Abstract
The authors report the characterization of a novel cyclic adenosine monophosphate (cAMP)–responsive luciferase (Luc) reporter that exhibits optimal performance in high-throughput screens of agonist binding at G protein–coupled receptors (GPCRs). This reporter (RIP1-CRE-Luc) incorporates a nonpalindromic cAMP response element (CRE) originally identified within the 5′ promoter of the rat insulin 1 gene (RIP1). When multimerized and fused to the coding sequence of firefly luciferase, the CRE of RIP1 allows for the efficient activation of luciferase expression by cAMP-elevating agents or by cAMP itself. Of primary importance is the demonstration that RIP1-CRE-Luc does not exhibit the relatively high levels of basal luciferase activity inherent to reporters incorporating the palindromic CRE first identified in the somatostatin gene promoter. Furthermore, studies of HEK cells expressing class II GPCRs for the cAMP-elevating hormones GLP-1, GIP, and glucagon demonstrate that RIP1-CRE-Luc affords a much wider dynamic range of activation upon exposure to agonist. Such properties of RIP1-CRE-Luc indicate its usefulness as a new and powerful tool for the identification of small-molecule compounds with receptor-stimulating actions or for the identification of constitutively active orphan receptors with cAMP-signaling properties.
Keywords: cAMP response element, luciferase reporter, G protein–coupled receptor, drug discovery, high-throughput screen
INTRODUCTION
Reporter constructs incorporating the coding sequence of luciferase (Luc) are convenient tools with which to perform high-throughput screens of multiple cellular functions.1 Typically, these reporters are introduced into mammalian cell lines by transfection so that the level of Luc activity may be determined in whole-cell lysates or living cells in which Luc-catalyzed oxidation of luciferin generates photons detected by a photomultiplier tube or an imaging photon detector. When the expression of Luc is placed under the control of a minimal promoter incorporating a cyclic adenosine monophosphate (cAMP) response element (CRE), cAMP-elevating agents act via protein kinase A (PKA) and the CRE-binding protein transcription factor (cAMP response element binding protein; CREB) to activate Luc gene expression. This technology is particularly applicable to the screening of cell lines expressing recombinant G protein–coupled receptors (GPCRs) positively linked to cAMP production. For example, small molecules with GPCR agonist activity or orphan GPCRs with constitutive activity can be identified on the basis of their ability to stimulate Luc expression.2,3
Here we report the characterization of a novel cAMP-responsive Luc reporter that exhibits optimal performance in high-throughput screens of agonist binding at GPCRs. This reporter (RIP1-CRE-Luc) incorporates a nonpalindromic CRE (5′-TGACGTCC-3′) originally identified within the 5′ promoter of the rat insulin 1 gene (RIP1).4 When compared with the palindromic CRE (5′-TGACGTCA-3′) first identified in the promoter of the somatostatin (SOM) gene,5 the nonpalindromic CRE of RIP1 has a reduced binding affinity for CREB.6 Consistent with this prior finding, we now demonstrate that RIP1-CRE-Luc exhibits substantially reduced basal activity, a feature that is likely to be of particular importance when performing high-throughput screens in which false-positive hits are to be avoided. Of added benefit, we also find that when compared with SOM-CRE-Luc, the RIP1-CRE-Luc reporter affords a much wider dynamic range of activation (>340 fold-stimulation) upon exposure to GPCR agonists with cAMP signaling properties.
MATERIALS AND METHODS
Cell culture
HEK, COS, and CHO cells were obtained from the American Type Culture Collection (Rockville, MD). HEK cells stably expressing the human GLP-1 receptor were a gift of Novo Nordisk A/S (Bagsvaerd, Denmark).7 HEK cells stably expressing the rat glucose-dependent insulinotropic peptide (GIP) receptor8 or rat glucagon receptor9 were provided by Drs. T. J. Kieffer (University of British Columbia) and C. G. Unson (Rockefeller University), respectively. HEK and COS cells were cultured in DMEM containing 10% fetal bovine serum (FBS), whereas CHO cells were cultured in Ham’s F-12 containing 10% FBS. All ingredients for cell culture were from GIBCO BRL (Carlsbad, CA). The cells were maintained at 37 °C in a humidified incubator gassed with 5% CO2 and were passaged by trypsinization once per week.
