SUMMARY
RIG-I detects invading viral RNA and activates the transcription factors NF-κB and IRF3 through the mitochondrial protein MAVS. Here we show that RNA bearing 5′-triphosphate strongly activates the RIG-I–IRF3 signaling cascade in a reconstituted system composed of RIG-I, mitochondria and cytosol. Activation of RIG-I requires not only RNA, but also polyubiquitin chains linked through lysine-63 (K63) of ubiquitin. RIG-I binds specifically to K63 polyubiquitin chains through its tandem CARD domains in a manner that depends on RNA and ATP. Mutations in the CARD domains that abrogate ubiquitin binding also impair RIG-I activation. Remarkably, unanchored K63 ubiquitin chains, which are not conjugated to any target protein, potently activate RIG-I. These ubiquitin chains function as an endogenous ligand of RIG-I in human cells. Our results delineate the mechanism of RIG-I activation, identify CARD domains as a new ubiquitin sensor, and demonstrate that unanchored K63 polyubiquitin chains are signaling molecules in antiviral innate immunity.
INTRODUCTION
Innate immunity is an evolutionarily conserved mechanism that detects and defends against invasion by microbial pathogens, including viruses, bacteria and parasites. Detection of pathogen-associated molecular patterns (PAMPs) is carried out by pattern recognition receptors (PRRs) that include membrane-bound Toll-like receptors (TLRs) and cytosolic receptors such as RIG-I-like receptors (RLRs) (Akira et al., 2006). In the case of infection by RNA viruses and some DNA viruses, viral RNAs are the major PAMPs, which are detected by some TLRs located on the endosomal membrane (e.g, TLR7) in specialized cells (e.g, plasmacytoid dendritic cells) and RLRs in many types of cells directly infected by viruses. Recognition of viral RNAs by TLRs and RLRs triggers signal transduction cascades that culminate in the production of type-I interferons (i.e, IFNα and IFNβ) and other molecules that collectively suppress viral replication and assembly.
RLRs include RIG-I, MDA5 and LGP2, all of which contain a DExD-box RNA helicase domain (Fujita, 2009; Yoneyama et al., 2004). RIG-I and LGP2 also contain a C-terminal regulatory domain (RD) that binds to RNA bearing 5′-triphosphate (5′-pppRNA) (Hornung et al., 2006; Pichlmair et al., 2006). The N-termini of RIG-I and MDA5, but not LGP2, contain tandem CARD domains that mediate downstream signaling. Signaling by both RIG-I and MDA5 requires the essential adaptor protein MAVS (also known as IPS-1, VISA or CARDIF) (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). MAVS contains a C-terminal transmembrane domain that targets it to the mitochondrial membrane and an N-terminal CARD domain that is exposed to the cytosol (Seth et al., 2005). This CARD domain interacts with the CARD domains of RIG-I, resulting in the activation of MAVS through an unknown mechanism. MAVS then activates the cytosolic protein kinases IKK and TBK1, which in turn activate the transcription factors NF-κB and IRF3, respectively. NF-κB, IRF3 and other transcription factors function together in the nucleus to induce the expression of type-I interferons and other antiviral molecules.
Recent studies have revealed a pivotal role of protein ubiquitination in diverse cell signaling pathways, including those in the immune system (Bhoj and Chen, 2009). In the RIG-I pathway, polyubiquitination involving lysine-63 (K63) of ubiquitin is important for signaling at steps both upstream and downstream of MAVS. Upstream of MAVS, K63-linked polyubiquitination of RIG-I by the RING domain ubiquitin ligase TRIM25 has been proposed as an important mechanism of RIG-I activation (Gack et al., 2007). Downstream of MAVS, the activation of TBK1 requires the E2 Ubc5 and K63 polyubiquitination of an unknown target (Zeng et al., 2009). Several additional ubiquitin ligases (e.g, Riplet/REUL, RNF125) have been shown to positively or negatively regulate the RIG-I pathway (Nakhaei et al., 2009). However, the biochemical mechanisms by which RIG-I and TBK1 are activated by ubiquitination remain poorly understood.
To dissect the signaling mechanisms in the RIG-I pathway, we establish a cell-free system that mimics viral infection in intact cells. This in vitro reconstitution system contains recombinant RIG-I protein, mitochondria, cytosol, RNA and ubiquitination enzymes including TRIM25. We demonstrate that 5′-pppRNA and dsRNA, which resemble viral RNA, can activate the entire RIG-I signaling cascade in vitro, leading to activation of IRF3. Interestingly, we find that upon incubation with both RNA and unanchored K63 polyubiquitin (polyUb) chains, RIG-I is fully activated to cause IRF3 dimerization. We show that the N-terminus of RIG-I, termed RIG-I(N), which contains tandem CARD domains, binds specifically to K63 polyUb chains, and that this binding is both necessary and sufficient to activate RIG-I(N). Furthermore, we devise a protocol to isolate endogenous ubiquitin chains associated with RIG-I in a human cell line, and demonstrate that these are unanchored K63 polyUb chains with potent ability to activate RIG-I. Our results support a model in which RIG-I binds to two ligands, viral RNA and endogenous ubiquitin chains, in a sequential manner through the C-terminal RD and N-terminal CARD domains, respectively. The binding of both ligands is required for full activation of RIG-I.
RESULTS
In vitro Reconstitution of a RIG-I Signaling Cascade from RNA to IRF3 Activation
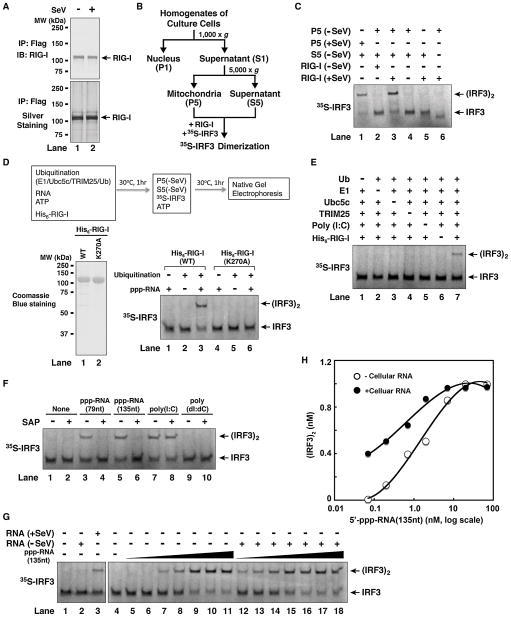
To reconstitute the RIG-I pathway in vitro, we first examined whether RIG-I isolated from virus infected cells could cause IRF3 activation in the presence of mitochondria, which contain MAVS, and cytosolic extracts, which contain TBK1 and many other components. A hallmark of IRF3 activation is its dimerization, which depends on its phosphorylation by TBK1 and can be measured by native gel electrophoresis (Yoneyama et al., 2002). To set up the assay, HEK293T cells stably expressing full-length RIG-I with a C-terminal Flag tag were infected with Sendai virus (SeV) or uninfected, then RIG-I was affinity purified (Figure 1A). Crude mitochondria (P5) and cytosolic extracts (S5) were prepared from uninfected HEK293T cells by differential centrifugation (Figure 1B), and 35S-labeled IRF3 protein was synthesized by in vitro translation. As shown in Figure 1C, dimerization of IRF3 was observed when RIG-I from virus-infected cells was incubated with mitochondria (P5) and cytosolic extracts (S5) in the presence of ATP (lane 3). In contrast, RIG-I from mock-treated cells did not promote IRF3 dimerization (lane 2). The activation of IRF3 required both cytosol and mitochondria (lanes 5 & 6). Mitochondria isolated from cells depleted of MAVS by RNAi could not support IRF3 dimerization (Supplementary Figure S1A), confirming that MAVS is essential for activating the downstream pathway in this in vitro assay. When RIG-I was isolated from cells depleted of TRIM25 by RNAi, its ability to promote IRF3 dimerization in the in vitro assay was greatly reduced (Supplementary Figure S1B), supporting an important role of TRIM25 in RIG-I activation. As we have shown recently, mitochondria isolated from virus-infected cells activated IRF3 in the absence of RIG-I (Figure 1C, lane 1) (Zeng et al., 2009).
Figure 1. In Vitro Reconstitution of the RIG-I Pathway and Regulation of RIG-I by RNA- and Ubiquitination.
