Abstract
Phosphoinositide kinases such as PI3-kinase synthesize lipid second messengers that control diverse cellular processes. Recently, these enzymes have emerged as an important class of drug targets, and there is significant interest in discovering new lipid kinase inhibitors. We describe here a procedure for the high-throughput determination of lipid kinase inhibitor IC50 values. This assay exploits the fact that phosphoinositides, but not nucleotides such as ATP, bind irreversibly to nitrocellulose membranes. As a result, the radiolabeled lipids from a kinase assay can be isolated by spotting the crude reaction on a nitrocellulose membrane and then washing. We show that diverse phosphoinositide kinases can be assayed using this approach and outline how to perform the assay in 96-well plates. We also describe a MATLAB script that automates the data analysis. The complete procedure requires 3–4 h.
INTRODUCTION
Signaling by lipid kinases
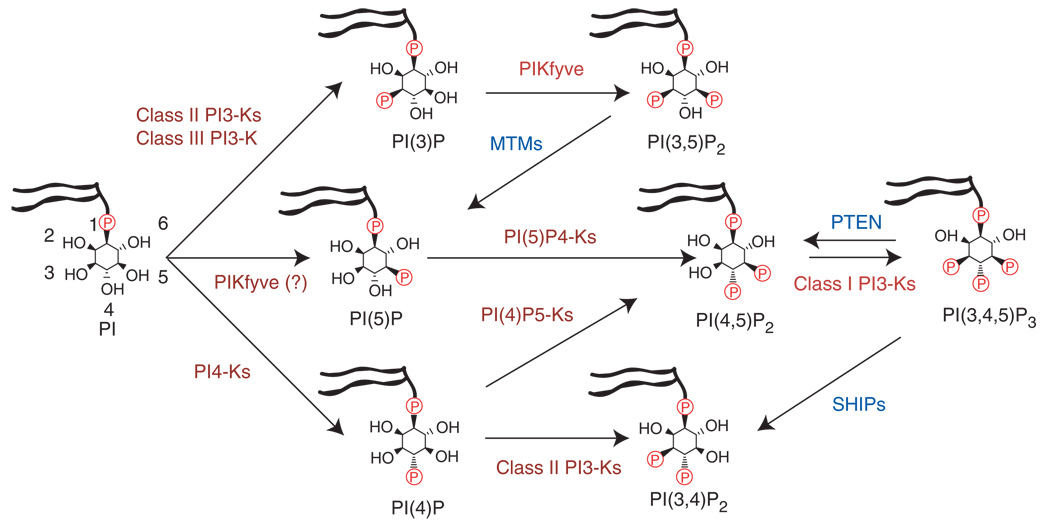
Phosphatidylinositol (PI) is an abundant component of all eukaryotic cell membranes. In response to upstream signals, lipid kinases phosphorylate PI at specific positions on the inositol head group to generate a spectrum of lipid second messengers (Fig. 1). These differentially phosphorylated lipids bind to and regulate the activity of diverse effector proteins, including protein kinases, ion channels, guanine-nucleotide exchange factors, phospholipases and adaptor proteins. In this way, lipid kinases are able to control a wide range of cellular processes.
Figure 1.
Routes for the synthesis of phosphoinositides in mammalian cells. Lipid kinases are shown in red and lipid phosphatases are shown in blue.
Recent interest in phosphoinositide signaling has been driven by studies of the class I PI3-kinases (p110α, p110β, p110δ and p110γ). These enzymes are activated by receptor tyrosine kinases and G-protein-coupled receptors to phosphorylate phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2), thereby generating the second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3; Fig. 1). PIP3 acts as a docking site at the plasma membrane for signaling proteins that include the protein kinases 3-phosphoinositide-dependent protein kinase-1 (PDK1) and Akt; these kinases are activated by PIP3 to promote nutrient uptake, suppress apoptosis and drive cell proliferation1. PI3-kinase signaling is antagonized by PTEN2, a lipid phosphatase that dephosphorylates PIP3 to generate PI(4,5)P2. The PI3-kinase pathway is frequently activated in solid tumors, and mutations in p110α, Ras (an upstream PI3-kinase activator3–5) and PTEN are among the most common genetic alterations in cancer6–9. In addition, there is considerable evidence that p110δ and p110γ may be useful targets for the treatment of inflammation and autoimmune diseases10–15. Together, these observations have stimulated widespread interest in identifying selective PI3-kinase inhibitors16.
In addition to the well-characterized class I PI3-kinases, several other families of lipid kinases have been identified (Fig. 1). These include the class II and III PI3-kinases, which synthesize phosphatidylinositol-3-phosphate (PI(3)P) and phosphatidylinositol-3,4-bisphosphate (PI(3,4)P2); two families of PI4-kinases (designated type II and type III) that phosphorylate PI to generate phosphatidylinositol-4-phosphate (PI(4)P); and a family of phosphatidylinositol phosphate kinases (PIP-kinases) that synthesize primarily PI(4,5)P2. Furthermore, these lipid kinases are antagonized by phosphatases that dephosphorylate specific positions on the inositol ring (Fig. 1). Together, these lipid kinases and phosphatases coordinate the synthesis of a complex pool of second messengers that regulate diverse aspects of cell biology.
Lipid kinase assays
Purified lipid kinases are assayed in vitro to characterize their enzymatic properties or identify small molecule inhibitors. Traditionally, lipid kinase activity has been assayed by monitoring phosphate transfer to lipid via thin-layer chromatography (TLC). In this approach, the kinase is allowed to phosphorylate lipid in the presence of [γ-32P]ATP; radiolabeled lipids are separated from [γ-32P]ATP by extracting into organic solvent followed by TLC; and the radioactivity in isolated TLC spots is detected by phosphorimaging or scintillation counting. This approach is highly sensitive, as phospholipid isomers are chromatographically separated from each other as well as from residual ATP. A limitation of this assay, however, is that it is labor intensive and of low-throughput due to the need for extraction and TLC steps. Recently, fluorescence-based assays for the class I PI3-kinases have been developed that overcome some of these difficulties through the use of protein domains that bind to PIP3 (ref. 17).
