Abstract
Previous work has demonstrated that the copper (Cu) transporters Ctr1, Atp7a and Atp7b regulate the cellular pharmacology of cisplatin (CDDP) by mediating its uptake and efflux. It was also shown that, in the process of uptake by Ctr1, CDDP triggers the rapid proteasomal degradation of its own transporter. The current study examined the role of the metallochaperone Atox1 in the regulation of uptake, efflux and subcellular distribution of CDDP by using a pair of fibroblast cell lines established from Atox1+/+ and Atox1-/- mice. Atox1 is a metallochaperone that is known to play a central role in distributing Cu within the cells and was recently shown to act as a Cu-dependent transcription factor. Loss of Atox1 increased Cu accumulation and reduced efflux. In contrast, loss of Atox1 reduced the influx of CDDP and subsequent accumulation in vesicular compartments and in DNA. Loss of Atox1 was found to block the CDDP-induced down regulation of Ctr1. Ctr1 was found to be polyubiquitinated in an Atox1-dependent manner during CDDP exposure. In conclusion, Atox1 is required for the polyubiquitination of Ctr1 and the Ctr1-mediated uptake of CDDP.
Keywords: Atox1, cisplatin, copper, Ctr1, degradation
1. Introduction
The mechanism of cellular accumulation of the anticancer drug cisplatin (CDDP) is often impaired in cells with acquired resistance to CDDP. Recent data has implicated the transporters of the copper (Cu) homeostasis system in the process of uptake and efflux of CDDP. In particular, a role for the Cu importer Ctr1 has been demonstrated by the observation that the absence of Ctr1 impairs the ability of cells to accumulate CDDP and thus increases the degree of resistance of cells to the cytotoxic effects of this drug [1-4]. A large body of evidence has also documented the involvement of the two Cu exporters, Atp7a and Atp7b, in the efflux or vesicular sequestration of CDDP and shown them to be regulators of the cytotoxic effect of CDDP [5-8]. While the exact mechanism by which these Cu transporters control the intracellular levels of CDDP remains to be discovered, available data confirm a role for the cysteine, histidine and methionine rich metal binding motifs which are found in these and several other Cu homeostasis proteins such as the metallochaperone Atox1 (antioxidant protein 1) (reviewed in [9]).
The metal binding domain of Ctr1 consists of a methionine rich motif (mets motif) that binds Cu and propels it into the central pore of a homotrimeric Ctr1 complex at the plasma membrane [10]. The metal binding domains of Atp7a and Atp7b consist of 6 cysteine-containing motifs with a classical ferredoxin-like βαββαβ fold [11-16] and a core sequence of CxxC, similar to the one found in the metallochaperone Atox1 [17]. Atox1 is a key metallochaperone that receives Cu from the importer Ctr1 and delivers it to Cu exporters Atp7a and Atp7b in the secretory compartment. Atox1 has 69 amino acids and is found in the cytosol and nucleus [18]. The single metal binding motif of Atox1 has a CggC sequence that can selectively bind a single Cu1+ [19] in a dimeric form [17]. It is believed that the exchange of Cu between the Atox1 and Ctr1 [20, 21], Atp7a and Atp7b [12, 22] occurs through direct protein-protein interactions and that these interactions are required for the delivery of Cu to the secretory cuproenzymes and detoxification of excess Cu via efflux. Loss of expression of Atox1 in transgenic mice and in yeast is accompanied by an increase in intracellular levels of Cu [23, 24], a finding which is consistent with its proposed function as a specific Cu chaperone for delivery of Cu to the secretory compartment [25]. However, in light of recent findings that Atox1 can also bind DNA and regulate transcription [26] it is likely that this protein plays an even more complex role in the regulation of cellular physiology in response to changing levels of intracellular Cu.
In this study, we examined the role of Atox1 in the regulation of the cellular pharmacology of CDDP using a pair of fibroblast cell lines from wild type (Atox1+/+) and knockout (Atox1-/-) mice. We show that Atox1 regulates the influx of CDDP by controlling the CDDP-induced down regulation of Ctr1through ubiquitination.
2. Materials and methods
2.1. Reagents
Cell culture media and sera were purchased from HyClone (Logan, UT). Antibodies to Ctr1 were from Novus Biologicals (Littleton, CO), tubulin, from Sigma Co. (St. Louis, MO) and polyubiquitin conjugates (FK1 and FK2), from BIOMOL (Exeter, UK). 64Cu was purchased from Isotrace Technologies, Inc. (O'Fallon, MO). Other chemicals were purchased from Sigma Co. (St. Louis, MO) and Fisher Scientific Co. (Tustin, CA). CDDP (PLATINOL-AQ) was received as a gift from Bristol Laboratories (Princeton, NJ.).
2.2. Cell culture and assay of sensitivity
Immortalized fibroblasts from Atox1+/+ and Atox1-/- mice were generously provided by Dr. JD Gitlin (Washington University, St. Louis, MO) [24]. These cells were maintained and propagated in DMEM supplemented with 10% fetal bovine serum and 250 μg/mL of G418. Sensitivity to the growth inhibitory effects of CDDP and Cu was determined by measuring total cellular protein using a Bio-Rad assay dye reagent (Bio-Rad, Richmond, CA) at the end of a 5 day drug exposure. Inoculation of 6,000 cells per well of 24-well plates and a growth period of 5 days in the presence of various concentrations of Cu or CDDP was found to yield the optimal dynamic range for the Atox1+/+ and Atox1-/- cells in this assay. After 5 days of growth, cells were washed with PBS (phosphate buffered saline) and then dissolved in situ in 100 μL of lysis buffer (150 mM NaCl, 5 mM EDTA, 1 % Triton-X 100, 10 mM Tris, pH 7.4), which was subjected to protein determination. Protein levels were measured at a wavelength of 595 nm using a Benchmark Micro-Plate Reader (BioRad, Richmond, CA).
2.3. Accumulation and efflux of 64Cu
Whole cell accumulation of Cu was measured by incubating 5 × 105 of Atox1+/+ or 7.5 × 105 Atox1-/- cells/well in 35 mm plates using basal medium containing 2 μM Cu traced with 1- 8 × 106 cpm 64Cu. After a 24 h exposure to 64Cu, cells were washed three times with ice-cold PBS, lysed with PBS containing 1% SDS and analyzed for 64Cu accumulation by using a Beckman 5500B Gamma Counter.
For efflux, cells were loaded with 2 μM Cu traced with 1- 8 × 106 cpm of 64Cu for 24 h and then medium was removed and cells were allowed to efflux for 0, 5, 10, 15, 30 and 60 min after which the levels of radioactivity were determined as described above. The Cu levels were normalized to the protein levels in each lysate sample.
2.4. Accumulation of CDDP in whole cell, subcellular fractions and assay of efflux of CDDP
For whole cell accumulation of CDDP 5 × 105 of Atox1+/+ and 7.5 × 105 Atox1-/- cells were inoculated l in 35 mm plates. CDDP was added to the culture medium to the final concentration of 2 μM. At different time points following CDDP exposure cells were rinsed quickly with three changes of ice-cold PBS and then lysed with 250 μL of lysis buffer (0.25% Nonidet P-40 in 100 mM Tris HCl, pH 8.0). An aliquot was set aside for protein determination and another 200 μL aliquot was mixed with 125 μL of 70% analytical grade nitric acid, heated to 65 °C overnight, diluted with 3 mL of distilled water containing 1 part per billion indium (Acros Organics, Tustin, CA) and subjected to Pt measurements using a Thermo Finnigan ICP-MS (inductively coupled plasma mass spectrometry, Element 2 at the Analytical Facility at the Scripps Institute of Oceanography). Pt values were normalized to protein levels, measured with a Bio-Rad protein assay dye reagent (Bio-Rad, Richmond, CA).
