SUMMARY
The yeast pheromone pathway consists of a canonical heterotrimeric G protein and MAP kinase cascade. To identify new signaling components we systematically evaluated 870 essential genes using a library of repressible-promoter strains. Quantitative transcription-reporter and MAPK activity assays were used to identify strains that exhibit altered pheromone sensitivity. Of the 92 newly identified essential genes required for proper G protein signaling, those involved with protein degradation were most highly-represented. Included in this group are members of the SCF (Skp-Cullin-F-Box) ubiquitin ligase complex. Further genetic and biochemical analysis reveals that SCFCdc4 acts together with the Cdc34 ubiquitin conjugating enzyme at the level of the G protein, promotes degradation of the G protein α subunit, Gpa1, in vivo and catalyzes Gpa1 ubiquitination in vitro. These new insights to the G protein signaling network reveal the essential-genome as an untapped resource for identifying new components and regulators of signal transduction pathways.
INTRODUCTION
The budding yeast Saccharomyces cerevisiae is an established model for investigating fundamental biological processes including cell division, cell growth, and intracellular communication. One unique attribute of the yeast system is the availability of several thousand isogenic gene-deletion strains, which allows for unbiased genome-scale analysis of cellular functions (Giaever et al., 2002). However, of the approximately 6,000 genes in the yeast genome, nearly 1,100 are essential for viability and difficult to study using standard gene-deletion mutants. This limitation has led to a poor understanding of a substantial fraction of the yeast genome (Mnaimneh et al., 2004). Notably, these essential genes are more likely to have a human ortholog, as compared with non-essential genes (38% vs. 20%) (Hughes, 2002). Here we describe the identification of new components and new regulators of the G protein signaling apparatus. Our approach was to conduct a systematic analysis of the “essential genome”, identify components required for efficient signal transduction, and establish their mode of action.
In yeast, a canonical heterotrimeric G protein signaling pathway regulates the process of cell mating. Yeast exists as one of two haploid cell types, a and α, that secrete peptide pheromones (a factor and α factor). These ligands bind to cell surface receptors, consequently promoting new gene transcription, morphological changes, cell cycle arrest, cell fusion, and the creation of an a/α diploid cell (Dohlman and Thorner, 2001).
As in other G protein pathways, agonist stimulation of the α factor receptor (Ste2) promotes exchange of GDP for GTP on the G protein α subunit (Gpa1). GTP-bound Gα undergoes conformational changes and dissociates from the Gβγ subunit dimmer (Ste4/18). Dissociated Gβγ can then signal through effector proteins including a mitogen-activated protein kinase (MAPK) cascade (Ste20, Ste11, Ste7, and Fus3). Inactivation of G protein signaling results from the slow intrinsic GTPase activity of Gα, hydrolyzing GTP to GDP, and the re-association of Gα and Gβγsubunits. GTP hydrolysis is further accelerated by the RGS (Regulator of G protein Signaling) protein Sst2. Therefore, Gpa1 functions primarily to sequester Gβγ in the absence of receptor stimulation (Dohlman and Thorner, 2001).
Many components of the yeast pheromone pathway were identified genetically, by isolating mutants that exhibit a mating-deficient (sterile) phenotype (Hartwell, 1980). Recent efforts to identify new components of G protein signaling have employed more systematic, genome-scale approaches (Slessareva et al., 2006). For example, a library of gene-deletion strains (representing almost all of the non-essential genes) was used to identify direct effectors of Gα signaling. Consequently, it was shown that Gpa1Gα modulates pheromone signaling through a direct interaction with phosphatidylinositol (PtdIns) 3-kinase, resulting in elevated production of the second messenger PtdIns 3-P (Slessareva et al., 2006).
While the non-essential genes have been thoroughly studied, the essential genes are inherently less tractable and have therefore been poorly characterized. Previous approaches to investigating essential gene function have included the isolation of temperature-sensitive (ts) alleles, or fusion to a heat-inducible degron sequence (Dohmen et al., 1994; Kanemaki et al., 2003). However, the use of temperature-sensitive alleles requires growth at sub-optimal temperatures, and introduces destabilizing mutations that could alter enzyme function or protein-protein interactions. Recently a new resource for studying essential genes has been developed. Hughes and colleagues have constructed a library of repressible-promoter strains representing 870 of the yeast essential genes (Mnaimneh et al., 2004). These strains employ the tetracycline-regulatable promoter (TetO7 promoter) system, allowing for precise control of gene expression, with no change in protein sequence or function. This TetO7 promoter library has been used previously to identify new components of the cell division cycle (Yu et al., 2006), translation, and mitochondria import machinery (Mnaimneh et al., 2004). However, the role of essential genes in signal transduction has not been explored in any systematic manner.
While signal transduction networks, such as those mediated by heterotrimeric G proteins, are not typically thought of as essential for cell viability, they can share components with essential processes such as control of cytoskeletal rearrangements and the cell division cycle (Dohlman and Thorner, 2001). In fact, GPA1 is an essential gene because when it is deleted, Gβγ is free to activate downstream effectors resulting in permanent cell cycle arrest (Miyajima et al., 1987).
Here, we systematically characterized 870 essential genes for participation in the yeast G protein signaling pathway. Our results show that proper G protein signaling requires the Cdc34 E2 ubiquitin conjugating enzyme and the SCFCdc4 E3 ubiquitin ligase (Feldman et al., 1997; Skowyra et al., 1997). Ubiquitin ligases, such as the SCF, promote covalent modification of specific substrate proteins with ubiquitin, which can, in turn, target them for degradation by the 26S proteasome (Deshaies, 1999). Previous work has showed that Cdc34 and the SCF complex are involved in regulating the cell cycle and the mating-associated cell cycle arrest (Henchoz et al., 1997; Skowyra et al., 1997). Here we show that SCF also regulates signal initiation, through ubiquitination of the G protein α subunit. More generally, these findings reveal considerable overlap among genes required for cell viability and signal propagation.
RESULTS
Screen of essential genes for new regulators of G protein signaling
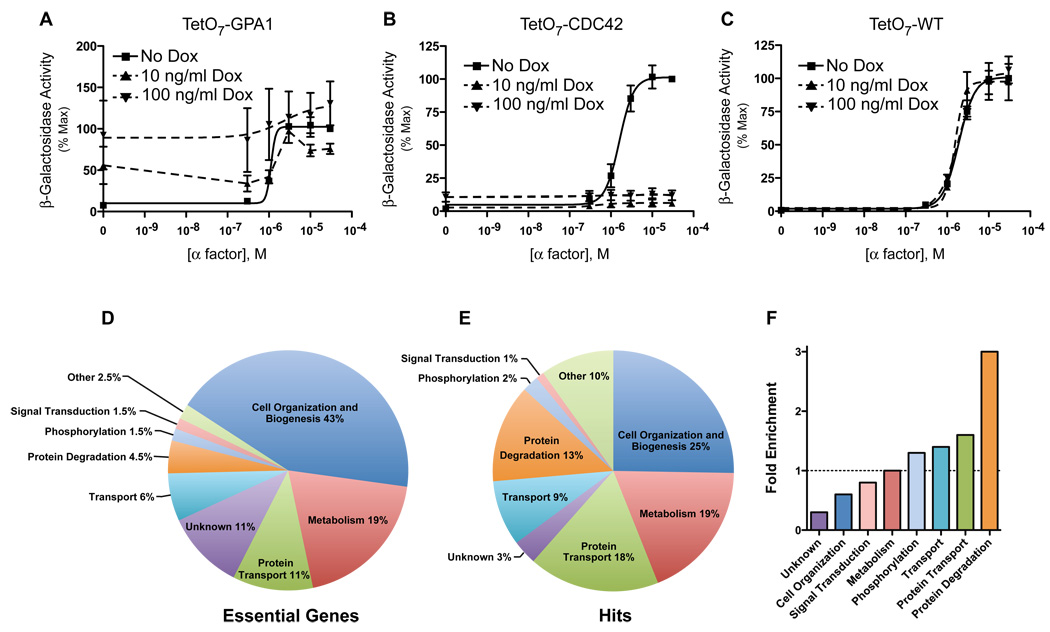
To identify new regulators of G protein signaling, we monitored the pheromone response in 870 TetO7 promoter strains. This strain collection represents nearly all genes essential for viability. Pathway activation was measured initially using a pheromone-inducible promoter from the FUS1 gene fused to the β-galactosidase (lacZ) gene. To validate this approach, we tested the effects of inactivation of two essential genes known to function in the pheromone response pathway: GPA1 (Miyajima et al., 1987) and CDC42 (Simon et al., 1995). GPA1 encodes a negative regulator of the pheromone pathway that functions to sequester Gβγ, and thereby prevents it from activating effectors. As expected, doxycycline treatment of the TetO7-GPA1 strain resulted in constitutive activation of the pathway, and a higher dose of doxycycline exacerbated this effect (Figure 1A). Conversely, CDC42 encodes a positive regulator required for full activation of the signaling cascade. Doxycycline treatment of the TetO7-CDC42 strain resulted in complete loss of the pheromone response (Figure 1B). As expected, treatment of the TetO7 Wild-Type strain with doxycycline had no effect (Figure 1C). These results validate our screening method and demonstrate that the reporter assay is sufficiently sensitive to identify bona fide signaling components.