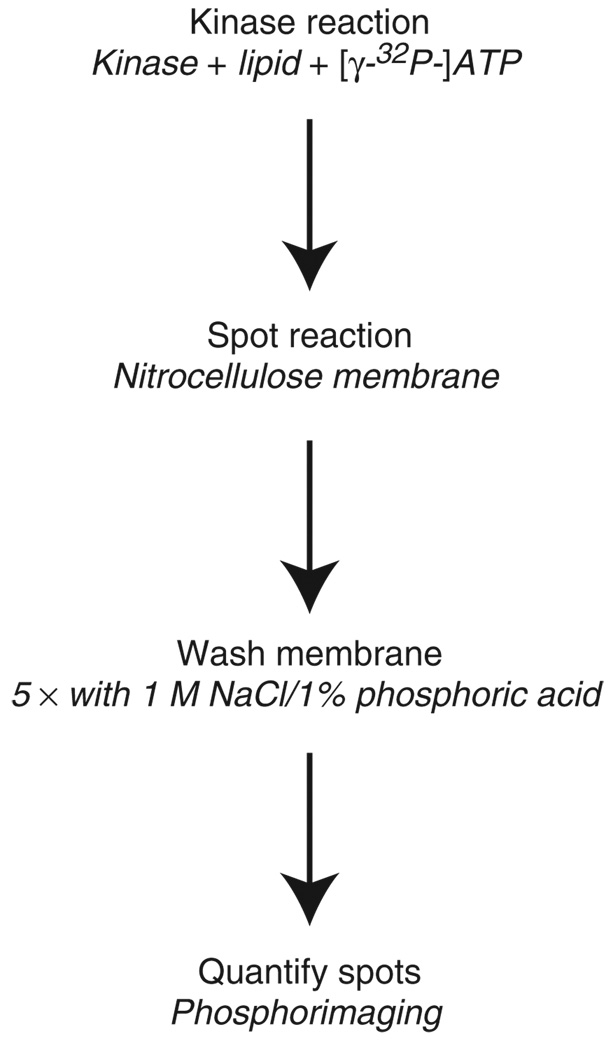
We have initiated an effort to discover pharmacological inhibitors of specific lipid kinases and characterize the selectivity of these molecules across the lipid kinome18,19. To this end, we have developed a simple lipid kinase assay to facilitate the high-throughput determination of lipid kinase inhibitor IC50 values on a laboratory scale. This assay exploits the fact that phosphoinositides, but not nucleotides such as ATP, bind irreversibly to nitrocellulose membranes through hydrophobic interactions. As a result, it is possible to isolate the radiolabeled lipids from a kinase assay by spotting the crude reaction on a nitrocellulose membrane and then washing (Fig. 2). Using this approach, we have assayed diverse lipid kinases in a single format and determined several thousand inhibitor IC50 values.
Figure 2.
Outline of the membrane capture lipid kinase assay.
We also describe here a MATLAB script (‘Spot’) that automates the analysis of phosphorimager data from this assay. By using this script, hand-spotted radioactivity can be counted quickly, uniformly and with minimal user intervention. The script, which utilizes MATLAB’s image analysis and statistics toolboxes, is available as MATLAB source code or as compiled executables for mac and pc platforms from http://www.ucsf.edu/shokat/SPOT.htm.
Assay validation
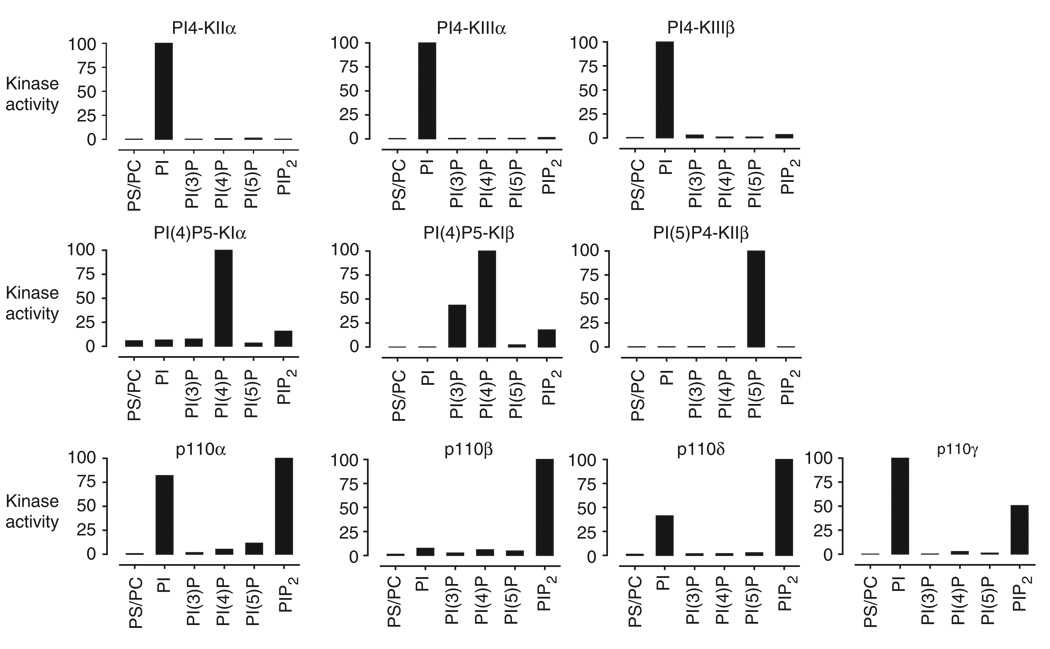
We have previously reported the profiling of lipid kinase inhibitors using this assay18. We describe here representative data illustrating features of this approach. Ten lipid kinases were assayed against a panel of phosphoinositides to characterize their lipid substrate specificity (Fig. 3). The observed substrate preferences in this assay closely mirror the reported biochemical selectivities of these enzymes. The three PI4-kinases each utilize exclusively PI as a substrate20–23. The PIP-kinases PI(4)P5-KIα and PI(4)P5-KIβ preferentially phosphorylate PI(4)P, whereas PI(5)P4-KIIβ phosphorylates PI(5)P24–26. The class I PI3-kinases (p110α, p110β, p110δ and p110γ) phosphorylate either PI or PI(4,5)P2 in a ratio that varies across the four isoforms. PI(4,5)P2 is the primary substrate of these enzymes in vivo, but PI is used preferentially in vitro under many assay conditions. The relative phosphorylation of these two substrates in vitro is known to depend on the identity and composition of other lipids in the membrane bilayer27. Overall, these data show that diverse lipid kinases that preferentially utilize at least four different phosphoinositide substrates can be assayed using the membrane capture approach.
