Summary
Gonadotropin-releasing hormone-1 (GnRH-1) is essential for mammalian reproduction, controlling release of gonadotropins from the anterior pituitary. GnRH-1 neurons migrate from the nasal placode into the forebrain during development. Although first located within the nasal placode, the embryonic origin/lineage of GnRH-1 neurons is still unclear. The migration of GnRH-1 cells is the best characterized example of neurophilic/axophilic migration, with the cells using a subset of olfactory-derived vomeronasal axons as their pathway and numerous molecules to guide their movement into the forebrain. Exciting work in this area is beginning to identify intersecting pathways that orchestrate the movement of these critical neuroendocrine cells into the CNS both spatially and temporally through a diverse and changing terrain. Once within the forebrain, little is known about how the axons target the median eminence and ultimately secrete GnRH-1 in a pulsatile fashion.
Keywords: GnRH-1, Kallman Syndrome, nasal placode, neuronal migration, olfactory development, reproduction
Introduction
The mammalian gonadotropin hormone-releasing hormone-1 (GnRH-1) system is essential for reproduction with cells residing within the forebrain postnatally but arising outside the CNS prenatally (Figure 1; 1, 2). In vertebrates, a maximum of three GnRH forms has been detected in any given species; each encoded by a separate GnRH gene - GnRH-1 [human chromosome 8], GnRH-2 [human chromosome 20] and GnRH-3 (primarily in teleosts). These 3 paralogous GnRH genes arose from two rounds of genome duplication early in evolution (3). GnRH-2 is the most ancient form of GnRH. During evolution, the preproGnRH-2 gene as well as the GnRH-2 receptor has been deleted or inactivated from the genome of many mammals (4). Thus, a physiological role for GnRH-2 in mammals remains controversial. GnRH-3 is prevalent in fish. Many fish express GnRH-3 and GnRH-1. GnRH-1 and not GnRH-3 is expressed in ancient teleosts, but modern teleosts (zebrafish) lost the mammalian GnRH-1 ortholog with GnRH-3 adopting its role in reproduction (4). In mammals, cells within this system express GnRH-1 and synthesize and secrete GnRH-1 into the portal capillary system (6, 7). GnRH-1 controls reproduction by binding to GnRH-1 receptors on gonadotropes of the anterior pituitary and triggering the release of luteinizing hormone and follicle stimulating hormone. These pituitary hormones subsequently act on the gonads (2). The importance of this system was demonstrated in experiments on the hypogonadal mouse (8). This mouse was shown to have a mutation in the mammalian GnRH-1 gene. In these mice, reproduction was rescued by GnRH-1 gene therapy. Thus, GnRH-1 cells are an integral component of the hypothalamic-pituitary-gonadal axis (HPG axis, Figure 1).
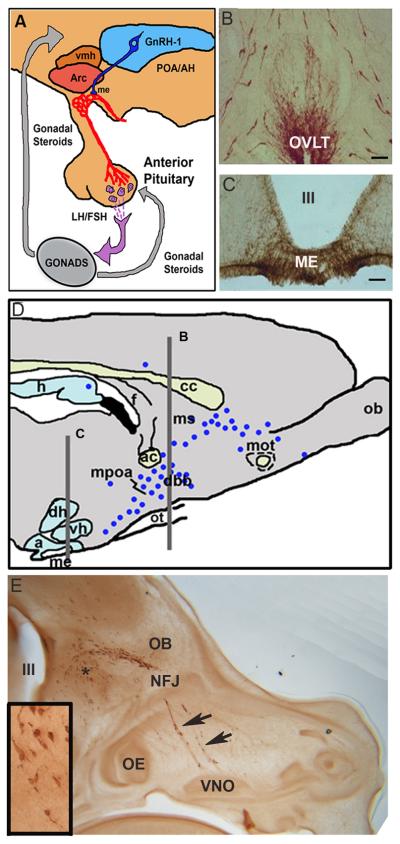
Figure 1. GnRH-1 neuroendocrine system.
A. Hypothalamic-Pituitary-Gonadal axis. GnRH-1 cells send axons to the median eminence (ME) where GnRH-1 is secreted into the portal capillary system. It is via this system that GnRH-1 cells affect gonadotropes of the anterior pituitary and subsequently gonadal function. POA/AH=preoptic area/anterior hypothalamus, vmh= ventromedial hypothalamus, arc=arcuate nucleus. C and D. Photomicrographs of sections immunocyto-chemically stained for GnRH-1 at the level of the OVLT (B) and ME (C). D. Parasagittal section of a rodent brain indicating the location of GnRH-1 cells (black dots, with levels in B and C shown by lines). GnRH-1 cells are distributed in a continuum from the olfactory bulbs to the caudal hypothalamus. h=hippocampus, f=fornix, ob=olfactory bulb, mot=medial olfactory tract, ms=medial septum, db=diagonal band of Broca, mpoa=medial preoptic area, d=dorsomedial hypothalamus, ot=optic tract, v=ventromedial hypothalamus, a=arcuate nucleus, me=median eminence. E. Parasagittal section of an E14.5 mouse embryo head immunocytochemically stained for GnRH-1. GnRH-1 cells can be found migrating from the VNO through the nasal forebrain junction (NFJ) into the forebrain. Asterisk shows location of inset, III= third ventricle.
Twenty years ago, two groups independently reported that in mice, the forebrain GnRH-1 cells migrate from the nasal placode into the CNS (Figure 1 and 2; 9-11). The migration of GnRH-1 cells or GnRH-3 cells into the CNS has been now documented by many groups in a number of species (1, 3, 6, 7). Despite their importance, the location of the precursors of the neuroendocrine GnRH cells has eluded characterization and has been hypothesized to be 1) nasal placode, 2) adenohypophyseal and/or 3) neural crest. However, much work has been done on the migration of GnRH-1 cells. The cells migrate in association with a subset of vomeronasal olfactory-derived axons in nasal regions, migrate through the cribriform plate with large numbers of olfactory sensory axons as well as blood vessels and then turn caudally on a transient pathway into the developing forebrain (1, 6, 12). The journey of the GnRH-1 cells is long in distance and one that is continually expanding as the embryo develops, i.e in mice the first GnRH-1 cells leave the nasal placode around E11 with cells continuing to migrate through the cribriform plate at E16.5. During migration of the GnRH-1 cells, the terrain through which they move as well as surrounding cell types are changing. Once within the brain, GnRH-1 neurons form bilateral continua distributed in areas that reflect the extension of the pathway that the GnRH-1 cells associate with during migration. GnRH-1 cells are often found rostrally from the olfactory bulbs to the caudal hypothalamus, but their final location varies across species (i.e. in fish cells are more rostrally located as compared to primates). This variation likely is the result of the pathway trajectory in each species as well as species' specific brain development during GnRH-1 cell movement. Independent of final location, the majority of GnRH-1 axons project to the median eminence where they access (via fenestrated capillaries) the pituitary portal capillary system (Figure 1). Little is known about the molecules used to guide GnRH axons (or other neuroendocrine axons for that matter) to the median eminence. But certainly it is one of the final steps in assuring a functional HPG axis.
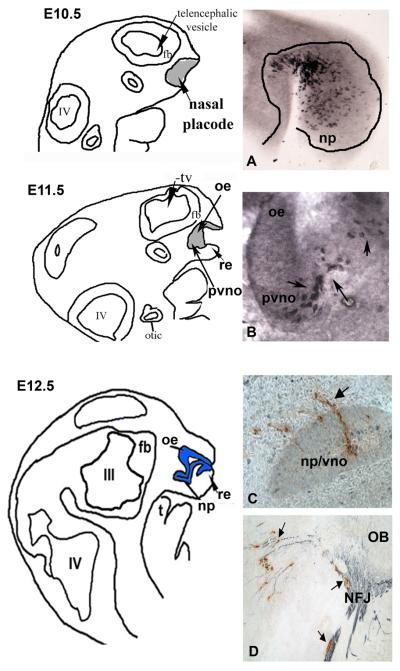
Figure 2. Development of the nasal placode/GnRH-1 neurons.
