Abstract
Cell migration and invasion are two critical cellular processes that are often deregulated during tumorigenesis. To identify factors that contribute to oncogenic progression by stimulating cell migration, we conducted a powerful retroviral based migration screen using an MCF7 cDNA library and the immortalized human breast epithelial cell line MCF-10A. We identified prostate derived Ets factor (PDEF), an Ets transcription factor that is overexpressed in both prostate and breast carcinoma, as a candidate promigratory gene from this screen. Whereas PDEF induced limited motility of MCF-10A cells, coexpression of PDEF with the receptor tyrosine kinases (RTK) ErbB2 and colony-stimulating factor receptor (CSF-1R)/CSF-1 significantly enhanced MCF-10A motility. Furthermore, cells coexpressing PDEF with either ErbB2 or CSF-1R/CSF-1 induced a dramatic invasive phenotype in three-dimensional cultures. Constitutive activation of the extracellular signal–regulated kinase (ERK) pathway also enhanced PDEF-induced motility and invasion, suggesting that activation of the ERK/mitogen-activated protein kinase by ErbB2 and CSF-1R/CSF-1 can cooperate with PDEF to promote motility and invasion. Furthermore, PDEF promoted anchorage-independent growth of ErbB2 and CSF-1R/CSF-1–expressing cells. Using laser capture microdissection, we also found that PDEF mRNA is overexpressed in breast tumor epithelia throughout tumor progression. Taken together, these findings suggest that the transcription factor PDEF may play an important role in breast tumorigenesis and that PDEF overexpression may be particularly significant in tumors that exhibit activation of oncogenic RTKs such as ErbB2 and CSF-1R.
Introduction
The progression of a focal lesion, such as ductal carcinoma in situ (DCIS), to a more aggressive tumor, such as invasive ductal carcinoma, is accompanied by the up-regulation of several key cellular processes, most notably cell migration and invasion. Although these processes are not sufficient for metastatic activity, motility and invasion are believed to be critical for tumor cell metastasis.
The Ets family of transcription factors regulate a number of biological processes including cell proliferation, differentiation, and invasion and are thought to play an important role in oncogenesis. Several Ets factors including Ets1, Ets2, and ESE-1 are overexpressed in both murine and human mammary tumors and are thought to be predictors of poor prognosis (1–5). In addition, overexpression of an inhibitory mutant of Ets2 was sufficient to revert Ras transformation of NIH 3T3 cells and to block anchorage-independent growth and invasion in various breast tumor cell lines (6–10). Notably, both Ets1 and Ets2 are primarily detected in the stromal compartment of tumors and thought to alter the tumor microenvironment by regulating matrix-remodeling proteins (3, 11–13). However, the mechanisms by which these and other Ets factors influence tumorigenesis are not clearly understood.
Ets family proteins share a unique DNA binding domain, known as the Ets domain that binds to a consensus GGA(A/T) sequence within the promoters of target genes. Ets targets include other transcription factors such as Fos and matrix remodeling proteins such as collagenase, stromelysin, and urokinase-type plasminogen activator receptor (14, 15). Prostate derived Ets factor (PDEF) is a recently identified Ets factor with homologues in both mouse (mPSE) and Drosophila (D-Ets), respectively (16). Domain analysis of PDEF revealed a COOH-terminal Ets DNA binding domain and an NH2-terminal regulatory region that includes the Pointed domain, which is present in a subset of Ets proteins and is thought to mediate target specificity (16, 17). PDEF also contains two PEST motifs that render the PDEF protein highly unstable as well as an optimal mitogen-activated protein kinase (MAPK) phosphorylation site homologous to those of Ets1 and Ets2 (16, 18, 19).
Unlike the majority of Ets factors, PDEF is expressed exclusively in tissues with a high epithelial content such as the prostate and breast (16, 20, 21). Furthermore, several studies showed PDEF to be one of the most highly overexpressed mRNAs in human and mouse mammary tumors (5, 20–22). Because the majority of human cancers are epithelial in origin, it is important to better understand the role of such epithelial-specific transcription factors in tumor development and progression.
Here we report the identification of PDEF from a genetic screen for factors that stimulate growth factor–independent migration of MCF-10A cells. In addition, we found that PDEF can cooperate with activated growth factor receptors including ErbB2 and colony-stimulating factor receptor (CSF-1R) to significantly enhance MCF-10A cell motility. Furthermore, coexpression of PDEF with ErbB2 or hyperactivated CSF-1R provoked a dramatic change in the morphology of structures formed by MCF-10A cells in three-dimensional cultures, converting spheroid-like structures into protrusive cords that invade into the basement membrane gel. Constitutive activation of the extracellular signal–regulated kinase (ERK)/MAPK pathway induced a similar morphologic conversion of PDEF-expressing cells. In addition, we found that PDEF promoted anchorage-independent growth of MCF-10A cells expressing ErbB2 or CSF-1R/CSF-1 cells. Finally, we found PDEF to be overexpressed in the epithelial cells isolated from a large percentage of breast tumors. Collectively, our findings suggest that PDEF may play an important role during breast tumorigenesis, particularly in the presence of constitutively activated receptor tyrosine kinases (RTK) such as ErbB2 and CSF-1R. To our knowledge, this is the first report supporting a role for PDEF as a promigratory and invasive cDNA in human mammary epithelial cells and its cooperation with the growth factor receptors ErbB2, CSF-1R, and the ERK/MAPK signaling pathway.
Materials and Methods
Cell culture and reagents
MCF-10A cells were cultured as described (23). The α-PDEF and α-actin antibodies were obtained from Zymed (San Francisco, CA) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. pBabe-MEK2DD and pBabe-ErbB2 were generously provided by S. Meloche (University of Montreal, Montreal, Canada) and Danielle Lynch (Harvard Medical School, Boston, MA). HMECs were obtained from Clonetics (East Rutherford, NJ) and the A375 and Lovo cell lines were kind gifts from S. Mani and R. Weinberg (Massachusetts Institute of Technology, Cambridge, MA). All other cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured according to the guidelines provided.
Library construction
Total RNA was isolated from MCF7 cells using RNA-STAT-60 (Tel-Test, Inc., Friendswood, TX). Poly(A) RNA was isolated using the Oligotex kit (Qiagen, Valencia, CA). The Superscript Choice System (Invitrogen, Carlsbad, CA) was used for cDNA synthesis. BstX1/EcoR1 adaptors (Invitrogen) were ligated to the cDNA using T4 DNA ligase (New England Biolabs, Ipswich, MA). The cDNA was then size fractionated on a low-melt agarose gel (American Bioanalytical, Natick, MA) into two fractions, >3 and 1 to 3 kb. Each fraction was purified with agarose treatment according to the protocol of the manufacturer (Roche, Palo Alto, CA). The cDNA was ligated into the nonpalindromic BstXI sites in the purified BstXI-digested pEYK3.1 vector. The ligation mixture was purified with phenol-chloroform extraction and electroporated into Electromax DH10B (Invitrogen). For amplification of the library, the bacteria were plated on LB plates containing 100 µg/mL of ampicillin and 50 mg/mL of zeocin (Invitrogen). After incubating at 37°C for 16 to 18 hours, the plates were scraped and the plasmid DNA was isolated using Qiagen Megaprep columns (Qiagen).
Retroviral vectors and constructs
The PDEF mutant T50A was created by site-directed mutagenesis using the primers 5′-AGTCCACCCGCCGCGCCCGAGCAGGGC-3′ and 5′-GCCCTGCTCGGGCGCGGCGGGTGGACT-3′ and cloned into the EcoRI site of pEYK3.1. Full-length PDEF, the T50A mutant, the NH2-terminal region (1–248 amino acids), and the COOH-terminal DNA binding domain (245–335 amino acids) of PDEF were cloned into pBabe-puro and PMX-N-GFP by PCR amplification with primers including a 5′ EcoRI site and a 3′XhoI or EcoRI site. pBabe-CSF-1R and pMSCV-CSF-1 were created as previously described (24).