Figure 3.
Substrate specificity of lipid kinases. Kinases were incubated in the appropriate reaction buffer with [γ-32P]ATP (5 µCi per each 50 µl reaction), nonradioactive ATP (10 µM), BSA (0.5 mg ml−1) and each phosphoinositide (0.1 mg ml−1) in the presence of the carrier lipids PS/PC (0.1 mg ml−1). Reactions were spotted on nitrocellulose, washed and assayed by phosphorimaging. Kinase activity was normalized to 100% for the preferred lipid substrate. PIP2 denotes PI(4,5)P2.
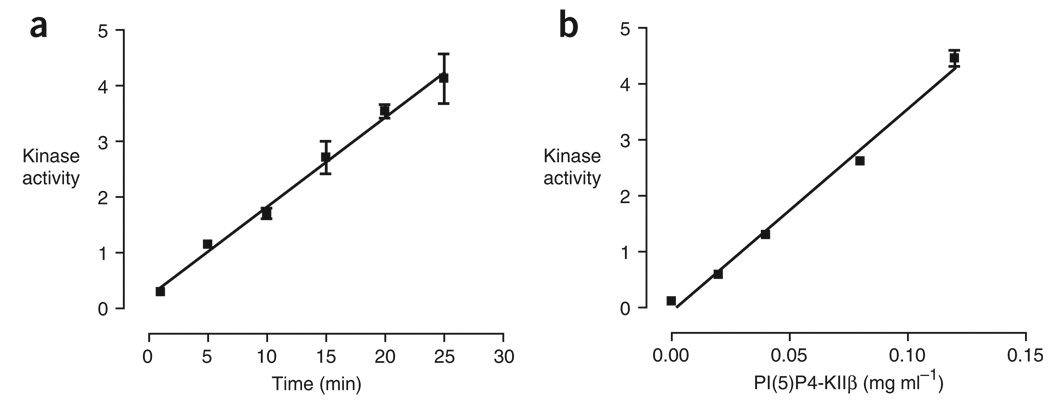
Signal strength in this assay depends on length of time that the kinase reaction is allowed to proceed and the amount of enzyme used (Fig. 4). We have not observed saturation of the phosphoinositide binding capacity of nitrocellulose under the assay conditions described here, although, as in any enzymatic assay, it is possible to deplete substrate at sufficiently high enzyme concentrations. The optimal concentration of kinase for this assay depends on the specific activity of the enzyme under the conditions used, and this varies greatly across lipid kinases (see General assay considerations).
Figure 4.
The membrane capture assay of PI(5)P4-KIIβ is linear with respect to time and enzyme concentration. (a) PI(5)P4-KIIβ (0.12 mg ml−1) was assayed in 25 mM HEPES (pH 7.4), 10 mM MgCl2 with [γ-32P]ATP (5 µCi per reaction), nonradioactive ATP (10 µM), BSA (0.5 mg ml−1) and PI(5)P (0.1 mg ml−1) in the presence of the carrier lipids PS/PC (0.1 mg ml−1). Reactions were terminated at the indicated times by spotting on nitrocellulose, followed by washing and quantification by phosphorimaging. Y axis represents arbitrary relative kinase activity. (b) Kinase reactions were performed as described in a with varying concentrations of PI(5)P4-KIIβ. The reactions were allowed to proceed for 15 min. Y axis represents arbitrary relative kinase activity.