To prepare nuclear, cytosolic and microsomal fractions, cells were harvested with trypsin, centrifuged at 1000 × g and the pellet was rinsed once with ice-cold PBS and resuspended in a hypotonic buffer (10 mM Tris, pH 7.4. 1 mM MgCl2, 10 mM NaCl, 5 mM CaCl2) containing protease inhibitors (Complete tablets from Sigma, St. Louis MO). Cells were then homogenized with a glass homogenizer and centrifuged at 1000 × g at 4 °C for 5 min to pellet the nuclei. Nuclei were further purified by two additional rounds of re-suspension in hypotonic buffer, homogenization and centrifugation as above. Microsomal fractions were prepared by centrifugation of the post-nuclear fractions at 130,000 × g for 1 h as previously described [27]. The cytosolic fractions were prepared from the post-nuclear fraction by sedimenting all particulate organelles with two rounds of centrifugation at 130,000 × g for 1h each. Microsomal and nuclear fractions were dissolved in lysis buffer (0.25% Nonidet P-40 in 100 mM Tris HCl, pH 8.0). Aliquots were taken from each fraction for protein assay and the remainder of the fraction was used for Pt determination by ICP-MS.
Efflux was measured in cells that had been loaded with 2 μM CDDP for 24 h; cells were quickly rinsed, three times with warm medium and then incubated with fresh medium for 0, 1, 5, 10, 15, 30, and 60 min; cells were then harvested and processed for protein and Pt determination as described above.
2.5. Pt-DNA adducts
Cells were grown in 150 cm2 flasks until 80% confluent and were then incubated in fresh medium containing 2 μM CDDP for 24 h. Cells were then rinsed three times with PBS and lysed by 3 mL of DNAZol (Invitrogen, Carlsbad, CA). The lysate was transferred to a 15 mL conical centrifuge tube and mixed with an equal volume of isopropanol prior to centrifugation at 20,000 × g for 5 min. The pellet was extracted 3 additional times with DNAZol and the final DNA product was assayed for purity with a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). DNA (100-300 μg) was dissolved in 70% nitric acid and heated to 65°C overnight, dissolved in indium-containing water and assayed for Pt content by ICP-MS.
2.6. Western blotting
Cells were harvested with trypsin, pelleted by centrifugation at 1000 x g at 4 °C, dissolved in lysis buffer (150 mM NaCl, 5 mM EDTA, 1 % Triton-X 100, 10 mM Tris, pH 7.4) containing protease inhibitors (Sigma Complete tablets, St. Louis, MO). The cells were then sonicated for 30 seconds and centrifuged at 500 × g for 10 min. The supernatant was assayed for protein concentration using the Bradford reagent (Bio-Rad, Richmond, CA) and aliquots of 25-75 μg protein/lane were electrophoresed on 4-20% gels using a Bio-Rad electrophoresis system. A Bio-Rad trans-blot system was used to transfer the proteins to Immobilin-P membranes (Millipore, Billerica, MA). Blots were incubated in 4% dry non-fat milk in TBST buffer (tris buffered saline, 150 mM NaCl, 300 mM KCl, 10 mM Tris, pH 7.4 and 0.01% Tween 20) overnight at 4 °C and then with 4% milk/TBST containing polyclonal antibodies to the N- or C-terminal regions of the Ctr1 for 12 h at 4 °C. A horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences, Piscataway, NJ) was dissolved in 4% milk in TBST buffer and incubated with the blot for 1h at room temperature. After three 15 min washes, the blots were exposed to the ECL chemiluminescence reagents (Amersham Biosciences, Piscataway, NJ) and detected on X-ray films (HyBlot CL, Denville Scientific, Inc. Metuchen, NJ). Each blot was stripped of antibodies using a the Restore™ Blot Stripping Buffer from Pierce Biotechnology, Inc. (Rockford, IL) and then probed with a monoclonal anti-tubulin antibody from Sigma (St. Louis, MO) according to the protocol provided by the vender. A ChemiImager™ 4400 instrument (Alpha Innotech, San Leandro, CA, USA) was used for determining the density of protein bands.
2.7. Confocal microscopy
Cells were cultured on coverslips and allowed to attach overnight. The cells were then exposed to either 10 μM CDDP or 300 μM Cu for 15 min after which they were quickly rinsed with cold PBS and fixed with 3.7% formaldehyde in PBS for 10 min. The cells were permeabilized with 0.3% Triton X 100 in PBS for another 10 min, blocked with 1 % (w/v) bovine serum albumin (BSA) in PBS for 1 h and then incubated with a polyclonal antibody against Ctr1 or monoclonal antibodies against polyubiquitin for 12 h. Following three 15 min rinses with PBS, cells were stained with an Alexa Fluor 488 conjugated anti-rabbit secondary antibody (Molecular Probes, Seattle, WA) or a Texas red conjugated anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) along with the nuclear marker Hoechst 33342 (Molecular Probes, Seattle, WA). Cells were then washed for 15 min three more times and then mounted on slides with Gelvatol and processed for microscopy with a DeltaVision deconvolution microscope system (Applied Precision, Inc., Issaquah, WA.) at the UCSD Cancer Center's Digital Imaging Shared Resource Facility as previously described [28].
2.8. Flow cytometry
Atox1+/+ and Atox1-/- cells were cultured on 150 mm flasks until 70% confluent and then exposed to 2 μM CDDP for 15 min. The cells were quickly rinsed with cold PBS before harvesting them with trypsin and centrifuging at 1000 × g for 10 min. The cells were resuspended in cold PBS, fixed for 10 min with 3.7% formaldehyde in PBS, permeabilized for 10 min with 0.3% Triton X 100 in PBS and then treated for 1 h with 1% BSA in PBS prior to incubation for 12 h with antibodies against Ctr1. After three 15 min washes with PBS, cells were incubated for 1 h with an Alexa Fluor 488-tagged secondary antibody (Molecular Probes, Seattle, WA). After washing the secondary antibody three times, 15 min each, with PBS, the cells were assayed for fluorescence using a BD FACSCalibur instrument at the Flow Cytometry Shared Resource of the UCSD Cancer Center.
2.9. Immunoprecipitation with antibodies to Ctr1 or polyubiquitin conjugates
Whole cell extracts were prepared using lysis buffer (150 mM NaCl, 5 mM EDTA, 1 % Triton-X 100, 10 mM Tris, pH 7.4) containing protease inhibitor tablets (Sigma, St Louis MO). The cell lysates were clarified by centrifugation at 10,000 × g for 10 min at 4 °C and then were diluted to the final concentration of 100 μg/mL with binding buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 10% glycerol; 2 mM β-mercaptoethanol, 0.1 % Triton X-100 and Roche Complete EDTA-free protease inhibitor tablet) [29] to which protein A-Sepharose (10μL/mL) was added and incubation continued for 1 h at room temperature. The samples were centrifuged at 1000 x g for 10 min at 4 °C and then 2 μg/mL of the primary antibody (anti-Ctr1, FK1 or FK2) was added and incubation continued overnight at 4 °C. The immune complexes were captured by adding 10 μL/mL of protein A-Sepharose (Thermo Scientific, Waltham, MA) for 1 h at 4 °C followed by washing five times in binding buffer. Proteins were eluted with elution buffer provided by Thermo Scientific and the samples were separated on a 4–15% polyacrylamide gels and electroblotted as described above.