Figure 1. Validation of the TetO7 Promoter Essential Gene Screen.
(A) Transcriptional activation (β-galactosidase activity) in response to α factor treatment was measured spectrofluorometrically in TetO7-GPA1 cells treated with doxycycline (Dox, 10ng/mL and 100ng/mL) or untreated control. Cells were transformed with a plasmid containing the pheromone-inducible reporter FUS1-lacZ. Data were analyzed by non-linear regression (sigmoidal-dose response, variable slope) using GraphPad Prism software. Results are the mean ± S.E. for three individual experiments each performed in triplicate.
(B) TetO7-CDC42 cells treated as in (A).
(C) TetO7-WT cells treated as in (A).
(D) Percentage of essential genes associated with the indicated GO Process.
(E) Percentage of essential gene hits associated with the indicated GO Process.
(F) Fold-enrichment of hits compared to all essential genes for each GO Process.
The 870 TetO7 strains were next arrayed in a 96-well format and transformed with the FUS1-lacZ reporter. Since gene product depletion varies depending on mRNA and protein half-life, we treated each strain with two doses of doxycycline (10ng/mL and 100ng/mL) for 15hrs. Each strain was then exposed to a range of six pheromone concentrations and β-galactosidase activity was measured using a spectrofluorometer. During the screening process, 61 essential genes repeatedly failed to either transform with the reporter plasmid or grow to a suitable cell density (A600nm = 0.8) required to conduct the reporter assay, and were not tested.
After the initial rounds of screening, we identified 92 genes required for normal pheromone response as measured by our transcriptional reporter (Figure 2). By using a highly specific reporter assay, we were assured that components of the mating pathway would be identified. However we excluded an additional 97 genes likely to have global effects on transcription and translation. The list of excluded genes is comprised of those involved in mRNA production, protein synthesis, DNA replication, RNA processing, or ribosome biogenesis (Table S1).
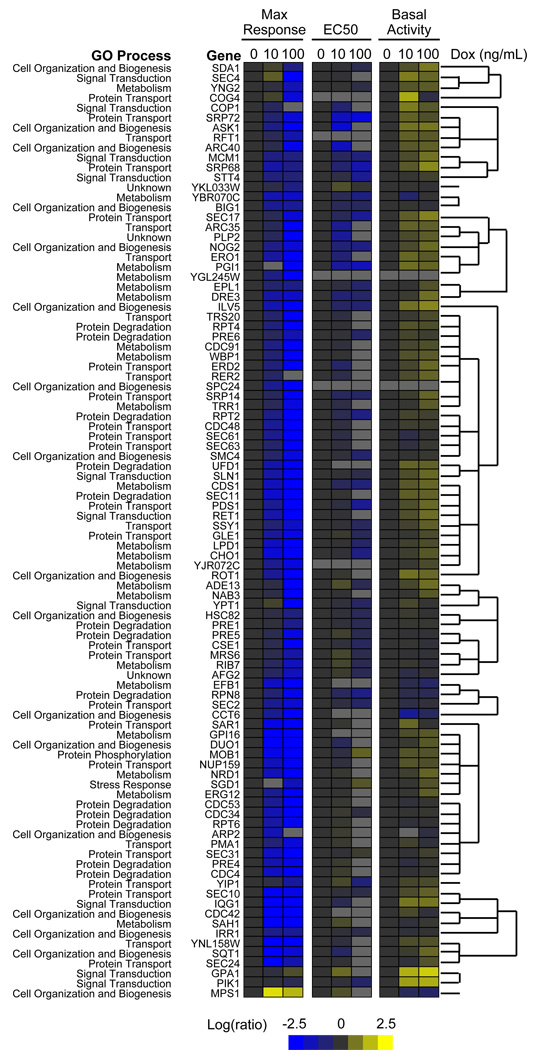
Figure 2. Phenotype Clustering Analysis.
Gene hits were analyzed by Cluster 3.0 software based on maximum response, EC50, and basal activity normalized to the untreated control and converted to Log2. Gene similarity was calculated using Pearson correlation (uncentered correlation) and clusters were generated using centroid linkage. Clustering data was visualized by Java TreeView (v 1.1.3). Genes were labeled by their involvement in the indicated GO Process. See also Figure S1 and Table S1.
We classified our hits by functional category using Gene Ontology (GO) annotations generated by Osprey, whereby each gene was classified according to one of 30 GO processes. When compared with all essential genes, our pheromone pathway hits were enriched for the following GO processes: protein degradation, cell cycle, protein transport, and phosphorylation (compare Figures 1D and 1E). Interestingly, genes involved in protein degradation were enriched 3-fold, by far the most highly represented group of functionally related genes (Figure 1F).
In order to further prioritize our investigations, we generated an interaction map using the Osprey Network Visualization System (Breitkreutz et al., 2002), which incorporates known physical and genetic interactions from the Biological General Repository for Interaction Datasets (BioGRID) (Stark et al., 2006) (Figure S1). We found several previously known interactions between pheromone pathway components and genes identified in our screen. In particular, genes involved in cell organization and biogenesis (dark blue), protein transport (green), and protein degradation (orange) were highly connected to known pheromone pathway components. We also used hierarchical clustering to organize genes into phenotypically similar groups based on changes in the maximal response, EC50, and basal activity (Figure 2). We found that genes involved with protein degradation tended to cluster more closely than genes involved in other cellular processes such as metabolism, cell organization and biogenesis. Given that genes involved in protein degradation were (i) over-represented in our screen, (ii) highly connected with known pathway components, and (iii) clustered more closely than any other functional group, we reasoned they must play a particularly important role in regulating G protein signaling.
Screen validation
Based on the above analysis, we selected six genes deemed likely to participate in cell signaling (Table 1). MPS1 is the yeast ortholog of mammalian TKK (Liu et al., 2003), and is known to encode a dual-specificity kinase (Lauze et al., 1995). According to our hierarchical analysis it was the only gene that failed to cluster with any of the other 91 hits. We speculate that Mps1 phosphorylates some component of the pheromone pathway. We also selected for further consideration two PtdIns 4-kinases, STT4 and PIK1 (Flanagan et al., 1993; Yoshida et al., 1994). Recently, components of the PtdIns 3-kinase, Vps15 and Vps34, were shown to bind directly to Gα and to be required for full activation of the pheromone signaling pathway (Slessareva et al., 2006). Given this precedent, we postulated a broad role for mono-phosphorylated inositides in G protein signaling. Validation of STT4 and PIK1 involvement in G protein signaling would strengthen the proposed model. Among genes involved in protein degradation, we were particularly interested in CDC4, CDC34, and CDC53 because they encode proteins that either form (CDC53, CDC4) or function with (CDC34) the SCFCdc4 ubiquitin ligase complex (Feldman et al., 1997; Skowyra et al., 1997). The Cdc34/SCF family of ubiquitin ligases regulates a variety of proteins, in most cases promoting their degradation by the proteasome. Among SCFCdc4 targets are proteins that play key roles in the regulation of cell growth and division, and the mating-associated cell cycle arrest. However, we were interested in determining a role of SCFCdc4 in regulating components of the G protein-coupled signaling cascade.