(A) Purification of RIG-I protein from Sendai virus-infected (+SeV) or untreated (−SeV) HEK293T cells stably expressing RIG-I containing a C-terminal Flag epitope. (B) Procedures for isolation of crude mitochondria (P5) and cytosol (S5) by differential centrifugation. (C) Virus-activated RIG-I induced IRF3 dimerization in vitro. The reconstitution reaction contained mitochondria (P5), RIG-I isolated from virus-infected or untreated cells, cytosolic extracts (S5) from uninfected cells, 35S-IRF3 and ATP. Dimerization of IRF3 was analyzed by native gel electrophoresis. (D) In vitro activation of RIG-I by 5′-pppRNA and ubiquitination. His6-tagged RIG-I (wild-type or WT) or its ATPase mutant (K270A) was purified from Sf9 cells (lower left panel), then incubated with 5′-pppRNA (79 nucleotides), ATP, and ubiquitination enzymes as outlined in the diagram. After incubation, aliquots of the reaction mixtures were further incubated with mitochondria (P5) and cytosol (S5) from uninfected cells together with 35S-IRF3 and ATP, and then IRF3 dimerization was analyzed by native gel electrophoresis. (E) In vitro activation of RIG-I by poly(I:C) and ubiquitination. Similar to (D), except that poly(I:C) was used and the dependency on ubiquitin and ubiquitination enzymes was tested. (F) The role of 5′-triphosphate for RNA to activate RIG-I in vitro. Similar to (D) except that the RNA was pretreated with or without shrimp alkaline phosphatase (SAP). (G) 5′-pppRNA and viral RNA are potent activators of the RIG-I pathway. Total RNA was extracted from HEK293T cells from viral-infected or untreated HEK293T cells, then incubated with RIG-I as in (D), followed by IRF3 dimerization assay (lanes 1–3). To measure the potency of 5′-pppRNA in RIG-I activation, increasing amounts of the RNA (135nt) (0.07 to 70 nM, at 3-fold increment) were incubated with RIG-I in the presence (lanes 12–18) or absence of cellular RNA from uninfected cells (lanes 5–11), then IRF3 dimerization assay was performed. (H) IRF3 dimer shown in (G) was quantified with ImageQuant, then plotted against the concentration of 5′-pppRNA. See also Figure S1.
Transfection of cells with synthetic RNAs, such as the dsRNA analogue poly(I:C) and 5′-pppRNA, potently induces IRF3 dimerization. To test if RNA could activate the entire RIG-I pathway in vitro, we expressed and purified full-length RIG-I from insect cells (Sf9; Figure 1D, lower left) and incubated it with ATP and a 79-nucleotide (79nt) 5′-pppRNA (Supplementary Figure S1C). This RNA strongly induced IFNβ when transfected into HEK293-IFNβ-luciferse reporter cells (Supplementary Figure S1D). However, incubation of this RNA with the recombinant RIG-I protein did not cause IRF3 dimerization in the reconstituted system (Figure 1D, lane 1 in lower right panel). As ubiquitination has been shown to be important in the RIG-I pathway, we incubated RIG-I with E1, Ubc5c (E2), TRIM25 (E3), and ubiquitin together with ATP and RNA. Remarkably, this condition caused RIG-I to activate the entire pathway, resulting in IRF3 dimerization (Figure 1D, lane 3 in lower right panel). A K270A mutation in the ATPase domain of RIG-I, which abrogates the ability of RIG-I to induce interferons in vivo (Yoneyama et al., 2004), also abolished its ability to induce IRF3 dimerization in vitro. Poly(I:C) stimulated IRF3 dimerization in this reconstituted system in a manner dependent on ubiquitin, E1, Ubc5c, TRIM25 and RIG-I (Figure 1E). Activation of RIG-I was also observed with another 5′-pppRNA containing 135 nucleotides (135nt) (Figure 1F). Dephosphorylation of the 5′-pppRNAs with shrimp alkaline phosphatase (SAP) destroyed their ability to activate IRF3 in vitro (Figure 1F) and IFNβ induction in HEK293T cells (Supplementary Figure S1D). In contrast, treatment of poly(I:C) with SAP did not inhibit its activity, consistent with the previous report that this dsRNA analogue activates RIG-I and MDA5 in a manner independent of 5′-triphosphate (Kato et al., 2008). As expected, the DNA poly(dI:dC) did not activate the RIG-I pathway in vitro or in cells (Figure 1F & Supplementary Figure S1D). In vitro reconstitution of the RIG-I pathway also led to site-specific phosphorylation of IRF3 and IκBα (Supplemental Results and Figures S1E, F & G).
To determine if RIG-I could be activated by naturally occurring viral RNA, we incubated purified RIG-I protein with total RNA from HEK293T cells infected with Sendai virus. Indeed, RNA from virus-infected but not mock-treated cells stimulated RIG-I to activate IRF3 (Figure 1G, lanes 1–3). To estimate the sensitivity of 5′-pppRNA detection by RIG-I, we incubated RIG-I with different amounts of 5′-pppRNA in the presence or absence of HEK293T cellular RNA (Figure 1G, lanes 4–18). The half maximal effective concentration (EC50) of the 5′-pppRNA was estimated at 1.2 nM and 0.4 nM in the absence and presence of the cellular RNA, respectively (Figure 1H). It is not clear how the cellular RNA enhances the potency of 5′-pppRNA, but one possibility is that these RNAs reduce the non-specific loss of very small amounts of 5′-pppRNA in the reactions. In any case, the fact that the presence of a large excess of cellular RNA does not interfere with the specific recognition of 5′-pppRNA by RIG-I underscores the remarkable specificity of viral RNA detection by RIG-I. Assuming that the cytoplasm of a human cell has a volume of ~500 μm3, we estimated that less than 20 molecules of viral RNA in a cell (equivalent to ~0.07 nM) are sufficient to trigger detectable IRF3 dimerization. Thus, our in vitro reconstitution recapitulates the entire RIG-I pathway with exquisite sensitivity and specificity for 5′-pppRNA (see Discussion).
Ubc5 and Ubc13 are Required for the Activation of RIG-I and MAVS
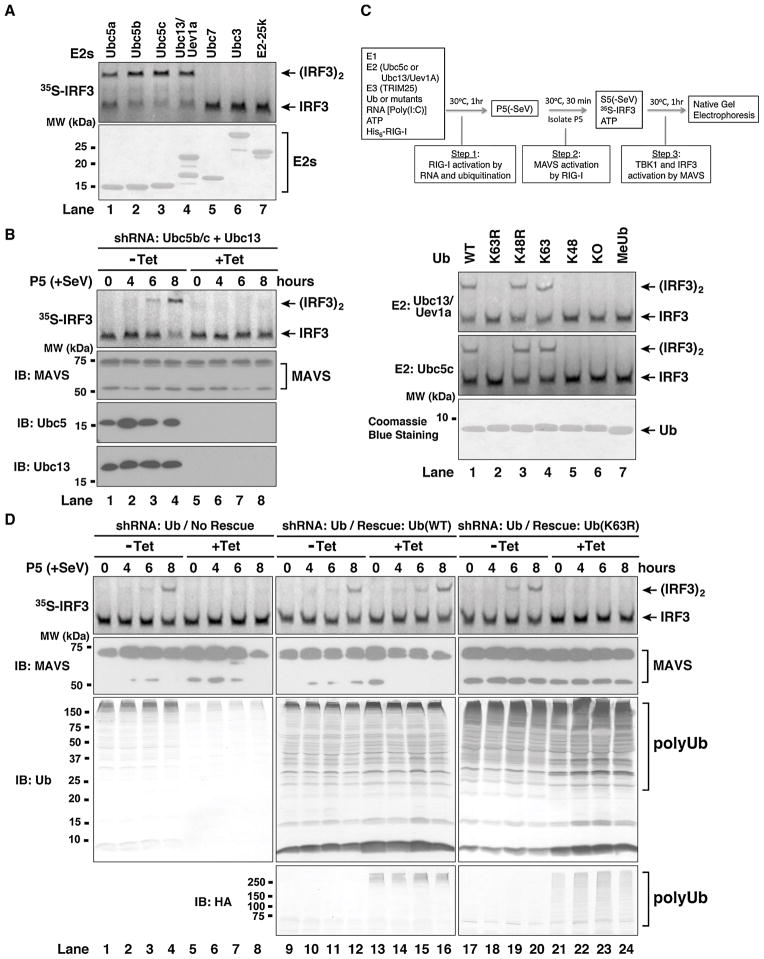
Ubc5 is a family of E2s comprising highly homologous isoforms (Ubc5a, b & c; putative Ubc5d in human) that catalyze the synthesis of polyUb chains linked through various lysines of ubiquitin, including K63 (Xu et al., 2009). In contrast to Ubc5, the E2 complex consisting of Ubc13 and Uev1A is highly specific in synthesizing K63 polyUb chains (Deng et al., 2000; Hofmann and Pickart, 1999). To determine the E2 involved in RIG-I activation, we examined a panel of E2s for their ability to stimulate RIG-I in the presence of TRIM25 and RNA (Figure 2A). The Ubc5 isoforms (Ubc5a, b & c) and Ubc13/Uev1A were capable of stimulating RIG-I to promote IRF3 dimerization. In contrast, Ubc3, Ubc7 and E2-25K had no activity (Figure 2A), despite the ability of these E2s to form thioesters with ubiquitin (data not shown; see also Zeng et al, 2009). To test if Ubc5 and/or Ubc13 are involved in the activation of MAVS by RIG-I, we established human osteosarcoma U2OS cell lines stably integrated with tetracycline-inducible shRNA vectors targeting both Ubc13 and two isoforms of Ubc5 (Ubc5b & c) (Xu et al., 2009). As shown in Figure 2B, the mitochondria from the cells depleted of Ubc13, Ubc5b and Ubc5c lost the ability to activate IRF3. These results suggest that Ubc5a and Ubc5d, which are not targeted by the Ubc5 shRNAs, play a minor role in IRF3 activation in U2OS cells. RNAi of Ubc13 or Ubc5 alone partially inhibited the IRF3-stimulatory activity of the mitochondria from virus-infected cells (Supplementary Figure S2A; Figure S3 in Zeng et al. 2009). These results suggest that both Ubc5 and Ubc13 are involved in the activation of MAVS in the mitochondria by RIG-I in response to viral infection.