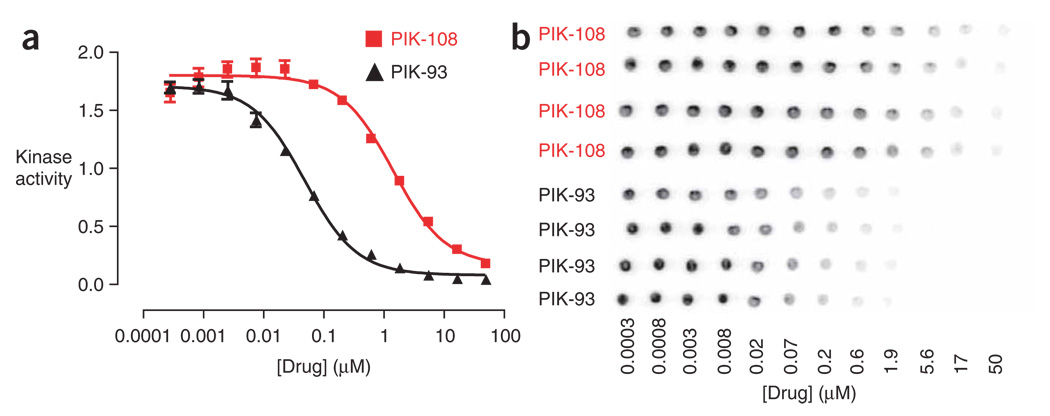
Figure 5 depicts typical data for the determination of IC50 values of two inhibitors (PIK-108 and PIK-93) against a lipid kinase (p110α). In this experiment, inhibitor was arrayed in 5 µl of 10% DMSO across eight rows of a 96-well PCR plate; PCR plates are used to facilitate handling small reaction volumes (25 µl), thereby minimizing the consumption of kinase and [γ-32P]ATP. The inhibitor was aliquoted in threefold dilutions to achieve final assay concentrations that range from 50 to 0.0003 µM, thereby spanning the predicted IC50 value for the drug. Kinase was then added to each well of the plate in 10 µl of a solution containing kinase reaction buffer, lipid and BSA. To initiate the kinase reaction, 10 µl of a solution containing 2 µCi [γ-32P]ATP was added to each well of the plate (adjusted to achieve a final ATP concentration of 10 µM in the assay). The kinase reaction was allowed to proceed for 30 min, at which point the reaction was terminated by spotting 4 µl from each well onto a nitrocellulose membrane. All liquid transfers were performed using a multi-channel pipettor to initiate and terminate the assay consistently. The membrane was then washed four times for 15 min each with 100–200 ml of wash solution (1 M NaCl/1% phosphoric acid). The last wash was allowed to proceed overnight. After allowing the membrane to dry, it was exposed to a phosphorimager screen to generate the raw data shown in Figure 5b. Quantification of these spots using the MATLAB script ‘Spot’ was performed as shown in Figure 6 to yield the dose–response data shown in Figure 5a. IC50 values for each inhibitor were obtained by fitting a sigmoidal dose–response curve to these data using the Prism software package.
Figure 5.
Determination of IC50 values for p110α with the PI3-kinase inhibitors PIK-108 and PIK-93 using the membrane capture assay. (a) Quantitated dose–response data for each compound. Y axis represents arbitrary relative PI3-K activity. IC50 (PIK-108) = 1.4 µM, IC50 (PIK-93) = 0.048 µM. (b) Raw dose–response data for each compound. Compounds were assayed in quadruplicate at threefold dilutions across the range 50–0.0003 µM.
Figure 6.
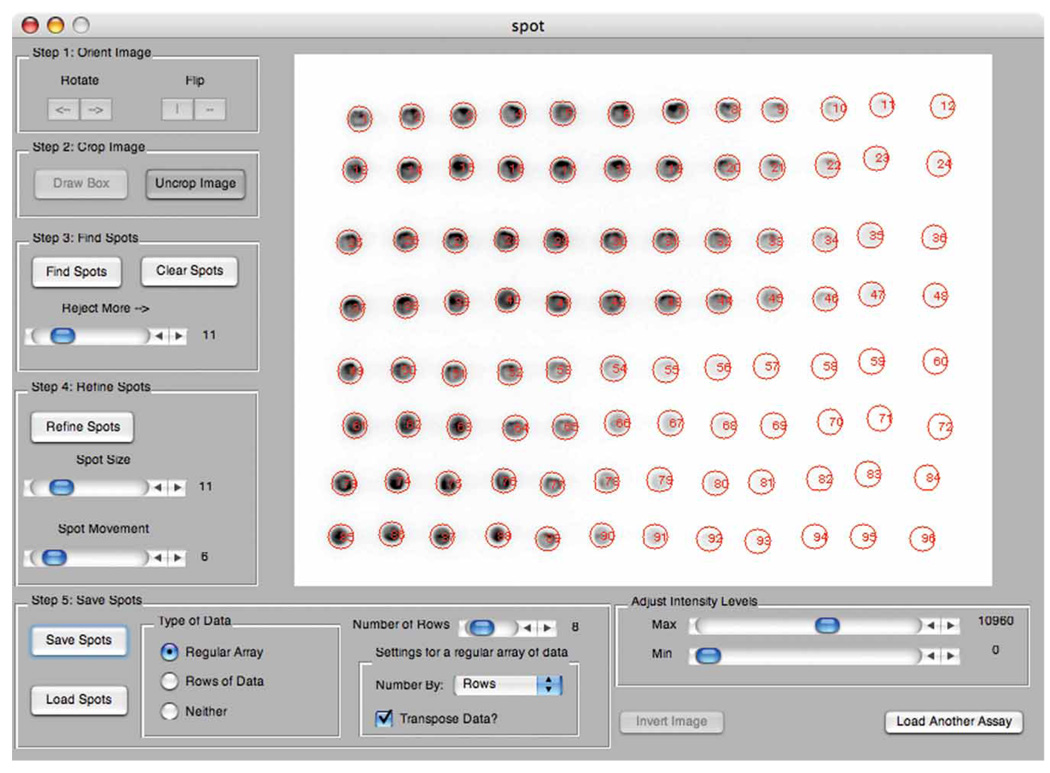
‘Spot’ image analysis tool. Screenshot depicting the analysis of dose–response data from Figure 5 using the MATLAB image analysis script Spot. Automatic location, refinement and integration of radioactive spots were accomplished by following the five steps indicated on the left panel of the graphical interface.