Table 1.
| Systematic Name |
Standard Name |
Functionb | Plasmid Rescue |
α factor Max Responsec,d |
α factor LogEC50c,d,e |
|---|---|---|---|---|---|
| YDL028C | MPS1 | Dual-specific kinase; required for spindle checkpoint function |
Yes | 178.7% ± 5.6% | −5.71 ± 0.04 |
| YLR305C | STT4 | Phosphatidylinositol-4- kinase; involved in the Pkc1 pathway |
Yes | 44.8% ± 0.7% | −6.10 ± 0.04 |
| YNL267W | PIK1f | Phosphatidylinositol-4- kinase; may control cytokinesis through actin cytoskeleton |
Yes | 88.3% ± 4.9% | −5.82 ± 0.05 |
| YFL009W | CDC4 | F-Box protein; component of the SCF ubiquitin ligase |
Yes | 46.1% ± 3.1% | −5.40 ± 0.03 |
| YDR054C | CDC34 | E2 ubiquitin conjugating enzyme; component of the SCF ubiquitin ligase |
Yes | 6.2% ± 0.2% | −5.98 ± 0.05 |
| YDL132W | CDC53 | Cullin; component of the SCF ubiquitin ligase |
Not Done | 36.1% ± 1.9% | −5.59 ± 0.06 |
The essential genes PMA1 and SLN1 were confirmed but were not characterized further.
Saccharomyces Genome Database (www.yeastgenome.org).
FUS1 reporter transcription was measured in the indicted TetO7 strains treated with doxycycline or untreated control (average ± SEM).
Transcriptional response with α factor pheromone; α factor Max Response and LogEC50 data derived from nonlinear regression analysis (sigmoidal dose-response).
LogEC50 for TetO7 Wild-Type strain is −5.60 ± 0.01
Showed 4× increase in basal activity when treated with doxycycline.
The six TetO7 strains described above were re-transformed with the FUS1-lacZ reporter and re-tested individually using a broader range of pheromone concentrations, as well as a higher dose of doxycycline (10µg/mL). Testing individual strains in this manner confirmed results obtained by the high-throughput screening method. For further validation, we transformed each TetO7 strain with a single-copy plasmid containing the absent wild-type gene, and showed this restored proper signaling (Table 1).
Transcriptional reporter assays are susceptible to false positives by proteins affecting overall gene expression. To determine if any of the six components have global effects on transcription or translation, we used a dual-reporter assay containing red fluorescent protein (RFP) under the control of a constitutive promoter (ADH1-RFP) and green fluorescent protein (GFP) under the control of a pheromone-responsive promoter (FUS1-GFP). None of the six genes exhibited any change in RFP abundance, despite clear differences in GFP expression. Investigation of five other randomly-selected genes revealed three that alter RFP as well as GFP expression. Thus each of the six genes of interest regulates signaling in a pathway-specific manner (Table 1 and Figure S2).
Analysis of pathway regulation by the newly identified essential genes
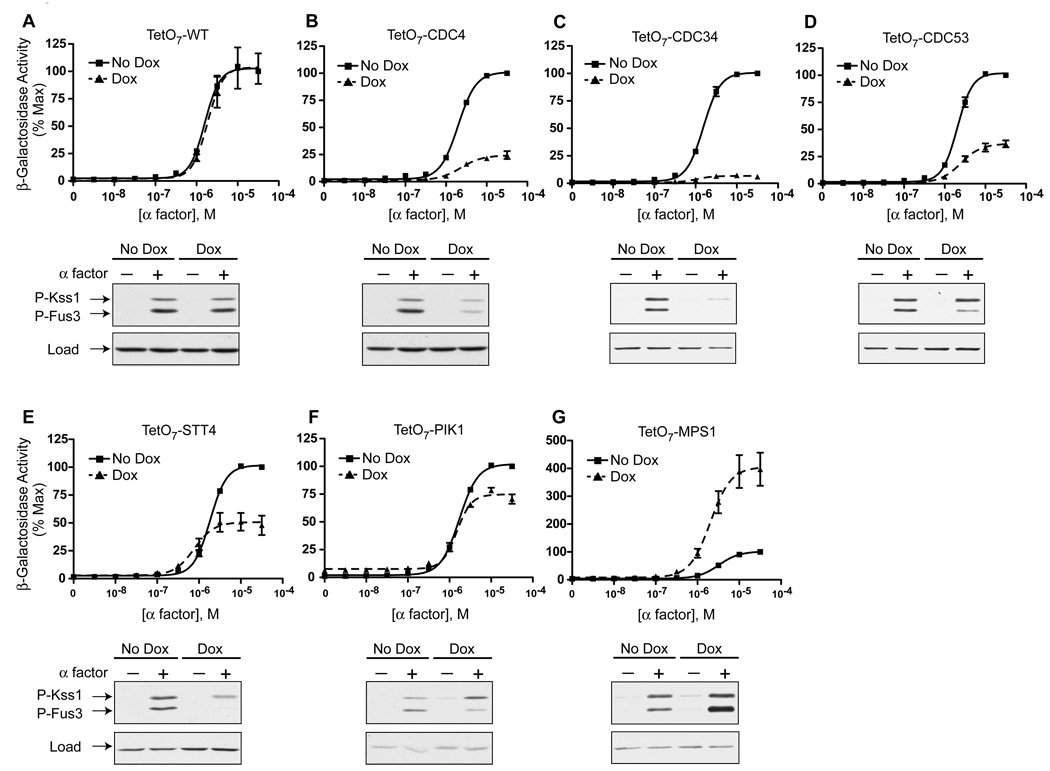
Information from dose-response curves can be used to infer function. For example, a gene mutation leading to an increase in maximal response typically indicates a negative role in signaling. Knockdown of five genes, CDC4, CDC34, CDC53, STT4 and PIK1, dampened the pheromone response (Figures 3 B–F), indicating a positive role in signaling. Conversely, knockdown of MPS1 resulted in an increase in maximal activation (Figure 3G), indicating a negative role in signaling. Knockdown of PIK1 resulted in substantial pathway activity even in the absence of pheromone addition (Figure 3F). These findings indicate that PIK1 (or its catalytic product PtdIns 4-P) serves to suppress basal signaling.
Figure 3. Verification of the Roles of Selected Essential Genes in Pheromone Signaling.
(A–G) The indicated essential genes were chosen for further validation and analysis. TetO7 strains expressing FUS1-lacZ were treated with 10µg/mL doxycycline for 15hrs and exposed to the indicated concentrations of α factor for 90min. Below each pheromone dose-response curve is a corresponding immunoblot probed using phospho-p42/44 (P-Fus3, P-Kss1) or G6PDH (load control) antibodies. TetO7 strains were treated with 10µg/mL doxycycline for 15hrs and then 3µM α factor for 30min. Results are the mean ± S.E. (n=5). See also Figure S3.
To further define the function of each candidate gene, we measured pathway activity upstream of transcriptional regulation. Phosphorylation of the MAPK is a prerequisite for transcription of pheromone-responsive genes. Therefore we measured MAPK phosphorylation using an antibody that recognizes the dually-phosphorylated, fully-activated form of Fus3 and the partially redundant MAPK Kss1. Compared to the TetO7 wild-type control strain (Figure 3A), knockdown of CDC4, CDC34, CDC53, STT4, or PIK1 resulted in a decrease in Fus3 phosphorylation (Figures 3B–F) in relation to total Fus3 levels (Figure S3). Conversely, knockdown of MPS1 resulted in an increase in Fus3 activation (Figure 3G). Thus the changes in Fus3 phosphorylation mirror the changes in β-galactosidase activity reported above. These results confirm a role for each gene in pheromone pathway regulation, and indicate they all function at a point upstream of the MAPK.