2.10. Assay of 20S proteasome
The chymotrypsin-like proteasome activity was measured using the 20S Proteasome Assay Kit from Calbiochem (Gibbstown, NJ). Briefly, triplicate assays were performed in a reaction mixture that contained 178 μL of reaction buffer (25 mM HEPES and 0.5 mM EDTA, pH 7.6), 10 μL of substrate (10 μM Suc-Leu-Val-Tyr-7-amino-4-methylcoumarin (AMC)), 2 μL of SDS (0.03%) and 10 μL of cell lysate (25 μg of protein). After incubation for 30 min at 37°C the fluorescence of liberated AMC was measured every 5 min for 30 min using excitation and emission wavelengths at 340 and 450 nm, respectively by a TECAN Infini M200 plate reader (Durham, NC). Estimates of the slope and curve fitting were made using Prism software (Prism Inc. Irvine, CA).
2.11. Statistical analysis
Groups were compared using the Student t test assuming unequal variance.
3. Results
3.1. Effects of the loss of Atox1 on the toxicity and cellular pharmacology of Cu
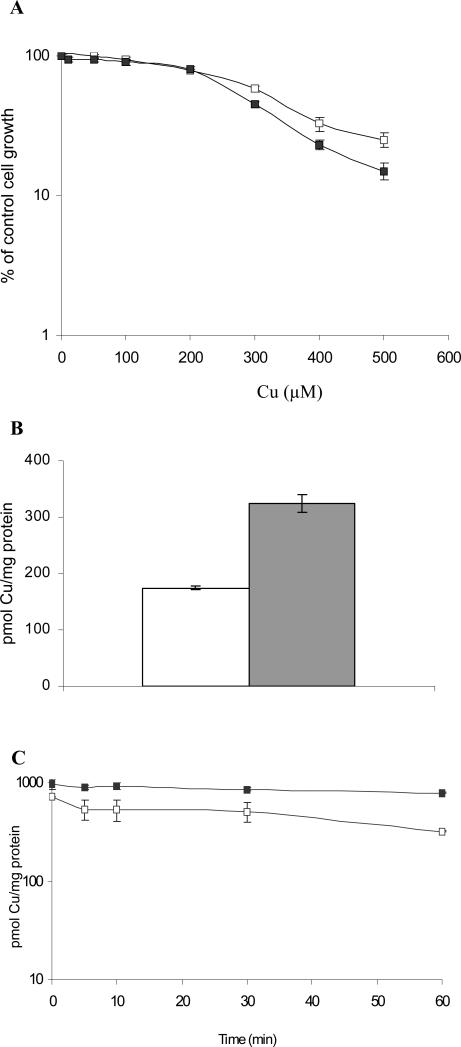
These studies utilized a pair of isogenic mouse embryo fibroblasts established from either wild type mice (Atox1+/+) or mice in which both alleles of Atox1 had been deleted (Atox1-/-). Sensitivity to the cytotoxic effect of Cu was assessed by examining the effect of increasing concentrations of Cu on the growth rate of the Atox1+/+ and Atox1-/- fibroblasts over a period of 5 days. The data presented in Figure 1A was obtained from 5 independent assays, each performed with triplicate cultures for each Cu concentration. The IC50 values were 337 ± 22 μM for the Atox1+/+ cells and 276 ± 9 μM for the Atox1-/- cells (p < 0.03) indicating that the loss of Atox1 was accompanied by a small increase in the sensitivity of cells to Cu.
Figure 1.
Effects of the loss of Atox1 on the cellular pharmacology of Cu. (A) Assay of sensitivity of Atox1+/+ (□) and Atox1-/- (■) cells to the growth inhibitory effects of Cu during a 5 day exposure period. (B) Whole cell accumulation of Cu after a 24 h incubation with 2 μM Cu traced with 64Cu; open bar, Atox1+/+ cells; gray bar, Atox1-/-cells. (C) Efflux of Cu after exposure of Atox1+/+ (□) and Atox1-/- (■) cells to 2 μM Cu traced with 64Cu for 24 h. In all assays error bars are SEM of at least 3 independent experiments, each performed with a minimum of 3 cultures per data point.
In order to investigate the effect of the loss of Atxo1 on the accumulation of Cu, Atox1+/+ and Atox1-/- cells were exposed to 2 μM 64Cu for 24 h, by which time uptake had reached steady-state; the amount of intracellular 64Cu was quantified by scintillation counting. As shown in Figure 1B, the 64Cu level in the Atox1-/- cells was 1.8-fold higher (324 ± 16) pmol/mg protein) than that in the Atox1+/+ cells (174 ± 4) pmol/mg protein) (p < 0.00001). This difference was similar to that previously reported [25]. Results of the study of the efflux of Cu in the Atox1+/+ and Atox1-/- cells were also consistent with previously reported data [25]; Atox1+/+ cells effluxed Cu at a significantly faster rate than the Atox1-/- cells (Figure 1C). At 30 min, the Atox1+/+ cells retained 72 ± 5 % of their initial load of Cu, while Atox1-/- cells retained 97 ± 3 %; at 60 min, Atox1+/+ cells only 33 ± 8 % of their initial load, while the Atox1-/- cells still retained 53 ± 8 % (p < 0.03). Thus, loss of Atox1 increased the steady-state level of Cu and this was associated with reduced efflux.
3.2. Effects of the loss of Atox1 on the cellular pharmacology of CDDP
As shown in Figure 2A, the Atox1+/+ cells were slightly more sensitive to the cytotoxic effects of CDDP than the Atox1-/- cells. The IC50 values for Atox1+/+ and Atox1-/- cells were respectively, 1.0 ± 0.1 and 1.5 ± 0.1μM (p < 0.02). Thus, although the difference in sensitivity was small, the loss of Atox1 had opposite effects on sensitivity to the growth inhibitory effects of Cu and CDDP, rendering cells more sensitive to Cu but more resistant to CDDP.
Figure 2.
Effect of the loss of Atox1 on the cellular pharmacology of CDDP. (A) Assay of sensitivity of Atox1+/+ (□) and Atox1-/- (■) cells to the growth inhibitory effects of CDDP during a 5 day exposure period. (B) Whole cell accumulation of CDDP during a 5 min to 24 h incubation with 2 μM CDDP; open bar, Atox1+/+ cells; gray bar, Atox1-/- cells. (C) Efflux of Cu after exposure of Atox1+/+ (□) and Atox1-/- (■) cells to 2 μM CDDP for 24 h. (D) Pt-DNA adducts formed per mg DNA; open bar, Atox1+/+ cells; gray bar, Atox1-/- cells. In all assays error bars are SEM of at least 3 independent experiments, each performed with a minimum of 3 samples per data point.
To determine whether the increased resistance of the Atox1-/- cells to CDDP was due to altered intracellular accumulation of the drug, the uptake of CDDP in the Atox1+/+ and Atox1-/- cells was measured after exposure of the cells to 2 μM CDDP for 1 and 5 min, and 1 and 24 h. As shown in Figure 2B, the level of Pt accumulation in the Atox1+/+ cells was significantly higher than in the Atox1-/- cells. The difference was already apparent at 1 min at which point the Atox1+/+ cells had accumulated 3.4 ± 0.1 pmol Pt/mg protein, whereas the Atox1-/-cells had accumulated 2.6 ± 0.3 pmol Pt/mg protein (p < 0.01). At 5 min the levels were 4.2 ± 0.1 and 2.4 ± 0.5 pmol Pt/mg protein for the Atox1+/+ and Atox1-/- cells, respectively (p < 0.004). The difference in accumulation persisted at 1 and 24 h. At 1 h the levels were 72.9 ± 12.8 and 45.1 ± 5.3 pmol/mg protein (p < 0.01), and at 24 h 167.0 ± 18.2 and 103.3 ± 23.4 pmol/mg protein (p = 0.03) for the Atox1+/+ and Atxox1-/- cells, respectively. At 24 h the Atox1-/- cells had accumulated only 62% as much CDDP as the Atox1+/+ cells. Thus, as in the case of sensitivity to the cytotoxic effects of Cu and CDDP, the loss of Atox1 had a different effect on the uptake of CDDP in comparison to that of Cu.