Plasmids
SOM-CRE-Luc was obtained from Stratagene (La Jolla, CA). It contains a 5′ minimal promoter incorporating 4 multimerized palindromic CREs with the sequence 5′-(AGCC[TGACGTCA]GAG)-3′ (CRE denoted in brackets). The RIP1-CRE-Luc generated in our laboratory consists of 4 multimerized nonpalindromic CREs of the sequence 5′-AGCC[TGACGTCC]GAG-3′. RIP1-CRE-Luc emulates SOM-CRE-Luc in all respects, with the exception of a single nucleotide substitution (C for A) within the CRE. RIP1-CRE-Luc was generated by the synthesis of complementary synthetic oligonucleotides. The positive strand contained a Hind-III overhang at its 5′ terminus and corresponds to 5′-AGCTTAGCC-[TGACGTCC]GAGAGCC[TGACGTCC]GAGAGCC[TGACGTCC]GAGAGCC[TGACGTCC]GAGA-3′. The (−) strand contained a Bgl-II overhang at its 5′ terminus and corresponds to 5′-GATCTCTC-[GGACGTCA]GGCTCTC[GGACGTCA] GGCTCTC[GGACGTCA]GGCTCTC[GGACGTCA]GGCTA-3′. The (+) and (−) strand oligos were annealed and ligated into Hind-III and Bgl-II digested pLuc-MCS Cis-Reporting Cloning Vector (Stratagene).
Transfection
Twenty-four hours prior to transfection, cells were trypsinized, washed in complete medium, and plated at 5000 cells/well using a 96-well culture plate (Becton Dickinson 3075; Franklin Lakes, NJ). On day 2, the medium was removed and replaced with a transfection cocktail containing plasmid DNA (200 ng/well) and Lipofectamine Plus transfection reagents (Invitrogen, Carlsbad, CA) dissolved in serum-free medium. After 4 h incubation in this medium, the transfection cocktail was removed and replaced with normal cell culture medium containing serum. After an overnight incubation, the cells were exposed for 4 h to serum-free culture medium containing 0.1% bovine serum albumin and the indicated test substances. Cells were then lysed and assayed for luciferase-catalyzed photoemissions using a luciferase assay kit (Promega, Madison, WI) and a luminometer (TR-717; Applied Biosystems, Foster City, CA). Plasmid DNA for all experiments was obtained from transformed JM-109 cells and purified using the HiSpeed Plasmid Midi Kit (Qiagen, Venlo, the Netherlands).
cAMP determination
After plating HEK cells stably expressing the GLP-1 receptor onto a 96-well culture plate, and after overnight equilibration of the cells in culture medium, the medium was substituted with Krebs Ringer bicarbonate buffer containing 10 mM HEPES and the GLP-1 receptor agonist exendin-4 (Ex-4). The duration of exposure to Ex-4 was 30 min. Measurements of total nonacetylated cAMP content were then performed using a rabbit polyclonal cAMP antiserum (Direct Biotrak EIA kit; Amersham, Buckinghamshire, UK), a plate-reading spectrophotometer (BenchMark Plus; BioRad, Hercules, CA), and synthetic cAMP as a standard.
Sources of reagents
Ex-4 and forskolin were from Sigma (St. Louis, MO). H-89 was from Calbiochem (San Diego, CA). All cAMP analogs were from BIOLOG (Hayward, CA). The constitutively active GαS plasmid in pcDNA1 was obtained through the American Type Culture Collection. The dominant-negative GαS plasmid in pcDNA1 was from Dr. C. H. Berlot.10 Dominant-negative A-CREB in pRc/CMV vector was from Dr. C. Vinson.11
Data analysis
Data that are presented graphically were plotted using the ligand-binding module of SigmaPlot 10.0 (Systat Software, San Jose, CA). For dose-response relationships, the curves were fit according to the Hill-slope equation and nonlinear regression analysis. From the best fit, the EC50 and Hill-slope parameters were determined. Each experiment was repeated a minimum of 3 times to establish the reproducibility of these findings. Data presented in each graph are the mean ± standard deviation of triplicate determinations (n = 3 wells per data point) for a single experiment. Error bars are not provided because the standard deviation was less than or equal to 0.01 of the mean for each determination.