Camera lucida drawing from E10.5-E12.5 mice embryos showing invagination of the nasal placode and the development of the respiratory epithelium (re), olfactory epithelium (oe) and presumptive VNO (pvno)/nasal pit (np). tv=telencephalon, III=third ventricle, IV=fourth ventricle, t=tongue, fb=forebrain. A. Parasagittal section immunocytochemically stained for Hu (early neuronal marker) shows many positive cells within the nasal placode at E10.5. B. Parasagittal section from an E11.5 GnRH-GFP mouse immunocytochemically stained for GFP shows positive cells within the presumptive VNO (pvno) C. Parasagittal section from an E12.5 mouse, immunocytochemically stained for GnRH-1 (C) shows GnRH-1 cells leaving the VNO (arrows) D. Parasagittal section from an E12.5 mouse, immunocytochemically stained for GnRH-1 (brown) and peripherin (blue) shows GnRH-1 cells associated with peripherin fibers crossing the nasal forebrain junction (NFJ), peripherin axons entering the developing olfactory bulb (OB) and GnRH-1 cells still associated with a subset of peripherin axons turning caudally towards the hypothalamus.
Origin of GnRH-1 cells (figure 2)
Several lines of data, in a variety of species, are consistent with GnRH-1 cells being derived from cells within the nasal placode. However, the possibility still exists that GnRH-1 cells are derived from a stage in development prior to placode formation and are incorporated into the area of the nasal placode were they differentiate and prepare for migration into the CNS. It should also be noted that in mammals, GnRH-1 expression appears in the embryo pre-implantation and immunoreactive GnRH1 is present in the morula and blastocyst stages (13). In chicken GnRH-1 expression was found along the primitive streak before gastrulation and in later stages, was seen associated with the anterior neural ridge. Subsequently, GnRH-1 expression was restricted to the presumptive nasal cavity and olfactory placode (14). The role of GnRH-1 in early development is not understood and, in some cases, may represent transitory expression or a role in non-neuronal cell types.
‘New GnRH-1 cells’ have also been identified at later stages within a) the forebrain that transiently express GnRH-1 transcript (15, 16), b) extrahypothalamic areas that express GnRH-1 transcript but truncated peptide (amino acids 1-5, 17), and c) the developing tooth ‘placode’ that express GnRH-1 (18). The population of forebrain neurons that express GnRH-1 transiently during development clearly express the correct combination of transcription factors for GnRH-1 production. The function, if any, of GnRH-1 in these brain cells is unknown. GnRH-1 expressing cells in rodent teeth may provide developmental information relevant to the neuroendocrine GnRH-1 population since the GnRH-1 cells are in epithelial derived structures and not in neural crest-derived structures (see below). Also noteworthy, are several independent reports documenting dental agenesis in patients with Kallman Syndrome (19, see section below). Whether these occurrences are due to overlaps in craniofacial factors and/or GnRH-1 factors is not known.
The olfactory placode was suggested as the source of CNS GnRH-1 cells since they were first detected in the vomeronasal organ (VNO)/olfactory pit (OP, fig. 2: 9-11). Classically, the olfactory placode was said to give rise to nonsensory respiratory and sensory olfactory epithelium (OE; fig. 2: 20, 21). The OE produces the main olfactory (chemosensory) as well as the VNO epithelia (pheromone reception). The location of the GnRH-1 cells, led to the hypothesis that GnRH-1 cells originate in an area of the olfactory placode associated with OE-derived structures. However, ablation studies in chick showed that removal of the posterior area of the nasal placode eliminated OE structures but spared the GnRH-1 nasal population. Conversely, removal of the anterior area eliminated GnRH-1 cells and respiratory epithelial cells, but spared OE structures (22-24). In addition, GnRH-1 cells were detected emerging from the respiratory epithelium in normal chick, although the majority of GnRH-1 cells were associated with the olfactory pit (25). Thus, data in chick suggested that the GnRH-1 lineage was developmentally associated with the respiratory area of the olfactory placode and distinct from OE progenitors.
Studies in mouse also argue that GnRH-1 progenitors are distinct from OE progenitors. These included 1) the absence of OE lineage markers Mash-1, Math4A, Math4C/neurogenin1 and NeuroD in early expressing GnRH-1 cells (26) and 2) ‘ectopic’ GnRH-1 cells emerging from the respiratory epithelium in mutant mice lacking Activator protein-2, which is expressed in respiratory epithelium but not OE (27). These studies suggest that GnRH-1 neurons diverge early from the OE lineage and that the progenitors of GnRH-1 cells are closely associated with anterior or respiratory regions during early stages of placode differentiation. Therefore a more generic name, the nasal placode (NP), is used by our laboratory to delineate the structure from which neuroendocrine GnRH-1 cells arise. An intermediate area between the anterior respiratory cells and the OE sensory cells was suggested as the location of GnRH-1 progenitor cells (27). However, the lineage and location of the precursors of neuroendocrine GnRH-1 cells still remains to be determined.
Multiple developmental origins for GnRH cells have been proposed in fish. If the fish has two rostral GnRH forebrain populations (both 1 and 3), then they may either share a nasal placode developmental origin or exhibit different developmental origins, nasal placode and telencephalon. An example of this is found in Medaka. Medaka, have a preoptic area population of GnRH-1 cells that projects to the anterior pituitary and regulates gonadotropin secretion and a population of GnRH-3 cells in the terminal nerve ganglion that regulates reproductive behavior. Within the forebrain POA area, Okuba et al (31) reported four GnRH-1 neuronal populations based upon development: 1) olfactory-derived ventral POA, 2) telencephalic-derived dorsal POA, 3) anterior telencephalic-derived medial ventral telencephalic population and 4) nonmigratory ventral hypothalamic population. The GnRH-3 population was shown to arise from the olfactory region. As in mammals, it is still unclear whether the precursors are located within the area of first detection or end up in this region as development progresses.
Studies using molecular techniques showed that in fish species with one rostral forebrain population (either GnRH-1 or GnRH-3), the cells are first detected in the nasal epithelium, similar to mammals. In zebrafish, GnRH lineage studies based on the expression of GnRH mRNAs and/or GFPs driven by specific GnRH-promoters reported that GnRH-3 cells were first detected in the nasal epithelium (5). However, in two zebrafish knockouts, you-too and detour, it was reported that when the adenohypophysis was missing or reduced, so were the hypothalamic GnRH neurons, yet olfactory structures were intact (32). Using tract tracing together with this mutant data, Whitlock (33) proposed that hypothalamic GnRH and terminal nerve/rostral telencephalon GnRH cell populations arise from two separate regions near the neural plate: the adenohypophyseal (anterior pituitary) region of the anterior neural plate and the cranial neural crest, respectively, and not the nasal placode.
Recent work in mice (34) addressed whether the GnRH-1 lineage in mammals is associated with the adenohypophyseal lineage, but end up residing in the nasal placode as the animal develops. These studies used mutant mouse lines Gli and Lhx3, that have complete deletion or reduction of the anterior pituitary. In mouse, pituitary organogenesis begins at E8.5 (35), prior to detection of GnRH-1 neurons. The oral ectoderm invaginates and forms Rathke's pouch which then moves toward the ventral diencephalon and matures to become the anterior and intermediate lobes of the pituitary gland (35). Gli factors are expressed in mouse from E7.5 - E11.5 in neural and other ectodermal and neural crest derivatives of the head, including Rathke's pouch and expression in nasal regions (36). Knockout of Gli2 (Gli2−/−), the equivalent of you-too in zebrafish, leads to variable loss of the pituitary (37). Knockout of Gli1 (Gli1−/−), the homologue of detour in zebrafish, leads to no pituitary defects in mice (37). However, Gli2−/− in conjunction with Gli1−/− causes complete pituitary ablation (38). Lhx3 is a LIM-homeobox transcription factor also expressed in Rathke's pouch (35). Knockout of Lhx3 (Lhx3−/−) leads to hypoplasia of the pituitary, with loss of most cells types (39). In all of the pituitary deficient mutant lines [Gli2−/−, Gli1−/−Gli2−/− and Lhx3−/−], GnRH-1 cells were detected within the developing VNO, across nasal regions, at the nasal forebrain junction and within the developing forebrain. In addition, the total number of GnRH-1 cells as well as the distribution of GnRH-1 cells was similar among genotypes (34). These results indicate that GnRH-1 progenitors are independent of the adenohypophyseal lineage and support the hypothesis that these cells are associated with the nasal placode.