To determine whether the lower level of CDDP accumulation in the Atox1-/- cells was due to a change in the ability of cells to efflux CDDP, we measured the efflux of CDDP from Atox1+/+ and Atox1-/- cells over a period of 1 h after the cells had been loaded with CDDP with 2 μM CDDP for 24 h and then washed and re-incubated in drug-free medium. As shown in Figure 2C, there was no clear difference in the ability of the Atox1+/+ and Atox1-/- cells to efflux CDDP. After 60 min of efflux the Atox1+/+ cells retained 85.2 ± 0.9 %, and the Atox1-/- cells retained 80.8 ± 2.4 %, of their respective initial loads of CDDP. Thus, the Atox1 protein appears to function primarily in the influx rather than the efflux of CDDP.
To assess the effect of the loss of Atox1 on the intracellular distribution of CDDP Atox1+/+ and Atox1-/- cells were exposed to 2 μM CDDP for 24 h and then subjected to subcellular fractionation using differential centrifugation. The cytosolic, microsomal and nuclear fractions were isolated and the percentage of the whole cell Pt found in each fraction was determined. As seen in Table 1, there were clear differences between the Atox1+/+and Atox1-/-cells in the intracellular distribution of Pt. When calculated as pmol Pt/mg protein of each fraction, the cytosolic fractions of the two cell types had similar levels of Pt but the microsomal and nuclear fractions of the Atox1+/+ cells had significantly higher levels of Pt than those of the Atox1-/- cells. However, when the data was calculated as a percentage of the whole cell Pt content, no significant differences between the Atox1+/+ and Atox1-/- and cells could be detected. Thus, the loss of Atox1 reduced the amount of Pt in the microsomal and nuclear compartment relative to their protein content but not the fractional distribution suggesting that loss of Atox1 had primarily affected the influx of CDDP.
Table 1.
Distribution of Pt in subcellular fractions of Atox1+/+ and Atoxl-/- cells.
| Cell Fraction | Percent of total Pt in fraction, mean ± SEM | pmol Pt/mg protein, mean ± SEM | |||
|---|---|---|---|---|---|
| Atox1+/+ cells | Atox1-/- cells | Atox1+/+ cells | Atox1-/- cells | p Value* | |
| Cytosol | 70.1 ± 4.4 | 73.9 ± 6.6 | 21.4 ± 0.4 | 18.6 ± 0.5 | |
| Microsomal | 5.9 ± 0.8 | 6.1 ± 0.9 | 221.8 ± 10.9 | 176.3 ± 12.5 | <0.05 |
| Nuclear | 24.0 ± 4.6 | 20.0 ± 4.8 | 318.5 ± 21.6 | 160.1 ± 21.5 | <0.04 |
For difference in Pt content.
Since the higher degree of resistance of Atox1-/- cells to treatment with CDDP was consistent with the lower accumulation of CDDP in DNA we measured the levels of Pt-DNA adducts in the two cell types following exposure to 2 μM CDDP for 24 h. As shown in Figure 2D, the loss of Atox1 was associated with a reduced level of Pt in DNA. The Atox1+/+ cells accumulated 34.4 ± 6.7 pmol Pt/mg DNA whereas the Atox1-/- cells accumulated only 18.8 ± 5.5 pmol Pt/mg DNA (p< 0.02). The magnitude of the difference in DNA adducts was in the same range as the difference in whole cell uptake of CDDP.
In summary, loss of Atox1 was associated with a small degree of resistance to CDDP, reduced influx and accumulation in the whole cell, the nuclear compartment and in the DNA but did not change the efflux of CDDP.
3.3. Effect of loss of Atox1 on Cu- and CDDP-induced down regulation of Ctr1
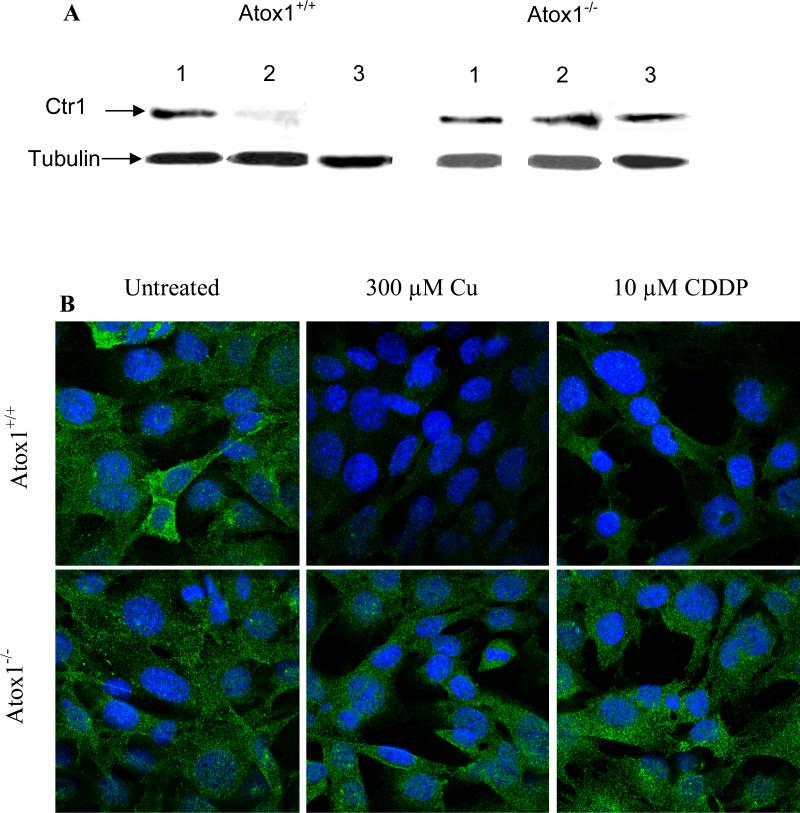
Since the primary consequence of the loss of Atox1 was a reduction in the uptake of CDDP, we examined the effect of the loss of Atox1 on the initial influx of CDDP mediated by Ctr1. Ctr1 is the major Cu influx transporter and has been shown to mediate the initial phase of CDDP influx in human ovarian carcinoma cells [2]. Exposure to either CDDP or Cu has been shown to rapidly down-regulate Ctr1 expression in several cell types [30]. We examined the levels of Ctr1 in the Atox1+/+ and Atox1-/- cells before and after exposure to CDDP. A Western analysis of the Atox1+/+ and Atox1-/- cells is shown in Figure 3A; in both cell types the monomeric form of Ctr1 was detected as a band migrating at ~ 28 kDa. In untreated cells, the levels of Ctr1 were similar relative to the lane loading controls. However, when the Atox1+/+ and Atox1-/- cells were exposed for 15 min to 10 μM CDDP or 300 μM Cu, only the Atox1+/+ cells exhibited a CDDP- or Cu-induced down-regulation of the Ctr1 (Figure 3A). Exposure of Atox1-/-cells to either compound resulted in Ctr1 expression levels that were unchanged from those found in the untreated cells. Therefore, while both CDDP and Cu triggered the down-regulation of Ctr1 only in the Atox1+/+ cells, neither reduced Ctr1 in the Atox1-/- cells.