Most genes involved in the pheromone response pathway regulate phosphorylation of both Fus3 and Kss1 in tandem. Interestingly, doxycycline treatment of both TetO7-PIK1 and TetO7-STT4 strains resulted in a reduction of Fus3 but not Kss1 activity (Figures 3E and 3F). Selective regulation of Fus3 has only been observed for a small number of gene deletion mutants, but this list includes the Fus3-binding protein Ste5 (Choi et al., 1994; Marcus et al., 1994) as well as the PtdIns 3-kinase Vps34 and its binding partner Vps15 (Slessareva et al., 2006). Pik1 and Stt4 are both PtdIns 4-kinases, but are present at different subcellular locations: Stt4 at the plasma membrane (Audhya and Emr, 2002) and Pik1 at the Golgi and the nucleus (Strahl et al., 2005). Taken together, these findings reveal a possible role for mono-phosphorylated inositides in Fus3 signaling.
SCFCdc4 regulates the pheromone pathway upstream of Ste4Gβ
Of the essential genes found to regulate pheromone signaling, we were particularly interested in those that promote protein degradation. As noted above, genes involved in degradation were enriched almost 3-fold compared to all the essential genes. Moreover they were clustered more closely than any other functional group in the hierarchical analysis. Among the genes identified in our screen, CDC4, CDC34, and CDC53 encode components of the Skp1, Cullin, F-box protein (SCF)-type ubiquitin ligase (Feldman et al., 1997; Skowyra et al., 1997). When treated with doxycycline, the TetO7-CDC4, TetO7-CDC34, and TetO7-CDC53 strains all exhibited a significant decrease in the maximal response to pheromone as well as diminished MAPK activation (Figures 3B, 3C, and 3D). The similarity of the responses of these strains, and the fact that the affected proteins exist as a complex in cells, suggests the SCFCdc4 has a particularly important role in signal regulation.
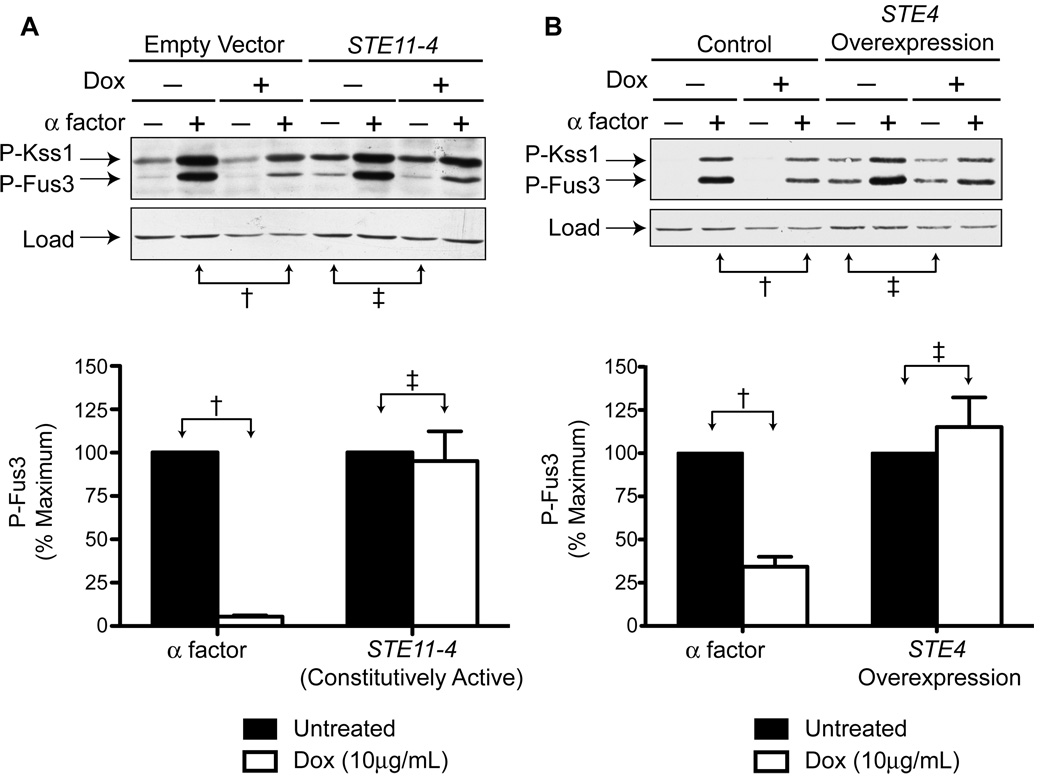
While each component of the SCF is necessary for ubiquitin-ligase function, the F-box protein, Cdc4, binds directly to the substrate and therefore defines substrate specificity of the SCF complex. For this reason we focused further efforts on characterizing the role of Cdc4. To narrow our search for Cdc4 substrates we activated the pathway at several points downstream of the pheromone receptor. First, we over-expressed the constitutively active STE11-4 mutant (Stevenson et al., 1992). STE11 encodes the MAPK kinase kinase (MAPKKK) that phosphorylates Ste7, which in turn phosphorylates and activates Fus3 and Kss1. TetO7-CDC4 cells expressing STE11-4 triggered MAPK phosphorylation in the absence of pheromone. However, doxycycline treatment of these cells had no effect on Fus3 activity, in contrast to the reduction seen in pheromone-stimulated cells (Figure 4A). These results indicate that SCFCdc4 acts on a protein component that is upstream of the MAPKKK.
Figure 4. The Cdc34 E2 and SCFCdc4 Regulate Signaling Upstream of Ste4Gβ.
(A) TetO7-CDC4 cells were transformed with a plasmid containing either STE11-4 (constitutively active mutant) or no insert. Cells were treated with 10µg/mL doxycycline for 15hrs and then 3µM α factor for 30min. Samples were analyzed by immunoblotting using phospho-p42/44 or G6PDH antibodies. Bar graphs represent quantification of the indicated bands. Results are the mean ± S.E. (n=3).
(B) TetO7-CDC4 cells were transformed with a plasmid containing STE4 under the control of a galactose-inducible promoter. Cells were treated with 10µg/mL doxycycline for 12hrs in medium containing either dextrose or switched to galactose (2% w/v final concentration) for 3hrs prior to α factor treatment (3µM for 30min). Samples were analyzed by immunoblotting using phospho-p42/44 or G6PDH antibodies. Bar graphs represent quantification of the indicated bands. Results are the mean ± S.E. (n=3).
Next, we over-expressed STE4Gβ using a galactose-inducible promoter. Since Gpa1 cannot sequester excess Ste4Gβ (Miyajima et al., 1987), the overproduced Ste4Gβ is free to activate effectors even in the absence of any stimulus. TetO7-CDC4 cells containing GAL-STE4 were grown in dextrose- or galactose-containing medium to induce protein expression. Overexpression of STE4 resulted in MAPK activation in the absence of pheromone. Once again, doxycycline treatment failed to dampen this signal (Figure 4B). Thus knock-down of Cdc4 attenuates signaling by pheromone but not the G protein subunit. These data indicate that SCF acts at the level of the G protein or the receptor.
