibility to nigrostriatal injury. Arch Neurol. 2002;59:999–1005. doi: 10.1001/archneur.59.6.999. [DOI] [PubMed] [Google Scholar]
- 159.Fasano M, Bergamasco B, Lopiano L. Modifications of the iron-neuromelanin system in Parkinson’s disease. J Neurochem. 2006;96:909–916. doi: 10.1111/j.1471-4159.2005.03638.x. [DOI] [PubMed] [Google Scholar]
- 160.Rasmussen RE, Painter RB. Radiation-stimulated DNA synthesis in cultured mammalian cells. J Cell Biol. 1966;29:11–19. doi: 10.1083/jcb.29.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Painter RB, Cleaver JE. Repair replication, unscheduled DNA synthesis, and the repair of mammalian DNA. Radiat Res. 1969;37:451–466. [PubMed] [Google Scholar]
- 162.Sega GA, Sotomayor RE, Owens JG. A study of unscheduled DNA synthesis induced by X-rays in the germ cells of male mice. Mutat Res. 1978;49:239–257. doi: 10.1016/0027-5107(78)90163-x. [DOI] [PubMed] [Google Scholar]