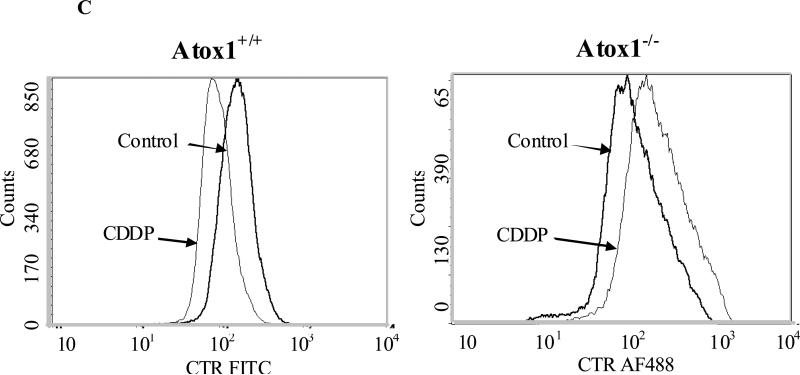
Figure 3.
Effect of the loss of Atox1 on the Cu- and CDDP-induced down-regulation of Ctr1. (A) Western blot analysis of cell lysates prepared from Atox1+/+ and Atox1-/- cells after exposure for 15 min to 10 μM CDDP or 300 μM Cu. Lane 1, no treatment; lane 2, 300 μM Cu; lane 3, 10 μM CDDP. (B) Confocal microscopy of untreated Atox1+/+ and Atox1-/-cells (left panel) or cells exposed to 300 μM Cu (middle panel) or 10 μM CDDP (right panel) for 15min. Ctr1 is detected with a rabbit polyclonal antibody; an Alexa Fluor 488 conjugated anti-rabbit antibody is used as secondary antibody (green); nuclei are stained blue with Hoechst 33342. (C) Flow cytometry of Atox1+/+ and Atox1-/- cells that were fixed and fluorescently labeled with antibodies against Ctr1 and subsequently with an Alexa Fluor 488 conjugated anti-rabbit antibody. Negative controls included unstained cells and cells stained only with the secondary antibody, treated with or without CDDP. Heavy line, untreated cells; light line, drug treated cells. Left panel, Atox1+/+cells treated with or without 2 μM CDDP; right panel, Atox1-/- cells treated with or without 2 μM CDDP.
To assess the subcellular localization of Ctr1 in Atox1+/+ and Atox1-/- cells before and after exposure to CDDP and Cu, we examined the cells by confocal fluorescent microscopy after exposing them to 10 μM CDDP or 300 μM Cu for 15 min prior to staining with an antibody to Ctr1. As shown in Figure 3B, while the overall subcellular distribution of Ctr1 was not visibly different in the two cell lines prior to drug treatment, a 15 min exposure to 10 μM CDDP or 300 μM Cu triggered down-regulation of Ctr1 only in the Atox1+/+ cells. This result was confirmed by a flow cytometric examination that demonstrated the reduction of Ctr1 levels in the Atox1+/+ but not in Atox1-/- cells following exposure to 2 μM CDDP; exposure to CDDP in fact produced a small increase in Ctr1 level in the Atox1-/- cells (Figure 3C). This result suggests that the reduced ability of Atox1-/- cells to accumulate CDDP may be due to the inability of CDDP to trigger the endocytotic process that putatively brings the CDDP into the cell and that accompanies the down regulation of Ctr1 during CDDP exposure.
A further assessment of the effect of the loss of Atox1 on the CDDP- and Cu-induced down regulation of Ctr1 was made by assaying the uptake of 64Cu following a 15 min treatment of cells with 10 μM CDDP. We reasoned that since Ctr1 is the major transporter of Cu, its down regulation by CDDP should diminish the levels of 64Cu uptake to a greater extent in the Atox1+/+cells than the Atox1-/- cells. Cu accumulation in the CDDP-treated Atox1+/+cells was 54.0 ± 1.3 % of that in untreated Atox1+/+ cells while Cu accumulation in the CDDP-treated Atox1-/- cells was 83.3 ± 2.4 % of that in untreated Atox1-/-cells (p < 0.01 in both cases). Thus, the functional down-regulation of Ctr1 by CDDP was less severe in the Atox1-/- than Atox1+/+ cells.
3.4. Effect of the loss of Atox1 on the proteasomal degradation of Ctr1
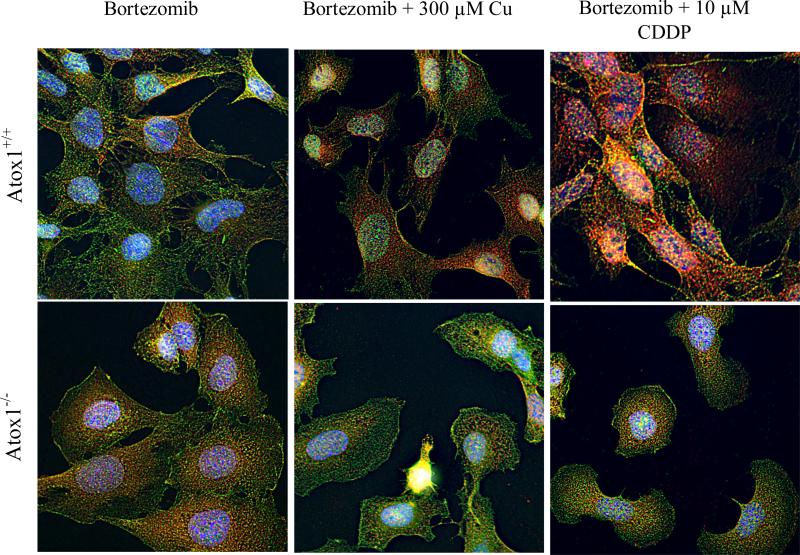
Current evidence suggests that Ctr1 transports Cu and CDDP by different mechanisms. In mouse fibroblasts and human ovarian carcinoma cells CDDP causes rapid down regulation of Ctr1 even at very low concentrations (1-2 μM) while Cu achieves this only at very high concentrations (>200 μM). Previous data indicates that the CDDP- and Cu-induced down-regulation of Ctr1 involves non-clathrin mediated endocytosis followed by degradation in the 26 S proteasome. Proteasomal degradation often involves the formation of ubiquitinated intermediates that serve to direct the molecule to the proteasome. To determine whether Ctr1 is ubiquitinated during CDDP- and Cu-induced degradation, Atox1+/+ and Atox1-/- cells were incubated for 4 h with the proteasome inhibitor bortezomib at a concentration of 40 nM and then exposed to 300 μM Cu or 10 μM CDDP for 15 min. The cells were then stained with antibodies to Ctr1 and polyubiquitin. As shown in Figure 4, confocal microscopic examination showed that the extent of co-localization of the Ctr1 and polyubiquitin signal, shown by the yellow color, was much greater in the Cu- and CDDP treated Atox1+/+cells than in the equivalently treated Atxo1-/-cells. In addition, in the Atox1+/+cells CDDP altered the patter of co-localization of Ctr1 and polyubiquitin more extensively than Cu. This analysis demonstrated a role for Atox1 in this process as very little co-localization of Ctr1 and the polyubiquitin signal was observed in the Atox1-/- cells under any of the conditions examined.
Figure 4.