Loss of Cdc4 stabilizes Gpa1 protein levels
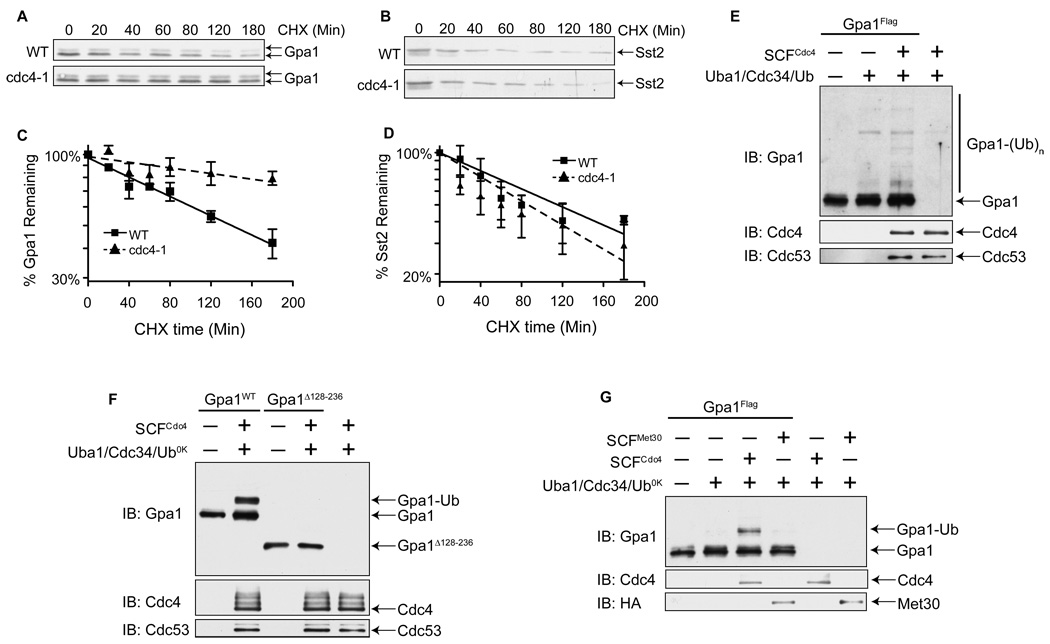
Our genetic epistasis analysis indicates that Cdc4 acts early in the pathway. We hypothesized further that Cdc4 must ubiquitinate a negative regulator, since knockdown of Cdc4 resulted in a decrease in signaling. There are only two well-characterized negative regulators that function upstream of Ste4Gβ, Gpa1 and the RGS protein Sst2, and both are known to be ubiquitinated (Hao et al., 2003; Madura and Varshavsky, 1994). To determine if Cdc4 affects Gpa1 or Sst2 stability, we tracked their rates of degradation in a temperature-sensitive cdc4-1 strain. This mutant strain has been used previously in similar experiments to identify Cdc4 substrates (Tang et al., 2005). The rate of Sst2 degradation was similar in both the wild-type and cdc4-1 cells (Figures 5B and 5D). In contrast, Gpa1 was considerably more stable in the absence of Cdc4 function. Whereas Gpa1 in wild-type cells exhibited an approximate half-life of 141min, in cdc4-1 cells the half-life was extended to 344min (Figures 5A and 5C). These data demonstrate that Cdc4 is required for proper turnover of Gpa1 in vivo. Interestingly, Wang et al. showed that a mutant Gpa1 that cannot be ubiquitinated in vivo produces a dampened pheromone response (Wang et al., 2005), comparable to that seen in the SCF mutant strains (Table 1). Taken together, these data indicate that SCFCdc4 regulates the function of Gpa1.
Figure 5. SCFCdc4 Ubiquitinates Gpa1 in vitro and Facilitates its Turnover in vivo.
(A) Gpa1 stability in wild-type vs. temperature-sensitive cdc4-1 mutant cells. Cultures were grown at 25°C, shifted to 37°C for 1hr, and then treated with cycloheximide (CHX) for the indicated times. Myristoylated (bottom band) and unmyristoylated (top band) Gpa1 detected by immunoblotting with Gpa1 antibodies.
(B) Samples from panel (A) analyzed with Sst2 antibodies.
(C and D) The intensity of bands from (A) and (B), analyzed by densitometry. Results are the mean ± S.E. (n=3).
(E) In vitro ubiquitination of Gpa1. Purified Gpa1-Flag was incubated with purified SCFCdc4 complex (Flag-Skp1/Cdc53/Myc-Rbx1/Cdc4), His6-Uba1, His6-Cdc34, and ubiquitin as indicated, followed by SDS-PAGE and immunoblotting. Unmodified (Gpa1) and ubiquitinated (Gpa1-(Ub)n) Gpa1 protein was visualized with Gpa1 antibodies. Membranes were also probed with Cdc4 antibodies and Cdc53 antibodies.
(F) In vitro ubiquitination using Lys-less ubiquitin (Ub0K). Reactions contain either Gpa1 or Gpa1Δ128-236, a mutant form of Gpa1 lacking the ubiquitinated subdomain.
(G) In vitro ubiquitination of Gpa1 using Ub0K and SCF complexes containing either Cdc4 or Met30 as indicated. Note that Met30 appears to bind weakly to Gpa1 but does not sustain Gpa1 ubiquitination (Figure S4B).
See also Figure S4.
We also considered whether the Cdc34/SCF complex regulates other components involved in G protein signaling. To this end we measured the abundance and stability of proteins downstream of the receptor but upstream of the transcription factor (Ste4, Ste20, Ste5, Ste11, Ste7, Fus3, and Kss1). To identify Cdc34/SCF substrates, including those that could be recruited by F-box proteins other than Cdc4, we used a temperature sensitive cdc34-2 mutant strain. Of the proteins tested, four (Ste4, Ste20, Ste7, and Ste5) were significantly stabilized in the cdc34-2 mutant as compared with wild-type cells (Figure S4A). These data corroborate reports that the Cdc34/SCF pathway promotes the degradation of Ste7 and Ste5 (Garrenton et al., 2009; Wang and Dohlman, 2002), and suggest that it may likewise act on Ste20 and Ste4. Of these proteins at least one binds to Cdc4 (Ste5) and another is clearly ubiquitinated (Ste7) (Garrenton et al., 2009; Wang and Dohlman, 2002). Strikingly, despite stabilized expression of multiple components that propagate the signal, loss of Cdc34 or SCFCdc4 function leads to a reduction in pheromone signaling. Furthermore, loss of CDC4 gene expression has no effect when the pathway is activated at the level of Ste4Gβ (Figure 4B). Taken together, these findings highlight the importance of a negative regulator as the most functionally significant target for Cdc34/SCFCdc4-dependent degradation, and demonstrate that the dampened pheromone signaling in SCF-depleted cells is due primarily to the stabilization of Gpa1.
Cdc34 poly-ubiquitinates purified Gpa1 in vitro in an SCFCdc4-dependent manner
Protein turnover is often dependent upon polyubiquitination. Gpa1 was shown previously to undergo ubiquitination in vivo (Madura and Varshavsky, 1994). To test the possibility that SCFCdc4 is directly responsible for the modification, we sought to establish whether SCFCdc4 could interact with Gpa1. We affinity purified Skp1-GST/Cdc4-HA from insect cell lysates and added purified Gpa1-Flag. As shown in Figure S4B, Gpa1 copurified with Cdc4. Additionally, we tested a closely related F-box protein, Met30. Both Met30 and Cdc4 contain a substrate-binding domain comprised of WD repeats (Chandrasekaran et al., 2006). Gpa1 bound to Met30 (Figure S4B), but with a lower affinity than Cdc4. These results suggest that the Cdc4 component of the SCFCdc4 ubiquitin ligase complex binds Gpa1, and as a consequence may promote its ubiquitination by Cdc34.
To establish whether Gpa1 is ubiquitinated by Cdc34/SCFCdc4 we sought to reconstitute Gpa1 ubiquitination in vitro. We purified Gpa1-Flag from yeast (to maintain post-translational modifications) and purified the SCFCdc4 complex (comprised of yeast Flag-Skp1, Cdc53, Rbx1, and Cdc4) from insect cells. As shown in Figure 5E, Gpa1 is polyubiquitinated in reaction mixtures composed of purified SCFCdc4, Cdc34, Uba1, Ubiquitin and ATP, but not in the absence of SCFCdc4 or Cdc34. To determine if the SCF polyubiquitinates Gpa1 at a single site, or instead monoubiquitinates Gpa1 at multiple sites, we performed the in vitro ubiquitination reactions with a mutant form of ubiquitin (Ub0K) that, due to the replacement of all the lysines with arginine, cannot be incorporated into poly-ubiquitin chains. Under such conditions, we observed only a single 8kD shift, suggesting Gpa1 is modified at a single lysine residue (Figure 5F).
In order to determine if Gpa1 is ubiquitinated by the SCFCdc4 at the previously identified site, we conducted an in vitro ubiquitination assay using a mutant form of Gpa1 that lacks the ubiquitinated subdomain (Gpa1Δ128-236). As shown in Figure 5F, Gpa1Δ128-236 is not ubiquitinated in vitro, indicating the SCFCdc4 ubiquitinates Gpa1 at the known site of ubiquitination. Interestingly, Gpa1Δ128-236 is not ubiquitinated, and the response to pheromone is partially abrogated. This gain-of-function phenotype is likely due to stabilized expression of the protein. However we cannot rule out the possibility that there are other functional differences between the mutant and wild-type protein. As an additional control, we conducted an in vitro reaction with the F-box protein Met30. We show above that Met30 was able to bind Gpa1. However, SCFMet30 complexes were not able to ubiquitinate Gpa1 (Figure 5G).