Generation of stable cell lines and analyses of protein lysates
MCF-10A cells expressing stable levels of the above genes were generated by infection with retrovirus as described (23). Populations of MCF-10A cells stably expressing CSF-1R/CSF-1 were obtained as described (24). MCF-10A cells overexpressing ErbB2/PDEF and CSF-1R/PDEF were created by sequential infection of ErbB2 and PDEF or CSF-1R/CSF-1 and PDEF. Cells were selected with 2 µg/mL puromycin or 200 µg/mL G418 after each round of infection. Protein lysates from MCF-10A cells were prepared in NP40 buffer, separated by SDS-PAGE, and analyzed by immunoblot as previously described (25).
Transwell migration assay
All MCF-10A–derived cell lines (ErbB2, CSF-1R/CSF-1, and MEK2DD) were starved overnight in assay medium (MCF-10A growth medium containing 2% serum and no EGF). The starved cells were trypsinized, 1 × 105 cells were added to the top chambers of the transwell (8 µm pore size; BD Bioscience, Bedford, MA), and assay medium was added to the bottom chambers and incubated for 16 to 20 hours. For all other cell lines (HMEC, SKBR3, MDA-MB-231, and MDA-MB-435), the appropriate growth medium as recommended by ATCC was used instead of assay medium. After overnight incubation, the migratory cells were fixed, stained with 4′,6-diamidino-2-phenylindole, and quantified as previously described (25). Each cell line was assayed in duplicate per experiment and repeated at least thrice. Paired Student’s t test analysis was done to determine significance of PDEF activity over control cells for each cell line over multiple experiments.
For the migration screen, 1 × 106 MCF-10A cells expressing the cDNA library were plated onto six-well transwell chambers in assay medium. After a 16-hour incubation, the migratory cells on the bottom of the filters were trypsinized, propagated, and rescreened twice.
Rescue of prostate epithelium–derived Ets transcription factor from migratory cells
To rescue pEYK3.1 PDEF, genomic DNA was isolated (Puregene, Gentra Systems Inc., Minneapolis, MN) from MCF-10A cells harvested after the third round of migration, digested with NotI, and self-ligated to create a provirus encoding PDEF (26, 27). The resulting proviral DNA was amplified in bacteria, purified by Maxi prep (Qiagen), and sequence verified.
Wound healing assays
MCF-10A cells were seeded onto six-well dishes at 1 × 105 per well in growth medium. Confluent monolayers were starved overnight in assay medium and a single scratch wound was created using a micropipette tip. Cells were washed with PBS to remove cell debris, supplemented with assay medium, and monitored. Images were captured by phase microscopy using a 10× objective at 0, 12, and 24 hours post wounding. To quantify the wounded area, we measured the area of the wound at the time of wounding (0 hours) and at 12 (MEK) or 24 hours (10A, ErbB2, and CSF) post wounding to calculate the area of wound healing (Metamorph image analysis software). This area is represented as a percentage of the initial wound. The percentage of wound healing for each of the cell lines expressing vector control or PDEF was averaged over four experiments and statistical significance was calculated using Student’s t test.
Three-dimensional cell culture
Cells were cultured in basement membrane gels composed of a 50:50 mixture of growth factor-reduced Matrigel (BD Bioscience) and bovine dermal collagen I (Vitrogen, Cohesion Technologies, Palo Alto, CA) as described (25). MCF-10A cells expressing vector control or PDEF alone were supplemented with 5 ng/mL EGF to allow proliferation whereas ErbB2, CSF-1R/CSF-1, and MEK2DD cells were cultured in the absence of EGF.
Soft agar assay
A mixture of 5,000 cells in assay medium and 0.3% agarose was seeded on to a solidified bed of 0.6% agarose on six-well plates. The plates were allowed to solidify at 4°C and incubated at 37°C. The cultures were fed once a week with assay medium containing 0.3% agarose. Each cell line was seeded in duplicate per experiment and was repeated at least thrice. The cultures were imaged and the number of colonies was counted after 15 days. The experiment was repeated thrice and the statistical significance was calculated using Student’s t test.
Laser capture microdissection
All laser capture microdissection– and tissue section–derived RNA samples were prepared and amplified as described (28). RNA samples were subjected to DNase treatment before cDNA synthesis and RNA amplification. Briefly, 2 µg of amplified RNA were converted into double-stranded cDNA and quantified with Picogreen (Molecular Probes, Eugene, OR) using a spectrofluorometer (Molecular Devices, Sunnyvale, CA). For each case, 12 ng of cDNA in triplicates were used for real-time PCR with an ABI 7900HT (Applied Biosystems, Foster City, CA). The sequences of the PCR primer pairs and fluorogenic probe (5′→3′) used for PDEF analysis are as follows: forward primer, TCCATCCGCCAGTATTACAAGA; reverse primer, GGTGCACGAACTGGTAGACGA; probe, CATCCGGAAGCCAGACATCTCCCA.
Results
Identification of PDEF in a screen for promigratory genes
To identify genes that induce migration of nontransformed breast epithelial cells, we screened an MCF7 cDNA library using the pEYK3.1 retroviral system. We chose MCF-10A cells for this study because they are stationary in the absence of exogenous motogenic factors. MCF-10A cells were infected with a pool of pEYK3.1 retroviral vectors encoding a library of MCF7 cDNAs and screened for motogen-independent motility using transwell assays. The resulting motile cells were subjected to two additional rounds of transwell screens, followed by the rescue of candidate promigratory cDNAs (Fig. 1A).
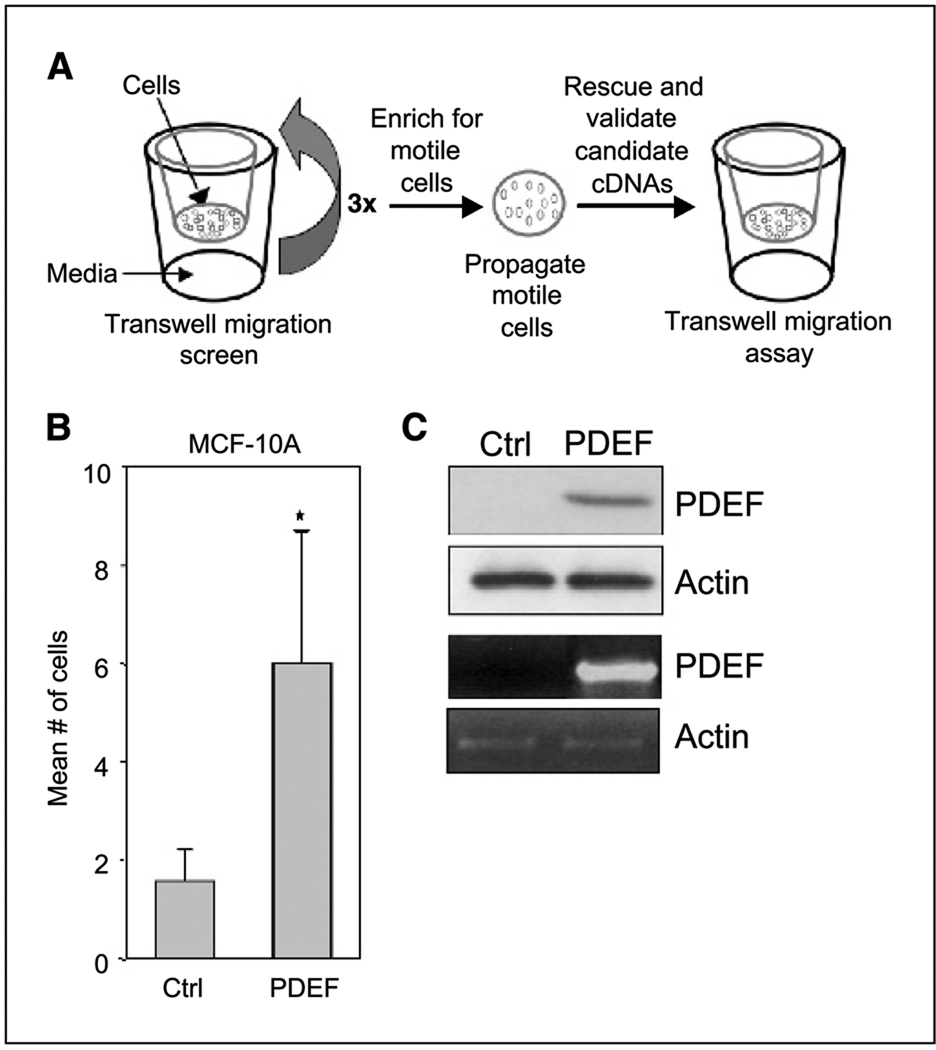
Figure 1.