Figure 2. K63 polyubiquitination is Essential for RIG-I Activation.
(A) Ubc5 and Ubc13/Uev1a activate RIG-I in vitro. RIG-I was incubated with E1, different E2s as indicated, TRIM25, ubiquitin, RNA and ATP, followed by IRF3 dimerization assay as described in Figure 1D. The E2 proteins (2 μg) were analyzed by Coomassie Blue staining (lower panel). (B) Ubc5 and Ubc13 are required for viral activation of MAVS in the mitochondria. U2OS cells stably integrated with tetracycline-inducible shRNA against Ubc5b/c and Ubc13 were treated with or without tetracycline (Tet). After viral infection for the indicated time, mitochondrial fraction (P5) was prepared and the MAVS activity was measured by IRF3 dimerization assay. (C) K63 of ubiquitin is essential for RIG-I activation in vitro. RIG-I and IRF3 activation assays were performed using Ubc5c (upper panel) or Ubc13/Uev1a (lower panel) as the E2, and various ubiquitin mutants as indicated. KO, lysine-less mutant; MeUb, methylated ubiquitin. (D) K63 polyubiquitination is essential for viral activation of MAVS in the mitochondria. U2OS cells stably integrated with tetracycline-inducible shRNA against endogenous ubiquitin genes and a rescue expression vector for wild-type or K63R mutant of ubiquitin were grown in the presence or absence of tetracycline. After infection by Sendai virus, mitochondria (P5) were prepared to measure MAVS activation. The expression of MAVS and ubiquitin was analyzed by immunoblotting (lower panels). The HA antibody detects HA-Ub(WT) or HA-Ub(K63R) expressed from the transgene rescue vector. See also Figure S2.
K63 Polyubiquitination is Essential for the Activation of RIG-I and MAVS
To determine if K63 of ubiquitin is required for RIG-I activation in the in vitro system, we incubated various ubiquitin lysine mutants with RIG-I in the presence of RNA, ATP, E1, TRIM25 and Ubc5c or Ubc13/Uev1A. The reaction mixture was then incubated with the mitochondrial fraction (P5) to activate MAVS. The activated mitochondria were isolated and then tested for their ability to stimulate IRF3 dimerization in the presence of cytosolic extracts (Figure 2C). The ubiquitin proteins containing a lysine at position 63 (wild-type, K48R and K63-only) were capable of activating RIG-I, whereas those containing a substitution at K63 (K63R, K48-only, KO and methylated ubiquitin) had no activity (Figure 2C; see Supplementary Figure S2C for an illustration of ubiquitin mutants). Interestingly, although Ubc5c and TRIM25 catalyze the synthesis of polyUb chains from K63R (Supplementary Figure S2D, lane 2), these chains did not stimulate RIG-I (Figure 2C, lane 2), indicating that K63 polyUb chain synthesis is specifically required for RIG-I activation in vitro.
To investigate the role of K63 polyubiquitination in the activation of MAVS in cells, we used recently developed U2OS cell lines in which endogenous ubiquitin is replaced with wild-type or K63R mutant of ubiquitin through a tetracycline-inducible strategy (Xu et al., 2009). As shown in Figure 2D, depletion of endogenous ubiquitin severely impaired the ability of the mitochondria to promote IRF3 dimerization, but this activity was rescued by the expression of the wild-type ubiquitin transgene. In contrast, when the endogenous ubiquitin was replaced with the K63R mutant, the mitochondria isolated from these cells failed to activate IRF3 in the vitro assay, strongly suggesting that K63 polyubiquitination is essential for viral activation of MAVS in the mitochondria.
K63 Polyubiquitin Chains Activate RIG-I through its N-terminal CARD Domains
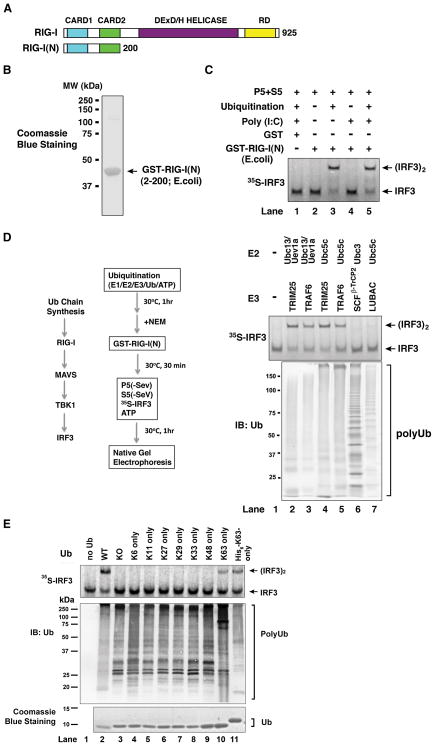
The N-terminus of RIG-I contains tandem CARD domains, which, when overexpressed in mammalian cells, constitutively activate IRF3 (Yoneyama et al., 2004). To determine if this N-terminal fragment [RIG-I(N), see Figure 3A] could activate IRF3 in vitro, we expressed the protein in E. coli and purified it to near homogeneity (Figure 3B). When the protein was incubated with mitochondria and cytosolic extracts, it did not promote IRF3 dimerization (Figure 3C, lane 2). However, when the incubation mixtures contained ubiquitination components, including E1, Ubc5c, TRIM25 and ubiquitin, robust IRF3 dimerization was detected.
Figure 3. Activation of RIG-I N-terminus by K63 Polyubiquitin Chains.
(A) Depiction of RIG-I functional domains. (B) Purification of recombinant GST-RIG-I(N) protein from E. coli. (C) Activation of RIG-I(N) by ubiquitination. GST or GST-RIG-I(N) was incubated with ubiquitination reaction (E1, Ubc5, TRIM25 and ubiquitin) and/or poly(I:C) as indicated, then an aliquot of the reaction was further incubated with mitochondria (P5) and cytosol (S5) to measure IRF3 dimerization. (D) K63 polyUb chain synthesis, but not ubiquitination of RIG-I(N), is required for RIG-I activation in vitro. Ubiquitination reactions containing different combinations of E2s and E3s as indicated were carried out before N-ethylmaleimide (NEM) was added to inactivate E1 and E2s. An aliquot of the reaction mixture was incubated with GST-RIG-I(N), then further incubated with mitochondria (P5) and cytosol (S5) to measure IRF3 dimerization. Another aliquot of the reaction was analyzed by immunoblotting with a ubiquitin antibody (lower panel). SCFβ-TrCP2: an E3 complex containing Skp1, Cul1, Rbx1 and β-TrCP2; LUBAC: an E3 complex containing HOIL-1L and HOIP. (E) Similar to D, except that ubiquitination reactions were carried out in the presence of Ubc5c, TRIM25 and various ubiquitin mutants as indicated. See also Figure S3.
To determine if ubiquitination of RIG-I(N) is required for IRF3 activation in the in vitro system, we carried out a ubiquitination reaction in the presence of different pairs of E2 and E3, and then treated the reaction mixture with the chemical N-ethylmaleimide (NEM), which alkylates the active site cysteine of E1 and E2. The reaction mixtures were then incubated with GST-RIG-I(N) before further incubation with the mitochondrial and cytosolic fractions to measure IRF3 dimerization (Figure 3D). Under these conditions, RIG-I(N) was not ubiquitinated because E1 and E2 had been inactivated by NEM. Remarkably, when TRIM25 or another RING domain E3 TRAF6 was incubated with either Ubc13/Uev1A or Ubc5c, robust IRF3 dimerization was detected (lanes 2–5). When K48 polyUb chains were synthesized by Ubc3 and an E3 complex consisting of Skp1, Cul-1, Roc1 and βTrCP2 (SCF-βTrCP2), these chains did not activate the RIG-I pathway. Similarly, linear polyUb chains generated by Ubc5c and an E3 complex consisting of HOIP-1 and HOIL-1L (termed LUBAC) led to very weak IRF3 dimerization. To further test if ubiquitin chains of other linkages could support IRF3 dimerization, we used a panel of ubiquitin mutants harboring a single lysine (Figure 3E). Although all the ubiquitin mutants were capable of forming polyUb chains in the presence of Ubc5c and TRIM25, the only mutants capable of supporting IRF3 activation were those containing a lysine at position 63 (K63-only and His6-K63 only). Collectively, these results clearly demonstrate that polyUb chains containing the K63 linkage, but not other linkages, specifically activate the RIG-I pathway. We also found that unanchored polyUb chains, but not ubiquitinated TRIM25, mediate RIG-I activation (Supplementary Results and Figure S3).