Effects of loss of Atox1 on the distribution of Ctr1 (green) and polyubiquitin (red) in Atox1+/+ (top panel) and Atox1-/- (lower panel) cells. All cells were treated with 40 nM bortezomib for 4 h and then with either 300 μM Cu or 10 μM CDDP for 15 min. A rabbit polyclonal antibody to Ctr1 and a mouse monoclonal antibody to polyubiquitin conjugates were used. Nuclei were stained with Hoechst 33342. Yellow depicts co-localization of Ctr1 and polyubiquitin signals.
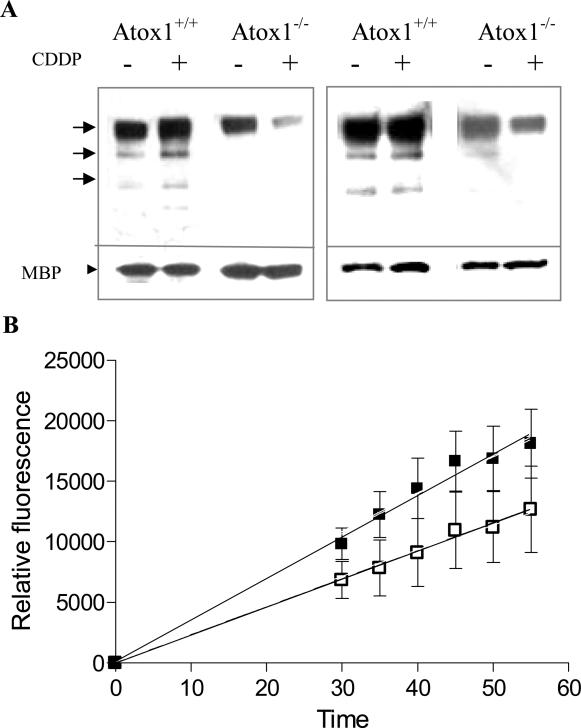
Western blot analysis of the total lysates from Atox1+/+ and Atox1-/- cells treated with bortezomib prior to exposure to 10 μM CDDP using antibodies against polyubiquitin conjugates indicated that the levels of a wide range of polyubiquitinated proteins were much higher in the Atxo1+/+ than the Atox1-/- cells following CDDP exposure (data not shown). To demonstrate that Ctr1 becomes ubiquitinated during exposure to CDDP, and provide further evidence for a difference in ubiquitination between the Atox1+/+ and Atox1-/- cells, the two cell lines were treated for 4 h with bortezomib and then with 10 μM CDDP for an additional 15 min prior to preparing lysates for immunoprecipitation with antibodies against Ctr1 or polyubiquitin conjugates. As shown in the left panel of Figure 5A, when equal amounts of protein (1.7 μg) immunoprecipitated by the Ctr1 antibody were resolved on SDS gels and probed with anti-polyubiquitin antibody a poly-ubiquitinated protein of ~ 130 kDa and several lower molecular weight forms were detected. The intensity of these bands was lower in the Atox1-/- cells. Their intensity increased in response to CDDP exposure in the Atox1+/+ cells but not in the Atox1-/-cells. Similar results were obtained when cell lysates from bortezomib and CDDP-treated cells were immunoprecipitated with anti-polyubiquitin antibodies and probed on Western blots with anti-Ctr1 antibodies (Figure 5A, right panel). Thus, the level of polyubiquitinated Ctr1 increased in Atox1+/+ cells following exposure to CDDP whereas it decreased in the Atox1-/-cells under similar conditions. This result establishes that Ctr1 does become polyubiquitinated, and that this is defective when Atox1 is deficient.
Figure 5.
Effect of loss of Atox1 on the levels of polyubiquitinated conjugates following exposure of Atox1+/+ and Atox1-/- cells to CDDP and on endogenous proteasomal activity. (A) Western blots of immunoprecipitated Ctr1 (left panel) or polyubiquitin conjugates (right panel) were analyzed for polyubiquitination and presence of Ctr1 respectively. Left panel: Western blot of Ctr1 immunoprecipitated from Atox1+/+ and Atox1-/- cells treated for 4 h either with 40 nM bortezomib alone or 40 nM bortezomib and a 15 min exposure to 10 μM CDDP probed with antibody to polyubiquitinated proteins. Each lane contained 1.7 μg of immunoprecipitated Ctr1 and 100 ng of purified maltose binding protein (MBP). Right panel, Western blot of polyubiquitin conjugates immunoprecipitated from Atox1+/+ and Atox1-/- cells treated for 4 h either with 40 nM bortezomib alone or 40 nM bortezomib and a 15 min exposure to 10 μM CDDP probed with antibodies against Ctr1. Each lane contained 8 μg of polyubiquitinated protein and 100 ng of the maltose binding protein (MBP). MBP was used to document equal lane loading. (B) Effect of loss of Atox1 on the activity of the 20S proteasome. Chymotrypsin-like proteasome activity was measured by determination of fluorescence during a 1 h assay using 25 μg of whole cell lysates from Atox1+/+ (□) and Atox1-/- (■) cells. Error bars are SEM of at least 3 independent experiments, each performed with a minimum of 3 samples per data point.
3.5. Effect of the loss of Atox1 on the activity of the 20 S proteasome
The activity of the 20S proteasome in the lysates from the Atox1+/+ and Atox1-/- cells was assayed by measuring chymotrypsin-like proteasomal activity using a fluorochrome-labeled substrate. The activity was linear with time in lysates at concentrations between 10 - 100 μg/mL. Final assays were performed with 25 μg/mL protein for 1 h during which the activity was recorded every 5 min following the first 30 min of incubation (Figure 5B). The mean slope of the plot of relative fluorescence obtained for the Atox1+/+ cells was 231 ± 52 (SD) whereas that for the Atox1-/- cells was 396 ± 14 (SD) (p < 0.03). Thus, the proteasome activity of the Atox1-/-cells was significantly higher than that of the Atox1+/+ cells, perhaps due to the lack of competition from adequately ubiquitinated endogenous substrates.
4. Discussion
The results of this study demonstrate that, like the Cu transporters Ctr1, Atp7a and Atp7b, the metallochaperone Atox1 also regulates the cellular pharmacology of CDDP. Key findings from this study are that Atox1 regulates the intracellular accumulation, compartmentalization and cytotoxicity of CDDP by mechanisms that are distinct from those that are involved in Cu homeostasis, as is evidenced by the effects of the loss of Atox1 on the uptake rather than efflux of CDDP.
Atox1 is known to regulate Cu levels by facilitating its efflux through a process that involves receiving Cu from Ctr1 and handing it to the exporters Atp7a/b in the trans-Golgi compartment via intimate protein-protein interactions. The effect of loss of Atox1 on the cellular pharmacology of Cu observed in this study is consistent with this prior understanding of the role of this protein as an acceptor and donor of Cu [25]. Thus, loss of Atox1 resulted in greater accumulation of Cu, reduced efflux and had a small increase in the sensitivity of the cells to Cu. These results provided excellent validation that the pair Atox1+/+ and Atox1-/- cells used in this study can serve as a reliable model system from which to assess the influence of Atox1 on the cellular pharmacology of CDDP.