Evidence now indicates that FGF signaling is important for the formation of GnRH-1 neurons via the neurotrophic molecule Fgf8 and one of its receptors FgFR1 (40). FGFR1 is expressed by GnRH-1 cells (41) and FGFR1 hypomorphs have a dramatic reduction in GnRH-1 neurons (40). Fgf8 is involved in induction and differentiation of the mouse nasal placode (42), with a loss of Fgf8 resulting in the absence of the VNO and absence of GnRH-1 neurons (40). Since GnRH-1 neurons were absent at E11.5 in these mice, a role in GnRH-1 neuron fate specification has been suggested. Interestingly, the anatomical region of the nasal placode from which the GnRH-1 cells are known to emerge, was missing in the homozygous Fgf8 hypomorphs. Thus, this system acting early, disrupts formation/specification of the GnRH-1 cell type. However, since an entire anatomical region in the nasal placode is missing in these mice, the full impact of these molecules on the development of GnRH-1 neurons and/or their precursors remains to be elucidated.
Neural crest can contribute to placodal areas. The neural crest is a cell population characteristic of vertebrates that gives rise to a variety of neural and nonneural derivatives, including peripheral neurons and glia, craniofacial skeleton and neuroendocrine cells (43-46). GnRH-1 cells in mouse do share some markers common to neural crest cells (e.g. AP-2; 47). However, to date, a direct lineage has not been shown in mammals. In zebrafish, where one population of GnRH cells was proposed to originate from the neural crest (33), the studies involved labeling of cells that lie close to one another – neural crest cells lie adjacent to the posterolateral boundary of the nasal placode. Due to the close apposition of these structures when the experiments were performed, inadvertent labeling of nasal placode cells cannot be ruled out. Gene expression domains have been used to delineate the anterior border of the neural plate; the region that gives rise to the sensory placodes (48) and it is clear that the olfactory organs differentiate from the nasal placodes, transient paired structures lying laterally at the anterior end of the forming neural tube. Whether GnRH-1 cells differentiate from precursors in the nasal placode is difficult to address since GnRH-1 is not detectable in mouse until 1-2 days after their final division (~ E9.5-E10.5, 11). However, it is also now known that many cells types emerge from nasal placode and move toward the CNS. These cells express neuronal markers including (depending on the species examined) GnRH-1, galanin, Hu, NPY, and TH (1, 49). All of these cells, unlike olfactory sensory cells, actually migrate across nasal regions toward the forebrain. Thus, like GnRH-1 cells, other non-sensory neuronal cell populations exist within this structure or become associated with it early in development. It is still unclear whether these other neuronal cells types, like the GnRH-1 cells, a) arise from the panplacodal primordium located at the neural plate border (48) and b) migrate across the cribriform plate into the CNS. Thus while the exact precursors of GnRH-1 (and-3) are still elusive, consensus is that GnRH-1 cells are associated with the nasal placode as they turn on GnRH-1 expression. Thereafter these cells migrate into the CNS by traversing several ‘distinct’ domains - the nasal area, the nasal forebrain junction and the forebrain. This rest of this review will concentrate on movement of GnRH-1 neurons primarily during migration within nasal areas.
Migration of GnRH-1 Cells into the CNS
Newly born CNS neurons use two main strategies to arrive at their final location, radial or tangential migration. Radial migration involves a trajectory that is perpendicular to the ventricular/pial surface and utilizes radial glial fibers (gliophilic). Pyramidal cortical cells have been studied extensively as a model for gliophilic migration. This movement relies on membrane-bound cell adhesion molecules as well as secreted extracellular molecules. Tangential migration involves a trajectory that is parallel to the ventricular surface and does not involve a pathway support per se, e.g. cells from the ganglion eminence to the cortex or cells in the olfactory migratory stream. This movement relies on interactions with substrate (extracellular matrix and/or surface of other cells). Interactions can be homotypic, with cells of the same type often creating chains as well as a specific microenvironment that facilitates migration, or heterotypic. Chemotactic gradients are also frequently utilized to achieve directional movement. GnRH-1 neurons migrate in association with a subset of olfactory axons. Thus, although not on radial glia, GnRH-1 cell migration does involve a pathway support. The orientation of the GnRH-1 migratory route cannot be easily defined by a single term, but when in the forebrain, tangential movement is more applicable than radial movement. GnRH-1 neurons migrate in clusters that resemble the chains seen in homotypic neuronal migration. Thus, GnRH-1 neurons exhibit characteristics common to both of the categories routinely used to described migration of CNS neurons. It is this mixture of traits that 1) makes classification of molecules modulating GnRH-1 neuronal migration into a hierarchy difficult and 2) helps insure that entry of GnRH-1 cells into the CNS is successful.
Cellular movement of GnRH-1 Cells
Migrating neurons within the CNS, generally bipolar in shape, undergo three synchronized steps; extension of the leading process, translocation of the nucleus into the leading process (nucleokinesis) and elimination of the trailing process. Nucleokinesis typically occurs in a saltatory pattern thought to result from coupling of nuclear movement to leading process dynamics (50). GnRH-1 neuronal migration has been analyzed using time-lapse microscopy in slices (51) and nasal explants (52). Similar to migrating cells in the CNS, migrating GnRH-1 neurons show extension of a leading process, nucleokinesis with somal translocation and exhibit saltatory movement (52). GnRH-1 neurons, like pyramidal cortical cells that utilize radial glia, had a neuronal speed of 13-20um/hr (51-52). Thus, GnRH-1 cell migration in nasal regions, with respect to speed and movement, resembles cells within the CNS.
Olfactory System/Substrate Pathway
The developmental relationship of the GnRH-1 system to the olfactory system is illustrated by Kallmann syndrome and has been extensively reviewed. Humans with the X-linked form of this disease have a monogenetic mutation that results in anosmia (lack of smell) and hypogonadism (failure to undergo pubertal development). Examination of Kallmann fetal material revealed lack of olfactory axon ingrowth into the developing forebrain and GnRH-1 cells ‘stuck’ on the CNS–side of the cribriform plate in a tangle of olfactory axons in one case (53) and GnRH-1 cells in only nasal regions in a second case (54). Thus, molecules can alter migration of GnRH-1 neurons by disrupting normal growth of olfactory sensory axons. In addition, altering the GnRH-1 specific migratory route and/or the signaling of molecules directly on the GnRH-1 cells themselves can prevent entrance of GnRH-1 cells into the CNS or lead to abnormal distribution and/or numbers of GnRH-1 cells within the brain.
Prior to GnRH-1 migration, ‘pioneer’ olfactory axons cross the nasal region and target the base of the developing telencephalon (55). The cues that direct this group of axons may be critical for the guidance and targeting of all future olfactory/vomeronasal axons and subsequently appropriate migration of GnRH-1 cells to the developing forebrain. In addition, migratory neuronal progenitor cells and neuronal precursors have been identified leaving the nasal placode prior to pioneer axonal outgrowth in nasal regions (49). What early guidance cues participate in these processes is unknown. Once guidance cues are in place, GnRH-1 cells use a subset of vomeronasal axons as their pathway across the nasal region (55-58). In mammals, the majority of peripherin-positive olfactory axons project to the main and accessory olfactory bulbs. However, in the embryonic mouse, peripherin has been shown to highlight a pathway that enters the CNS and turns caudally toward the developing hypothalamus. GnRH-1 cells are associated with this caudally projecting peripherin-positive axonal pathway. This pathway [often termed the nervus terminalis or the caudal branch of the vomeronasal nerve] also expresses TAG-1 [an axonal surface glycoprotein that is transiently expressed; 56] and DCC [deleted in colorectal cancer; receptor for the guidance molecule netrin-1; 57-58]. These two markers are expressed on olfactory/vomeronasal axons in the nasal region but not on the branches of these nerves that enter the forebrain and course rostrally into the olfactory bulb/accessory regions. Guidance of this caudal axonal pathway is important for the establishment of the adult-like GnRH-1 cell distribution. In DCC knockout mice, the caudal branch of the vomeronasal nerve turns towards the cerebral cortex instead of the hypothalamus. GnRH-1 cells follow this pathway and migrate into the cortex instead of the hypothalamus. Thus, GnRH-1 cells, probably via multiple membrane-bound cell adhesion molecules, are tightly coupled to their pathway substrate. With respect to movement of GnRH-1 cells in either nasal or forebrain regions, to my knowledge, no report has documented the majority of GnRH-1 cells using (directed to) another pathway for migration, i.e. following the olfactory sensory axons into the olfactory bulb. In fact, at the nasal/forebrain junction, it appears that both membrane-bound molecules as well as extracellular factors (see below) guide the GnRH-1 neurons into the developing forebrain. With respect to membrane bound molecules, they can be expressed on the rostral olfactory/vomeronasal axons and absent from the caudal hypothalamic projection. In addition, several molecules have been shown to be down-regulated on GnRH-1 neurons as they enter the forebrain. Such spatiotemporal events, suggest that a combination of molecules some turning on while others turn off play an important role in establishing the appropriate GnRH-1 adult-like distribution. In addition, multiple regulatory domains exist (each potentially expressing a new molecule/factor) through which GnRH-1 (and GnRH-3) neurons migrate from their origin to their final destination within the CNS (figure 3).