RESULTS AND DISCUSSION
Characterization of a novel luciferase reporter incorporating the CRE of RIP1
We compared the basal and agonist-stimulated activities of 2 Luc constructs in HEK cells stably expressing a GPCR for the cAMP-elevating hormone glucagon-like peptide-1-(7-36)-amide (GLP-1).7 These cells, referred to here as HEK-GLP-1-R cells, were exposed for 4 h to Ex-4, a synthetic peptide that is a stable, high-affinity agonist of the GLP-1 receptor.12 The 2 Luc constructs tested were the conventional SOM-CRE-Luc incorporating the palindromic CRE of the somatostatin gene promoter5 or RIP1-CRE-Luc, which incorporates the nonpalindromic CRE of the rat insulin 1 gene promoter.4
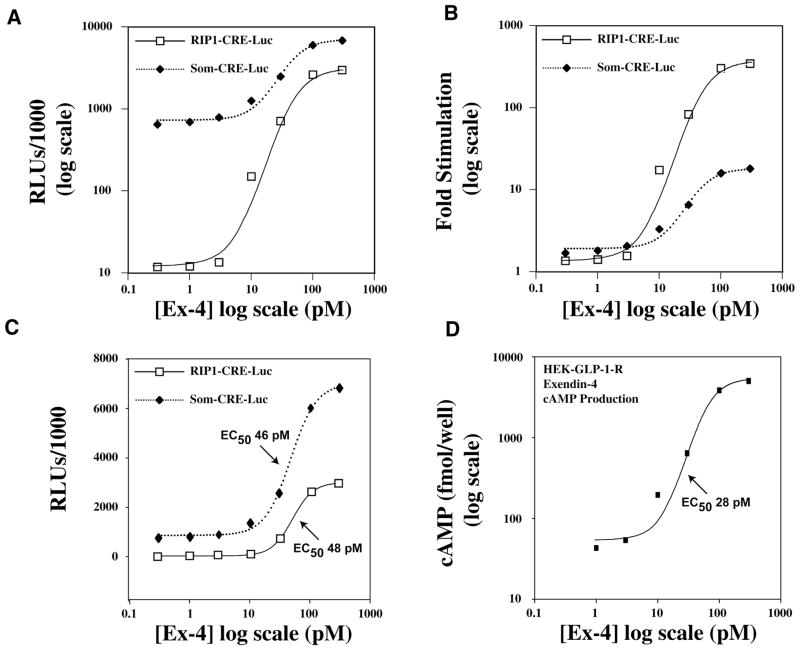
When HEK-GLP-1-R cells were exposed to a nonstimulatory concentration of Ex-4 (0.3 pM), the basal activity of SOM-CRE-Luc in whole-cell lysates was 55-fold higher than that of RIP1-CRE-Luc (Fig. 1A; Luc activities expressed as relative light units [RLUs]; note the log-log scales). Exposure of HEK-GLP-1-R cells to progressively higher concentrations of Ex-4 produced a dose-dependent increase of Luc activity, such that the response saturated at a concentration of about 300 pM (Fig. 1A). The maximal Luc activity determined for SOM-CRE-Luc was 2.3-fold greater than that of RIP1-CRE-Luc (6.8 × 106 and 2.9 × 106 RLUs, respectively). Thus, SOM-CRE-Luc exhibits a dramatically higher basal activity when compared with RIP1-CRE-Luc yet yields only a modestly higher peak effect under conditions of agonist stimulation. This conclusion is particularly evident when the action of Ex-4 is quantified as the fold stimulation of Luc activity relative to basal values. A saturating concentration of Ex-4 (300 pM) produced a 345-fold stimulation of RIP1-CRE-Luc activity but only an 18-fold stimulation of SOM-CRE-Luc activity (Fig. 1B ). The EC50 and Hill-slope values for Ex-4–stimulated RIP1-CRE-Luc activity in HEK-GLP-1-R cells were 48 pM and 2.5, respectively, values similar to those determined for the stimulatory action of Ex-4 at SOM-CRE-Luc (46 pM and 2.0, respectively; Fig. 1C).
FIG. 1.
Exendin-4 stimulates RIP1-CRE-Luc activity, SOM-CRE-Luc activity, and cyclic adenosine monophosphate (cAMP) production in HEK-GLP-1-R cells. (A) The log-log dose-response relationship describing the stimulatory action of exendin-4 (Ex-4) is illustrated over a concentration range of 0.3 to 300 pM of the peptide (x axis). Luciferase activity determined by luminometry of whole-cell lysates is quantified as relative light units (RLUs; y axis). (B) Same as in A except that the action of Ex-4 is quantified as the fold stimulation (y axis) of luciferase activity relative to basal levels of luciferase activity, as measured in the absence of Ex-4. (C) Illustrated is the semilogarithmic plot of the dose-response relationship for the stimulatory action of Ex-4 at RIP1-CRE-Luc and SOM-CRE-Luc. (D) The log-log plot describing the dose-response relationship for Ex-4–stimulated cAMP production in HEK-GLP-1-R cells. The duration of exposure to Ex-4 was 4 h in A through C and 30 min in D.