The effect of the loss of Atox1 on the cellular pharmacology of CDDP was quite distinct from the effect on Cu suggesting that the Atox1 functions differently with respect to the two metalloids. Whereas loss of Atox1 primarily affected the efflux of Cu, it significantly impaired the accumulation of CDDP. The reduced ability of the Atox1-/- cells to accumulate CDDP was reflected in the higher degree of resistance of these cells to CDDP and their lower accumulation of Pt-DNA adducts. It is surprising, however, that the degree of resistance of Atox1-/- cells was quite small; one reason may be that Atox1 has several and perhaps opposing regulatory functions in cells as is evidenced by the involvement of this protein in transcription [31] and oxido-redox [32, 33] processes. Loss of Atox1 is known to affect the expression of Atp7a [25] and SOD3 [34] and can be reasonably expected to affect the level of Cu-regulated proteins such as XIAP [35] and p53 [36]. In addition, Atox1 can potentially affect the cellular responses to CDDP through Cu-dependent processes; the role of Cu-containing antioxidants in conferring CDDP resistance to cells is well documented [37].
The higher levels of CDDP accumulation found in the Atox1+/+ cells, particularly in the nuclei and in the vesicular compartment which are known to associate with the Atox1, suggests that Atox1 may be a CDDP-binding protein. However, since the microsomes and nuclei of both cell types accumulated similar fractions of the total cellular CDDP, it appears that the main role of Atox1 is in the influx of CDDP rather than in subcellular compartmentalization of the drug.
Previous data from this laboratory have demonstrated that Ctr1 regulates the initial uptake of CDDP through a mechanism that involves macropinocytosis linked to subsequent rapid degradation of Ctr1 [38]. The prior studies also demonstrated that the rapid disappearance of Ctr1 from the plasma membrane and its subsequent degradation following exposure to CDDP can be blocked when macropinocytosis and proteasomal activity are pharmacologically or biologically disabled [30]. The observation that loss of Atox1 impairs the ability of CDDP to down-regulate Ctr1 is novel and indicates that Atox1 plays a central role in the process of CDDP-induced Ctr1 internalization and degradation. The reduced uptake of CDDP in the Atox1-/- cells is consistent with the concept that Ctr1 mediates CDDP uptake by binding the drug extracellularly and internalizing it by macropinocytosis. The fact that the down-regulation of Ctr1 produced by pre-treatment of cells with CDDP resulted in a lower uptake of 64Cu confirms that the down regulation of Ctr1 by CDDP is physiologically significant. Furthermore, the fact that pre-treatment of cells with CDDP produced a greater effect on 64Cu uptake in the Atox1+/+ than the Atox1-/- cells confirms that Atox1 plays a role in this internalization process. How then does Atox1 regulate the influx of CDDP via Ctr1? Previous studies have demonstrated that Atox1 interacts with the C-terminal end of Ctr1 [21]. It is likely that this interaction facilitates the internalization, ubiquitination and subsequent degradation of Ctr1 by the proteasome. Evidence that Atox1-/- cells are defective in the ubiquitination and degradation of Ctr1 confirms this view. The interaction of Atox1 with the Ctr1 may not be required for the influx of Cu which seems to be unimpaired in Atox1-/- cells; however, Ctr1 degradation in response to Cu nevertheless occurs when the Cu concentration is very high and this suggests that, through Atox1, Ctr1 serves as a sensor of toxic metals in the cellular environment. It is intriguing to speculate that the absence of Atox1 may also influence the function of the Ctr1 in the FGF signaling mechanisms which were recently demonstrated in Xenopus [39].
The observation that a mammalian Ctr1 is ubiquitinated is new and confirms previous data from yeast [40]. The mouse Ctr1 has two lysine residues at the C-terminal end (residues 185 and 186, accession # CH466527.2) that may serve as the interaction site for ubiquitination enzymes. The observation that Atox1 is required for the poly-ubiquitination of Ctr1 is also novel and provides evidence that Atox1 has other functions in addition to its role as a Cu chaperone. Whether the effect of Atox1 on the ubiquitination of Ctr1 is due to its direct interaction with Ctr1 [21] or through secondary mechanisms such as transcription remains unknown. However, the failure of the Atox1-/- cells to down regulate Ctr1 during exposure to CDDP is not related to a reduction in overall proteasomal activity which we found to have increased in these cells. In the absence of CDDP the untreated Atox1+/+ and Atox1-/- cells ubiquitinate Ctr1 at similar levels, but in the presence of CDDP there is a failure of ubiquitination in the Atox1-/- cells. Since CDDP has previously been shown to bind ubiquitin [41], it is possible that Atox1 competes with ubiquitin for binding to CDDP and thus protects the available ubiquitin pool from inactivation by CDDP. Another possibility is that the higher Cu levels in the Atox1-/- cells inhibits the proteasomal activity [42] or sequesters ubiquitin or both. This later possibility, however, is unlikely as pretreatment of cells with BCS 24 h prior to the application of CDDP failed to alter the rate of Ctr1 degradation or the levels of polyubiquitination of this protein.
It is interesting to note that the Atox1-/- cells have higher levels of chymotrypsin-like proteasome activity than the Atox1+/+ cells. The increase in the level of proteasome activity may be an adaptive response to the inhibitory effects of the higher intracellular Cu found in the Atox1-/- cells [42], or a reflection of the reduced abundance of appropriately poly-ubiquitinated proteins in the Atox1-/- cells. It is also possible that Atox1 acts as a transcriptional regulator of the proteasome subunits many of which contain the GAAAGA, the putative binding site for Atox1 within their proximal or distal promoter regions [31].
5. Acknowledgements
We thank Dr. J. Gitlin for his generosity in providing the mouse embryo fibroblast cell lines and Ms. Angela Robles for assistance with the preparation of this manuscript. This work was supported by the NIH grant CA095298, the DOD grant USAMRAA W81XWH-08-0135 and a grant from the Clayton Medical Research Foundation, Inc.