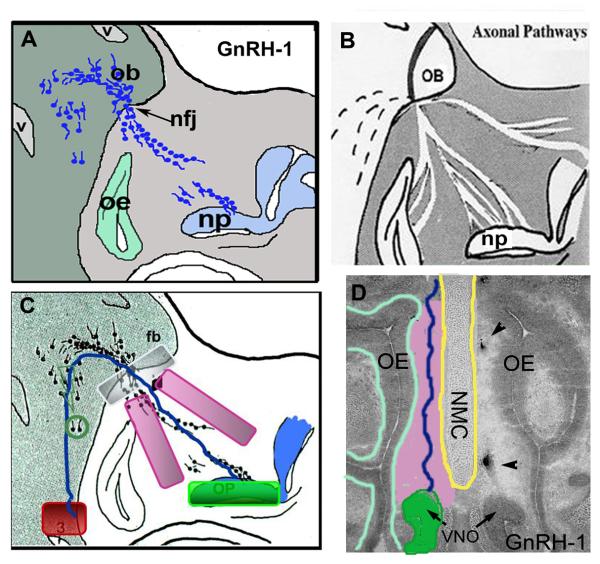
Figure 3. Multiple Domains Along the GnRH-1 neuronal migratory route.
A. Schematic of GnRH-1 neuronal migration at E12.5-E14.5. Blue dots = GnRH-1 neurons. Np=nasal pit, oe=olfactory epithelium, ob = olfactory bulb, nfj = nasal forebrain junction. B. Schematic of axons in nasal area at E12.5-E14.5. Axons form pathways to NFJ, through the cribriform plate and predominately enter olfactory bulbs (OB), but a small subset of axons form pathway caudally on which the GnRH-1 neurons migrate (dotted line). C and D. During their movement GnRH-1 neurons are exposed to multiple regulatory domains along the migratory route (light green; nasal pit, blue; olfactory epithelium, pink; nasal mesenchyme, white; nasal forebrain junction, blue line; transient pathway, red; basal forebrain, dark green; GnRH-1 cells themselves or other migrating cells along pathway. C= parasagittal; D=coronal, nmc = nasal midline cartilage.
Cues Regulating GnRH-1 Neuronal Migration in Nasal Regions
Within the CNS, a number of secreted extracellular molecules regulate radial and/or tangential neuronal migration including netrins, slits, Reelin, and semaphorins. In addition, tangentially migrating neurons are influenced by growth factors and chemokines that often form chemotactic gradients (either attractive or repellent) to give directional information to the migrating cells. Finally, neural activity through activation of ligand-gated and voltage-dependent ion channels can also regulate neuronal migration (59) and it is now clear that a variety of neurotransmitters are involved in the regulation of migration of olfactory bulb neurons, cerebellar neurons, hippocampal neurons, GABAergic interneurons and cortical principal neurons (references see 60).
Extracellular molecules
Semaphorins (61, 62)
Semaphorins constitute a large family of guidance cues. A role for Semaphorin3F as a repellent for the migration of GnRH-1 cells has been reported. The semaphorin-4D receptor, PlexinB1, is highly expressed in the developing nasal placode and is expressed on olfactory axons and GnRH-1 cells during prenatal development. Semaphorin-4D is present in the nasal mesenchyme though appears stronger at the nasal forebrain junction as well as developing forebrain. PlexinB1-deficient mice exhibit a defect in migration of GnRH-1 neurons that results in a reduction of GnRH-1 neurons in the adult brain. Examination of postnatal day 3 tissue revealed a decrease in GnRH-1 neurons in the caudal region of forebrain and an increase in GnRH-1 neurons in rostral brain regions and a delay in movement of the GnRH-1 cells into the brain was already observed at E14.5. In addition, Semaphorin-4D promoted directional migration in immortalized GnRH-1 cells by coupling PlexinB1 with activation of the HGF receptor Met tyrosine kinase (62). In this study no abnormalities were found in the development or organization of the olfactory axons, suggesting that the migratory defect might be cell autonomous or that the subpopulation of axons that the GnRH-1 cells associate with were not specifically identified. Interestingly, brains from Semaphorin-4D-deficient adult mice show no changes in GnRH-1 cell number, suggesting compensation by another ligand and/or compensation by other morphogenetic signals such as HGF (see below).
Prokineticin (Prok; 63-65)
Proks are multifunctional secreted proteins with Prok2 shown to function as an output molecule from the suprachiasmatic circadian clock. Studies in Prok2 receptor−/− mice and Prok2 −/− mice showed olfactory bulb defects and disrupted GnRH neuron migration, resulting in a dramatic decrease in GnRH neuron population in the hypothalamus as well as hypogonadotropic hypogonadism.
Receptor Tyrosine kinases/cytokines
Ephrins (66) and Hepatocyte growth factor (HGF; 67)
GnRH-1 neurons express the receptor tyrosine kinases EphA5 and Met. Ephrins represent short-range repulsive cues for migration of of neural stem cells and progenitors such as neural crest cells, and of differentiated neurons such as cerebellar granule cells. GnRH-1 neurons express the receptor tyrosine kinases EphA5. The vomeronasal organ and olfactory epithelium express ephrin A3 at E11.5, while ephrin A3 and A5 where present in the vomernasal organ at E12.5. Overexpression of EphA5 in transgenic mice resulted in slower migration of GnRH-1 cells and the formation of cell clusters along the migratory route. HGF is a cytokine expressed in the nasal compartment and GnRH-1 neurons express its receptor Met and tissue-type plasminogen activator (tPA), a known HGF activator. Met was also expressed by the olfactory axons. Studies showed that activation of this pathway promotes migration of GnRH-1 neurons and that a gradient of HGF is required for proper GnRH-1 cell migration.
Chemokines
Stromal cell-derived factor-1 (SDF-1; 68-70)
In the nervous system, the chemokine SDF-1 and one of its receptors, CXCR4 (a G protein-coupled seven-transmembrane receptor) are highly expressed during development in the cerebellum, hippocampus and neocortex (references see 69). Deletion of the gene for either CXCR4 or SDF-1 results in developmental abnormalities in brains areas characterized by altered cell survival, axonal pathfinding and migration. At E12.5-E14.5, the time when the olfactory/GnRH-1 systems are developing, a prominent signal is detected in the olfactory epithilum for CXCR4, while SDF-1 is present in the meninges and in mesodermal components surrounding the olfactory epithelium. Three studies verified a function of SDF-1 and CXCR4 in the developing GnRH-1/olfactory systems. CXCR4 and SDF-1 were expressed in migratory GnRH-1 neurons and olfactory nerve fibers that were associated with GnRH-1 neurons and SDF-1 expression appears in cells localized to the nasal forebrain junction, on the nasal side. Disruption of SDF- 1/CXCR4 signaling affects olfactory axon outgrowth, inhibits GnRH-1 neuronal migration, and results in loss of GnRH-1 cells as they move away from the nasal pit and/or approach the developing forebrain.
Neurotransmitters/Neuromodulators
Cholecystokinin (CCK; 71)
CCK is expressed in sensory cells in the developing OE and VNO, with both ligand and receptors (CCK-1R and CCK-2R) found on olfactory axons throughout prenatal development. In addition, migrating GnRH-1 neurons in nasal regions express CCK-1R. Exogenous application of CCK via CCK-1R reduced both olfactory axon outgrowth and GnRH-1 cell movement. Mice carrying a genetic deletion of CCK-1R, but not CCK-2R, had more GnRH-1 neurons in the forebrain, consistent with an accelerated migratory process. Thus, during development CCK provides an inhibitory influence on GnRH-1 neuronal migration, contributing to appropriate entrance of these cells into the brain.