PDEF induces migration of MCF-10A cells. A, schematic representation of the growth factor-independent migration screen. B, validation of PDEF as a promigratory gene in MCF-10A cells. Transwell migration assay of MCF-10A cells overexpressing the vector control or PDEF in the absence of motogenic factors is shown. Columns, mean number of motile cells per 20× field from six independent experiments; bars, SD. Each experiment was conducted in duplicate where ten 20× fields (five per duplicate) were counted and the means calculated. A paired Student’s t test (*, P < 0.005) was done on the means from six experiments. C, protein and RNA analysis of PDEF levels in MCF-10A cells. Top, immunoblot analysis of MCF-10A cells stably expressing the vector control or PDEF using antibodies against PDEF. Actin was used as a loading control. Bottom, RT-PCR analysis of total RNA isolated from MCF-10A cells overexpressing the vector control or PDEF using primers against PDEF.
We identified PDEF as one of several candidate promigratory cDNAs from this migration screen. To verify that PDEF overexpression is sufficient to stimulate migratory activity, MCF-10A cells were infected with a pEYK3.1 retrovirus encoding a full-length PDEF cDNA and subjected to transwell migration assays. PDEF overexpression induced a 2- to 5-fold increase in migration of MCF-10A cells in the absence of motogens, suggesting that PDEF can independently stimulate motility (Fig. 1B). Both PDEF mRNA and protein were readily detected in the retrovirus-transduced MCF-10A cells but absent in control cells, indicating that MCF-10A cells do not express endogenous PDEF (Fig. 1C). The identification of PDEF as a promigratory factor from this screen validates the use of this unique retroviral system as a valuable tool for gain-of-function screens.
PDEF induces migration in normal and cancer cell lines
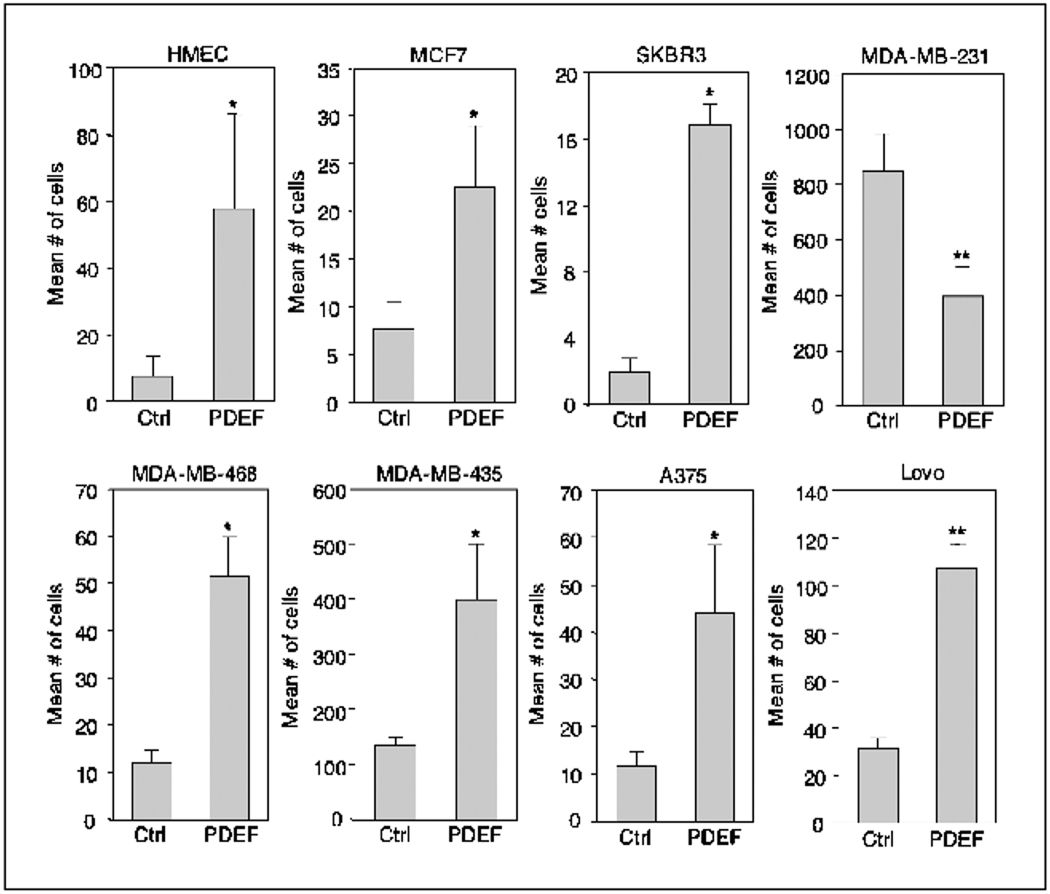
To determine if PDEF can also induce migration in other nontumorigenic breast epithelial cells, we introduced PDEF by retroviral infection into human mammary epithelial cells (HMECs). PDEF overexpression caused an ~8-fold increase in HMEC motility in transwell migration assays (Fig. 2). Furthermore, PDEF likewise increased motility in the nonmetastatic breast carcinoma cell lines MCF7 and SKBR3 and the metastatic cell line MDA-MB-468 (Fig. 2). In contrast, PDEF overexpression decreased motility of the invasive breast cancer cell line MDA-MB-231 as previously described (29). However, we found that PDEF also promoted motility in melanoma (A375 and MDA-MB-435) as well as colon carcinoma (Lovo) cell lines (Fig. 2). Taken together, these findings show that, with the exception of MDA-MB-231 cells, PDEF can promote cell motility in multiple cell lines derived from both normal and tumorigenic epithelia.
Figure 2.
PDEF enhances migration in other normal and tumorigenic epithelial cell lines. Transwell migration assays of HMECs, nonmetastatic breast carcinoma cell lines MCF7 and SKBR3, metastatic breast carcinoma cell lines MDA-MB-231 and MDA-MB-468, melanoma cell lines MDA-MB-435 and A375, and colon carcinoma cell line Lovo. All cell lines expressing vector control or PDEF were assayed as described in Fig. 1B. Columns, mean number of motile cells per 20× field from four independent experiments; bars, SD. *, P < 0.05; **, P < 0.001.
