Summary
Genetic analysis of DNA polymerases that are essential for progression through the S phase of the cell division cycle in budding yeast suggests that two separate DNA polymerases copy different strands of the DNA double helix, with a third polymerase involved in priming.
The entire genome of organisms must be copied accurately to ensure the inheritance of the genetic information over many generations. A large and complex cellular machinery has evolved to accomplish this remarkable feat. The machinery consists of the core proteins that copy DNA that coordinate with many other proteins that repair mistakes, handling damage to the DNA, recover stalled replication forks and hold the two daughter helices together until they are evenly segregated during mitosis. Well over 200 polypeptides are devoted to these processes.
The DNA double helix has its two strands intertwined with opposite polarity. Since all known DNA polymerases must use a 3′-OH of a nucleoside as a primer and synthesize DNA in the 5′ → 3′ direction, both strands of the DNA helix are copied differently, with one strand synthesized continuously (leading strand) and the opposite strand copied in short ~200 base pair Okazaki fragments that are then joined together (lagging strand). Both the leading strand and every Okazaki fragment on the lagging strand are primed by a short RNA that is synthesized de novo by a specialized RNA polymerase called primase that in eukaryotes in part of the DNA polymerase α enzyme.
Of the 15 known eukaryotic cell DNA-templated DNA polymerases (Weill and Reynaud, 2008), many are auxiliary and handle the repair of DNA that can be coupled to progression of the DNA replication fork itself. Three polymerases, α , δ and ε are at the core of the coping process and work in concert to do duplicate the bulk of the genome. Recent genetic analyses have clarified how these polymerases coordinate the process (Nick McElhinny et al., 2008; Pursell et al., 2007).
The general mechanism of how DNA replication occurs in eukaryotes was first determined by studying the duplication of the Simian Virus 40 (SV40) genome. Biochemical studies revealed that two essential DNA polymerases, α and δ, cooperated to replicate DNA along with proteins that unwound the helix, loaded the polymerases, kept them attached to the template and processed Okazaki fragments on the lagging strand (Waga and Stillman, 1998). The four-subunit polymerase α /primase is initially loaded onto the origin of DNA replication by interaction with the origin recognition protein, T antigen that also functions as a DNA helicase to unwind the DNA double helix. Together with the single stranded DNA binding protein RPA (replication protein A) polymerase α /primase makes short RNA primers and the polymerase activity extends these primers to about 30 bases.
The RNA-DNA primer attached to the single stranded template DNA provides a unique DNA structure to which an ATP-dependent machine called RFC binds specifically, expelling the polymerase α /primase enzyme. The bound RFC loads the ring shaped proliferating cell nuclear antigen (PCNA) onto the double stranded DNA near the primer and hands of the primer to another DNA polymerase called polymerase δ. PCNA is a DNA polymerase clamp that is topologically linked to the double stranded, replicated DNA and binds to polymerase δ, thereby ensuring that the polymerase synthesizes DNA in a processive manner. Indeed PCNA bound polymerase δ can copy RPA coated single stranded templates as long as 10 kb or more without disassociating and re-loading. The same loading mechanism that primes leading strand replication primes leading strand synthesis (figure, left panel). Whether PCNA stimulates polymerase ε is still unclear.
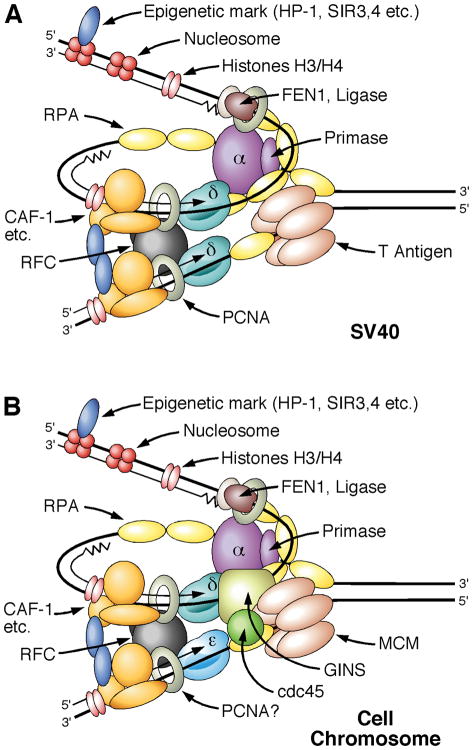
Figure.
Left panel: DNA replication fork showing the proteins that are required for replication of SV40 DNA. The SV40 T antigen functions to recruit polymerase α/primase and also as a DNA helicase to unwind the DNA. Primers made by polymerase α/primase are then recognized by RFC and PCNA to load polymerase δ that completes the Okazaki fragments on the lagging strand. Polymerase δ also synthesizes the leading strand. Right panel: Replication forks that emerge from cellular origins of DNA replication are established by priming by polymerase α/primase priming, but in this case polymerase ε synthesizes the leading strand and polymerase δ the lagging strand. Instead of T antigen to unwind the DNA, helicase activity is provided by the Cdc45-MCM-GINS (CMG) complex. Polymerase δ is activated by the PCNA polymerase clamp, but whether this occurs on the leading strand for polymerase ε remains to be determined. How polymerase ε is loaded is under investigation.