GnRH (72-74)
A recent study examined the role of GnRH signaling in the proper development of GnRH neurons. Using the hypogonadal mouse crossed to GFP targeted to GnRH neurons and the GnRH receptor mutant mouse, a normal GnRH population was found within the forebrain, distributed similar to WT mice and fibers were present in the median eminence. These studies indicate that in mice, autocrine-paracrine GnRH-signals are not necessary for the developmental migration of GnRH neurons into the brain or for the projection of GnRH neurosecretory neurons. However, in zebrafish, morpholino-modified antisense oligonucleotides directed against GnRH3 disrupted the establishment of the GnRH3 fiber network and altered GnRH3 perikarya localization, suggesting an autocrine-paracrine role of the secreted peptide in establishment of the final system. Whether, early disruption in mice is compensated at later developmental time points or whether correction of the zebrafish system might have occurred at later time points is unknown. In chick, using an in vitro olfactory nerve bundle explant, a GnRH antagonist was found to significantly inhibit GnRH neurite outgrowth and GnRH neuronal migraton. These effects were attenuated by the addition of chicken GnRH-1. These experiments suggest that in some species GnRH-1 itself may autoregulation the development of the GnRH-1 system, but likely is not essential.
Ligand-gated and voltage-dependent ion channels
Pioneer studies performed on cerebellar granule cells showed that neural activity through activation of ligand-gated and voltage-dependent ion channels was involved in the regulation of neuronal migration (59). In particular, calcium transients via L-type and/or N-type voltage-gated calcium channels (VGCCs) altered neuronal migration and axon growth. Neurotransmitter effects on neuronal migration mediated by changes in intracellular calcium have been shown for glutamate, GABA and serotonin (59). Activation of each of these neurotransmitters has also been linked to activation of VGCCs. N- and L-type VGCGs are in GnRH-1 cells during prenatal development (60) and perturbation studies showed that activation of N-type VGCCs has a role in facilitating GnRH-1 cell movement in nasal areas.
Unknown class
Nasal embryonic LHRH Factor (Nelf; 75-78)
Nelf was identified in a single cell library screen comparing cDNAs from mouse migrating GnRH-1 cells to nonmigrating GnRH-1 cells (75). It is expressed in olfactory axons and GnRH-1 cells during development and in vitro studies implicated Nelf in the development of the GnRH-1 system in mice (75-77). During embryonic development in zebrafish, nelf is expressed in GnRH-3 neurons and in target sites of GnRH3 projections and perikarya prior to the initiation of GnRH-3 cell migration. Nelf knockdown resulted in a disruption of the GnRH-3 system (78), including absence or misguidance of the GnRH-3 axons and incorrect or arrested migration of the GnRH-3 cells.
Nasal/Forebrain Junction
The crossover from nasal region to CNS is a dramatic change in environment. Chemokines, growth factors and neurotransmitters localized to, or forming gradients at, the nasal forebrain junction, have been shown to alter movement of GnRH-1 cells into the forebrain/out of nasal regions. At the nasal forebrain junction, multiple tissue types exist including midline nasal cartilage, nasal mesenchyme, the cribriform plate as well as meninges and developing CNS tissue. At this region, GnRH-1 cells appear to pause. The reason for this pause is unclear, but it may ensure 1) maturation of GnRH-1 neurons, 2) establishment/targeting of migratory pathway to appropriate brain regions, and/or 3) changes in the extracellular millieu composition. A molecular gradient towards the nasal-forebrain junction or away from olfactory/vomeronasal epithelium would be important for developing nasal systems, including GnRH-1 neurons. To date, two molecules [discussed earlier] have been shown with an increasing gradient pattern towards the frontonasal mesenchyme, HGF (67) and SDF-1 (68-70).
Previous in vitro and in vivo studies demonstrated that GABA regulates CNS neuronal migration, acting as a chemoattractant, regulator of cell mobility and mediator of the migration process (references see 79). It is now well established that GABA is excitatory during early development, depolarizing rather than hyperpolarizing the cell membrane. Since migrating neurons do not form chemical synapses, extracellular GABA activates the postsynaptic receptors in a paracrine manner. GABA also appears to be important at the nasal/forebrain junction (cribriform plate) for the migration of GnRH-1 neurons into the CNS. Both in vivo and in vitro experiments indicate that GABA inhibits migration of GnRH-1 neurons (1,12, 79-81). In GAD 67 KO mice, attenuated levels of GABA accelerated GnRH-1 cell migration in nasal areas as well as movement into the CNS. One of the hypothesized functions of the GABA-induced pause at the nasal/fore-brain junction is that it causes a developmental change in GnRH-1 neurons prior to their entrance into the CNS (1). The data from GAD67 KO mice (81) suggest that faster movement into the CNS is also associated with an apparent ‘loss’ of GnRH-1 cells at the junction. In situ hybridization histochemistry for GnRH-1 mRNA positive cells was performed and KO mice had fewer cells expressing GnRH-1 mRNA in brain areas. The changes observed in KO mice are consistent with the idea that premature entrance of GnRH-1 neurons into CNS regions down-regulates GnRH-1 expression at the level of transcription. Necdin, a gene whose mutation is associated with Prader-Willi syndrome (82) has recently been shown to alter GnRH-1 migration into brain regions, with GnRH-1 cells again reaching the cribriform plate but lower numbers if GnRH-1 cells in the forebrain. A decrease of GnRH-1 cells as the cells enter the forebrain, or shortly thereafter, has also been documented in AP-2 (30) mutant mice, Reelin (83) and Nhlh2 mutant mice (84, 85). Attenuated expression of both GnRH-1 protein and mRNA may result in these mice because the GnRH-1 cells encountered a cellular environment for which they were not temporally ready for, and/or that the GnRH-1 cells missed a developmental step preparing them for entrance into the CNS.
Kallman Syndrome
Human Kallmann Syndrome patients exhibit anosmia and reproductive dysfunction, specifically hypogonadotropic hypogonadism. Genetic work on Kallmann patients identified the Kal gene (86, 87) that encodes anosmin-1. Mutations in this gene result in the failure of GnRH-1 cells to migrate from nasal areas into the brain. The Kal gene has been lost from the mouse/rat genome, but antisense knockdown of Medaka KAL1.1 a) inhibited GnRH-1 cell migration into the POA, leading to accumulation of GnRH-1 neurons in the olfactory region and dorsal telencephalon, and b) suppressed migration of GnRH-3 neurons from the olfactory region into the forebrain. The other GnRH-1 populations appeared unaffected (31). In zebrafish, knockdown of Kal1.1 disrupted GnRH cell migration into the hypothalamus, but alterations in hypothalamic development were also noted while GnRH cells remaining in the terminal nerve were not affected (88). The C. elegans Kal-1 has also been cloned and shown to interact with multiple heparan sulfate proteoglycans to promote neuroblast migration (89).
Several other monogenic mutations have been identified in Kallman patients including NELF (90), FGFR1 (91) and Prokineticin 2 and its receptor (92). These mutations alter the development of the GnRH-1/olfactory systems. Where examined, Anosmin-1, FGFR1, NELF and Prokinectin, are not limited to nasal regions during development, often being found in areas actively showing cell/neuronal migration. In patients with Kallman syndrome, other deficits occur, both within the CNS as well as other organs. Whether deficits occur in other regions in vertebrates in general, after knock-down on these genes, remains to be determined. As genetic details emerge from work in human patients who come to the clinic because of pubertal delay, one can expect other genes involved in the development/migration of GnRH-1 neurons to be discovered. In fact recent work has begun to address the heterogeneity of phenotypes in Kallmann families, as well as the fact the defects in the above mentioned genes only account for a small percentage of the cases. These studies show that digenic mutations (e.g. in FGFR1 and NELF [93]) can synergize to produce a given phenotype.
The list of molecules expressed by GnRH-1 neurons during their migration into the forebrain has grown quite long (see reviews 1, 6, 7, 12, 40, 94). The large number of molecules however, most likely still underestimates the complexity of possible interactions. One must remember that these cells spatially and temporally travel across areas (nasal region, nasal-forebrain junction, forebrain) that have very different composites of guidance molecules and factors. Thus, although the GnRH-1 cells may express a given receptor throughout their journey, it may be activated once in a specific region or multiple times in different regions. In addition, many molecules will defy anatomical boundaries working in multiple areas and may even in the end, produce different responses depending on the relative ‘dose’ to which the GnRH-1 cells are exposed. To put all the pieces in their appropriate sequence is what makes the development of the GnRH-1 system challenging and exciting.