Finally, the SCF complex is typically recruited to substrates in response to substrate phosphorylation (Ardley and Robinson, 2005). To determine if Gpa1 ubiquitination by the SCFCdc4 is regulated by phosphorylation we conducted in vitro ubiquitination assays with Gpa1 from E. coli, which should lack all post-translational modifications. As shown in Figure S4C, Gpa1 expressed from E. coli is ubiquitinated poorly, indicating that Gpa1 ubiquitination by SCFCdc4 may be regulated by a post-translational modification.
Notably, the Gpa1Δ128-236 mutant, which is not ubiquitinated in vivo or in vitro (Wang et al., 2005) (Figure 5F), exhibits a dampened pheromone response comparable to that seen following knock-down of SCFCdc4 expression (Table 1). The ability of SCFCdc4 to ubiquitinate Gpa1 in vitro, and to accelerate Gpa1 turnover in vivo, reveals Gpa1 as a critical target of SCF regulation. To date, only a handful of SCFCdc4 substrates have been verified by direct ubiquitination using purified components. Indeed, of the mating pathway components only the cell cycle regulatory protein Far1 has been characterized in this manner (Figure 6) (Blondel et al., 2000). The identification of Gpa1 as an SCFCdc4 substrate could explain the critical role of SCFCdc4 in pheromone signaling, and expands the repertoire of regulatory SCFcdc4 functions in the pheromone pathway (Figure 6).
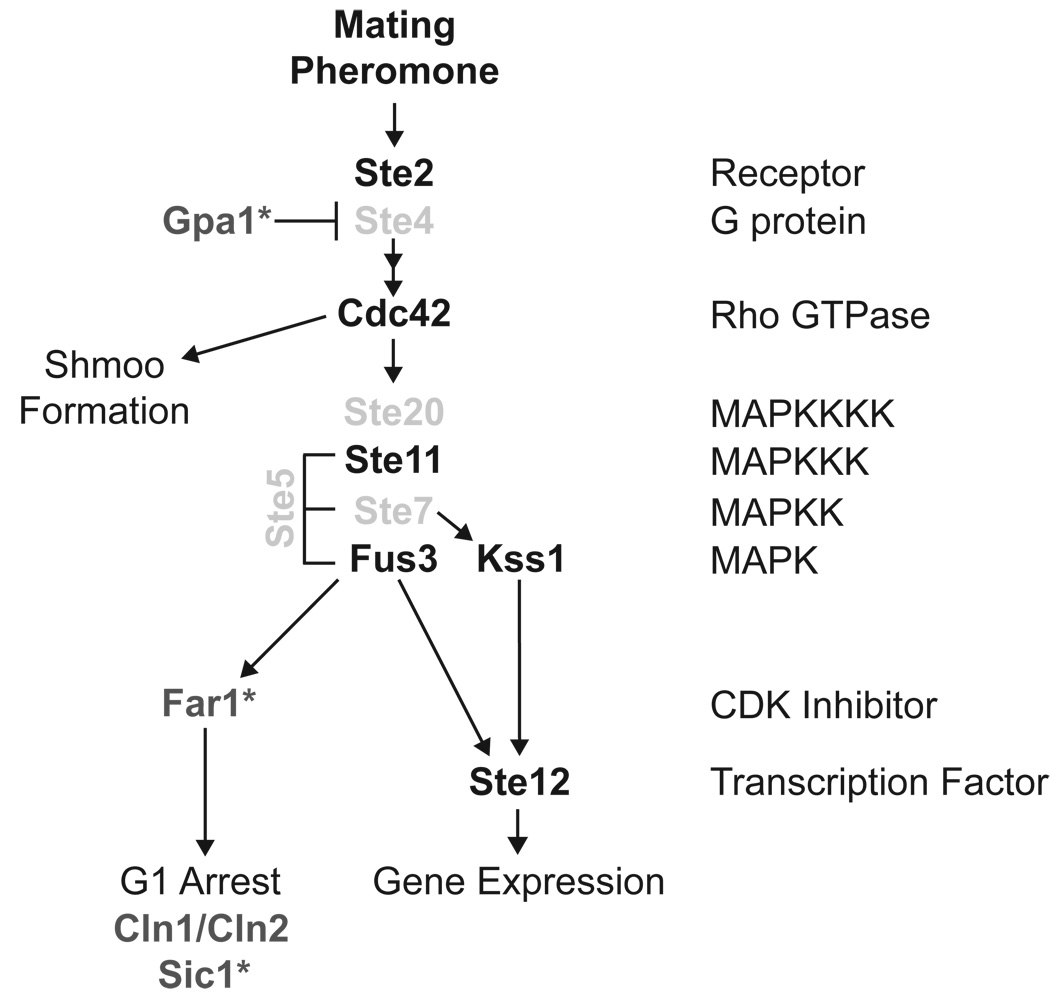
Figure 6. The Pheromone Response Pathway.
The Cdc34/SCF complex targets several known substrates in the pheromone response pathway. Known substrates of Cdc34/SCF are shown in dark grey. Likely substrates of the SCF are shown in light grey. SCFCdc4 substrates that have been verified using in vitro ubiquitination assays are designated with an asterisk (*).
DISCUSSION
The yeast pheromone response system is perhaps the most thoroughly characterized of any signaling pathway. The contributions of non-essential gene products have been exhaustively and systematically characterized over much of the past three decades. Much less is known about genes that are essential to cell viability, but which nevertheless contribute to effective signal transduction. Until now, only two major components of the pheromone signaling cascade, Gpa1 and Cdc42, were known to be encoded by essential genes. To address this deficit, we conducted a systematic analysis of the essential yeast genome. Our results implicate 92 essential genes in the regulation of G protein signaling, verify the involvement of 6 selected genes, and define their mode of action. These findings suggest a level of complexity in G protein signaling that has not been fully appreciated.
Protein Degradation
Among the genes required for proper pheromone responses, those involved in protein degradation (13 of 92 genes) were over-represented in our screen, indicating that they may have a particularly important role in signal regulation. Included in this group are four components of the 20S proteasome, four components of the 19S regulatory particle (together forms the 26S proteasome complex), as well as the Cdc34 E2 ubiquitin conjugating enzyme and the SCFCdc4 E3 ubiquitin ligase. These data are consistent with the observation that many of the core components of the pheromone pathway are ubiquitinated, including the receptor, G protein, RGS protein, and components of the MAP kinase cascade (Esch and Errede, 2002; Garrenton et al., 2009; Hao et al., 2003; Henchoz et al., 1997; Hicke and Riezman, 1996; Madura and Varshavsky, 1994; Wang and Dohlman, 2002). Further, there is indirect evidence that SCF complexes promote ubiquitination of several proteins that propagate the signal, including Ste5 (Garrenton et al., 2009), Ste7 (Wang and Dohlman, 2002), Ste4 and Ste20 (Figure S3). Strikingly, despite stabilized expression of these positive regulators, loss of SCFCdc4 function results in a reduction in pheromone signaling. A negative regulator is thus the most functionally significant target of the SCFCdc4 in signal transduction. Our analysis indicates that Gpa1 is this critical target of the SCFCdc4.
These findings expand the classic roles of SCF complexes in orchestrating the events leading to mating (Figure 6). Arrest of cell division in the G1 phase is needed to ensure cell cycle synchrony prior to cell fusion. Essential to G1 cell cycle arrest is inactivation of the cyclin-dependent kinase (CDK) Cdc28. Several mechanisms contribute to the G1-specific functions of Cdc28; these include degradation of the G1 cyclins Cln1 and Cln2, and direct inhibition of Cln/Cdc28 activity by Far1 (Peter and Herskowitz, 1994), a protein that is upregulated in a Ste12-dependent manner during mating (Chang and Herskowitz, 1990). Additionally, these changes prevent proteolysis of the Sic1 S-phase CDK inhibitor, which normally depends on G1 CDK activity, thereby ensuring that the S-phase Cdc28 functions are not activated in mating cells. Each of these processes is regulated by SCF complexes. Whereas SCFCdc4 promotes the ubiquitination and degradation of Far1 (Henchoz et al., 1997) and Sic1 (Feldman et al., 1997; Skowyra et al., 1997), SCFGrr1 promotes the degradation of Cln1 and Cln2 (Barral et al., 1995; Schweitzer et al., 2005).