General assay considerations
The specific activity of the lipid kinase is the major determinant of the signal-to-background ratio in this assay. Extended washing removes the vast majority of radioactive ATP and phosphate from the nitrocellulose membrane, but trace amounts of radioactive material will adhere to the membrane independent of kinase activity. For relatively active lipid kinases such as the class I PI3-kinases or the PI4-kinases, we have found that a final kinase concentration in the assay of ~ 1 µg ml−1 (~ 5 nM) typically yields a signal-to-background ratio greater than 10. At this concentration, ~ 2 µg of enzyme is sufficient to assay a 96-well plate, which is cost efficient for most purposes. Other lipid kinases, such PI(5)P4-KIIβ, have very low specific activity under all assay conditions we have tested, and for these kinases it may be necessary to use 50- to 100-fold more enzyme to achieve similar results. Similar considerations apply to the analysis of kinase activity from immunoprecipitates. However, as the 96-well assay format described below requires a homogenous source of enzyme, immunoprecipitated kinases must be eluted from the beads before assaying in 96-well plates.
For each kinase preparation, it is necessary to empirically determine the appropriate enzyme concentration before starting high-throughput assays. As a general guideline, one should aim for a radioactive signal that is at least tenfold greater than negative controls in which either the kinase or the lipid has been omitted. During the optimization of enzyme concentration, it is also advisable to test several different reaction lengths (e.g., 30, 60 and 120 min) to assess the stability of the enzyme and the potential for enhanced signal-to-background ratio at longer reaction times.
The signal-to-background ratio in this assay is largely independent of the concentration of radioactive ATP, because the signal and background are both expected to increase as the concentration of radioactive ATP is increased (the background is caused by radioactivity adhering nonspecifically to the nitrocellulose membrane). The concentration of radioactive ATP primarily determines the overall signal strength of the assay and, therefore, the length of time that the membrane must be exposed to the phosphorimager screen. We typically use a radioactive ATP concentration of 0.1 Ci µl−1 with a total ATP concentration of 10 µM. If desired, the radioactive ATP concentration can be decreased at least fivefold without impairing the assay. [γ-33P]ATP can also be substituted for [γ-32P]ATP.
The ATP concentration used in this assay influences the IC50 value that is measured for a kinase inhibitor. For an ATP competitive kinase inhibitor, the IC50 value is related to the Ki of the inhibitor, the KM,ATP of the kinase and the ATP concentration by the Cheng–Prusoff equation28,29: IC50 = Ki(1 + [ATP]/KM,ATP). At ATP concentrations below the KM,ATP of the kinase, the IC50 value approximates the Ki. At ATP concentrations above the KM,ATP of the kinase, the measured IC50 value exceeds the Ki. The KM,ATP has been reported for many protein and lipid kinases28. Because the intracellular ATP concentration is 1–5 mM30,31, most kinase inhibitors appear less potent in cells than when assayed at lower ATP concentrations (typically 10–100 µM) in vitro.
The source and physical form of the lipids used for the assay can be important in some cases. Commercially available phosphoinositides may be purified from natural sources or synthesized chemically. We have found that both natural and synthetic lipids are compatible with the membrane capture assay, although in some cases natural lipids are less expensive. Synthetic lipids are often available in several hydrocarbon lengths (e.g., C8 and C16); we recommend using the longer hydrocarbons to ensure irreversible binding to the nitrocellulose membrane. The purity of commercial phosphoinositides is variable, and impurities in commercial preparations have led to the misassignment of lipid kinase substrate specificity26. For experiments in which the purity of the phosphoinositide is critical, commercial phosphoinosites should be independently characterized by HPLC or TLC. In this respect, the membrane capture assay does not distinguish between different phosphoinositides that may be generated by a kinase that phosphorylates a single lipid at multiple sites. These considerations are less relevant for the determination of inhibitor IC50 values using well-characterized lipid kinases.
The protocol outlined below describes how to perform lipid kinase assays in either individual microcentrifuge tubes or 96-well plates. The former protocol is appropriate for determining the optimal enzyme concentration and troubleshooting the assay. The 96-well plate procedure facilitates the rapid determination of inhibitor IC50 values.
MATERIALS
REAGENTS
Purified lipid kinase. Class I PI3-kinases are commercially available (Upstate). Other lipid kinases can be expressed and purified using published techniques
Purified phosphoinositides (Avanti Polar Lipids, Sigma)
1:1 dioleoyl phosphatidylserine/dioleoyl phosphatidylcholine (DOPS/DOPC) carrier lipid (Avanti Polar Lipids)
Tris (Sigma)
MgCl2 (Sigma)
Nitrocellulose (Bio-Rad)
Phosphoric acid (Sigma)
 Phosphoric acid is corrosive.
Phosphoric acid is corrosive.NaCl (Sigma)
ATP (Sigma)
[γ-32P]ATP (Perkin-Elmer)
 Radioactive. Suitable screening should be used.
Radioactive. Suitable screening should be used.96-well assay plates (VWR, cat. no. 82006-636)
96-well plate covers (Eppendorf, cat. no. 3211-360)
PCR tube strips as reagent reservoirs (VWR, cat. no. 20170-0074)
PCR tube holder (VWR, cat. no. 80086-084)
EQUIPMENT
Multichannel pipettors (Finnpipette; Thermo Fisher)
Water bath sonicator (VWR, cat. no. 50HT)
Phosphorimager (GE Healthcare)
Rocking platform (VWR, cat. no. 40000-300)
Heat lamp (optional) (Fisher, cat. no. 11-504-50)
REAGENT SETUP
10× kinase assay buffer (all kinases except PI4-Ks and VPS34) 250 mM HEPES (pH 7.4) and 100 mM MgCl2.
10× kinase assay buffer for VPS34 500 mM Tris (pH 8.0) and 200 mM MnCl2. Add MnCl2 immediately before use.
10× kinase assay buffer for PI4-Ks 500 mM Tris (pH 7.4), 200 mM MgCl2 and 4% (vol/vol) Triton X-100.
10× lipid stock solution for PI Suspend the solid PI in water to a final concentration of 1 mg ml−1. Sonicate in a water bath sonicator for 20 s to generate lipid vesicles. This solution can be stored at 4 °C for at least 1 month. PI can also be assayed in the presence of DOPS/DOPC (below) if desired.