ErbB2 and CSFIR activation enhances PDEF–induced motility
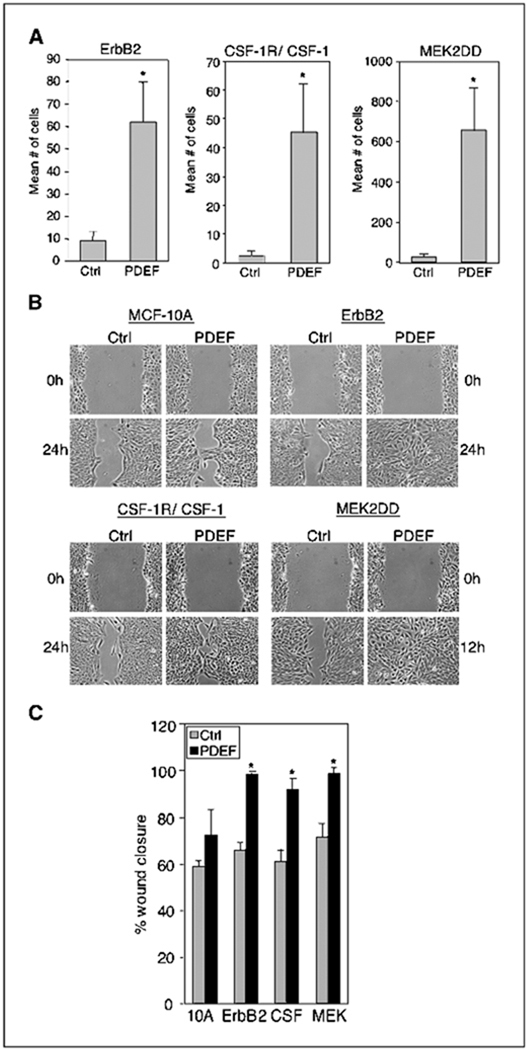
Breast tumors commonly exhibit overexpression or activation of growth factor receptors such as ErbB2 and CSF-1R. To determine whether ErbB2 or CSF-1R activity influences the biological function of PDEF, we expressed PDEF in MCF-10A cells expressing wild-type ErbB2 or hyperactivated CSF-1R and assayed for cell motility. Expression of ErbB2 alone did not induce significant motility of MCF-10A cells in transwell assays (Fig. 3A). However, cells expressing both ErbB2 and PDEF (ErbB2/PDEF) showed a 5- to 10-fold enhancement in motility (Fig. 3A) whereas cells expressing PDEF alone only stimulated 2- to 5-fold enhancement in motility (Fig. 1B). We also analyzed PDEF-induced motility in wound healing assays. At 24 hours post wounding, ErbB2/PDEF cells had achieved near-complete wound closure (98%) whereas control ErbB2 cells did not (Fig. 3B and C). To determine whether PDEF would have a similar effect on CSF-1R hyperstimulated cells, we introduced PDEF into MCF-10A cells coexpressing both CSF-1R and CSF-1 ligand (CSF-1R/CSF-1). We found that coexpression of PDEF with CSF-1R/CSF-1 (CSF-1R/PDEF) resulted in a robust increase in cell motility in both transwell (10- to 30-fold) and wound healing assays (Fig. 3A–C). In contrast, expression of PDEF alone did not significantly enhance wound closure of MCF-10A cells [Fig. 3B (top left) and C]. Taken together, these results show that both ErbB2 and CSF-1R/CSF-1 can enhance PDEF-induced motility of MCF-10A cells.
Figure 3.
ErbB2, CSF-1R/CSF-1, and MEK2DD enhance PDEF induced migration. A, transwell migration assays of MCF-10A cells coexpressing ErbB2, CSF-1R/CSF-1, or MEK2DD with vector control or PDEF as described in Fig. 1B. Columns, mean number of motile cells per 20× field from six independent experiments; bars, SD. *, P < 0.005. B, wound healing assays of the same cell lines expressing vector control (Ctrl) or PDEF. Representative images captured with a 10× objective at the time of wounding (t = 0) and 12 hours (MEK2DD) or 24 hours after wounding. All experiments were repeated at least thrice. C, quantification of wound healing. Columns, percentage of wound closure at t = 24 hours [MCF-10A (10A), ErbB2, and CSF-1R/CSF-1 (CSF)] or t = 12 hours MEK2DD (MEK) averaged over four separate experiments (described in Material and Methods); bars, SD. *, P < 0.01.
To determine if the ERK/MAPK signaling pathway, which is common to both ErbB2 and CSF-1R/CSF-1 signaling, would influence PDEF-induced motility, we expressed a constitutively activated form of MEK2 (MEK2DD) in MCF-10A cells. MEK2DD expression results in constitutive activation of phosphorylated ERK (data not shown) and induces a low level of motility in MCF-10A cells (Table 1, 10A versus MEK). Coexpression of PDEF with MEK2DD (MEK2DD/PDEF) resulted in a robust increase in cell motility in both transwell (20- to 30-fold) and wound healing assays (Fig. 3A–C). These findings show that constitutive activation of the Ras/ERK MAPK pathway results in a similar enhancement of PDEF-induced motility as overexpression of ErbB2 or CSF-1R/CSF-1. In contrast, coexpression of PDEF with activated AKT, the kinase downstream of the phosphatidylinositol-3 kinase signaling pathway, or cyclin D1, an oncogene and an effector common to both RTKs, did not enhance PDEF-induced motility of MCF-10A cells (data not shown).
Table 1.
PDEF synergizes with ErbB2, CSF-1R/CSF-1, and MEK2DD to enhance MCF-10A cell motility
| Experiment 1 | Experiment 2 | Experiment 3 | |
|---|---|---|---|
| 10A + Ctrl | 0 | 1.1 ± 0.1 | 0 |
| 10A + PDEF | 5.5 ± 0.4 | 9.1 ± 1.3 | 7 ± 1.4 |
| ErbB2 + Ctrl | 2.9 ± 0.1 | 12.2 ± 0.3 | 2.3 ± 0.1 |
| ErbB2 + PDEF | 26.9 ± 1 | 72 ± 1.4 | 30.1 ± 1 |
| 10A + Ctrl | 0 | 2 ± 0.6 | 2.4 |
| 10A + PDEF | 5.5 ± 0.4 | 8.1 ± 1 | 8.2 ± 0.3 |
| CSF + Ctrl | 1.5 ± 0.1 | 4.7 ± 0.7 | 3.3 ± 0.1 |
| CSF + PDEF | 48 ± 4.2 | 56.8 ± 8.2 | 31.5 ± 0.1 |
| 10A + Ctrl | 2 ± 0.6 | 2.1 ± 0.4 | 2.4 ± 0.4 |
| 10A + PDEF | 8.1 ± 1 | 8.7 ± 1.6 | 8.2 ± 0.3 |
| MEK + Ctrl | 31.5 ± 2.4 | 24.9 ± 6.4 | 33.4 ± 0.3 |
| MEK + PDEF | 788 ± 79 | 753 ± 52 | 795 ± 49 |
NOTE: The average number of migratory cells per 20× field from three separate experiments is listed. Each experiment was conducted in duplicate (± SD) where vector control (10A + Ctrl) or PDEF-expressing MCF-10A cells (10A + PDEF) were assayed alongside ErbB2, CSF-1R/CSF-1 (CSF), or MEK2DD (MEK) cells.
Direct comparison of the motility of parental MCF-10A cells expressing vector control or PDEF with MCF-10A cells coexpressing ErbB2, CSF-1R/CSF-1, or MEK2DD, alone or together with PDEF, showed that the enhanced motility observed with PDEF is not additive, but synergistic, with ErbB2, CSF-1R, and MEK2DD (Table 1).