Short, Unanchored, K63-Linked Ubiquitin Chains Activate RIG-I
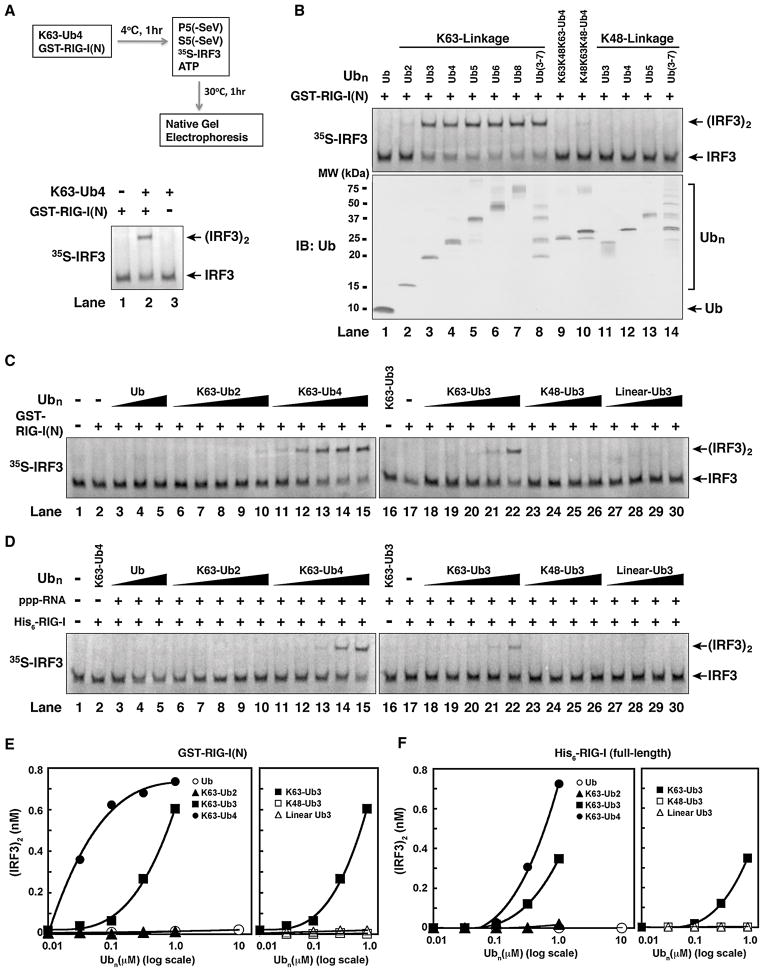
To determine if short ubiquitin chains are capable of activating RIG-I, we incubated GST-RIG-I(N) with a ubiquitin polymer containing four units of ubiquitin linked through K63 (K63-Ub; Figure 4A). Strikingly, incubation of GST-RIG-I(N) with K63-Ub4 led to robust activation of IRF3 in this assay (lane 2), whereas GST-RIG-I(N) or K63-Ub4 alone had no activity (lanes 1 & 3). We then tested a panel of ubiquitin chains of different lengths and linkages for their ability to activate RIG-I(N) (Figure 4B). Interestingly, K63 Ub chains containing more than two ubiquitin moieties potently activated RIG-I(N) (Figure 4B, lanes 3–8). K63-Ub2 had very weak activity (lane 2), whereas monomeric ubiquitin and K48-linked ubiquitin chains were inactive (lane 1; lanes 11–14). Ub4 containing mixed linkages of K48 and K63 also had greatly reduced activity (lanes 9–10). We then carried out titration experiments to quantify the relative potency of different ubiquitin chains in the activation of RIG-I(N) (Figure 4C and 4E). Similar titration experiments were also carried out using full-length RIG-I in the presence of 5′-pppRNA (Figure 4D and 4F). Among the ubiquitin chains tested, K63-Ub4 was the most potent activator, with an EC50 of ~25 nM for RIG-I(N) activation. The potency of K63-Ub3 was about 27 fold less (EC50 ≈ 680 nM), whereas K63-Ub2 was nearly inactive even at the highest concentration (1 μM). Ubiquitin, K48-Ub3 and linear-Ub3 were inactive at all the concentrations tested. These results demonstrate that short, unanchored K63 ubiquitin chains containing at least three ubiquitin moieties are potent and specific activators of RIG-I.
Figure 4. Short, Unanchored, K63-Ubiquitin Chains Potently Activate RIG-I.
(A) K63-Ub4 activates RIG-I(N). GST-RIG-I(N) (0.2 μM) was incubated with or without K63-Ub4 (0.3 μM), followed by IRF3 dimerization assay as outlined. (B) K63, but not K48, ubiquitin chains activate RIG-I(N). Ubiquitin chains of defined lengths and linkages were incubated with GST-RIG-I(N), then IRF3 dimerization assay was carried out as in (A). The quality of the ubiquitin chains was evaluated by immunoblotting (lower panel) or silver staining (see Figure S4). (C) K63 ubiquitin chains potently activate RIG-I(N) in a chain length- and linkage-dependent manner. Different concentrations of Ub chains (0.01–1 μM) or ubiquitin (0.1–10 μM) were tested for RIG-I(N) activation using the IRF3 dimerization assay. (D) Similar to (C), except that full-length His6-RIG-I and 5′-pppRNA were used in lieu of RIG-I(N) in the reactions. (E & F) IRF3 dimer in C & D, respectively, was quantified using ImageQuant, then plotted against the concentration of Ub or Ub chains.
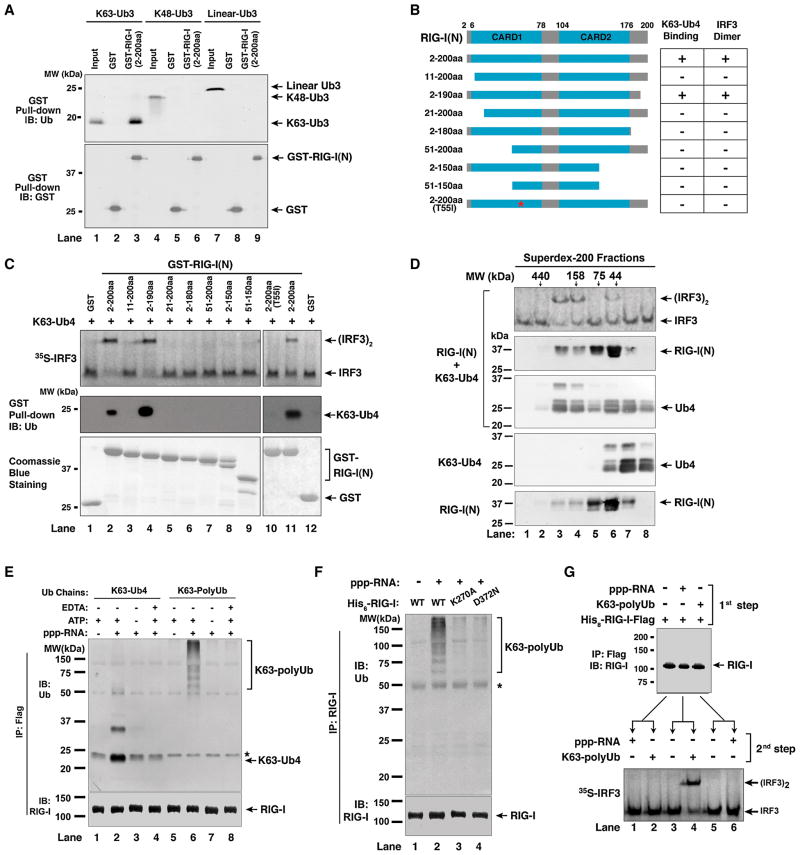
The Tandem CARD Domains of RIG-I Bind to K63 Ubiquitin Chains
The potent activation of RIG-I(N) by K63 ubiquitin chains implies that the N-terminus of RIG-I binds to these chains. Indeed, GST-RIG-I(N) was able to pull down K63-Ub3, but not K48-Ub3 or linear Ub3 (Figure 5A). Both CARD domains are required for ubiquitin binding, because the fragments containing residues 11–200 or 2–180 of RIG-I failed to bind K63-Ub4 or activate IRF3 (Figure 5B & 5C; see also Supplementary Figure S5A & S5B). We also generated a RIG-I(N) protein containing a T55I mutation, which was previously shown to render RIG-I defective in interferon induction (Sumpter et al., 2005). Interestingly, T55I mutation severely impaired the ability of RIG-I(N) to bind K63-Ub4 and activate IRF3 (Figure 5C, lane 10), suggesting that the signaling defect of this mutant may be due to its failure to bind K63 ubiquitin chains.
Figure 5. RIG-I CARD Domains Bind K63 Ubiquitin Chains and This Binding in Full-Length RIG-I is Regulated by RNA and ATP.
(A) RIG-I(N) binds specifically to K63 ubiquitin chains. GST or GST-RIG-I(N) was incubated with Ub3 containing K63, K48, or linear linkage, then pulled down with glutathione-Sepharose, followed by immunoblotting. Input represents 10% of Ub3 used in the pull-down experiments. (B) Diagram of RIG-I N-terminus containing the tandem CARD domains and various deletion and point mutants. The table on the right summarizes the results in panel C. (C) Both CARD domains of RIG-I are required for polyUb binding and IRF3 activation. GST-RIG-I(N) and various mutants were incubated with K63-Ub4. 1-μl aliquot of each mixture was used for IRF3 dimerization assay (upper panel), and the remainder was pulled down with glutathione-Sepharose followed by immunoblotting with a Ub antibody (middle panel). The GST-RIG-I(N) and the mutant proteins (2 μg each) were analyzed by Coomassie Blue staining (lower panel). See also Figure S5. (D) RIG-I(N) and K63-Ub4 form active high molecular weight complex. RIG-I(N) was incubated with K63-Ub4 and then the mixture was fractionated on Superdex-200. Aliquots of the fractions were assayed for their ability to stimulate IRF3 dimerization, whereas other aliquots were subjected to immunoblotting with antibodies against RIG-I and ubiquitin, respectively. K63-Ub4 or RIG-I(N) alone was also analyzed by gel filtration on the same column (lower two panels). (E) Full-length RIG-I binds to ubiquitin chains in a manner that depends on 5′-pppRNA and ATP. His8-RIG-I-Flag was incubated with K63-Ub4 or K63-polyUb chains in the presence or absence of 5′-pppRNA (135nt), ATP or EDTA. Following immunoprecipitation with a Flag antibody, the precipitated proteins were detected with an antibody against Ub or RIG-I. The asterisk indicates a non-specific band. (F) Similar to (E), except that RIG-I ATPase mutants (K270A and D372N) were also tested for binding to K63 polyUb chains. The asterisk indicates a non-specific band. (G) Sequential binding of RIG-I to RNA and polyUb leads to IRF3 activation. His8-RIG-I-Flag was incubated with 5′-pppRNA or K63-polyUb in the first step, then immunopurified using a Flag antibody. The purified RIG-I was incubated with K63-polyUb or 5′-pppRNA in the second step, followed by IRF3 dimerization assay.