10× lipid stock solution for all other phosphoinositides Prepare a 1mg ml−1 solution of the carrier lipids DOPS/DOPC (1:1). Sonicate this solution in a water bath sonicator for 20 s. Suspend the solid phosphoinositide in this solution to a final concentration of 1 mg ml−1. Sonicate this solution in a water bath sonicator for 20 s to generate lipid vesicles. This solution can be stored at 4 °C for at least 1 month.
100× BSA stock solution Prepare a 50 mg ml−1 solution of BSA in water. This can be stored in frozen aliquots at −20 °C indefinitely.
1,000× ATP stock solution Prepare a 10 mM solution of ATP in 25 mM Tris (pH 7.4) and 10 mM EDTA. This can be stored in frozen aliquots at −20 °C for at least 1 year.
2.5× buffer–substrate solution On the day of the assay, prepare a 2.5 × solution containing kinase buffer, lipid and BSA by mixing 10× kinase assay buffer, 10× lipid stock solution and water in a 1:1:2 ratio by volume. Dilute the 100× BSA stock solution 40-fold into this solution. Store this buffer–substrate on ice.
2.5× ATP solution On the day of the assay, prepare a 2.5× ATP solution by diluting 1,000× ATP stock solution 400-fold into water. Add [γ-32P]ATP to a final concentration of 0.1–0.25 µCi µl−1. Store this diluted ATP solution on ice.
 This solution is radioactive. Follow appropriate safety procedures for handling 32P.
This solution is radioactive. Follow appropriate safety procedures for handling 32P.Membrane wash solution 1 M NaCl/1% phosphoric acid.
PROCEDURE
Preparing the assay  1–2 h
1–2 h
Prepare the stock solutions described above.
Cut a rectangular piece of nitrocellulose to a size that will fit in a washbasin. Lids from P-1000 pipette tip boxes make convenient washbasins for this assay and accommodate nitrocellulose membranes of approximately 12 cm × 8.5 cm.
-
Mark the nitrocellulose membrane with a pencil to indicate where the kinase reactions will be spotted. Follow option A for assays in individual microcentrifuge tubes and option B for assays in 96-well plates.
-
Assays in individual microcentrifuge tubes
Place numbered marks on the membrane at least 1 cm apart. A total of 12–15 marks can be conveniently placed on a 12 cm × 8.5 cm membrane.
-
Assays in 96-well plates
Mark the upper left-hand corner of the membrane with a pencil. Use this mark to orient the membrane with respect to the 96-well assay plate after the kinase reactions have been spotted.
-
-
Aliquot inhibitor to each kinase reaction. The final concentration of DMSO in the assay should not exceed 2%. Follow option A for assays in individual microcentrifuge tubes and option B for assays in 96-well plates.
-
Assays in individual microcentrifuge tubes
Label the appropriate number of microcentrifuge tubes.
For a final reaction volume of 50 µl, aliquot 10 µl of inhibitor in 10% DMSO to each tube in this step. If not testing any inhibitors, aliquot 10 µl of water instead.
-
Assays in 96-well plates
Prepare a 5× 96-well stock plate containing serial dilutions of the inhibitors to be tested in 10% DMSO. These plates can be stored at −20 °C.
For a final reaction volume of 25 µl, transfer 5 µl of inhibitor in 10% DMSO to each well of a 96-well assay plate. Use a 96-well PCR plate as the assay plate to facilitate handling 25 µl reaction volumes.
-
Prepare the 2.5× buffer–substrate and ATP solutions described above. Determine the appropriate volume of each by multiplying the number of kinase reactions by 20 µl (for assays in microcentrifuge tubes) or 10 µl (for assays in 96-well plates) and then adding at least 10% extra to account for sample losses during pipetting. Store these solutions on ice.
Dilute the appropriate amount of kinase into freshly made 2.5× diluted kinase assay buffer. Store on ice. For the class I PI3-kinases, a final enzyme concentration of ~ 1 µg ml−1 in the assay is a good starting point.
-
Aliquot the enzyme–buffer–substrate solution to each reaction. Follow option A for assays in individual microcentrifuge tubes and option B for assays in 96-well plates.
-
Assays in individual microcentrifuge tubes
For a final reaction volume of 50 µl, aliquot 20 µl of enzyme–buffer–substrate solution to each tube. Mix by tapping the tube.
-
Assays in 96-well plates
Place a strip of 12 PCR tubes in a PCR tube holder. This will serve as a reservoir for transferring the enzyme–buffer–substrate solution to the assay plate with a multichannel pipettor.
Aliquot enzyme–buffer–substrate solution to each of the PCR tubes. Calculate the amount of solution needed by multiplying the number of rows being assayed by 10 µl. For example, a single 96-well plate requires at least 80 µl of solution per PCR tube. Always include at least 10% extra solution in the reservoir to account for sample loss during transfer.
Transfer 10 µl of solution to each well of the assay plate using a multichannel pipettor. Mix by pipetting up and down five times.
-
Performing the kinase reaction  ~ 1 h
~ 1 h
-
8
Start the assay by adding the 2.5× ATP solution to each reaction. Allow the reaction to proceed for 30 min or for designated length of time before terminating in Step 11. Follow option A for assays in individual microcentrifuge tubes and option B for assays in 96-well plates.
-
Assays in individual microcentrifuge tubes
-
Add 20 µl of the diluted ATP solution to each tube and mix by tapping.
 From this point onward, the procedure involves radioactive components. Take appropriate precautions.
From this point onward, the procedure involves radioactive components. Take appropriate precautions.