PDEF promotes invasive activity in three-dimensional basement membrane cultures
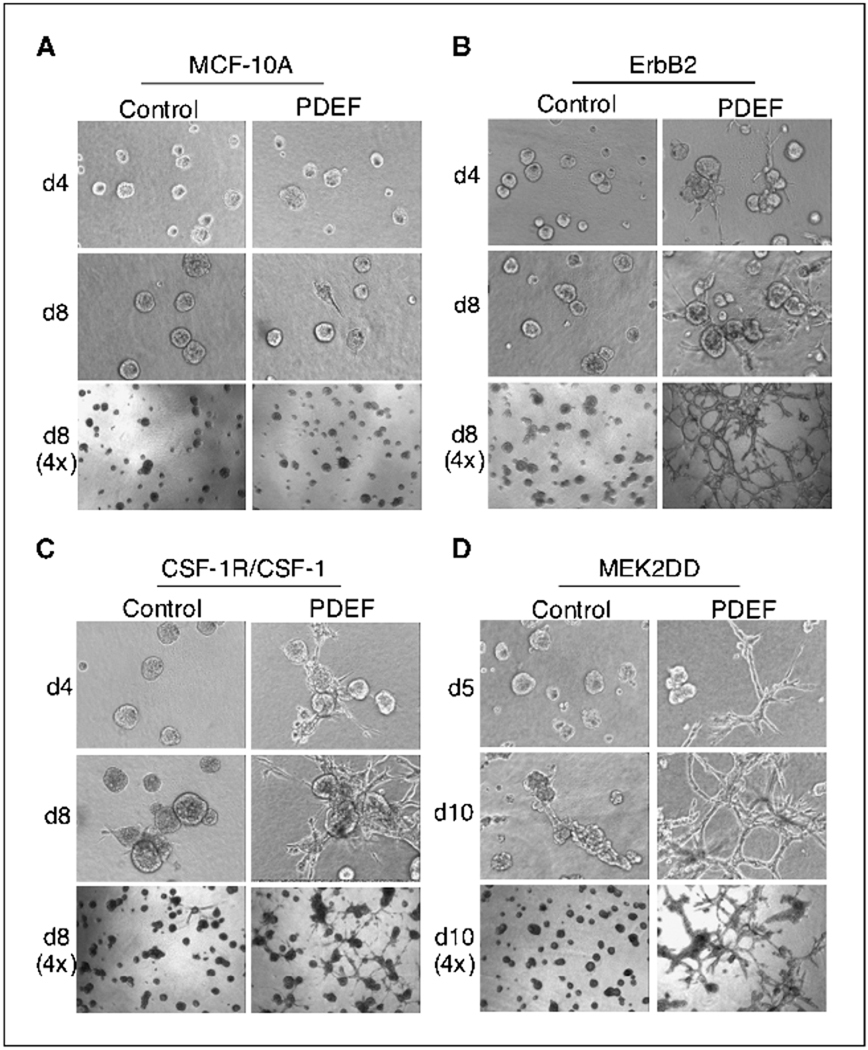
To examine the effects of PDEF activity in a context that more closely resembles in vivo mammary architecture, we assessed the consequences of PDEF overexpression on the formation of MCF-10A–based acini. To this end, we cultured PDEF-expressing cells in a mixture of Matrigel and collagen I. The majority of MCF-10A cells overexpressing PDEF formed normal acini in three-dimensional (3D) cultures; a subset of these acini (22%) contained short projections but did not form highly invasive structures at later days in culture (Fig. 4A and data not shown). Overexpression of ErbB2 or CSF-1R/CSF-1 in MCF-10A cells promotes the formation of hyperproliferative acini (24). 5 In addition, overexpression of CSF-1R/CSF-1 also causes a disruption in cell-cell adhesion leading to a discohesive phenotype (24). However, neither of these receptors (ErbB2 or CSF-1R/CSF) stimulated invasive activity in MCF-10A cells when cultured in 3D (Fig. 4B and C, left). In contrast, ErbB2/PDEF and CSF-1R/PDEF cells formed highly invasive acini on basement membrane (Fig. 4B and C, right). By day 8, the invasive projections emanating from these acini formed a network of multicellular cords that penetrated the basement membrane (Fig. 4B and C). By day 12 in culture, invasive structures were detected in ~100% of ErbB2/PDEF acini and 75% of CSF-1R/PDEF acini (Fig. 4). Taken together, these results show that PDEF profoundly affects the behavior of cells expressing activated RTKs such as ErbB2 and CSF-1R, leading to a highly invasive phenotype in 3D culture.
Figure 4.
MCF-10A cells coexpressing PDEF with ErbB2, CSF-1R/CSF-1, or MEK2DD form invasive acini in three-dimensional cultures. A, three-dimensional Matrigel and collagen cultures of MCF-10A acini expressing vector control or PDEF in the presence of EGF. B to D, MCF-10A cells coexpressing either vector control (left) or PDEF (right) with ErbB2 (B), CSF-1R/CSF-1 (C), or MEK2DD (D) cultured in the absence of EGF. Representative phase images obtained at various days in culture using a 10× (top and middle rows) and a 4× (bottom rows) objective are shown.
To determine if constitutive MEK activity could similarly confer invasive activity to PDEF-expressing cells, we cultured cells expressing either MEK2DD or MEK2DD/PDEF three-dimensionally as described above. Control cells expressing MEK2DD alone were hyperproliferative and adopted a spheroid morphology but lacked invasive activity in 3D culture (Fig. 4D). Strikingly, MEK2DD/PDEF acini displayed robust invasive activity forming a lattice-like network of invasive cords similar to those formed by ErbB2/PDEF and CSF-1R/PDEF cells (Fig. 4B–D). Consistent with the increased motility described above, the finding that MEK2DD/PDEF cells exhibit invasive activity on basement membrane suggests that the ERK-MAPK pathway may collaborate with PDEF to induce invasion.
Domain analysis of PDEF in motility and invasion
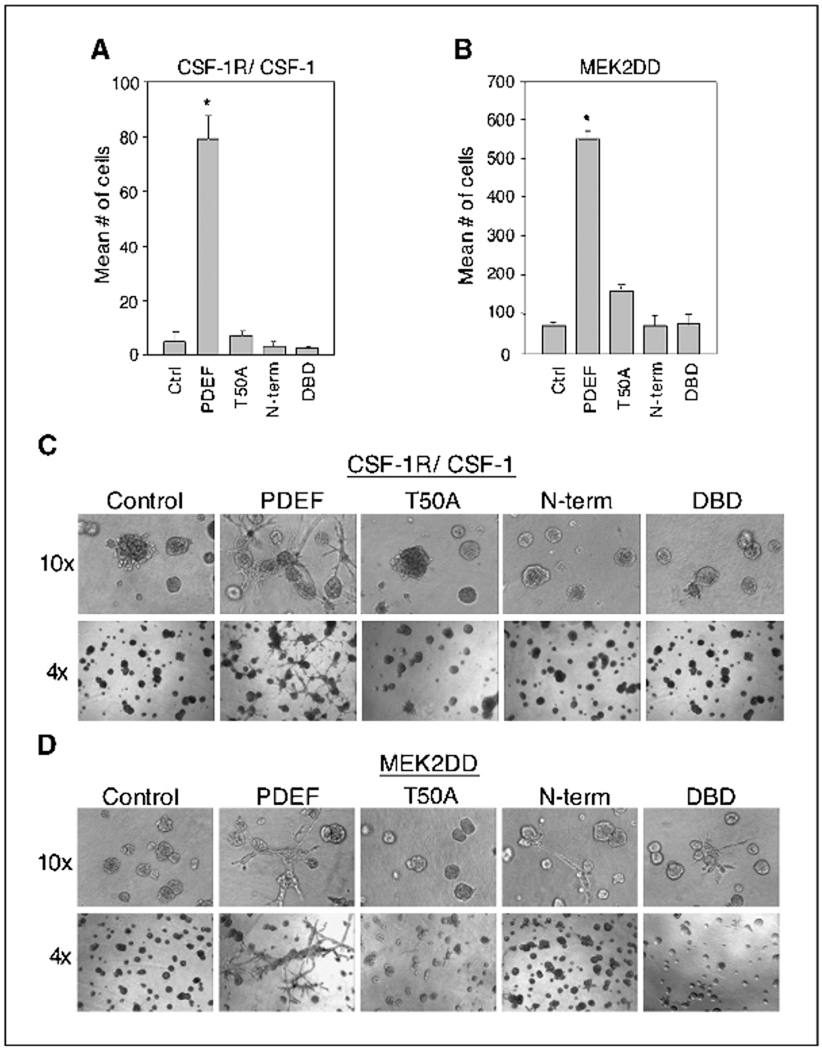
PDEF consists of a highly conserved COOH-terminal Ets DNA binding domain and an NH2-terminal regulatory region. Whereas the activity of most Ets family transcription factors is dependent on the DNA binding domain, ESE-1, which belongs to the same subgroup as PDEF, functions in the cytoplasm independent of this domain (30). Therefore, we examined whether the NH2-terminal region of PDEF (1–248 amino acids), which contains the Pointed domain as well as the aforementioned consensus MAPK phosphorylation site, or the COOH-terminal region of PDEF (245–335 amino acids), comprising the DNA binding domain, would be sufficient to induce motility and invasion. In addition, we created a mutant PDEF protein that contains a threonine-to-alanine substitution at the consensus MAPK phospho-acceptor site (T50A) to determine whether this site is required for PDEF activity. We found that both deletion constructs and the T50A mutant failed to induce migration or invasion in MCF-10A cells coexpressing ErbB2, CSF-1R/CSF-1, or MEK2DD (Fig. 5 and data not shown). These results indicate that neither the NH2-terminal regulatory domain nor the DNA binding domain is sufficient for PDEF function. In addition, these findings suggest that the optimal MAPK phosphorylation site at T50 is essential for PDEF-induced motility and invasion.