Validation that RIP1-CRE-Luc is an authentic cAMP-responsive luciferase reporter
To test for a positive correlation between Ex-4–stimulated Luc activity and the expected stimulatory action of Ex-4 on cAMP levels, cAMP production was determined using an enzyme immunoassay protocol that employs a cAMP-specific antiserum (see the Materials and Methods section). Ex-4 stimulated cAMP production in a dose-dependent manner in HEK-GLP-1-R cells not transfected with the luciferase reporter (Fig. 1D). The concentration-response relationship in terms of the threshold, EC50 (28 pM), and peak effect was close to that describing the stimulatory action of Ex-4 at RIP1-CRE-Luc (cf. Figs. 1C–D). Although not shown, a dose-response relationship nearly identical to that illustrated in Figure 1D was determined when examining the ability of Ex-4 to stimulate cAMP production in HEK-GLP-1-R cells transfected with RIP1-CRE-Luc (n = 2 experiments).
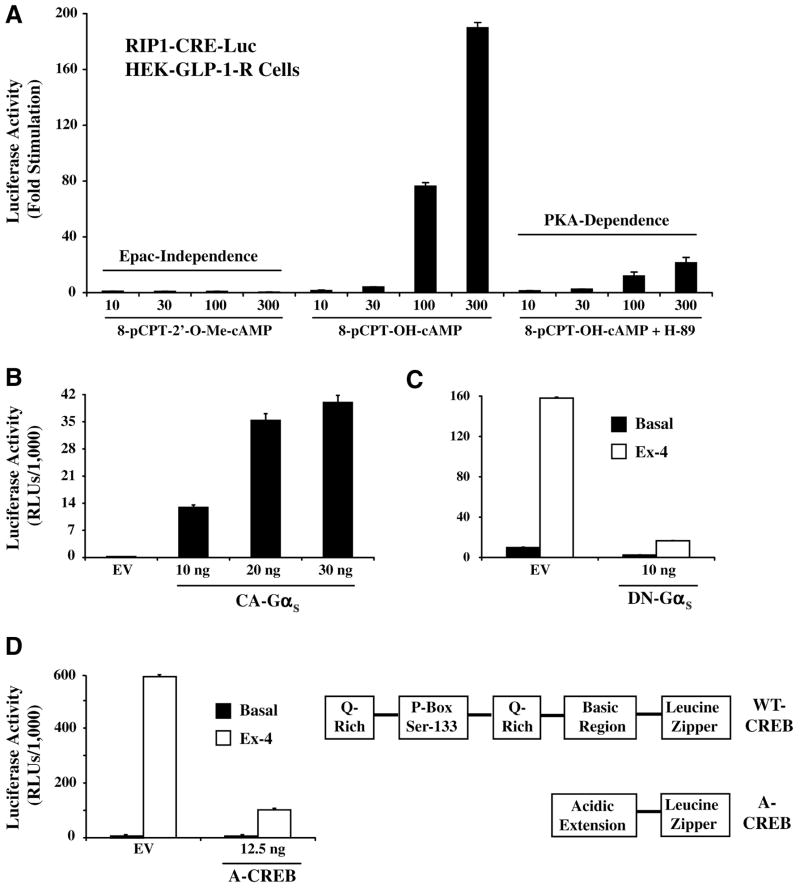
To more directly assess the cAMP responsiveness of RIP1-CRE-Luc, the stimulatory actions of cell-permeant cAMP analogs were evaluated. For this purpose, 2 structural analogs of cAMP were tested. 8-pCPT-2′-O-Me-cAMP is a selective activator of the Epac family of cAMP-regulated guanine nucleotide exchange factors (cAMPGEFs) that are expressed in HEK cells, whereas 8-pCPT-OH-cAMP activates not only Epac but also PKA. A 4-h exposure of HEK-GLP-1-R cells to 8-pCPT-OH-cAMP (10–300 μM) produced a dose-dependent stimulation of RIP1-CRE-Luc activity, whereas 8-pCPT-2′-O-Me-cAMP was entirely without effect (Fig. 2A). In these same cells, the stimulatory action of 8-pCPT-OH-cAMP was nearly abolished by H-89, an inhibitor of PKA activity (Fig. 2A). It may be concluded that the stimulatory action of cAMP at RIP1-CRE-Luc is PKA mediated and is unlikely to be dependent on the activation of Epac.