To determine if ubiquitin chain binding induces oligomerization of RIG-I(N), we incubated RIG-I(N) with K63-Ub4, and then performed gel filtration analysis using Superdex-200. K63-Ub4 and RIG-I(N) have predicted molecular masses of ~30 and ~24 kDa, respectively; however, they formed an active complex with an apparent molecular mass of approximately 200 kDa (Fig. 5D). When each protein alone was fractionated on the same column, K63-Ub4 and RIG-I(N) eluted at positions corresponding to approximately 25 and 50 kDa, respectively. The elution profile of RIG-I(N) suggests that it forms a dimer, and the binding of K63-Ub4 to RIG-I(N) apparently promotes the formation of an oligomeric complex (e.g, a tetramer).
Full-length RIG-I Binds to K63 Ubiquitin Chains in a Manner that Depends on RNA and ATP
As full-length RIG-I is regulated by RNA binding and ATP hydrolysis, we tested its binding to K63 ubiquitin chains in the presence or absence of RNA and ATP. Immunoprecipitation of His8-RIG-I-Flag led to co-precipitation of K63-Ub4 and long K63 polyUb chains in the presence of 5′-pppRNA and ATP (Figure 5E, lanes 2 & 6). This binding was lost when the RNA was absent or when EDTA was added to chelate Mg2+, which is required for ATP binding. Two different mutations that abrogate the ATPase activity of RIG-I, K270A and D372N, also disrupted the ability of RIG-I to bind K63 polyUb (Figure 5F). To determine if RIG-I binds to RNA and K63 polyUb in a sequential manner, we incubated RIG-I with 5′-pppRNA or K63 polyUb in reciprocal orders (Figure 5G). If RIG-I was pre-incubated with 5′-pppRNA in the first step and then with K63 polyUb in the second step, it was capable of activating IRF3 dimerization in the reconstitution assay (lane 4). In contrast, if RIG-I was pre-incubated with K63 polyUb first, then immunoprecipitated to remove unbound polyUb, its subsequent incubation with 5′-pppRNA failed to activate it (lane 6). Thus, the binding of RIG-I to RNA precedes its binding to K63 polyUb.
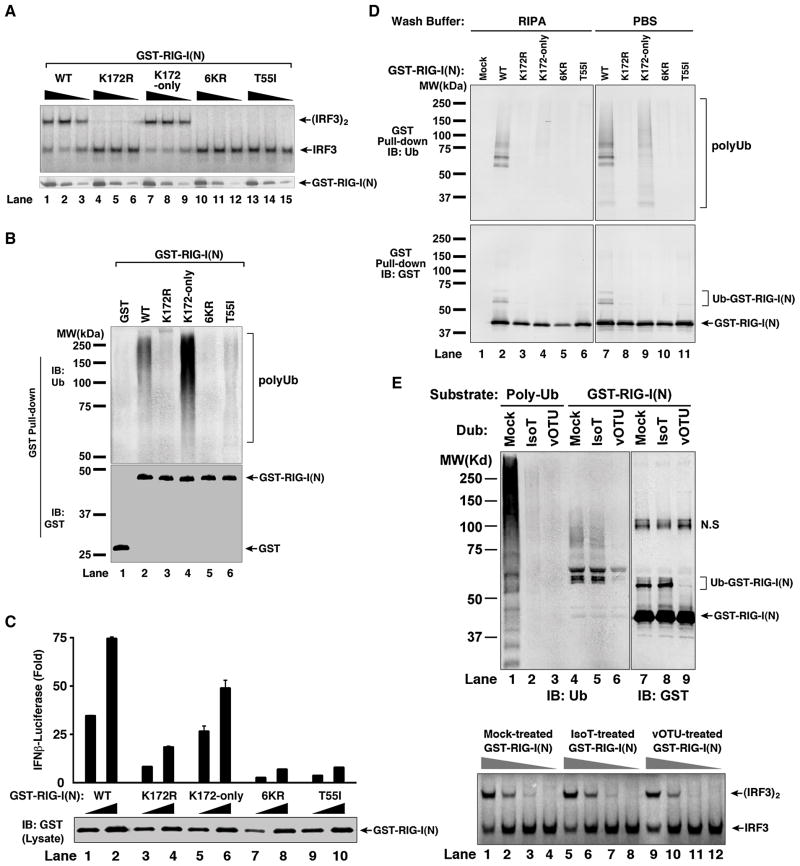
Binding of K63 Ubiquitin Chains is Necessary for RIG-I Activation
Previous studies have shown that RIG-I is ubiquitinated at K172 and that a mutation of this residue (K172R) impairs its ability to induce IFNβ (Gack et al., 2007). Consistent with an important role of K172 in RIG-I activation, we found that GST-RIG-I(N) containing the K172R mutation failed to activate IRF3 in the in vitro reconstitution assay (Figure 6A, lanes 4–6). A GST-RIG-I(N) protein containing mutations at six lysines (K99, 169, 172, 181, 190, 193; herein referred to as 6KR) was also inactive (lanes 10–12), but this activity was rescued by keeping the lysine at position 172 (K172-only; lanes 7–9). Interestingly, the K172R, 6KR and T55I mutants were greatly compromised in their ability to bind K63 polyUb, whereas K172-only was fully capable of binding to these chains (Figure 6B). These results show that the IRF3-activating function of RIG-I correlates well with its ubiquitin binding activity.
Figure 6. Polyubiquitin Binding is Required for RIG-I Activation.
(A) Mutations at K172 and T55 impair RIG-I(N) activation in vitro. GST-RIG-I(N) and the indicated mutants were expressed and purified from E. coli, then incubated with K63-Ub4, followed by IRF3 dimerization assay (upper panel). Aliquots of the reaction mixtures were analyzed by immunoblotting with a GST antibody (lower panel). 6KR: GST-RIG-I(N) containing six K>R mutations, including K172R. K172-only: similar to 6KR except that K172 is not mutated. (B) Mutations at K172 and T55 impair polyUb binding by RIG-I(N) in vitro. GST-RIG-I(N) and mutant proteins were incubated with K63 polyUb chains, then pulled down with glutathione-Sepharose and analyzed by immunoblotting. (C) Mutations at K172 and T55 impair RIG-I(N)’s ability to induce IFN-β in cells. Different amounts (30 ng and 100 ng) of mammalian expression vectors for GST-RIG-I(N) and mutants were transfected into HEK293-IFNβ-Luc cells, then luciferase activity was measured. Error bars represent the variation range of duplicate experiments. (D) GST-RIG-I(N) and mutant proteins as described in (C) were pulled down with glutathione-Sepharose beads, washed with PBS or RIPA buffer, then analyzed by immunoblotting. (E) GST-RIG-I(N) was expressed in HEK293T cells, then purified using glutathione-Sepharose. The purified protein was treated with isopeptidase T (IsoT) or viral OTU (vOTU) or mock-treated, then immunoblotted with an antibody against ubiquitin or GST (upper panel; N.S indicates a non-specific band). Aliquots of the treated GST-RIG-I(N) were tested for its ability to promote IRF3 dimerization in the reconstitution assay (lower panel). See also Figure S6.
To determine if ubiquitination or ubiquitin-binding of RIG-I is important for its activation in cells, we expressed GST-RIG-I(N) and various mutants in HEK293-IFNβ-luciferase reporter cells (Figure 6C), then pulled down these proteins with glutathione-Sepharose (Figure 6D). To distinguish ubiquitin chains covalently conjugated to RIG-I from those non-covalently associated with RIG-I, the Sepharose beads were washed with either phosphate-buffered saline (PBS) or a more stringent buffer containing 0.1% SDS and 1% deoxycholate (RIPA; Figure 6D). Immunoblotting with a GST antibody showed that while the wild-type GST-RIG-I(N) was conjugated by ubiquitin chains, no apparent ubiquitination of the mutants, including K172R and K172-only, was detected (Figure 6D, lower panel). Immunoblotting of the pull-down proteins with a ubiquitin antibody showed an interesting difference depending on the wash buffers (upper panel). While GST-RIG-I(N) containing K172-only was able to pull down polyUb chains from the cells when the beads were washed with PBS (lane 9), these chains were largely washed away with the RIPA buffer (lane 4), indicating that this mutant was defective in ubiquitination, but retained the ability to bind polyUb chains. Because GST-RIG-I(N)-K172-only was active in inducing IFNβ (Figure 6C), these results suggest that polyUb binding by RIG-I is responsible for its function. The other RIG-I mutants, including K172R, 6KR and T55I, were unable to bind ubiquitin chains in cells, consistent with their inability to induce IFNβ.