Figure 5.
Mutational and domain analyses of PDEF. A and B, transwell migration assays of CSF-1R/CSF-1 or MEK2DD cells overexpressing vector control, PDEF, T50A, NH2-terminal domain (N-term), or DNA binding domain (DBD) of PDEF. Columns, mean number of motile cells per 20× field from four independent experiments; bars, SD. *, P < 0.005. C and D, CSF-1R/CSF-1 and MEK2DD cells expressing the same constructs as above were seeded on basement membrane and grown in the absence of EGF. Phase images of these cultures taken at day 8 using a 10× (top row) and a 4× (bottom row) objective are shown.
PDEF enhances transformation activity
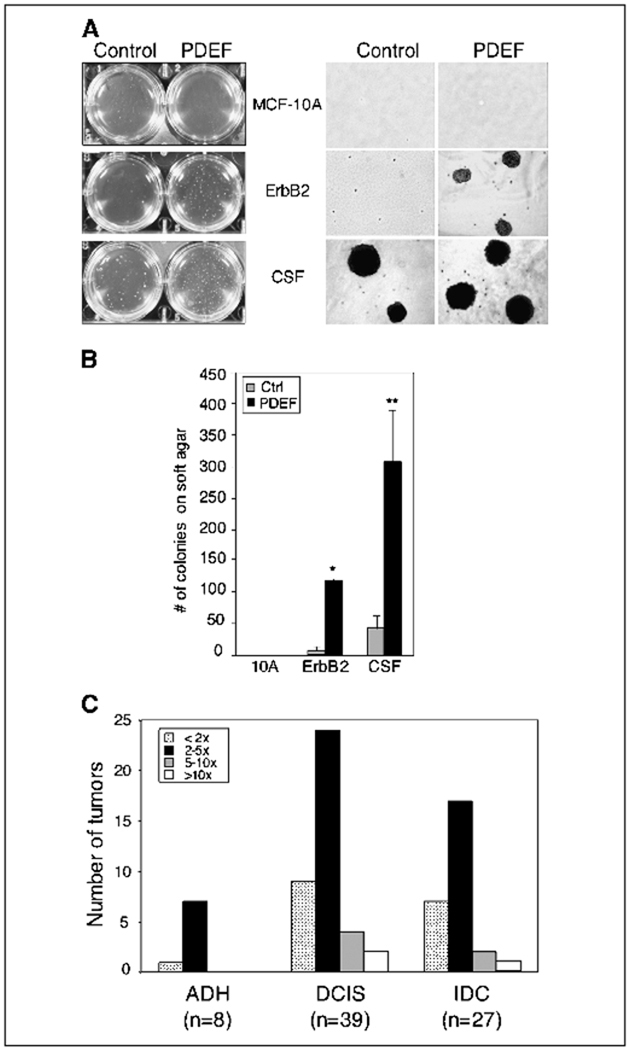
Although the migratory and invasive activities of PDEF suggest that this transcription factor may play a role in late stages of tumor progression, we wanted to determine if PDEF induces other phenotypic alterations associated with oncogenic transformation. Therefore, we assessed whether PDEF expression would result in anchorage-independent growth of MCF-10A cells in soft agar. MCF-10A cells expressing PDEF alone did not form colonies in soft agar. As reported previously, control cells expressing ErbB2 alone did not form colonies in soft agar (31). In contrast, ErbB2/PDEF cells gave rise to a significant number of colonies (~ 100) after 15 days in culture, suggesting that the synergy between ErbB2 and PDEF allows these cells to grow in soft agar. Whereas control CSF-1R/CSF-1 cells formed a few colonies in soft agar, CSF-1R/PDEF cells formed ~ 10-fold more colonies (Fig. 6A and B). These results show that PDEF promotes anchorage-independent growth of both ErbB2 and CSF-1R/CSF-1 cells.
Figure 6.
PDEF enhances transformation activity and is overexpressed in breast tumors. A, PDEF enhances transformation activity of MCF-10A cells expressing ErbB2 or CSF-1R/CSF-1. Cells were grown in soft agar and photographed on day 15 (A, left). Representative phase images (4×) from each cell line are shown (A, right). B, columns, mean number of soft agar colonies from three independent experiments; bars, SD. *, P < 0.001; **, P < 0.05. C, quantification of tumors overexpressing PDEF mRNA. Columns, number of tumors from each stage categorized according to their levels of PDEF overexpression. The fold difference (see legend) for each tumor was calculated by dividing the expression level of PDEF mRNA in the tumor by the levels in the normal breast epithelia from the same patient.
PDEF is overexpressed in breast carcinomas
PDEF has previously been shown to be overexpressed in both human and mouse mammary tumors. To further investigate the association of PDEF in human breast tumors, we were interested in investigating its expression in different stages of breast cancer. Because most Ets factors are expressed ubiquitously or are restricted to tumor stroma, it was also important to determine whether PDEF is expressed in the epithelial cells within breast tumors. To address these questions, we used laser capture microdissection technology to isolate epithelial cells from both normal breast tissue and tumor samples [ranging from atypical ductal hyperplasia (ADH) to invasive ductal carcinoma (IDC)] from a previously described cohort of breast cancer patients (32). Quantitative reverse transcription-PCR (RT-PCR) analysis revealed that PDEF is overexpressed in the epithelial cells from a majority of these breast tumors in comparison with normal breast epithelia from the same patient (Fig. 6C). Furthermore, a 2- to 10-fold increase in PDEF levels was detected in >75% of samples derived from ADH, DCIS, and IDC tumor samples (Fig. 6C). Taken together, these results suggest that PDEF activity is induced early in tumorigenesis and is elevated in tumor epithelial cells through the progression of breast carcinoma.