-
-
Assays in 96-well plates
Place a strip of 12 PCR tubes in a PCR tube holder. This will serve as a reservoir for transferring the 2.5× ATP solution to the assay plate with a multichannel pipettor.
-
Aliquot 2.5× ATP solution to each of the PCR tubes. Calculate the amount of solution needed by multiplying the number of rows being assayed by 10 µl. For example, a single 96-well plate requires at least 80 µl of solution per PCR tube. Always include at least 10% extra solution in the reservoir to account for sample loss during transfer.
 From this point onward, the procedure involves radioactive components. Take appropriate precautions.
From this point onward, the procedure involves radioactive components. Take appropriate precautions. Transfer 10 µl of solution to each well of the assay plate using a multichannel pipettor. Mix by pipetting up and down five times. Tap the plate gently on the bench to ensure that all the liquid collects at the bottom of each well.
-
-
9
Place a sheet of plastic wrap on the bench adjacent to the kinase reaction. Place several paper towels on top of the plastic wrap, and then place the marked nitrocellulose membrane on top of the paper towels. These will absorb any radioactivity that passes through the nitrocellulose.
-
10
Terminate the assay by spotting the reactions onto the nitrocellulose membrane. Follow option A for assays in individual microcentrifuge tubes and option B for assays in 96-well plates.
-
Assays in individual microcentrifuge tubes
Spot 4 µl from each reaction onto the designated mark on the nitrocellulose membrane. When spotting is complete, allow lipids to adhere to the membrane for an additional 2 min. Discard the remainder of the kinase reactions and the paper towels in an appropriate container for solid 32P radioactive waste.
-
Assays in 96-well plates
Spot 4 µl from each well onto the nitrocellulose membrane using a multichannel pipettor. Allow approximately 1 cm between rows of spots. When spotting is complete, allow lipids to adhere to the membrane for an additional 2 min. Discard the remainder of the kinase reactions and the paper towels in an appropriate container for solid 32P radioactive waste.
-
Washing the membrane  2 h to overnight
2 h to overnight
-
11
Place the membrane in its own washbasin and add 100–200 ml of wash solution to the basin. Mix with gentle rocking for 30 s and then discard the wash solution in an appropriate container for liquid 32P radioactive waste.
-
12
Add 100–200 ml of wash solution to the basin and mix with gentle rocking for 5 min. Discard the wash solution in an appropriate container for liquid 32P radioactive waste, and then repeat for at least four additional washes. After the third wash, the residual radioactivity removed in the wash is often sufficiently low that it can be disposed of down the sink rather than as radioactive waste.
 The final wash can be allowed to proceed overnight if desired.
The final wash can be allowed to proceed overnight if desired.
Imaging the membrane  ~ 1–2 h
~ 1–2 h
-
13
Remove the membrane and allow it to dry for at least 1 h. Drying can be accelerated by placing the membrane under a heat lamp.
-
14
Wrap the membrane in plastic wrap to avoid transferring radioactivity to the phosphor screen. Tape the wrapped membrane to the phosphor screen cassette and expose to the screen. The optimal length of exposure will depend on the signal in the assay. Class I PI3-kinases assayed using this procedure typically require approximately 15-min exposures.
-
15
Scan the phosphor screen using a phosphorimager such as the Storm or Typhoon instruments (GE Healthcare). The intensity of each spot can be quantified by densitometry using the software supplied with the phosphorimager. Alternatively, follow Steps 16–22 to quantify the data using the MATLAB script Spot.
 The data can be analyzed later if desired.
The data can be analyzed later if desired.
Using Spot to quantify the phosphorimager data  ~ 10 min
~ 10 min
-
16
At the MATLAB command, prompt type ‘spot’ to start the software. Use the dialog box to choose an image file for analysis. Typhoon Gel files as well as Tiffs are accepted.
-
17
The graphical interface is organized by steps to help guide the user. Each step is a set of software buttons and options grouped together within a box. Use the ‘Rotate’ and ‘Flip’ button within ‘Step 1’ to adjust the image orientation.
-
18
Click on ‘Draw Box’ within ‘Step 2’ and draw a box on the image that only contains the spots to be analyzed. Click on ‘Crop Image’ to focus on the spots to be analyzed.
-
19
Click on ‘Find Spots’ within ‘Step 3’. If too few spots are found, adjust the rejection slider to a lower value, and if too many are found, move the slider to a higher value. Left click on the image to manually identify a spot. Right click on the edge of an already identified spot to delete it. Left click and hold on a spot identifier to move it.
-
20
Use the ‘Spot Size’ slider in ‘Step 4’ to adjust the spot size so that the darkest spots are just contained within the spot indicators. Adjust the intensity level of the image to confirm that low-intensity pixels are contained within the spot indicators. Click ‘Refine Spots’ to optimize the location of each spot. If the spots move too far, adjust the ‘Spot Movement’ slider to a lower value and return to Step 19.
-
21
Before saving the spots, click on the button that best indicates the type of data (Regular Array, Rows of Data or Neither). For data in rows or in a regular array, specify the number of rows. For data in a regular array, also specify whether to number the data by rows or columns and whether to transpose the data before writing it to file. Click ‘Save Spots’ and assign a file name in the dialog box. A tab-delimited text file will be written. If the spots are misnumbered, see the troubleshooting advice in Table 1. After saving the file, the data will also be available on the clipboard.
-
22
To view previously saved spots, first perform Steps 16 and 17 and then press ‘Load Spots’. At the file selection dialog, choose a file created previously at Step 21.

TABLE 1.