To further evaluate whether RIG-I ubiquitination is required for its function, we treated GST-RIG-I(N) isolated from HEK293T cells with a deubiquitination enzyme (DUB), which contains the OTU domain of the Crimean Congo hemorrhagic fever virus (CCHFV) large (L) protein (Figure 6E) (Frias-Staheli et al., 2007; Xia et al., 2009). The viral OTU (vOTU) can cleave polyUb chains from target proteins as well as unanchored polyUb chains. In contrast, another DUB, isopeptidase T (IsoT) only cleaves unanchored polyUb chains (Reyes-Turcu et al., 2006). Following treatment with vOTU or IsoT, GST-RIG-I(N) was tested for its ability to activate IRF3 in the in vitro reconstitution assay. Unlike GST-RIG-I(N) expressed in E. coli, which does not have the ubiquitin system, the same protein expressed in HEK293T cells was able to activate IRF3 without preincubation with ubiquitin chains (Figure 6E, bottom panel), probably because RIG-I(N) isolated from human cells is already bound to endogenous polyUb chains (e.g., lane 9 in Figure 6D). Interestingly, although vOTU removed most of the ubiquitin chains from GST-RIG-I(N) (Figure 6E, lanes 6 & 9 in upper panel), the deubiquitinated RIG-I protein was as active in causing IRF3 dimerization as the protein that was mock treated (lanes 1–4 and 9–12, lower panel). These results suggest that ubiquitination of the RIG-I CARD domains may be dispensable for its activity.
Unanchored K63 Polyubiquitin Chains Isolated from Human Cells are Potent Activators of RIG-I
In Figure 6E, we noted that IsoT treatment did not remove much of the polyUb chains from GST-RIG-I(N) (lane 5 in upper panel), indicating that RIG-I(N) protects the ubiquitin chains from degradation by IsoT (see Supplemental Results and Figure S6). The protection of polyUb by RIG-I(N) offers an opportunity to detect the otherwise labile ubiquitin chains in cells, but poses another challenge of how to release these chains while preserving their functions. We devised a protocol to isolate the RIG-I-bound ubiquitin chains from HEK293T cells by employing two strategies (Figure 7A). First, we lysed the cells in the presence of NEM to inactivate most of the deubiquitination enzymes. Ubiquitin contains no cysteine, therefore evading modification by NEM. Second, as ubiquitin is known to be relatively stable at high temperatures, we heated the RIG-I(N):polyUb complex at different temperatures to identify a condition that denatures RIG-I(N) but preserves the structure and function of ubiquitin chains. Following centrifugation that precipitates denatured protein aggregates, the supernatant is expected to contain polyUb chains and can be tested for its ability to activate IRF3 in the presence of fresh RIG-I(N). Because GST-RIG-I(N)-K172-only appears to capture more polyUb chains, we expressed this protein in HEK293T cells (Figure 7B). In control experiments, we incubated GST-RIG-I(N)-K172-only protein with K63 polyUb chains in vitro, carried out GST pull-down and then heated the complex (lanes 1–6). Five minutes of heating at 70–80°C was sufficient to inactivate the RIG-I protein (lane 1, 3, 5) and release polyUb chains into the supernatant, which were capable of activating IRF3 when supplied with fresh GST-RIG-I(N) (lane 2, 4, 6). When the GST-RIG-I(N) protein expressed in HEK293T cells was heated at 70°C, it was still capable of activating IRF3 in the absence of fresh RIG-I(N) (lane 7), indicating that the activated RIG-I(N) complex was resistant to NEM and 70°C treatment. However, raising the temperatures to 75°C and 80°C inactivated the GST-RIG-I(N) protein (lanes 9 & 11). Strikingly, the supernatant containing polyUb chains released from the RIG-I(N) complex at the high temperatures were capable of activating IRF3 when supplied with fresh GST-RIG-I(N) (lanes 10 & 12).
Figure 7. Endogenous Unanchored K63 Polyubiquitin Chains Activate the RIG-I Pathway.
(A) A protocol for isolating functional endogenous polyUb chains in human cells. (B) Endogenous polyUb chains bound to RIG-I can be released by heat treatment and remained functional. GST-RIG-I(N)-K172-only was expressed in HEK293T cells and isolated as described in (A), then heated for 5 minutes at the indicated temperatures. After centrifugation, the supernatant containing heat-resistant ubiquitin chains was incubated with GST-RIG-I(N) expressed and purified from E. coli (lanes 7–12). The activity of GST-RIG-I(N) was then measured by IRF3 dimerization assay. As positive controls, K63 polyUb chains were incubated with GST-RIG-I(N)-K172-only, which was then pulled down and heated in parallel experiments (lanes 1–6). endo. polyUb: endogenous polyUb. (C) Endogenous unanchored polyUb chains activate RIG-I(N). PolyUb chains associated with GST-RIG-I(N)-K172-only were captured and released at 75°C as in (B). The heat-resistant supernatant was incubated with GST-RIG-I(N) followed by IsoT treatment (lane 9), or in reverse order (lane 8). As positive controls, unanchored K63 polyUb chains were incubated with GST-RIG-I(N) and IsoT in sequential orders as indicated. In the right panel, the heat supernatant containing endogenous polyUb from HEK293T cells was incubated with or without IsoT, then analyzed by immunoblotting with a ubiquitin antibody. The arrow denotes a ~40 kDa band that is likely K63-Ub6 (see Figure S7B). (D) Similar to (C), except that the supernatant containing endogenous polyUb chains were treated with CYLD. The ubiquitin chains were detected with a ubiquitin antibody or another antibody specific for the K63 linkage of ubiquitin chains. (E) siRNA oligos targeting GFP (control), TRIM25 or CYLD were transfected into HEK293T cells, which were subsequently transfected with an expression vector encoding GST-RIG-I(N)-K172-only. Endogenous polyUb chains associated with the GST-RIG-I(N) protein were isolated as described in (A), then tested in IRF3 dimerization assay and visualized by immunoblotting with a ubiquitin antibody. The efficiency of RNAi was also confirmed by immunoblotting. (F) Potent activation of RIG-I by endogenous polyUb chains. Different amounts of heat-resistant supernatant containing endogenous polyUb were incubated with GST-RIG-I(N) to measure IRF3 dimerization. The concentration of the ubiquitin chains was estimated by semi-quantitative immunoblotting (Figure S7B). Error bars represent the variation range of duplicate experiments. (G) A proposed mechanism of RIG-I activation by RNA and polyUb (see Results and Discussion).
To determine whether the supernatant of the heated GST-RIG-I(N) complex contained unanchored polyUb chains and whether these chains were responsible for activating the RIG-I pathway, we performed two sets of experiments. First, we incubated the supernatant with E1 to determine if the endogenous ubiquitin chains could form thioesters with E1. Indeed, in the presence of E1 and ATP, substantial fractions of both synthetic and endogenous ubiquitin chains formed thioesters that were sensitive to reduction by β-mercaptoethanol, indicating that they contained unanchored C-termini (Supplementary Figure S7A). Second, we incubated the endogenous ubiquitin chains with IsoT and then measured their activity in the IRF3 dimerization assay (Figure 7C). Importantly, the IsoT treatment completely abolished the ability of the supernatant to activate IRF3 in the presence of GST-RIG-I(N) (lane 8). However, if we reversed the order by incubating the supernatant with GST-RIG-I(N) first and then with IsoT, IRF3 dimerization was observed (lane 9). Parallel control experiments show that unanchored K63 polyUb chains were sensitive to IsoT treatment but became resistant after pre-incubation with GST-RIG-I(N) (lanes 2–5). To directly visualize unanchored polyUb chains associated with the GST-RIG-I(N) complex isolated from HEK293T cells, the heated supernatants treated with and without IsoT were analyzed by immunoblotting with a ubiquitin antibody (Figure 7C, right panel). Several bands in the range of 40–75 kDa, most prominently the 40 kDa band, disappeared in the IsoT treated sample, indicating that these are unanchored ubiquitin polymers containing approximately 5–10 ubiquitin moieties. However, most of the high molecular weight bands remained resistant to IsoT treatment, suggesting that they represent ubiquitin chains conjugated to some protein targets. Because the IsoT treatment completely abolished the activity of the heat supernatant to promote IRF3 dimerization (Figure 7C, lane 8 on left panel), the high molecular weight ubiquitin conjugates remaining after IsoT treatment apparently did not contribute to RIG-I activation.
To determine if the endogenous unanchored polyUb chains are linked through K63 of ubiquitin, we treated the heat-resistant supernatant with the K63-specific deubiquitination enzyme CYLD. This treatment markedly diminished the ability of the supernatant to activate RIG-I (Figure 7D, left panel). CYLD also destroyed the ubiquitin chains with molecular masses in the range of 40–75 kDa, which could be detected with an antibody specific for the K63 ubiquitin linkage (Figure 7D, right panel).