Discussion
Here we report the identification of an Ets family transcription factor, PDEF, from a screen designed to isolate genes that stimulate motility of normal human breast epithelial cells. Ectopic expression of PDEF promoted limited migration of MCF-10A cells in the absence of motogens and strongly enhanced motility induced by hyperstimulation of the RTKs ErbB2 and CSF-1R. Furthermore, PDEF expression induced a dramatic invasive phenotype in MCF-10A acini coexpressing ErbB2 or CSF-1R/CSF-1 in three-dimensional basement membrane cultures. The migratory and invasive activities of PDEF were dependent on the presence of a highly conserved ERK-MAPK phosphoacceptor site, T50, and mutation of this site abolished PDEF-induced migration and invasion. Furthermore, PDEF enhanced anchorage-independent growth of ErbB2 or CSF-1R/CSF-1 cells. These results suggest that PDEF can promote motility, invasion, and anchorage independence in breast epithelial cells, particularly in the context of activated RTKs.
In addition to stimulating motility of normal mammary epithelial cell lines such as MCF-10A and HMECs, PDEF expression also enhanced motility in several tumorigenic epithelial cell lines with the exception of MDA-MB-231 cells (29). These findings suggest that PDEF, similar to many other transcription factors, may play differing roles depending on cell context. Our finding that PDEF enhances motility of several epithelial cell lines also shows that the promigratory activity of PDEF is not restricted to MCF-10A cells.
PDEF induced a 2- to 5-fold increase in motility of parental MCF-10A cells and a robust 10- to 30-fold increase in motility when coexpressed with the growth factor receptors ErbB2 or CSF-1R. Furthermore, ErbB2/PDEF and CSF-1R/PDEF cells formed highly invasive structures in three-dimensional cultures. The ability of PDEF to induce invasive activity is consistent with previous reports that other Ets factors including Ets1, Ets2, and ESE-1 also promote invasion (1, 7, 10, 33–35). Because expression of ErbB2 or CSF-1R/CSF-1 alone does not stimulate motility or invasion of MCF-10A cells, our findings suggest that PDEF can synergize with hyper-stimulated RTKs, such as ErbB2 or CSF-1R/CSF-1, to promote these cellular processes. Coexpression of PDEF with other oncogenes, such as AKT and cyclin D1, did not enhance motility or invasion, indicating the specificity of this synergy. Our finding that MEK2DD/PDEF cells are highly motile and invasive suggests a similar synergy between PDEF and MEK2DD and a role for the Ras/MAPK pathway in PDEF-induced motility and invasion.
Previous work has shown that both Ras and ErbB2 regulate other Ets factors by promoting Ras/MAPK phosphorylation (17). Ets1, Ets2, and Pointed each contain a highly conserved MAPK phosphorylation site (PxTP) that is required for the transcriptional activity of these proteins (17, 18, 33, 36, 37). Furthermore, MMTV-PyMT transgenic mice expressing Ets2 with a mutation at the MAPK phosphorylation site formed fewer and smaller breast tumors with decreased levels of stromal matrix metalloproteinase 3 (MMP3) and MMP9 (13). In our studies, a PDEF variant bearing a similar mutation at an optimal MAPK phosphorylation site (T50A) failed to promote migratory and invasive activity, suggesting that the MAPK pathway may likewise regulate PDEF activity. The T50A mutant was able to localize to the nucleus and activate basal transcription of an Ets promoter element (data not shown), suggesting that this mutation does not interfere with the subcellular targeting or basal transactivation capacity of PDEF. Whereas our results show that this putative MAPK phosphorylation site is essential for PDEF-induced invasive activity, further studies will be required to confirm the regulation of PDEF activity by the Ras-ERK pathway.
Multiple studies have shown that Ets proteins, including Ets1, Ets2, and ESE-1, are overexpressed in breast tumors (2–5). Although a recent study reported that PDEF protein levels were reduced in several breast tumors (29), previous studies have shown that PDEF mRNA is overexpressed in a large number of breast cancer cell lines and tumors (20, 21). For instance, a SAGE analysis of a collection of breast tumor and cell line cDNA showed that a majority of breast tumor cells express 32–240 PDEF tags/200,000 tags compared with <2 in normal epithelial cells (http://cgap.nci.nih.gov/SAGE/FreqsOfTag?ORG=Hs&METHOD=SS10,LS10&FORMAT=html&TAG=GTGCAGGGAG&TISS=mammary+gland&-HIST=neoplasia&CELL=2&NOT=fetus). Furthermore, murine PDEF (mPSE) was found to be significantly elevated in mammary tumors in both PyMT and ErbB2 tumor models (5). Because most Ets factors, including Ets1 and Ets2, are mainly expressed in the stromal cells, we used laser capture microdissection and quantitative RT-PCR to determine PDEF expression in the epithelial cells of human breast tumor samples and found that PDEF is overexpressed in the epithelial cells in a majority (>75%) of breast tumors. Our findings are supported by a recent SAGE analysis (http://cgap.nci.nih.gov/SAGE/BreastCellViewer?TAG=GTGCAGGGAG&ORG=Hs&METHOD=SS10,LS10) showing that luminal epithelial cells in breast tumors express PDEF whereas tumor-associated myoepithelial and stromal cells do not (22). Although most tumors displayed only a 2- to 5-fold increase in PDEF mRNA levels, analysis of epithelial cells from a number of advanced tumors revealed a 5- to 10 or >10-fold enhancement of PDEF expression compared with the adjacent normal epithelium (Fig. 6C). However, a larger sample size will be required to determine if there is a significant correlation between tumor stage and PDEF expression.
Overall, our studies reveal that PDEF can promote migration and, in the context of hyperstimulation of the ERK/MAPK pathway, can induce invasion and anchorage independence of breast epithelial cells. The absence of invasive activity in MCF-10A cells expressing PDEF alone and the presence of elevated levels of PDEF in the epithelia of early, noninvasive stages of breast carcinoma suggest that PDEF expression alone is insufficient to induce invasive activity. However, elevated levels of PDEF may initiate transforming activity and/or sensitize these cells to additional insults, such as the amplification or mutational activation of a RTK, to promote tumor progression. Consistent with an early role for Ets factors in tumorigenesis, we found that PDEF supports anchorage-independent growth when coexpressed with ErbB2 or CSF-1R/CSF-1. This finding suggests that PDEF may regulate phenotypic effects other than motility and invasion in human tumors. Therefore, we speculate that PDEF could be important in promoting distinct phenotypic effects at different stages of tumorigenesis, particularly in tumors with genetic alterations that can lead to the hyperstimulation of RTKs, such as ErbB2 or CSF-1R, which regulate the ERK pathway. The dramatic morphologic changes and invasive activity of ErbB2/PDEF and CSF-1R/PDEF cells in three-dimensional cultures may reflect some aspects of tumor invasion during later stages of tumor progression in vivo.
Collectively, our data suggest that PDEF is a promigratory gene that synergizes with the oncogenes ErbB2 and CSF-1R to promote various aspects of tumor progression including enhanced cell motility, invasion, and anchorage-independent growth of mammary epithelia. Currently, we are identifying downstream targets of PDEF to determine the mechanism by which PDEF stimulates these activities in mammary epithelial cells.
Acknowledgments
Grant support: Department of Defense Breast Cancer Research Program DAMD17-03-1-0408 (R.N. Gunawardane); the Avon Foundation, the Susan G. Komen Breast Cancer Foundation, and the Department of Defense Grants DAMD17-03-1-0428 (D.C. Sgroi); and the Cell Migration Consortium NIH/NIGMS U54 GM64346-04 and NIH Specialized Program of Research Excellence (J.S. Brugge).