References
- 1.Wray S. Development of gonadotropin-releasing hormone-1 neurons. Frontiers in Neuroendocrinology. 2002;23:292–316. doi: 10.1016/s0091-3022(02)00001-8. [DOI] [PubMed] [Google Scholar]
- 2.Herbison AE. Physiology of the GnRH neuronal network. In: Neill JD, editor. Knobil and Neill's Physiology of Reproduction. Academic Press; San Diego, CA: 2006. pp. 1415–1482. [Google Scholar]
- 3.Okubo K, Nagahama Y. Structural and functional evolution of gonadotropin-releasing hormone in vertebrates. Acta Physiol (Oxf) 2008;193:3–15. doi: 10.1111/j.1748-1716.2008.01832.x. [DOI] [PubMed] [Google Scholar]
- 4.Stewart AJ, Katz AA, Millar RP, Morgan K. Retention and Silencing of PreproGnRH-II and Type II GnRH Receptor Genes in Mammals. Neuroendocrinology. 2009;90:416–432. doi: 10.1159/000233303. [DOI] [PubMed] [Google Scholar]
- 5.Palevitch O, Kight K, Abraham E, Wray S, Zohar Y, Gothilf Y. Ontogeny of the GnRH systems in zebrafish brain: in situ hybridization and promoter-reporter expression analyses in intact animals. Cell Tissue Res. 2007;327:313–322. doi: 10.1007/s00441-006-0279-0. [DOI] [PubMed] [Google Scholar]
- 6.Wray S. Developmental of Luteinizing Hormone Releasing Hormone Neurones. J. of Neuroendocrinology. 2001;13:3–11. doi: 10.1046/j.1365-2826.2001.00609.x. [DOI] [PubMed] [Google Scholar]
- 7.Wray S. Gonadotropin-Releasing Hormone: GnRH-1 System. In: Squire LR, editor. Encyclopedia of Neuroscience. Vol. 4. Academic Press; Oxford: 2009. pp. 967–973. [Google Scholar]
- 8.Mason AJ, Pitts SL, Nikolics K, Szonyi E, Wilcox JN, Seeburg PH, Stewart TA. The hypogonadal mouse: reproductive functions restored by gene therapy. Science. 1986;234:1372–8. doi: 10.1126/science.3097822. [DOI] [PubMed] [Google Scholar]
- 9.Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 10.Wray S, Nieburgs A, Elkabes S. Spatiotemporal cell expression of luteinizing hormone-releasing hormone in the prenatal mouse: evidence for an embryonic origin in the olfactory placode. Dev. Brain Res. 1989;46:309–318. doi: 10.1016/0165-3806(89)90295-2. [DOI] [PubMed] [Google Scholar]
- 11.Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tobet SA, Schwarting GA. Minireview: Recent Progress in Gonadotropin-releasing hormone neuronal migration. Endocrinology. 2006;147:1159–1165. doi: 10.1210/en.2005-1275. [DOI] [PubMed] [Google Scholar]
- 13.Casañ EM, Raga F, Polan ML. GnRH mRNA and protein expression in human preimplantation embryos. Mol Hum Reprod. 1999;5(3):234–9. doi: 10.1093/molehr/5.3.234. [DOI] [PubMed] [Google Scholar]
- 14.Witkin JW, Dao D, Livne I, Dunn IC, Zhou XL, Pula K, Silverman AJ. Early expression of chicken gonadotropin-releasing hormone-1 in the developing chick. J Neuroendocrinol. 2003;15:865–870. doi: 10.1046/j.1365-2826.2003.01073.x. [DOI] [PubMed] [Google Scholar]
- 15.Wu TJ, Gibson MJ, Silverman A-J. Gonadotropin-releasing hormone (GnRH) neurons of the developing tectum of the mouse. J. Neuroendocrinol. 1995;7:899–902. doi: 10.1111/j.1365-2826.1995.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 16.Skynner MJ, Slater R, Sim JA, Allen ND, Herbison AE. Promoter transgenics reveal multiple gonadotropin-releasing hormone-I-expressing cell populations of different embryological origin in mouse brain. J. Neurosci. 1999;19:5955–5966. doi: 10.1523/JNEUROSCI.19-14-05955.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terasawa E, Busser BW, Luchansky L, Sherwood NM, Jennes L, Millar RP, Glucksman MJ, Roberts JL. Presence of luteinizing hormone releasing hormone fragments in the Rhesus monkey forebrain. JCN. 2001;439:491–504. doi: 10.1002/cne.1364. [DOI] [PubMed] [Google Scholar]
- 18.Tiong JDR, Pakiam JG, Wray S. GnRH-1 (Gonadotropin Releasing Hormone–1) Expression in Incisors of Mice. Endocrinology. 2004;145:3608–3612. doi: 10.1210/en.2004-0387. [DOI] [PubMed] [Google Scholar]
- 19.de Zegher F, Lagae L, Declerck D, Vinckier F. Kallmann syndrome and delayed puberty associated with agenesis of lateral maxillary incisors. J Craniofac Genet Dev Biol. 1995;15(2):87–9. [PubMed] [Google Scholar]
- 20.Farbman AI. Cell biology of olfaction. Cambridge University Press; New York: 1992. [Google Scholar]
- 21.Halpern M. The organization and function of the vomeronasal system. Ann. Rev. Neurosci. 1987;10:325–362. doi: 10.1146/annurev.ne.10.030187.001545. [DOI] [PubMed] [Google Scholar]
- 22.Akutsu S, Takada M, Ohki-Hamazaki H, Murakami S, Arai Y. Origin of luteinizing hormone-releasing hormone (LHRH) neurons in the chick embryo: effect of the olfactory placode ablation. Neurosci.Lett. 1992;142:241–244. doi: 10.1016/0304-3940(92)90382-h. [DOI] [PubMed] [Google Scholar]
- 23.El Amrauoui A, Dubois PM. Experimental evidence for an early commitment of gonadotropin-releasing hormone neurons, with special regard to their origin from the ectoderm of nasal cavity presumptive territory. Neuroendocrinology. 1993;57:991–1002. doi: 10.1159/000126490. [DOI] [PubMed] [Google Scholar]
- 24.Arai Y, Murakami S, Seki T. Removal of olfactory placode prevents the development of LHRH neurons in the forebrain of the chick embryo: possible interaction between migrating LHRH neurons and highly polysialylated form of neural cell adhesion molecule (NCAM-H) Acta Biologica Hungarica. 1994;45:155–168. [PubMed] [Google Scholar]
- 25.Hilal EM, Chen JH, Silverman A-J. Joint migration of gonadotropin-releasing hormone (LHRH) and neuropeptide Y (NPY) neurons from olfactory placode to central nervous system. J.Neurobiol. 1996;31:487–502. doi: 10.1002/(SICI)1097-4695(199612)31:4<487::AID-NEU8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Kramer PK, Wray S. Midline nasal tissue influences nestin expression in nasal placode derived-Luteinizing Hormone-Releasing Hormone (LHRH) neurons during development. Dev. Biol. 2000;227:343–357. doi: 10.1006/dbio.2000.9896. [DOI] [PubMed] [Google Scholar]
- 27.Kramer PK, Guerrero G, Krishnamurthy R, Mitchell PJ, Wray S. Ectopic expression of LHRH and peripherin in the respiratory epithelium of mice lacking transcription factor AP-2a. Mech. Dev. 2000;94:79–94. doi: 10.1016/s0925-4773(00)00316-6. [DOI] [PubMed] [Google Scholar]
- 28.Kramer PK, Wray S. Novel gene expressed in nasal regions influences outgrowth of olfactory axons and migration of luteinizing hormone releasing hormone (LHRH) neurons. Genes & Development. 2000;14:1824–1834. [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer PK, Wray S. Nasal embryonic LHRH factor (NELF) expression within the CNS and PNS of the rodent. Gene Expression Patterns. 2001;1:23–26. doi: 10.1016/s1567-133x(01)00004-7. [DOI] [PubMed] [Google Scholar]
- 30.Kramer PK, Krishnamurthy R, Mitchell PJ, Wray S. Transcription factor AP-2 is required for continued Luteinizing hormone releasing hormone (LHRH) expression in the forebrain of developing mice. Endocrinology. 2000;141:1823–1908. doi: 10.1210/endo.141.5.7452. [DOI] [PubMed] [Google Scholar]
- 31.Okubo K, Sakai F, Lau EL, Yoshizaki G, Takeuchi Y, Naruse K, Aida K, Nagahama Y. Forebrain gonadotropin-releasing hormone neuronal development: insights from transgenic medaka and the relevance to X-linked Kallmann syndrome. Endocrinology. 2006;147:1076–1084. doi: 10.1210/en.2005-0468. [DOI] [PubMed] [Google Scholar]
- 32.Whitlock KE, Wolf CD, Boyce ML. Gonadotropin-releasing hormone (GnRH) cells arise from cranial neural crest and adenohypophyseal regions of the neural plate in the zebrafish, Danio rerio. Dev Biol. 2003;257:140–152. doi: 10.1016/s0012-1606(03)00039-3. [DOI] [PubMed] [Google Scholar]
- 33.Whitlock KE. A new model for olfactory placode development. Brain Behav Evol. 2004;64(3):126–40. doi: 10.1159/000079742. [DOI] [PubMed] [Google Scholar]