TRIM25 and CYLD have previously been shown to be important for the activation and inactivation of the RIG-I pathway, respectively (Friedman et al., 2008; Gack et al., 2007; Zhang et al., 2008). To determine if these enzymes are involved in the synthesis and disassembly of endogenous K63 ubiquitin chains, we used siRNA to knock down the expression of TRIM25 or CYLD in HEK293T cells, then isolated the endogenous ubiquitin chains as outlined in Figure 7A. Immunoblotting experiments showed that depletion of TRIM25 diminished, whereas depletion of CYLD enhanced, the production of endogenous polyUb chains, which in turn activated IRF3 (Figure 7E).
To evaluate the potency of endogenous polyUb chains in activating RIG-I, we carried out titration experiments using different amounts of heat-resistant supernatants isolated from HEK293T cells (Figure 7F). The amount of endogenous K63-Ub6 was estimated by comparing to synthetic K63-Ub6 using semi-quantitative immunoblotting (Supplementary Figure S7B). Because K63-Ub6 is the dominant species of endogenous polyUb chains, its concentration was used to calculate the EC50 for stimulating RIG-I. Remarkably, the EC50 for the endogenous ubiquitin chains was estimated to be approximately 50 picomolar (pM), indicating that these ubiquitin chains are highly potent ligands of RIG-I.
Finally, we examined whether viral infection induces the formation of polyUb chains associated with full-length RIG-I in cells (Supplementary Figure S7C). HEK293T cells expressing RIG-I-Flag were infected with Sendai virus or mock treated, and then the RIG-I complex was immunoprecipitated and heated at 76°C to release polyUb chains. The heat supernatant prepared from the virus-infected cells, but not mock-treated cells, stimulated IRF3 dimerization in the presence of RIG-I(N). This activity was diminished by IsoT treatment (Supplementary Figure S7D), indicating that the supernatant contained unanchored polyUb chains. It should be noted that the activity associated with full-length RIG-I was much lower than that associated with RIG-I(N), because the activation of full-length RIG-I is limited by its accessibility to viral RNA (see Discussion). Nevertheless, given the potency of 5′-ppp-RNA and unanchored polyUb chains in activating RIG-I, small amounts of viral RNA and endogenous ubiquitin chains are likely to be sufficient to trigger the RIG-I signaling cascade.
In sum, the results presented herein demonstrate that: a) the unanchored K63 ubiquitin chains are present in human cells; b) they are bound and protected by RIG-I; c) they are potent activators of the RIG-I pathway. We propose that sequential binding of RIG-I to RNA and unanchored K63 polyUb chains leads to full activation of RIG-I, which in turn activates MAVS and downstream signaling cascades (Figure 7G).
DISCUSSION
In this report, we demonstrate that RNA with characteristics of viral RNA (5′-pppRNA and dsRNA) can activate the RIG-I pathway in vitro in a reconstitution system with exquisite specificity and sensitivity. To our knowledge, this is the first in vitro reconstitution of an immune signaling cascade from a microbial ligand (viral RNA) to activation of a transcription factor (IRF3). Our success in reconstituting the RIG-I pathway in vitro is beneficiary of at least three previous discoveries that: a) RNAs containing 5′-triphosphate are direct functional ligands of RIG-I (Hornung et al., 2006; Pichlmair et al., 2006); b) mitochondria containing MAVS are essential for RIG-I signaling (Seth et al., 2005); c) ubiquitination is important for RIG-I activation (Gack et al., 2007). The in vitro reconstitution of the RIG-I pathway sets the stage for delineating the biochemical mechanism of RIG-I activation and subsequent steps of downstream signaling. Testimonial to the power of this in vitro system are the discoveries revealed in this study that the tandem CARD domains of RIG-I represent a new class of ubiquitin-binding domain with specificity for K63 polyUb chains and that unanchored K63 polyUb chains are potent intracellular signaling molecules that activate the RIG-I pathway.
Specificity and Sensitivity of Viral RNA Detection by RIG-I
A major determinant that distinguishes viral RNA from the host RNA in mammalian cells is the presence of 5′-triphosphate in viral RNA but not in host RNA (Fujita, 2009). 5′-pppRNA is generated in the course of RNA replication by viral RNA polymerases. However, RIG-I may have very limited access to viral 5′-pppRNA for the following reasons: 1) viral RNA replication normally occurs within a membrane compartment (e.g, vesicles) that is sequestered from the cytosolic immune sensors such as RIG-I; 2) some viruses such as influenza replicate in the nucleus rather than in the cytosol; 3) the majority of viral RNA is in complex with viral capsid proteins to form ribonucleoprotein particles (RNPs), and the 5′ end of viral RNA can be blocked by viral proteins; 4) like their mammalian counterparts, most viral mRNAs also contain 5′-cap or other 5′-modifications. Thus, RIG-I must be capable of detecting very tiny amounts of 5′-pppRNA that may be inadvertently or transiently exposed during the course of viral replication and intracellular trafficking. By reconstituting RNA-mediated activation of the RIG-I cascade in vitro, we demonstrate that RIG-I can detect a few 5′-pppRNA molecules (<20 in a cell) in the presence of abundant cellular RNA and trigger a signaling cascade leading to IRF3 activation. Thus, our reconstitution system recapitulates the specificity and sensitivity of viral RNA recognition by the RIG-I pathway in cells.
RIG-I CARD Domains Represent a New Type of Ubiquitin Sensor
CARD domains are protein interaction domains that play key roles in many cellular processes including inflammation and apoptosis (Park et al., 2007). Unexpectedly, we found that the tandem CARD domains of RIG-I bind specifically to K63-linked polyUb chains. We also found that the tandem CARD domains of MDA5, but not the CARD domains of NOD2 or MAVS, bind to K63 polyUb chains (Jiang, X., and Chen, Z., unpublished; Supplementary Figure S5C). Thus, a subset of CARD domains appears to have the specialized ability to bind K63 polyUb chains. Importantly, mutations that disrupt ubiquitin-binding of RIG-I also impair its ability to activate IRF3, indicating that polyUb binding plays a key role in RIG-I activation. It will be of significant interest to identify other CARD domains capable of ubiquitin binding and determine whether proteins harboring these CARD domains require ubiquitin binding for their functions.
Polyubiquitin Binding is Required for RIG-I Activation
An important role of RIG-I ubiquitination in its activation was suggested based on the observations that K172R mutation of RIG-I diminishes its ubiquitination and its ability to induce IFNβ and that the K172-only mutant can induce IFNβ (Gack et al., 2007). Further supporting this conclusion was the finding that the T55I mutation, which was known to abrogate IFNβ induction, also impairs RIG-I ubiquitination (Gack et al., 2008). However, closer examination of these mutants reveals that K172R and T55I mutations also impair the ability of RIG-I to bind K63 polyUb chains (Figure 6B). Moreover, the K172-only mutation reinstalls polyUb binding by RIG-I, but not ubiquitination of RIG-I (Figure 6D), implying that polyUb binding but not ubiquitin modification is required for RIG-I’s function. In support of this notion, removal of ubiquitin chains from RIG-I with a DUB (vOTU) had virtually no effect on RIG-I’s activity (Figure 6E). Most importantly, we demonstrated that unanchored K63 polyUb chains exist in human cells and that these endogenous chains are potent ligands of RIG-I (Figure 7). Interestingly, although the RIG-I complex isolated from HEK293T cells contains both IsoT-sensitive unanchored ubiquitin chains and IsoT-resistant ubiquitin conjugates, the IsoT treatment completely abolished the ability of these ubiquitin chains to activate RIG-I (Figure 7C), strongly suggesting that unanchored polyUb chains are responsible for RIG-I activation. Although we cannot rule out the possibility that ubiquitination of some signaling proteins may contribute to RIG-I activation, our data show that ubiquitinated TRIM25 is incapable of activating RIG-I (Supplementary Figure S3).
Regulation of RIG-I by Endogenous Polyubiquitin Chains During Viral Infection
We estimated that approximately 10 ng of endogenous K63-Ub6 could be isolated from 20 million HEK293T cells using our protocol (Supplementary Figure S7B). This is equivalent to approximately 6000 molecules of K63-Ub6 per cell. These Ub chains activate RIG-I with an EC50 of approximately 50 pM, which is equivalent to ~15 molecules per cell. This estimate, although imprecise, suggests that there should be abundant endogenous Ub chains to activate RIG-I(N), explaining why overexpression of RIG-I(N) is sufficient to induce interferons in the absence of viral infection. However, in normal cells containing endogenous full-length RIG-I, which likely adopts an auto-inhibited conformation, the ubiquitin chains cannot gain access to the CARD domains of RIG-I and are rapidly disassembled by high levels of cellular DUB activity. Viral infection or introduction of 5′-pppRNA into the cytosol presumably induces a conformational change of RIG-I that exposes the CARD domains, which then capture and stabilize unanchored K63 polyUb chains synthesized by TRIM25. We speculate that local and transient accumulation of these ubiquitin chains in the vicinity of RNA-bound RIG-I allows the CARD domains to bind these chains, resulting in full activation of RIG-I. However, as discussed above, only a very small fraction of RIG-I is activated by viral infection due to limited access to viral RNA in the cytosol. Nevertheless, we obtained evidence that Sendai virus infection led to association of IsoT-sensitive ubiquitin chains with full-length RIG-I (Supplementary Figure S7C & S7D). Given the high potency of unanchored K63 ubiquitin chains in RIG-I activation, accumulation of just a few molecules of these chains in a cell are likely to activate endogenous RIG-I, provided that a few molecules of viral RNA are also present.