We thank Laura Selfors for guidance on the statistical analysis of the data in this manuscript as well as past and present members of the Brugge lab for stimulating discussions and helpful technical advice during the preparation of this article; Jason Smith, Tobias Schmelze, and Kaylene Simpson for critical reading of the manuscript; T. Libermann (BIDMC) for helpful discussions; D. Lynch and S. Meloche for plasmids; R. Mulligan for VSV-GPG viral packaging of cells; M. Roussel for the CSF-1R; and S. Mani and R. Weinberg for cancer cell lines.
Footnotes
R.N. Gunawardane, E. Lin, and D. Lynch, unpublished observation.
References
- 1.Gilles C, Polette M, Birembaut P, Brunner N, Thompson E. Expression of c-ets-1 mRNA is associated with invasive, EMT-derived phenotype in breast carcinoma cell lines. Clin Exp Metastasis. 1997;15:519–526. doi: 10.1023/a:1018427027270. [DOI] [PubMed] [Google Scholar]
- 2.Chang C, Scott GK, Kuo WL, et al. ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene. 1997;14:1617–1622. doi: 10.1038/sj.onc.1200978. [DOI] [PubMed] [Google Scholar]
- 3.Behrens P, Rothe M, Wellmann A, Krischler J, Wernert N. The Ets-1 transcription factor is up-regulated together with MMP 1 and MMP 9 in the stroma of pre-invasive breast cancer. J Pathol. 2001;194:43–50. doi: 10.1002/path.844. [DOI] [PubMed] [Google Scholar]
- 4.Span P, Manders P, Heuvel JJ, et al. Expression of the transcription factor Ets-1 is an independent prognostic marker for relapse-free survival in breast cancer. Oncogene. 2002;21:8506–8509. doi: 10.1038/sj.onc.1206040. [DOI] [PubMed] [Google Scholar]
- 5.Galang C, Mullers WJ, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–11292. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- 6.Wasylyk CMS, Sobieszczuk P, Wasylyk B. Reversion of Ras transformed cells by Ets trans-dominant mutants. Oncogene. 1994;9:3665–3673. [PubMed] [Google Scholar]
- 7.Delannoy-Courdent A, Mattot v, Fafeur v, et al. The expression of an Ets1 transcription factor lacking its activation domain decreases uPA proteolytic activity and cell motility, and impairs normal tubulogenesis and cancerous scattering in mammary epithelial cells. J Cell Sci. 1998;111:1521–1534. doi: 10.1242/jcs.111.11.1521. [DOI] [PubMed] [Google Scholar]
- 8.Sapi E, Flick MB, Rodoc S, Kacinski BM. Ets-2 trans-dominant mutant abolishes anchorage-independent growth and macrophage colony-stimulating factor-stimulated invasion by BT20 breast carcinoma cells. Cancer Res. 1998;58:1027–1033. [PubMed] [Google Scholar]
- 9.Foos G, Garcia-Ramirez JJ, Galang CK, Hauser CA. Elevated expression of Ets2 or distinct portions of Ets2 can reverse Ras-mediated cellular transformation. J Biol Chem. 1998;273:18871–18880. doi: 10.1074/jbc.273.30.18871. [DOI] [PubMed] [Google Scholar]
- 10.Foos G, Hauser CA. Altered Ets transcription factor activity in prostate tumor cells inhibits anchorage-independent growth, survival, and invasiveness. Oncogene. 2000;19:5507–5516. doi: 10.1038/sj.onc.1203946. [DOI] [PubMed] [Google Scholar]
- 11.Wernert N, Raes MB, Lassalle P, et al. C-ets1 proto-oncogene is a transcription factor expressed in endothelial cells during tumor vascularization and other forms of angiogenesis in humans. Am J Pathol. 1992;140:119–127. [PMC free article] [PubMed] [Google Scholar]
- 12.Wernert NGF, Fafeur v, Bouali F, et al. Stromal expression of c-Ets1 transcription factor correlates with tumor invasion. Cancer Res. 1994;54:5683–5688. [PubMed] [Google Scholar]
- 13.Man A, Young LJT, Tynan JA, et al. Ets2-dependent stromal regulation of mouse mammary tumors. Mol Cell Biol. 2003;23:8614–8625. doi: 10.1128/MCB.23.23.8614-8625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sementchenko v, Watson DK. Ets target genes: past, present, and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 15.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 16.Oettgen P, Finger E, Sun Z, et al. PDEF, a novel prostate epithelium-specific ETS transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem. 2000;275:1216–1225. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- 17.Wasylyk B, Hagman J, Gutierrez-Hartman A. Ets transcription factors: nuclear effectors of the Ras-MAP-Kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- 18.Yang B, Hauser CA, Henkel G, et al. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wasylyk C, Bradford AP, Gutierrez-Hartmann A, Wasylyk B. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets1 and Pointed P2. Oncogene. 1997;14:899–913. doi: 10.1038/sj.onc.1200914. [DOI] [PubMed] [Google Scholar]
- 20.Ghadersohi A, Sood AK. Prostate epithelium-derived Ets transcription factor mRNA is overexpressed in human breast tumors and is a candidate breast tumor marker and a breast tumor antigen. Clin Cancer Res. 2001;7:2731–2738. [PubMed] [Google Scholar]
- 21.Mitas M, Mikhitarian K, Hoover L, et al. Prostate-specific Ets (PSE) factor: a novel marker for detection of metastatic breast cancer in axillary lymph nodes. Br J Cancer. 2002;86:899–904. doi: 10.1038/sj.bjc.6600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 24.Wrobel C, Debnath J, Lin E, Beausoleil S, Roussel MF, Brugge JS. Autocrine CSF-1R activation promotes Srcdependent disruption of mammary epithelial architecture. J Cell Biol. 2004;165:263–273. doi: 10.1083/jcb.200309102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seton-Rogers S, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Cooperation of the ErbB2 receptor and transforming growth factor β in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci U S A. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh E, Chen T, Daley GQ. Novel retroviral vectors to facilitate expression screens in mammalian cells. Nucleic Acids Res. 2002;30:142–148. doi: 10.1093/nar/gnf142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh E, Chen T, Daley GQ. Genetic complementation of cytokine signaling identifies central role of kinases in hematopoietic cell proliferation. Oncogene. 2004;23:1214–1220. doi: 10.1038/sj.onc.1207209. [DOI] [PubMed] [Google Scholar]
- 28.Ma X, Wang Z, Ryan PD, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Feldman R, Sementchenko VI, Gayed M, Fraig MM, Watson DK. PDEF expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res. 2003;63:4626–4631. [PubMed] [Google Scholar]
- 30.Prescott J, Koto KSN, Singh M, Gutierrez-Hartmann A. The Ets transcription factor ESE-1 transforms MCF-12A human mammary epithelial cells via a novel cytoplasmic mechanism. Mol Cell Biol. 2004;24:5548–5564. doi: 10.1128/MCB.24.12.5548-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muthuswamy S, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X, Salunga R, Tuggle JT, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watabe T, Yoshida K, Shinodoh M, et al. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int J Cancer. 1998;77:128–137. doi: 10.1002/(sici)1097-0215(19980703)77:1<128::aid-ijc20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Schedin P, Eckel-Mahan KL, McDaniel SM, et al. ESX induces transformation and functional epithelial to mesenchymal transition in MCF-12A mammary epithelial cells. Oncogene. 2004;23:1766–1779. doi: 10.1038/sj.onc.1207391. [DOI] [PubMed] [Google Scholar]
- 35.Neznov N, Man AK, Yamamoto H, Hauser CA, Cardiff RD, Oshima RG. A single targeted Ets2 allele restricts development of mammary tumors in transgenic mice. Cancer Res. 1999;59:4242–4246. [PubMed] [Google Scholar]
- 36.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seidel J, Graves BJ. An Erk2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes Dev. 2001;16:127–137. doi: 10.1101/gad.950902. [DOI] [PMC free article] [PubMed] [Google Scholar]