Abbreviations
- Atox1
Antioxidant 1
- BSA
Bovine serum albumin
- Ctr1
Copper transporter 1
- CDDP
Cisplatin
- DMEM
Dulbecco's modified Eagle medium
- ICP-MS
inductively coupled plasma mass spectrometry
- PBS
Phosphate buffered saline
- TBST
Tris buffered saline with tween 20
6. References
- 1.Holzer AK, Manorek GH, Howell SB. Mol Pharmacol. 2006;70(4):1390–4. doi: 10.1124/mol.106.022624. [DOI] [PubMed] [Google Scholar]
- 2.Holzer AK, Samimi G, Katano K, Naerdemann W, Lin X, Safaei R, Howell SB. Mol. Pharmacol. 2004;66:817–23. doi: 10.1124/mol.104.001198. [DOI] [PubMed] [Google Scholar]
- 3.Ishida S, Lee J, Thiele DJ, Herskowitz I. Proc. Natl. Acad. Sci. USA. 2002;99:14298–14302. doi: 10.1073/pnas.162491399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin X, Okuda T, Holzer A, Howell SB. Mol. Pharmacol. 2002;62:1154–1159. doi: 10.1124/mol.62.5.1154. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu M, Sumizawa T, Mutoh M, Chen Z-S, Terada K, Furukawa T, Yang X-L, Gao H, Miura N, Sugiyama T, Akiyama S. Cancer Res. 2000;60:1312–1316. [PubMed] [Google Scholar]
- 6.Katano K, Kondo A, Safaei R, Holzer A, Samimi G, Mishima M, Kuo YM, Rochdi M, Howell SB. Cancer Res. 2002;62:6559–65. [PubMed] [Google Scholar]
- 7.Safaei R, Howell SB. Crit. Rev. Oncol. Hematol. 2005;53:13–23. doi: 10.1016/j.critrevonc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Samimi G, Katano K, Holzer AK, Safaei R, Howell SB. Mol. Pharmacol. 2004;66:25–32. doi: 10.1124/mol.66.1.25. [DOI] [PubMed] [Google Scholar]
- 9.Safaei R. Cancer Lett. 2006;234:34–39. doi: 10.1016/j.canlet.2005.07.046. [DOI] [PubMed] [Google Scholar]
- 10.Nose Y, Rees EM, Thiele DJ. Trends Biochem. Sci. 2006;31:604–7. doi: 10.1016/j.tibs.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Arnesano F, Banci L, Bertini I, Huffman DL, O'Halloran TV. Biochemistry. 2001;40:1528–39. doi: 10.1021/bi0014711. [DOI] [PubMed] [Google Scholar]
- 12.Banci L, Bertini I, Ciofi-Baffoni S, Chasapis CT, Hadjiliadis N, Rosato A. FEBS J. 2005;272:865–71. doi: 10.1111/j.1742-4658.2004.04526.x. [DOI] [PubMed] [Google Scholar]
- 13.Banci L, Bertini I, Ciofi-Baffoni S, Huffman DL, O'Halloran TV. J. Biol. Chem. 2001;276:8415–26. doi: 10.1074/jbc.M008389200. [DOI] [PubMed] [Google Scholar]
- 14.Rosenzweig AC, Huffman DL, Hou MY, Wernimont AK, Pufahl RA, O'Halloran TV. Struct. Fold Des. 1999;7:605–17. doi: 10.1016/s0969-2126(99)80082-3. [DOI] [PubMed] [Google Scholar]
- 15.Wimmer R, Herrmann T, Solioz M, Wuthrich K. J. Biol. Chem. 1999;274:22597–603. doi: 10.1074/jbc.274.32.22597. [DOI] [PubMed] [Google Scholar]
- 16.Poger D, Fuchs JF, Nedev H, Ferrand M, Crouzy S. FEBS Lett. 2005;579:5287–92. doi: 10.1016/j.febslet.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 17.Elam JS, Thomas ST, Holloway SP, Taylor AB, Hart PJ. Adv. Protein Chem. 2002;60:151–219. doi: 10.1016/s0065-3233(02)60054-3. [DOI] [PubMed] [Google Scholar]
- 18.Klomp LW, Lin SJ, Yuan DS, Klausner RD, Culotta VC, Gitlin JD. J. Biol. Chem. 1997;272:9221–6. doi: 10.1074/jbc.272.14.9221. [DOI] [PubMed] [Google Scholar]
- 19.Boultwood J, Strickson AJ, Jabs EW, Cheng JF, Fidler C, Wainscoat JS. Hum. Genet. 2000;106:127–9. doi: 10.1007/s004399900215. [DOI] [PubMed] [Google Scholar]
- 20.Hamza I, Schaefer M, Klomp L, Gitlin J. Proc. Natl. Acad. Sci. USA. 1999;96:13363–8. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao Z, Wedd AG. Chem. Commun. (Camb) 2002;6:588–9. doi: 10.1039/b111180a. [DOI] [PubMed] [Google Scholar]
- 22.Walker JM, Tsivkovskii R, Lutsenko S. J. Biol. Chem. 2002;277:27953–9. doi: 10.1074/jbc.M203845200. [DOI] [PubMed] [Google Scholar]
- 23.Lin SJ, Pufahl RA, Dancis A, O'Halloran TV, Culotta VC. J. Biol. Chem. 1997;272:9215–20. [PubMed] [Google Scholar]
- 24.Hamza I, Prohaska J, Gitlin JD. Proc. Natl. Acad. Sci. USA. 2003;100:1215–20. doi: 10.1073/pnas.0336230100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamza I, Faisst A, Prohaska J, Chen J, Gruss P, Gitlin JD. Proc. Natl. Acad. Sci. USA. 2001;98:6848–52. doi: 10.1073/pnas.111058498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoh S, Kim HW, Nakagawa O, Ozumi K, Lessner SM, Aoki H, Akram K, McKinney RD, Ushio-Fukai M, Fukai T. J. Biol. Chem. 2008;283:9157–9167. doi: 10.1074/jbc.M709463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katano K, Safaei R, Samimi G, Holzer A, Tomioka M, Goodman M, Howell SB. Clin. Cancer Res. 2004;10:4578–88. doi: 10.1158/1078-0432.CCR-03-0689. [DOI] [PubMed] [Google Scholar]
- 28.Safaei R, Katano K, Larson BJ, Samimi G, Holzer AK, Naerdemann W, Tomioka M, Goodman M, Howell SB. Clin. Cancer Res. 2005;11:756–67. [PubMed] [Google Scholar]
- 29.Safaei R, Larson BJ, Otani S, Rasmussen ML, Howell SB. Mol. Pharmacol. 2008;73:461–468. doi: 10.1124/mol.107.040980. [DOI] [PubMed] [Google Scholar]
- 30.Holzer AK, Howell SB. Cancer Res. 2006;66:10944–52. doi: 10.1158/0008-5472.CAN-06-1710. [DOI] [PubMed] [Google Scholar]
- 31.Itoh Y, Tamai M, Yokogawa K, Nomura M, Moritani S, Suzuki H, Sugiyama Y, Miyamoto K. Anticancer Res. 2002;22:1649–53. [PubMed] [Google Scholar]
- 32.Lin SJ, Culotta VC. Proc. Natl. Acad. Sci. USA. 1995;92:3784–8. doi: 10.1073/pnas.92.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelner GS, Lee M, Clark ME, Maciejewski D, McGrath D, Rabizadeh S, Lyons T, Bredesen D, Jenner P, Maki RA. J. Biol. Chem. 2000;275:580–4. doi: 10.1074/jbc.275.1.580. [DOI] [PubMed] [Google Scholar]
- 34.Jeney V, Itoh S, Wendt M, Gradek Q, Ushio-Fukai M, Harrison DG, Fukai T. Circ. Res. 2005;96:723–9. doi: 10.1161/01.RES.0000162001.57896.66. [DOI] [PubMed] [Google Scholar]
- 35.Mufti AR, Burstein E, Csomos RA, Graf PC, Wilkinson JC, Dick RD, Challa M, Son JK, Bratton SB, Su GL, Brewer GJ, Jakob U, Duckett CS. Mol. Cell. 2006;21:775–85. doi: 10.1016/j.molcel.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 36.Tassabehji NM, VanLandingham JW, Levenson CW. Exp. Biol. Med. (Maywood) 2005;230:699–708. doi: 10.1177/153537020523001002. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka-Kagawa T, Kitahara J, Seko Y, Toyoda H, Imura N, Naganuma A. Biochem. Pharmacol. 1999;57:545–8. doi: 10.1016/s0006-2952(98)00328-1. [DOI] [PubMed] [Google Scholar]
- 38.Holzer AK, Katano K, Klomp LW, Howell SB. Clin. Cancer Res. 2004;10:6744–9. doi: 10.1158/1078-0432.CCR-04-0748. [DOI] [PubMed] [Google Scholar]
- 39.Haremaki T, Fraser ST, Kuo YM, Baron MH, Weinstein DC. Proc. Natl. Acad. Sci. USA. 2007;104:12029–34. doi: 10.1073/pnas.0701413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Sitaram A, Burd CG. Traffic. 2007;8:1375–84. doi: 10.1111/j.1600-0854.2007.00616.x. [DOI] [PubMed] [Google Scholar]
- 41.Hartinger CG, Tsybin YO, Fuchser J, Dyson PJ. Inorg. Chem. 2008;47:17–9. doi: 10.1021/ic702236m. [DOI] [PubMed] [Google Scholar]
- 42.Milacic V, Chen D, Giovagnini L, Diez A, Fregona D, Dou QP. Toxicol. Appl. Pharmacol. 2008;231:24–33. doi: 10.1016/j.taap.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]