- 34.Metz H, Wray S. Use of mutant mouse lines to Investigate origin of GnRH-1 neurons: lineage independent of the adenohypophysis. Endocrinology. 2010;151:766–773. doi: 10.1210/en.2009-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X, Wang J, Ju BG, Rosenfeld MG. Signaling and epigenetic regulation of pituitary development. Curr Opin Cell Biol. 2007;19:605–611. doi: 10.1016/j.ceb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- 37.Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, Joyner AL, Hui C. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 38.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 39.Sheng HZ, Zhadanov AB, Mosinger B, Jr., Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang SP, Mahon KA, Westphal H. Specification of pituitary cell lineages by the LIM homeobox gene Lhx3. Science. 1996;272:1004–1007. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- 40.Chung WC, Tsai PS. Role of Fibroblast Growth Factor Signaling in Gonadotropin-Releasing Hormone Neuronal System Development. Front Horm Res. 2010;39:37–50. doi: 10.1159/000312692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gill JC, Moenter SM, Tsai PS. Developmental regulation of gonadotropin-releasing hormone neurons by fibroblast growth factor signaling. Endocrinology. 2004;145(8):3830–9. doi: 10.1210/en.2004-0214. [DOI] [PubMed] [Google Scholar]
- 42.Kawauchi S, Shou J, Santos R, Hébert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–23. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- 43.Crane JF, Trainor PA. Neural crest stem and progenitor cells. Annu Rev Cell Dev Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- 44.Kalcheim C, Burstyn-Cohen T. Early stages of neural crest ontogeny: formation and regulation of cell delamination. Int J Dev Biol. 2005;49:105–116. doi: 10.1387/ijdb.041949ck. [DOI] [PubMed] [Google Scholar]
- 45.Kalcheim C, Langley K, Unsicker K. From the neural crest to chromaffin cells: introduction to a session on chromaffin cell development. Ann N Y Acad Sci. 2002;971:544–546. doi: 10.1111/j.1749-6632.2002.tb04524.x. [DOI] [PubMed] [Google Scholar]
- 46.Le Douarin NM, Brito JM, Creuzet S. Role of the neural crest in face and brain development. Brain Res Rev. 2007;55:237–247. doi: 10.1016/j.brainresrev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 47.Kramer PR, Krishnamurthy R, Mitchell PJ, Wray S. Transcription factor activator protein-2 is required for continued luteinizing hormone-releasing hormone expression in the forebrain of developing mice. Endocrinology. 2000;141:1823–1838. doi: 10.1210/endo.141.5.7452. [DOI] [PubMed] [Google Scholar]
- 48.Schlosser G. Evolutionary origins of vertebrate placodes: insights from developmental studies and from comparisons with other deuterostomes. J Exp Zoolog B Mol Dev Evol. 2005;304:347–399. doi: 10.1002/jez.b.21055. [DOI] [PubMed] [Google Scholar]
- 49.Fornaro M, Geuna S, Fasolo A, Giacobini-Robecchi MG. Evidence of very early neuronal migration from the olfactory placode of the chick embryo. Neuroscience. 2001;107:191–197. doi: 10.1016/s0306-4522(01)00334-7. [DOI] [PubMed] [Google Scholar]
- 50.Tobet SA, Chickering TW, King JC, Stopa EG, Kim K, Kuo-Leblank V, Schwarting GA. Expression of gamma-aminobutyric acid and gonadotropin-releasing hormone during neuronal migration through the olfactory system. Endocrinology. 1996;137:5415–5420. doi: 10.1210/endo.137.12.8940365. [DOI] [PubMed] [Google Scholar]
- 51.Casoni F, Wray S. In situ visualization of GnRH-1 neuronal migration in mouse nasal explants: Perturbation by GABA. International Journal of Developmental Neuroscience. 2008;26:880–881. [Google Scholar]
- 52.Marin O, Valiente M, Ge X, Tsai L-H. Guiding Neuronal Cell Migration. Cold Spring Harb Perspect Biol. 2010:2a001834. doi: 10.1101/cshperspect.a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwanzel-Fukuda M, Bick D, Pfaff DW. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Mol.Brain.Res. 1989;6:311–326. doi: 10.1016/0169-328x(89)90076-4. [DOI] [PubMed] [Google Scholar]
- 54.Quinton R, Hasan W, Grant W, Thrasivoulou C, Quiney RE, Besser GM, Bouloux PM. Gonadotropin-releasing hormone immunoreactivity in the nasal epithelia of adults with Kallmann's syndrome and isolated hypogonadotropic hypogonadism and in the early midtrimester human fetus. J Clin Endocrinol Metab. 1997;82:309–14. doi: 10.1210/jcem.82.1.3673. [DOI] [PubMed] [Google Scholar]
- 55.Wray S, Key S, Qualls R, Fueshko SM. A subset of peripherin positive olfactory axons delineates the luteinizing hormone releasing hormone neuronal migratory pathway in developing mouse. Dev.Biol. 1994;166:349–354. doi: 10.1006/dbio.1994.1320. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida K, Tobet SA, Crandall JE, Jimenez TP, Schwarting GA. The migration of luteinizing hormone-releasing hormone neurons in the developing rat is associated with a transient, caudal projection of the vomeronasal nerve. J.Neurosci. 1995;15:7769–7777. doi: 10.1523/JNEUROSCI.15-12-07769.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deiner MS, Sretavan DW. Altered midline axon pathways and ectopic neurons in the developing hypothalamus of netrin-1 and DCC-deficient mice. J. Neuroscience. 1999;19:9900–9912. doi: 10.1523/JNEUROSCI.19-22-09900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarting GA, Kostek C, Bless EP, Ahmad N, Tobet SA. Deleted in colorectal cancer (DCC) regulates the migration of luteinizing hormone-releasing hormone neurons to the nasal forebrain. J. Neuroscience. 2001;21:911–919. doi: 10.1523/JNEUROSCI.21-03-00911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komuro H, Rakic P. Orchestration of neuronal migration by activity of ion channels, neurotransmitter receptors, and intracellular Ca2+ fluctuations. J Neurobiol. 1998;37:110–130. (1998) [PubMed] [Google Scholar]
- 60.Toba Y, Pakiam J, Wray S. Voltage-gated calcium channels in the developing GnRH-1 neuronal systems. Êuropean Journal of Neuroscience. 2005;22:79–92. doi: 10.1111/j.1460-9568.2005.04194.x. [DOI] [PubMed] [Google Scholar]
- 61.Cariboni A, Hickok J, Rakic S, Andrews W, Maggi R, Tischkau S, Parnavelas JG. Neuropilins and their ligands are important in the migration of gonadotropin-releasing hormone neurons. J Neurosci. 2007;27:2387–95. doi: 10.1523/JNEUROSCI.5075-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giacobini P, Messina A, Morello F, Ferraris N, Corso S, Penachioni J, Giordano S, Tamagnone L, Fasolo A. Semaphorin 4D regulates gonadotropin hormone-releasing hormone-1 neuronal migration through PlexinB1-Met complex. J Cell Biol. 2008;183:555–66. doi: 10.1083/jcb.200806160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou QY, Cheng MY. Prokineticin 2 and circadian clock output. FEBS J. 2005;272:5703–9. doi: 10.1111/j.1742-4658.2005.04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsumoto S, Yamazaki C, Masumoto KH, Nagano M, Naito M, Soga T, Hiyama H, Matsumoto M, Takasaki J, Kamohara M, Matsuo A, Ishii H, Kobori M, Katoh M, Matsushime H, Furuichi K, Shigeyoshi Y. Abnormal development of the olfactory bulb and reproductive system in mice lacking prokineticin receptor PKR2. Proc Natl Acad Sci U S A. 2006;103:4140–5. doi: 10.1073/pnas.0508881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng KL, Li JD, Cheng MY, Leslie FM, Lee AG, Zhou QY. Dependence of olfactory bulb neurogenesis on prokineticin 2 signaling. Science. 2005;308:1923–7. doi: 10.1126/science.1112103. [DOI] [PubMed] [Google Scholar]