FIG. 2.
Cyclic adenosine monophosphate (cAMP)–dependent regulation of RIP1-CRE-Luc in HEK-GLP1-R cells. (A) 8-pCPT-2′-O-Me-cAMP (10-300 μM; left x axis) failed to stimulate RIP1-CRE-Luc, whereas RIP1-CRE-Luc was stimulated in a dose-dependent manner by 8-pCPT-OH-cAMP (10–300 μM; middle x axis). The action of 8-pCPT-OH-cAMP was nearly abrogated by pretreatment of cells with H-89 (10 μM; right x axis), an inhibitor of protein kinase A (PKA). (B) The basal activity of RIP1-CRE-Luc was stimulated by transient transfection with a constitutively active α subunit of GS (CA-GαS), whereas transfection with empty vector (EV) was without effect. (C) The stimulatory action of Ex-4 at RIP1-CRE-Luc was diminished by transient transfection with a dominant-negative α subunit of GS (DN-GαS) but not by EV. (D) Transfection with A-CREB, a dominant-negative cAMP response element binding protein (CREB), inhibited the action of Ex-4 at RIP1-CRE-Luc, whereas EV was without effect. Illustrated on the right is the structure of A-CREB relative to that of wild-type (WT) CREB. A-CREB is a genetically engineered isoform of CREB that dimerizes with endogenous CREB via a leucine zipper and an acidic extension.11 It lacks the glutamine-rich (Q-Rich) and phosphorylation box (P-Box) domains found in WT CREB. Dimerization of A-CREB with WT-CREB prevents the interaction of WT-CREB with the CRE. RLU = relative light unit.
We also found that the basal activity of RIP1-CRE-Luc was stimulated upon transfection of HEK-GLP-1-R cells with a constitutively active GαS (Fig. 2B). This is significant because GαS couples GLP-1 receptors to the stimulation of adenylyl cyclase, thereby generating cAMP. Furthermore, the stimulatory action of Ex-4 at RIP1-CRE-Luc was nearly abrogated following transfection with a dominant-negative GαS (Fig. 2C). This dominant-negative GαS fails to bind guanyl nucleotides and does not activate adenylyl cyclase. However, it retains its ability to interact with GPCRs, thereby preventing their functional interaction with endogenous GαS. 10 Because the action of Ex-4 at RIP1-CRE-Luc was also diminished following transfection with a dominant-negative A-CREB (Fig. 2D),11 it is apparent that GαS, adenylyl cyclase, PKA, and CREB link GPCR activation to cAMP-dependent stimulation of RIP1-CRE-Luc.
Class II GPCRs for GIP and glucagon mediate the stimulation of RIP1-CRE-Luc
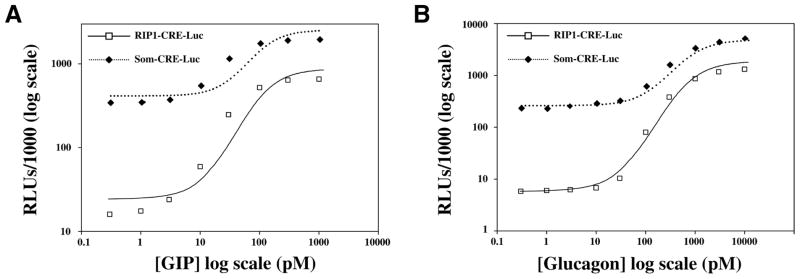
Agonist-dependent stimulation of RIP1-CRE-Luc was not simply a feature of HEK cells expressing the GLP-1-R. A dose-dependent stimulation of RIP1-CRE-Luc activity was also observed when HEK cells stably expressing GPCRs for GIP8 or glucagon9 were exposed to GIP or glucagon, respectively (Fig. 3A–B). The EC50 for GIP-stimulated RIP1-CRE-Luc activity was 38 pM, whereas that for glucagon was 135 pM. In contrast, the EC50 for GIP-stimulated SOM-CRE-Luc activity was 56 pM, whereas that for glucagon was 302 pM.
FIG. 3.