Troubleshooting table.
| Problem | Possible reason | Solution |
|---|---|---|
| Low signal-to-background ratio | Kinase concentration too low | Increase the concentration of kinase |
| Insufficient washing of membrane |
Wash at least six times and allow the final wash to proceed overnight | |
| Low kinase activity | Test the effect of additives such as DTT and BSA on kinase activity; allow kinase reaction to proceed longer than 30 min; confirm that you are using the correct phosphoinositide at a final assay concentration of at least 0.1 mg ml−1; test other sources of lipid kinase (e.g., commercial p110α) as a positive control |
|
| Membrane spots bleed into each other |
Spotting too large a volume | Spot a maximum of 4 µl when using a multichannel pipettor. Note that spots may appear to bleed to some extent when initially spotted, but this generally does not affect the assay, because phosphoinositides adhere to the nitrocellulose quickly and therefore do not migrate across the membrane |
| Spots on the right or left side of the membrane after transferring from a 96-well plate are consistently of lower intensity than spots in the middle |
Multichannel pipetting error | When loading a multichannel pipettor with tips, the tips on the right and left sides may not fully seal, such that they do not transfer the full volume. To avoid this, firmly roll the pipettor from side to side (rather than forcefully up and down) when loading tips |
| Spots are misnumbered by the Spot script |
Rows of spots are overlapping | Clear the current spot identifiers by clicking ‘Clear Spots’. Manually add new spot identifiers by left clicking on the image. Add the spot identifiers in the order they should be numbered. Click ‘Refine Spots’ to refine their positions. Before saving, specify ‘Neither’ as the data type to number the spots in the order they were selected |
Steps 1–7, preparing the assay: 1–2 h
Steps 8–10, performing the kinase reaction: ~ 1 h
Steps 11 and 12, washing the membrane: 2 h to overnight
Steps 13–15, imaging the membrane: ~ 1–2 h
Steps 16–22, using Spot to quantify the phosphorimager data: ~ 10 min
![]()
Troubleshooting advice can be found in Table 1.
ANTICIPATED RESULTS
The membrane capture procedure can be used to screen small-molecule inhibitors against many different lipid kinases using a single assay format. The assay is of relatively high-throughput, yet requires no automation. Following the procedure described here, a single technician can expect to comfortably assay approximately ten 96-well plates in a day. As the principles of the assay are simple, many variations are possible for different applications.
ACKNOWLEDGMENTS
Z.A.K. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. M.E.F. is an ARCS Foundation Fellow. We acknowledge funding from NIH training grant GM08284. K.M.S. received funding from the Howard Hughes Medical Institute. The research of T.B. and A.B. was supported by the Intramural Research Program of the National Institute of Child Health and Human Development of the National Institutes of Health. We thank James Hurley for the generous gift of PI(5)P4-KIIβ.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Katso R, et al. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 2.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 3.Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Viciana P, et al. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Viciana P, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 6.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 7.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br. J. Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow LM, Baker SJ. PTEN function in normal and neoplastic growth. Cancer Lett. 2006;241:184–196. doi: 10.1016/j.canlet.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 9.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 10.Okkenhaug K, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 11.Ali K, et al. Essential role for the p110delta phosphoinositide 3-kinase in the allergic response. Nature. 2004;431:1007–1011. doi: 10.1038/nature02991. [DOI] [PubMed] [Google Scholar]
- 12.Wymann MP, et al. Phosphoinositide 3-kinase gamma: a key modulator in inflammation and allergy. Biochem. Soc. Trans. 2003;31:275–280. doi: 10.1042/bst0310275. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch E, et al. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 14.Camps M, et al. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat. Med. 2005;11:924–955. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 15.Barber DF, et al. PI3Kgamma inhibition blocks glomerulonephritis and extends lifespan in a mouse model of systemic lupus. Nat. Med. 2005;11:933–935. doi: 10.1038/nm1291. [DOI] [PubMed] [Google Scholar]
- 16.Knight ZA, Shokat KM. Chemically targeting the PI3K family. Biochem. Soc. Trans. 2007;35:245–249. doi: 10.1042/BST0350245. [DOI] [PubMed] [Google Scholar]
- 17.Gray A, Olsson H, Batty IH, Priganica L, Peter Downes C. Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal. Biochem. 2003;313:234–245. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 18.Knight ZA, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight ZA, et al. Isoform-specific phosphoinositide 3-kinase inhibitors from an arylmorpholine scaffold. Bioorg. Med. Chem. 2004;12:4749–4759. doi: 10.1016/j.bmc.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Balla T, et al. Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J. Biol. Chem. 1997;272:18358–18366. doi: 10.1074/jbc.272.29.18358. [DOI] [PubMed] [Google Scholar]
- 21.Meyers R, Cantley LC. Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J. Biol. Chem. 1997;272:4384–4390. doi: 10.1074/jbc.272.7.4384. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa T, Goto K, Kondo H. Cloning and characterization of a 92 kDa soluble phosphatidylinositol 4-kinase. Biochem. J. 1996;320(Part 2):643–649. doi: 10.1042/bj3200643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa T, Goto K, Kondo H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J. Biol. Chem. 1996;271:12088–12094. doi: 10.1074/jbc.271.20.12088. [DOI] [PubMed] [Google Scholar]
- 24.Ishihara H, et al. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem. 1998;273:8741–8748. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- 25.Ishihara H, et al. Cloning of cDNAs encoding two isoforms of 68-kDa type I phosphatidylinositol-4-phosphate 5-kinase. J. Biol. Chem. 1996;271:23611–23614. doi: 10.1074/jbc.271.39.23611. [DOI] [PubMed] [Google Scholar]
- 26.Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 27.Carpenter CL, et al. Purification and characterization of phosphoinositide 3-kinase from rat liver. J. Biol. Chem. 1990;265:19704–19711. [PubMed] [Google Scholar]
- 28.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 30.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 31.Gribble FM, et al. A novel method for measurement of submembrane ATP concentration. J. Biol. Chem. 2000;275:30046–30049. doi: 10.1074/jbc.M001010200. [DOI] [PubMed] [Google Scholar]