- 66.Gamble JA, Karunadasa DK, Pape JR, Skynner MJ, Todman MG, Bicknell RJ, Allen JP, Herbison AE. Disruption of ephrin signaling associates with disordered axophilic migration of the gonadotropin-releasing hormone neurons. J Neurosci. 2005;25:3142–50. doi: 10.1523/JNEUROSCI.4759-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giacobini P, Messina A, Wray S, Giampietro C, Crepaldi T, Carmeliet P, Fasolo A. Hepatocyte growth factor acts as a motogen and guidance signal for gonadotropin hormone-releasing hormone-1 neuronal migration. J Neurosci. 2007;27:431–45. doi: 10.1523/JNEUROSCI.4979-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwarting GA, Henion TR, Nugent JD, Caplan B, Tobet S. Stromal cell-derived factor-1 (chemokine C-X-C motif ligand 12) and chemokine C-X-C motif receptor 4 are required for migration of gonadotropin-releasing hormone neurons to the forebrain. J Neurosci. 2006;26:6834–40. doi: 10.1523/JNEUROSCI.1728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toba Y, Tiong JD, Ma Q, Wray S. CXCR4/SDF-1 system modulates development of GnRH-1 neurons and the olfactory system. Dev Neurobiol. 2008;68:487–503. doi: 10.1002/dneu.20594. [DOI] [PubMed] [Google Scholar]
- 70.Palevitch O, Abraham E, Borodovsky N, Levkowitz G, Zohar Y, Gothilf Y. Cxcl12a-Cxcr4b signaling is important for proper development of the forebrain GnRH system in zebrafish. Gen Comp Endocrinol. 2010;165:262–8. doi: 10.1016/j.ygcen.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 71.Giacobini P, Kopin AS, Beart PM, Mercer LD, Fasolo A, Wray S. Cholecystokinin modulates migration of gonadotropin-releasing hormone-1 neurons. J Neurosci. 2004;24:4737–4748. doi: 10.1523/JNEUROSCI.0649-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palevitch O, Abraham E, Borodovsky N, Levkowitz G, Zohar Y, Gothilf Y. Nasal embryonic LHRH factor plays a role in the developmental migration and projection of gonadotropin-releasing hormone 3 neurons in zebrafish. Dev Dyn. 2009;238:66–75. doi: 10.1002/dvdy.21823. [DOI] [PubMed] [Google Scholar]
- 73.Gill JC, Wadas B, Chen P, Portillo W, Reyna A, Jorgensen E, Mani S, Schwarting GA, Moenter SM, Tobet S, Kaiser UB. The gonadotropin-releasing hormone (GnRH) neuronal population is normal in size and distribution in GnRH-deficient and GnRH receptor-mutant hypogonadal mice. Endocrinology. 2008;149:4596–604. doi: 10.1210/en.2008-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abraham E, Palevitch O, Gothilf Y, Zohar Y. Targeted gonadotropin-releasing hormone-3 neuron ablation in zebrafish: effects on neurogenesis, neuronal migration, and reproduction. Endocrinology. 2010;151:332–40. doi: 10.1210/en.2009-0548. [DOI] [PubMed] [Google Scholar]
- 75.Kramer PK, Wray S. Novel gene expressed in nasal regions influences outgrowth of olfactory axons and migration of luteinizing hormone releasing hormone (LHRH) neurons. Genes & Development. 2000;14:1824–1834. [PMC free article] [PubMed] [Google Scholar]
- 76.Kramer PK, Wray S. Nasal embryonic LHRH factor (NELF) expression within the CNS and PNS of the rodent. Gene Expression Patterns. 2001;1:23–26. doi: 10.1016/s1567-133x(01)00004-7. [DOI] [PubMed] [Google Scholar]
- 77.Xu N, Bhagavath B, Kim HG, Halvorson L, Podolsky RS, Chorich LP, Prasad P, Xiong WC, Cameron RS, Layman LC. NELF is a nuclear protein involved in hypothalamic GnRH neuronal migration. Mol Cell Endocrinol. 2010;319:47–55. doi: 10.1016/j.mce.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanaho Y, Enomoto M, Endo D, Maehiro S, Park MK, Murakami S. Neurotrophic effect of gonadotropin-releasing hormone on neurite extension and neuronal migration of embryonic gonadotropin-releasing hormone neurons in chick olfactory nerve bundle culture. J Neurosci Res. 2009;87:2237–44. doi: 10.1002/jnr.22051. [DOI] [PubMed] [Google Scholar]
- 79.Fueshko SM, Key S, Wray S. GABA Inhibits Migration of Luteinizing Hormone Releasing Hormone (LHRH) Neurons in Embryonic Olfactory Explants. J. Neuroscience. 1998;18:2560–2569. doi: 10.1523/JNEUROSCI.18-07-02560.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bless EP, Westaway WA, Schwarting GA, Tobet SA. Effects if g-aminobutyric acidA receptor manipulation on migrating gonadotropin-releasing hormone neurons through the entire migratory route in vivo and in vitro. Endocrinology. 2000;141:1254–1262. doi: 10.1210/endo.141.3.7348. [DOI] [PubMed] [Google Scholar]
- 81.Lee JM, Tiong J, Maddox DM, Condie BG, Wray S. Temporal migration of gonadotrophin-releasing hormone-1 neurones is modified in GAD67 knockout mice. J Neuroendocrinol. 2008;20:93–103. doi: 10.1111/j.1365-2826.2007.01623.x. [DOI] [PubMed] [Google Scholar]
- 82.Miller NL, Wevrick R, Mellon PL. Necdin, a Prader-Willi syndrome candidate gene, regulates gonadotropin-releasing hormone neurons during development. Hum Mol Genet. 2009;18:248–60. doi: 10.1093/hmg/ddn344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cariboni A, Rakic S, Liapi A, Maggi R, Goffinet A, Parnavelas Reelin provides an inhibitory signal in the migration of gonadotropin-releasing hormone neurons. Development. 2005;132:4709–4718. doi: 10.1242/dev.02033. [DOI] [PubMed] [Google Scholar]
- 84.Krüger M, Ruschke K, Braun T. NSCL-1 and NSCL-2 synergistically determine the fate of GnRH-1 neurons and control necdin gene expression. EMBO J. 2004;23:4353–64. doi: 10.1038/sj.emboj.7600431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cogliati T, Delgado-Romero P, Norwitz ER, Guduric-Fuchs J, Kaiser UB, Wray S, Kirsch IR. Pubertal impairment in Nhlh2 null mice is associated with hypothalamic and pituitary deficiencies. Mol Endocrinol. 2007;21:3013–27. doi: 10.1210/me.2005-0337. [DOI] [PubMed] [Google Scholar]
- 86.Legouis R, Hardelin JP, Levilliers J, Claverie JM, Compain S, Wunderle V, Millasseau P, Le Paslier D, Cohen D, Caterina D, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–35. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 87.Ballabio A, Camerino G. The gene for X-linked Kallmann syndrome: a human neuronal migration defect. Curr Opin Genet Dev. 1992;2:417–421. doi: 10.1016/s0959-437x(05)80152-2. [DOI] [PubMed] [Google Scholar]
- 88.Whitlock KE, Smith KM, Kim H, Harden MV. A role for foxd3 and sox10 in the differentiation of gonadotropin-releasing hormone (GnRH) cells in the zebrafish Danio rerio. Development. 2005;132:5491–502. doi: 10.1242/dev.02158. [DOI] [PubMed] [Google Scholar]
- 89.Rugarli EI, Di Schiavi E, Hilliard MA, Arbucci S, Ghezzi C, Facciolli A, Coppola G, Ballabio A, Bazzicalupo P. The Kallmann syndrome gene homolog in C. elegans is involved in epidermal morphogenesis and neurite branching. Development. 2002;129:1283–94. doi: 10.1242/dev.129.5.1283. [DOI] [PubMed] [Google Scholar]
- 90.Miura K, Acierno JS, Jr, Seminara SB. Characterization of the human nasal embryonic LHRH factor gene, NELF, and a mutation screening among 65 patients with idiopathic hypogonadotropic hypogonadism (IHH) J Hum Genet. 2004;49:265–8. doi: 10.1007/s10038-004-0137-4. [DOI] [PubMed] [Google Scholar]
- 91.Dodé C, Levilliers J, Dupont JM, De Paepe A, Le Dû N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pêcheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-van de Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–5. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 92.Dodé C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2(10) doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–63. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwarting GA, Wierman ME, Tobet SA. Gonadotropin-releasing hormone neuronal migration. Semin Reprod Med. 2007;25:305–12. doi: 10.1055/s-2007-984736. [DOI] [PubMed] [Google Scholar]