Assessment of agonist-stimulated luciferase reporter activity in HEK cells stably expressing class II G protein–coupled receptors (GPCRs) for glucose-dependent insulinotropic polypeptide (GIP) and glucagon. Illustrated are the dose-response relationships for GIP and glucagon-stimulated RIP1-CRE-Luc and SOM-CRE-Luc activities in HEK-GIP-R cells (A) and HEK-glucagon-R cells (B). The methods of experimental design were identical to those described for studies of HEK-GLP-1-R cells stimulated with exendin-4.
Cell line comparison of RIP1-CRE-Luc activation
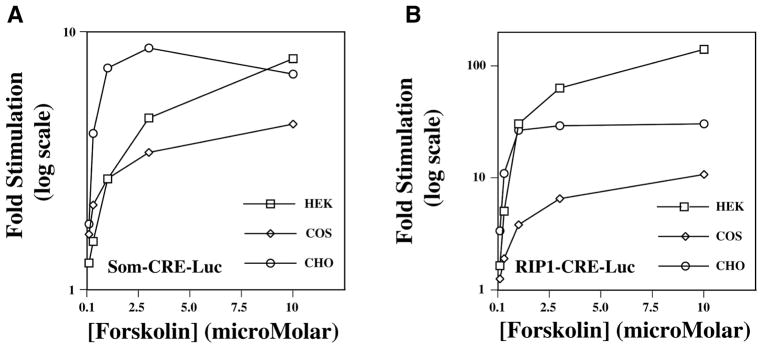
HEK, COS, and CHO cells are 3 cell lines frequently used for reporter gene assays. To compare the cAMP responsiveness of SOM-CRE-Luc and RIP1-CRE-Luc in each of these cell lines, the action of forskolin, a cAMP-elevating agent, was examined. For HEK cells, a 4-h exposure to forskolin produced a graded stimulatory effect at both SOM-CRE-Luc (Fig. 4A) and RIP1-CRE-Luc (Fig. 4B). This action of forskolin was observed over the full concentration range of 0.1 to 10 μM tested. A smaller but very similar graded response was also observed when examining the action of forskolin at either SOM-CRE-Luc or RIP1-CRE-Luc in COS cells (cf. Fig. 4A–B). However, for CHO cells, the activities of both reporters saturated at low concentrations of forskolin (cf. Fig. 4A–B). Importantly, for all 3 cell lines, the fold-stimulatory effect of forskolin at RIP1-CRE-Luc greatly exceeded that measured using SOM-CRE-Luc (Fig. 4A–B). For HEK cells, the action of forskolin at RIP1-CRE-Luc exceed that at SOM-CRE-Luc by a factor of >10 when the action of forskolin was expressed as the fold stimulation of luciferase activity relative to basal levels of activity. Taken together, these findings seem to indicate that for optimal assay performance, the screening of cAMP-elevating agents should be tailored to the use of HEK cells expressing RIP1-CRE-Luc. Given that considerable attention is presently focused on the identification of small molecules with agonist activity at specific subtypes of GPCRs, this RIP1-CRE-Luc–based assay should be of general utility for any project seeking to identity GPCR ligands with cAMP signaling potential.
FIG. 4.
Forskolin-stimulated luciferase activity in HEK, COS, and CHO cells. Illustrated is the dose-response relationship for forskolin-stimulated SOM-CRE-Luc activity (A) or RIP1-CRE-Luc activity (B).
A note of caution
It is interesting to note that prior studies of INS-1 insulin-secreting cells transfected with a luciferase reporter incorporating −410 bp of RIP1 demonstrate that GLP-1 and Ex-4 upregulate promoter activity in a CRE-mediated but PKA-independent manner.13,14 Evidently, the nonpalindromic CRE of RIP1 located between −177 and −184 bp of the promoter is regulated in a context-specific manner such that its sensitivity to PKA and CREB is diminished by adjacent regulatory elements.15 This allows for PKA-independent activation of the CRE by an as-yet-to-be identified transcription factor.14 In marked contrast, findings presented here demonstrate that this same nonpalindromic CRE, when isolated, multimerized, and fused to luciferase, is activated in HEK cells by Ex-4 in a PKA-dependent manner. A PKA-dependent stimulation of the nonpalindromic CRE is also observed when RIP1-CRE-Luc activity is measured in INS-1 cells, and in fact, SOM-CRE-Luc is also stimulated in a PKA-dependent manner in INS-1 cells (O. G. Chepurny, unpublished observations). Thus, when studied in isolation and devoid of adjacent regulatory elements, the nonpalindromic CRE of RIP1 behaves in a manner similar to that of the palindromic CRE, at least with respect to PKA sensitivity. Because this phenomenon of context specificity is not characteristic of the isolated CREs found within RIP1-CRE-Luc, it appears that RIP1-CRE-Luc is best suited for assays of GPCR function rather than for detailed mechanistic studies of CRE-dependent insulin gene expression.