The polymerase switching mechanism in which α hands off to δ explains how coordinated leading and lagging strand DNA replication could occur by the polymerase δ working on both strands of the DNA replication fork. It also explains why polymerase α does not need a proofreading exonuclease activity that polymerase δ contains since any DNA that polymerase α makes is eventually excised during Okazaki fragment maturation. In many ways, the polymerase switching mechanism and role of PCNA-polymerase δ on both strands of the fork explained how SV40 DNA is duplicated.
One problem in translating this mechanism to duplication of cell chromosomal DNA was that T antigen did not play a role and a third essential DNA polymerase, called polymerase ε was found in S. cerevisiae (Morrison et al., 1990). This enzyme has a large polymerase subunit encoded by the POL2 gene, but it was later shown that in both S. cerevisiae and S. pombe the polymerase catalytic activity was not essential, although genome replication was not normal (Dua et al., 1999; Feng and D'Urso, 2001; Kesti et al., 1999). Thus α and δ can replicate the genome. The essential part of POL2 was a domain of the protein involved in checkpoint signaling that handles DNA damage during S phase.
Polymerase ε, like polymerase δ has a proofreading exonuclease, suggesting that it synthesizes DNA that must be accurately inherited, but the question is what DNA? Studies in Kunkel’s laboratory over many years have elucidated the mechanism of fidelity of several DNA polymerases, including polymerases δ and ε. Based both on amino acid sequence conservation and structures of several DNA polymerases, they measured the contribution of selected amino acids in the DNA polymerase catalytic active site in vivo and in vitro. One particular amino acid was proposed to interact with a conserved tyrosine that guided the incoming dNTP. Mutations in these residues (pol3-L612M or pol2-M644G) produced DNA polymerases that had normal rates of DNA synthesis but miss-incorporated dNMPs, thereby causing mutations in vivo. These studies enabled the analysis of the relative contributions of the two DNA polymerases to the leading or lagging strand replication.
By placing a reporter gene in either orientation and at either side of a single origin of DNA replication, a given strand of the reporter gene would be replicated either as the leading or lagging strand, depending on the orientation of the gene. Because the DNA sequences of the two strands of the reporter gene are complementary, the altered DNA polymerases encounter different DNA sequences that are hot spots for mutations. Thus the accumulated mutations are characteristic of the strand being replicated. When the yeast harbored the altered polymerase δ, mutations occurred during lagging strand replication and the opposite was the case for the altered polymerase ε, which accumulated mutations when the leading strand was synthesized (Nick McElhinny et al., 2008; Pursell et al., 2007). These results argue that the two polymerases share the load during DNA replication, with polymerase ε on the leading strand and polymerase δ on the lagging strand (figure, right panel).
After establishment of pre-replicative complexes at origins of DNA replication, entry into S phase occurs by activation of the S phase cyclin-dependent kinase (CDK) and the Cdc7-Dbf4-dependent kinase (DDK). An essential role of the CDK is to phosphorylate the Sld2 and Sld3 proteins that bind to the Dbp11 protein that then promote loading of proteins such as Cdc45 and GINS at replication origins just when they are activated in S phase (Botchan, 2007; Labib and Gambus, 2007). The latter two proteins interact with the MCM to form a helicase complex that, with Dpb11 interacts with polymerase ε. Other proteins such as Mcm10, Cft4 and Pob1, all present in a larger protein complex, interact with polymerase α . The details, however, of how polymerases α and ε are loaded at origins of DNA replication needs to be established. Polymerase δ, on the other hand, is loaded by the RFC-PCNA dependent mechanism established for SV40 DNA replication.
On a more global genome level, remains to be determined about the relative contributions of the polymerases δ and ε during DNA replication, although the recent genetic analysis has clarified the situation significantly. For example, stalling of DNA replication forks during S phase may lead to dissociation of the polymerase and require re-loading. It is clear how polymerase δ can be re-loaded, but whether polymerase ε can be loaded at such sites remains to be determined. It would be therefore valuable to use yeast containing the altered polymerases in which the reporter gene is placed at a large distance from the origin, even down stream of DNA sequences that promote replication fork stalling, to determine whether polymerase ε or δ synthesizes the remaining leading strand in a replicon. Furthermore, both early and late replicating origins could be examined to see if there is a difference. For the future, it is clear that more biochemical information on DNA polymerase loading will fill in gaps in our knowledge about how the genome is duplicated.
References
- Botchan M. Cell biology: a switch for S phase. Nature. 2007;445:272–274. doi: 10.1038/445272a. [DOI] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- Feng W, D'Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol. 2001;21:4495–4504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 2007;17:271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PJ, Kunkel TA. Division of labor at the eukaryotic replication fork. Molecular Cell. 2008 doi: 10.1016/j.molcel.2008.02.022. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Weill JC, Reynaud CA. DNA polymerases in adaptive immunity. Nat Rev Immunol. 2008;8:302–312. doi: 10.1038/nri2281. [DOI] [PubMed] [Google Scholar]