As noted above, loss of Gpa1 leads to G1 cell cycle arrest. Thus an additional role for SCFCdc4 in ubiquitination and degradation of Gpa1 is likely to contribute to cell cycle regulation. Inhibition of SCFCdc4 could promote a timely recovery following pheromone stimulation, and also prevent pheromone signaling in non-G1 phases of the cell cycle when improper activation of cell mating could result in aneuploidy. The observation that many of the pheromone pathway components are not expressed in diploid cells suggests that SCF could also promote degradation of unnecessary (haploid-specific) signaling proteins and, as such, prevent aberrant pathway activation after mating.
Gpa1 as a target for proteasomal and vacuolar proteolysis
Gpa1 is a rare example of a protein that can be either mono- or poly-ubiquitinated (Madura and Varshavsky, 1994). Whereas polyubiquitinated Gpa1 is targeted to the proteasome, monoubiquitinated Gpa1 is internalized and degraded within the vacuole (Wang et al., 2005). Left unresolved was whether mono- and poly-ubiquitination of Gpa1 requires two distinct sets of E2 and E3 enzymes. Recently, we showed that Rsp5 monoubiquitinates Gpa1 (Torres et al., 2009). Here we show that the SCF complex polyubiquitinates Gpa1. Taken together these findings reveal that Gpa1 mono- and poly-ubiquitination occur by distinct pathways and/or mechanisms.
Although complex, there may be specific benefits to having two ubiquitinating pathways that can target the same protein for distinct proteolytic machineries. One such benefit would be that degradation could be triggered in response to different signals and/or functional states of the protein. In support of this model, we showed that the Rsp5 E3 ligase ubiquitinates only the fully myristoylated (fully mature) form of the G protein. Myristoylated Gpa1 would localize to the plasma membrane and its degradation in vacuoles could therefore be linked to endocytosis. Conversely, there may be another modification that directs Gpa1 to SCFCdc4. Indeed, many E3 ubiquitin ligases including the SCF, are recruited to substrates in response to substrate phosphorylation (Ardley and Robinson, 2005). Recently, two independent phospho-proteomic screens revealed that Gpa1 is phosphorylated at Thr-189 and Ser-200 (Li et al., 2007; Smolka et al., 2007), and it is noteworthy that both sites are located near a known Gpa1 ubiquitination site, Lys-165, established by mass spectrometry (Marotti et al., 2002). In this study, we show that Gpa1 purified from E. coli and lacking any post-translational modifications is not ubiquitinated in vitro. Thus phosphorylation of Gpa1 may serve to recruit the SCF ubiquitination machinery. It is also possible, however, that phosphorylation of a Gpa1-binding partner, rather than Gpa1 itself, is sufficient to direct the protein to the SCFCdc4 complex. Knowing when Gpa1 is phosphorylated, whether there are additional sites of phosphorylation, and the identity of the protein kinase(s) could further establish the role of Gpa1 ubiquitination in regulating the pheromone pathway.
Interestingly, depletion of Cdc4 does not result in increased steady-state levels of Gpa1 (data not shown). While any differences in Gpa1 abundance are small, even small differences in abundance could account for the 54% loss of signal observed in the TetO7-CDC4 strain. The SCFCdc4 likely targets a small pool of Gpa1 that is functionally important for signaling as has been shown for other proteins (Chandrasekaran et al., 2006; Tworkowski et al., 2002). In this case, small changes in protein levels, in a specific functional context, can result in large changes in signaling. We further expect that slower Gpa1 turnover in the TetO7-CDC4 strain would increase the proportion of protein that has had time to be correctly folded, fully modified, properly localized, and assembled into the heterotrimeric complex. Unfortunately, it is not currently feasible to distinguish Gpa1 that is folded and functional from protein that is non-functional but nevertheless expressed in the cell.
G protein ubiquitination may also be regulated by external stimuli. F-box proteins appear to serve as receptors for the plant hormones auxin and jasmonates; in each case binding to these hormones enhances the interaction between SCF and its substrates (Dharmasiri et al., 2005; Katsir et al., 2008; Kepinski and Leyser, 2005). Thus F-box proteins and ubiquitin ligases can function as hormone receptors. By extension, F-box proteins might also serve as targets for drugs that enhance or diminish signaling by G proteins and G protein-coupled receptors.
Non-proteolytic new essential regulators of G protein signaling
We also characterized three other essential genes required for proper G protein signaling. Of particular interest are the PtdIns 4-kinases Pik1 and Stt4. Whereas pheromone stimulation leads to activation of both Fus3 and Kss1, we found that knockdown of PIK1 or STT4 leads to a selective diminution of Fus3 activity. These observations suggest a positive role for PtdIns 4-P in signal transduction, and in particular for Fus3. Similarly, there is a selective loss of Fus3 activation in cells that lack the PtdIns 3-kinase components Vps34 and Vps15, or in cells treated with the PtdIns 3-kinase inhibitor Wortmannin (Slessareva et al., 2006). Taken together, these findings indicate a potential role for mono-phosphorylated phosphoinositides in Fus3 activation. Notably, Fus3 activation requires the MAPK scaffolding protein Ste5, while Kss1 activation does not. Moreover, Ste5 was shown to bind to PtdIns 4-P and PtdIns 4,5-P2 in vitro (Garrenton et al., 2006). We hypothesize that PtdIns 4-P interaction is required for Ste5 activity or proper localization. Additionally, PtdIns 3-P and 4-P are produced in different sub-cellular locations, and these differences could contribute further to signaling specificity and activity. In any event, these discoveries suggest an important and expanded role for phosphoinositides in the pheromone-response pathway.
Conclusions
We have systematically characterized 870 essential genes for participation in the yeast G protein signaling pathway, identified up to 92 new regulators of the pathway, and characterized six in detail. Our findings reveal considerable overlap among genes required for cell viability and signal propagation. More significantly, our work reveals that there are still many new pathway components to be found. While we focused on components for which there was a specific and rigorously-testable hypothesis, there are still dozens of others that will be pursued in the future. Based on these results, we regard the essential-genome as an under-utilized resource for the identification of new signal transduction factors. Further efforts to screen the essential genes in yeast and other model systems will undoubtedly lead to a more complete understanding of signal transduction networks. Given the conservation of G protein function across species, newly described functions in yeast are likely to extend as well to humans. Due to the established importance of G proteins in physiology and pharmacology, our findings may also reveal future opportunities for drug discovery.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
See Supplemental Methods.
Cell-Extract Preparation and Immuno-Blot Analysis
Preparation and Purification of Recombinant Proteins
See Supplemental Methods.
Screen of Essential Genes
852 TetO7 promoter strains (Open Biosystems, yTHC) were transformed with pRS423 FUS1-lacZ, and β-galactosidase activity was measured in a 96-well plate format as described previously (Chasse and Dohlman, 2004). Prior to pheromone stimulation, cells were grown for ~15hrs in either untreated medium or medium containing 10ng/mL, or 100ng/mL doxycycline hyclate (Sigma-Aldrich). β-galactosidase activity was measured in triplicate for each condition. Strains were considered for further analysis if doxycycline treatment resulted in a >50% increase or decrease in maximal response, at least a two standard deviation shift in the EC50, or at least a four-fold increase in the basal activity as determined by non-linear regression analysis (Graphpad Prism). Twenty-six candidate strains were re-tested individually using twelve concentrations of α factor pheromone and a higher dose of doxycycline (10µg/mL). Eleven confirmed strains were transformed with the pRS316-AR/FG dual reporter and tested for GFP and RFP expression by immunoblotting. While all eleven strains exhibited changes in GFP expression, three TetO7-strains representing the essential genes RIO2, KIC1, and SSY1 also exhibited detectable changes in RFP expression and were not considered further. The remaining eight TetO7-strains representing the essential genes MPS1, STT4, PIK1, CDC4, CDC53, CDC34, PMA1, and SLN1 showed no change in RFP expression. Of the remaining eight strains, six were selected for transformation with a single-copy plasmid containing the corresponding wild-type gene, and these were re-tested for restoration of normal β-galactosidase activity (Table 1).