Acknowledgments
G.G.H. acknowledges the support of the National Institutes of Health (R01-DK045817, R01-DK069575) and the American Diabetes Association (Research Grant Award).
References
- 1.Greer LF, III, Szalay AA. Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence. 2002;17:43–74. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- 2.Stables J, Scott S, Brown S, Roelant C, Burns D, Lee MG, et al. Development of a dual glow-signal firefly and Renilla luciferase assay reagent for the analysis of G-protein coupled receptor signalling. J Recept Signal Transduct Res. 1999;19:395–410. doi: 10.3109/10799899909036660. [DOI] [PubMed] [Google Scholar]
- 3.Chen G, Jayawickreme C, Way J, Armour S, Queen K, Watson C, et al. Constitutive receptor systems for drug discovery. J Pharmacol Toxicol Methods. 1999;42:199–206. doi: 10.1016/s1056-8719(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 4.Philippe J, Missotten M. Functional characterization of a cAMP-responsive element of the rat insulin I gene. J Biol Chem. 1990;265:1465–1469. [PubMed] [Google Scholar]
- 5.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci U S A. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oetjen E, Diedrich T, Eggers A, Knepel W. Distinct properties of the cAMP-responsive element of the rat insulin 1 gene. J Biol Chem. 1994;269:27036–27044. [PubMed] [Google Scholar]
- 7.Gromada J, Rorsman P, Dissing S, Wulff BS. Stimulation of cloned human glucagon-like peptide 1 receptor expressed in HEK 293 cells induces cAMP-dependent activation of calcium-induced calcium release. FEBS Lett. 1995;373:182–186. doi: 10.1016/0014-5793(95)01070-u. [DOI] [PubMed] [Google Scholar]
- 8.Kim SJ, Choi WS, Han JS, Warnock G, Fedida D, McIntosh CH. A novel mechanism for the suppression of a voltage-gated potassium channel by glucose-dependent insulinotropic polypeptide: protein kinase A-dependent endocytosis. J Biol Chem. 2005;280:28692–28700. doi: 10.1074/jbc.M504913200. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y, Cypess AM, Muse ED, Wu CR, Unson CG, Merrifield RB, et al. Glucagon receptor activates extracellular signal-regulated protein kinase 1/2 via cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 2001;98:10102–10107. doi: 10.1073/pnas.131200398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berlot CH. A highly effective dominant negative alpha s construct containing mutations that affect distinct functions inhibits multiple Gs-coupled receptor signaling pathways. J Biol Chem. 2002;277:21080–210805. doi: 10.1074/jbc.M201330200. [DOI] [PubMed] [Google Scholar]
- 11.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goke R, Fehmann HC, Linn T, Schmidt H, Krause M, Eng J, et al. Exendin-4 is a high potency agonist and truncated exendin-(9–39)-amide an antagonist at the glucagon-like peptide 1-(7–36)-amide receptor of insulin-secreting β-cells. J Biol Chem. 1993;268:19650–19655. [PubMed] [Google Scholar]
- 13.Skoglund G, Hussain MA, Holz GG. Glucagon-like peptide-1 stimulates insulin gene promoter activity by protein kinase A–independent activation of the rat insulin I gene cAMP response element. Diabetes. 2000;49:1156–1164. doi: 10.2337/diabetes.49.7.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chepurny OG, Hussain MA, Holz GG. Exendin-4 as a stimulator of rat insulin I gene promoter activity via bZIP/CRE interactions sensitive to serine/threonine protein kinase inhibitor Ro 31–8220. Endocrinology. 2002;143:2303–2313. doi: 10.1210/endo.143.6.8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggers A, Siemann G, Blume R, Knepel W. Gene-specific transcriptional activity of the insulin cAMP-responsive element is conferred by NF-Y in combination with cAMP response element-binding protein. J Biol Chem. 1998;273:18499–18508. doi: 10.1074/jbc.273.29.18499. [DOI] [PubMed] [Google Scholar]