Sequential Binding to RNA and Unanchored K63 Polyubiquitin Chains Leads to Full Activation of RIG-I
Our study reveals unanchored K63 polyUb chains as the second ligand of RIG-I and demonstrates that full activation of RIG-I requires an orderly binding of viral RNA followed by polyUb chains. Based on available evidence, we propose a multi-step model of RIG-I activation (Figure 7G). After viral infection, RIG-I binds to viral 5′-pppRNA through its C-terminal RD domain and may bind to the double-stranded segments of the RNA through the central helicase domain (Fujita, 2009). The RNA binding causes dimerization of RIG-I and activates its ATPase activity (Cui et al., 2008). The energy derived from ATP hydrolysis propels the translocation of RIG-I along the viral RNA (Myong et al., 2009). The translocation of RIG-I on viral RNA may help displace viral proteins from the RNA, facilitating further RIG-I binding. The translocating RIG-I dimer likely has an ‘open’ conformation that exposes the N-terminal CARD domains, which form a cluster consisting of four CARD domains. This cluster of CARD domains binds to K63 polyUb chains. This binding may cause additional conformational changes and/or oligomerization of RIG-I, forming a new signaling platform to activate MAVS. The dependency of RIG-I activation on binding to unanchored K63 ubiquitin chains may provide another level of regulation of antiviral immune responses.
Unanchored K63 Polyubiquitin Chains are Intracellular Signaling Molecules
Our previous study has shown that unanchored polyUb chains directly activate the protein kinases TAK1 and IKK by binding to the essential regulatory subunits TAB2 and NEMO, respectively (Xia et al., 2009). We now show that unanchored polyUb chains bind and activate RIG-I, suggesting that these ubiquitin chains may have more general functions in cell signaling. However, unlike TAK1 and IKK activation, which requires very long polyUb chains, RIG-I can be activated by short K63 chains consisting of only 3 or 4 ubiquitin moieties. Indeed, K63-Ub4 and endogenous Ub chains are very potent activators of RIG-I(N) (Figure 4E and Figure 7F). The clusters of multiple CARD domains at the N-termini of RIG-I may bind to K63 ubiquitin chains with strong affinity and high selectivity. This is in contrast to most ubiquitin-binding domains, which bind to ubiquitin chains with weak affinity and low selectivity (Hurley et al., 2006). Our finding that short ubiquitin chains associate with and potently activate RIG-I in cells strongly suggests that these are endogenous ligands of RIG-I. To our knowledge, this is the first example of the biological functions of short unanchored ubiquitin chains, and it deviates from the well-established paradigm that ubiquitin functions through covalent conjugation of another protein. It will be of enormous interest to investigate whether unanchored polyUb chains regulate other physiological functions besides the RIG-I and NF-κB pathways.
EXPERIMENTAL PROCEDURES
Biochemical Assay for RIG-I Activation in vitro
For standard assay with full-length RIG-I, each μl mixture contained 10 ng His6-E1, 15 ng Ubc5c, 50 ng His8-TRIM25, 1 μg ubiquitin, indicated amounts of RNA [200 ng poly (I:C), 30 ng 5′-pppRNA (79nt) or 50 ng 5′-pppRNA (135nt)], and 50 to 100 ng His8-RIG-I-Flag in Buffer A [20 mM Tris-HCl (pH 7.5), 2 mM ATP, 5 mM MgCl2, and 0.5 mM DTT]. After incubation at 30°C for 1 hour, 1 μl of reaction mixture was directly used in IRF3 activation assay as described in Supplemental Experimental Procedures (see also Zeng et al., 2009). For RIG-I activation assays containing mutant ubiquitin, 1 μl of reaction mixture was mixed with 10 μg of mitochondrial fraction (P5) in 10 μl Buffer B [20 mM HEPES-KOH (pH 7.0), 2 mM ATP, 5 mM MgCl2, and 0.25 M D-mannitol] at 30°C for 30 minutes. P5 was then pelleted at 10,000 g for 10 minutes, and washed twice with Buffer C [20 mM HEPES-KOH at pH 7.4, 0.5 mM EGTA, 0.25 M D-mannitol, and EDTA-free protease inhibitor cocktail] before it was used for IRF3 activation assay.
For standard assay with RIG-I (N), each μl mixture contained 100 ng GST-RIG-I (N) and 50 to 100 ng ubiquitin chains in a buffer containing 20 mM HEPES-KOH (pH 7.0) and 10% (v/v) glycerol. After incubation at 4°C for 10 minutes or up to 1 hour, the mixture was directly used in IRF3 activation assay.
Ubiquitin-Binding Assay
For ubiquitin-binding by full-length RIG-I, His8-RIG-I-Flag (1 μg) or the ATPase mutant (K270A or D372N) was incubated with K63-Ub4 or K63-polyUb (1 μg), and 5′-pppRNA (0.3 μg for 135nt RNA) in 10 μl Buffer A at 4°C for 1 hour. For reactions lacking ATP, EDTA (10 mM) was added. The mixture was then diluted 5 fold in Buffer D [20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.5% (w/v) NP-40 and 10% (v/v) glycerol], and immunoprecipitated with anti-Flag or anti-RIG-I antibody. The bound proteins were eluted with SDS sample buffer and subjected to immunoblotting analysis with antibodies specific for ubiquitin or RIG-I.
For ubiquitin-binding by RIG-I(N), 2 μg of wild-type or mutant GST-RIG-I(N) was incubated with 1 μg ubiquitin chains in 10 μl buffer containing 20 mM HEPES-KOH (pH 7.0) and 10% (v/v) glycerol at 4°C for 1 hour. The mixture was then diluted 5 fold in Buffer D before the GST proteins were pulled down with glutathione-Sepharose. The bound proteins were analyzed by Immunoblotting.
To analyze RIG-I(N):K63-Ub4 complex, GST-RIG-I(N) was cleaved with Tev protease and then RIG-I(N) was purified on Q-Sepharose and Superdex-200 columns. 5 μg each of purified RIG-I(N) and K63-Ub4 were incubated at 4°C for 60 minutes, and then the mixture was separated on Superdex-200 PC (3.2/30) using the SMART purification system. The elution buffer contained 20 mM Tris-HCl, pH7.5, 0.1 mM EGTA, 0.5 mM DTT and 0.02% CHAPS. Aliquots of the fractions were analyzed in the in vitro IRF3 dimerization assay and also by immunoblotting using antibodies against RIG-I or ubiquitin.
Isolation and Functional Analysis of Endogenous Polyubiquitin Chains
HEK293T cells were transfected in 10-cm dishes with 5 μg of pEF-GST-RIG-I(N)-K172 only. The cells were lysed 48 hours later in Buffer E [20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 1% (w/v) TritonX-100, 20 mM β-glycerol phosphate, 1 mM Na3VO4, 0.1 mM NaF, 0.1 mM phenylmethanesulfonyl fluoride, and EDTA-free protease inhibitor cocktail] supplemented with 10 mM NEM. After centrifugation at 20,000 g for 15 minutes, 5 mM DTT was added to the supernatant to quench NEM, then the GST proteins were isolated with glutathione-Sepharose. The proteins on the beads were incubated in 30 μl Buffer F (20 mM HEPES-KOH at pH 7.0, 10% (v/v) glycerol, and 0.02% (w/v) CHAPS) at 70°C, 75°C or 80°C for 5 minutes. Following a brief centrifugation, the supernatant containing the dissociated polyUb chains was incubated with 100 ng of bacterially expressed GST-RIG-I(N) at 4°C for 1 hour, followed by IRF3 dimerization assay. For deubiquitination enzyme treatment, the heated supernatant was incubated with 10 ng/μl IsoT or CYLD at 30°C for 1 hour. The concentration of endogenous polyUb chains was estimated by semi-quantitative immunoblotting using synthetic K63-Ub6 (Boston Biochem) as the standard (Supplementary Figure S7B).
Supplementary Material
Acknowledgments
We thank Francesco Melandri and Dr. Kal Lapan (Boston Biochem) for generously providing some of the ubiquitin chains, and Dr. Jae Jung (University of Southern California) for the mammalian expression plasmids of GST-RIG-I(N), K172R, K172-only and 6KR mutants. We also thank Brian Skaug for reading the manuscript. This work was supported by grants from NIH (RO1-AI09919 and RO1-GM63692) and the Welch Foundation (I-1389). Z. J. C is an Investigator of Howard Hughes Medical Institute.
Footnotes
Supplemental Data include Supplemental Results, Experimental Procedures and seven figures and can be found with this article online at http://www.cell.com
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CS, O’Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008 doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T. A nonself RNA pattern: tri-p to panhandle. Immunity. 2009;31:4–5. doi: 10.1016/j.immuni.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA Is an Adapter Protein Required for Virus-Triggered IFN-beta Signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A Ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fujita T. Control of IRF-3 activation by phosphorylation. J Interferon Cytokine Res. 2002;22:73–76. doi: 10.1089/107999002753452674. [DOI] [PubMed] [Google Scholar]
- Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu X, Lee AJ, Jin W, Chang M, Wright A, Imaizumi T, Sun SC. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008;283:18621–18626. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.