Protein Turnover Measurements
Cells were grown in 100mL of selective medium at room temperature to A600nm ~0.6, and then shifted to 37°C for 1hr. Cells were then treated with cycloheximide (final concentrations, 10µg/mL in 0.1% ethanol) for up to 3hrs. Growth was stopped by the addition of TCA (5% final concentration) and samples were normalized to the same A600nm. Cell pellets were washed and brought up directly in boiling SDS-PAGE sample buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 1% 2-mercaptoethanol, 0.0005% bromophenol blue) and lysed using glass beads as previously described (Wang et al., 2003). Protein extracts were resolved by 10% SDS-PAGE and immunoblotting with Gpa1 antibodies at 1:1000 (Dohlman et al., 1993) and Sst2 antibodies at 1:2000 (Dohlman et al., 1996).
In Vitro Ubiquitination Assay
Ubiquitination reactions were prepared with FlagSCF complexes containing Flag-Skp1/Cdc53/Myc-Rbx1/Cdc4 or Met30 purified from insect cells as described in Supplemental Methods, and in (Skowyra et al., 1997). 9µL of purified and eluted SCF complex (~2 pmol) was combined with 13.5 pmol HisCdc34, 1.0 pmol HisUba1, 120 pmol Ub or 0K Ub (Boston Biochem), 1.5 pmol Gpa1-Flag and supplemented with 1mM ATP, 5mM MgCl2, and 20µM GDP in a volume of 15µL. Reactions were allowed to proceed at 30°C and stopped after 90min with the addition of boiling SDS-PAGE sample buffer, followed by SDS-PAGE and immunoblotting with Gpa1 antibodies. Membranes were stripped and re-probed with Cdc4 antibodies at 1:1000 and Cdc53 antibodies at 1:1000 (provided by Mark Goebl, University of Indiana Medical School).
Supplementary Material
ACKNOWLEDGMENTS
We thank Mike Lee and Anh Nguyen for the construction of plasmids used in this research; Mike Tyers for supplying yeast strains; Johannes Hegemann for supplying the pUG35 vector; George Sprague for providing plasmids; Mark Goebl for providing antibodies; Kathryn Cappell for help with bioinformatic analysis; Agnieszka Lass for the preparation of purified Cdc34 and Uba1; Sarah Clement for providing purified Gpa1. We would like to specially thank Corinne Zeller and Chris Broda for their contributions to the early stages of this work. This work was supported by NIH grant RO1 GM059167.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- Blondel M, Galan JM, Chi Y, Lafourcade C, Longaretti C, Deshaies RJ, Peter M. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. Embo J. 2000;19:6085–6097. doi: 10.1093/emboj/19.22.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2002;2 PREPRINT0012. [PubMed] [Google Scholar]
- Chandrasekaran S, Deffenbaugh AE, Ford DA, Bailly E, Mathias N, Skowyra D. Destabilization of binding to cofactors and SCFMet30 is the rate-limiting regulatory step in degradation of polyubiquitinated Met4. Mol Cell. 2006;24:689–699. doi: 10.1016/j.molcel.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Chasse SA, Dohlman HG. Identification of yeast pheromone pathway modulators by high-throughput agonist response profiling of a yeast gene knockout strain collection. Methods Enzymol. 2004;389:399–409. doi: 10.1016/S0076-6879(04)89024-4. [DOI] [PubMed] [Google Scholar]
- Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Goldsmith P, Spiegel AM, Thorner J. Pheromone action regulates G-protein alpha-subunit myristoylation in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993;90:9688–9692. doi: 10.1073/pnas.90.20.9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Esch RK, Errede B. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc Natl Acad Sci U S A. 2002;99:9160–9165. doi: 10.1073/pnas.142034399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Schnieders EA, Emerick AW, Kunisawa R, Admon A, Thorner J. Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science. 1993;262:1444–1448. doi: 10.1126/science.8248783. [DOI] [PubMed] [Google Scholar]
- Garrenton LS, Braunwarth A, Irniger S, Hurt E, Kunzler M, Thorner J. Nucleus-specific and cell cycle-regulated degradation of mitogen-activated protein kinase scaffold protein Ste5 contributes to the control of signaling competence. Mol Cell Biol. 2009;29:582–601. doi: 10.1128/MCB.01019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrenton LS, Young SL, Thorner J. Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain. Genes Dev. 2006;20:1946–1958. doi: 10.1101/gad.1413706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Hao N, Yildirim N, Wang Y, Elston TC, Dohlman HG. Regulators of G protein signaling and transient activation of signaling: experimental and computational analysis reveals negative and positive feedback controls on G protein activity. J Biol Chem. 2003;278:46506–46515. doi: 10.1074/jbc.M308432200. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980;85:811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies RJ, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hughes TR. Yeast and drug discovery. Funct Integr Genomics. 2002;2:199–211. doi: 10.1007/s10142-002-0059-1. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–724. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Lauze E, Stoelcker B, Luca FC, Weiss E, Schutz AR, Winey M. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. Embo J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K, Varshavsky A. Degradation of G alpha by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci U S A. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotti LA, Jr., Newitt R, Wang Y, Aebersold R, Dohlman HG. Direct identification of a G protein ubiquitination site by mass spectrometry. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- Miyajima I, Nakafuku M, Nakayama N, Brenner C, Miyajima A, Kaibuchi K, Arai K, Kaziro Y, Matsumoto K. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell. 1987;50:1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- Schweitzer K, Cocklin R, Garrett L, Desai F, Goebl M. The ubiquitin ligase SCFGrr1 is necessary for pheromone sensitivity in Saccharomyces cerevisiae. Yeast. 2005;22:553–564. doi: 10.1002/yea.1234. [DOI] [PubMed] [Google Scholar]
- Simon MN, De Virgilio C, Souza B, Pringle JR, Abo A, Reed SI. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BJ, Rhodes N, Errede B, Sprague GF., Jr. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- Strahl T, Hama H, DeWald DB, Thorner J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol. 2005;171:967–979. doi: 10.1083/jcb.200504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Liu Q, Willems A, Sicheri F, Tyers M. Genome-wide surveys for phosphorylation-dependent substrates of SCF ubiquitin ligases. Methods Enzymol. 2005;399:433–458. doi: 10.1016/S0076-6879(05)99030-7. [DOI] [PubMed] [Google Scholar]
- Torres MP, Lee MJ, Ding F, Purbeck C, Kuhlman B, Dokholyan NV, Dohlman HG. G Protein Mono-ubiquitination by the Rsp5 Ubiquitin Ligase. J Biol Chem. 2009;284:8940–8950. doi: 10.1074/jbc.M809058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworkowski KA, Salghetti SE, Tansey WP. Stable and unstable pools of Myc protein exist in human cells. Oncogene. 2002;21:8515–8520. doi: 10.1038/sj.onc.1205976. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dohlman HG. Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J Biol Chem. 2002;277:15766–15772. doi: 10.1074/jbc.M111733200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ge Q, Houston D, Thorner J, Errede B, Dohlman HG. Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the Skp1-Cullin-F-box complex. J Biol Chem. 2003;278:22284–22289. doi: 10.1074/jbc.M301272200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Marotti LA, Jr., Lee MJ, Dohlman HG. Differential regulation of G protein alpha subunit trafficking by mono- and polyubiquitination. J Biol Chem. 2005;280:284–291. doi: 10.1074/jbc.M411624200. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ohya Y, Goebl M, Nakano A, Anraku Y. A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J Biol Chem. 1994;269:1166–1172. [PubMed] [Google Scholar]
- Yu L, Pena Castillo L, Mnaimneh S, Hughes TR, Brown GW. A survey of essential gene function in the yeast cell division cycle. Mol Biol Cell. 2006;17:4736–4747. doi: 10.1091/mbc.E06-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.