Abstract
Images produced by ultrasound systems are adversely hampered by a stochastic process known as speckle. A despeckling method based upon removing outlier is proposed. The method is developed to contrast enhance B-mode ultrasound images. The contrast enhancement is with respect to decreasing pixel variations in homogeneous regions while maintaining or improving differences in mean values of distinct regions. A comparison of the proposed despeckling filter is compared with the other well known despeckling filters. The evaluations of despeckling performance are based upon improvements to contrast enhancement, structural similarity, and segmentation results on a Field II simulated image and actual B-mode cardiac ultrasound images captured in vivo.
I. Introduction
AN accurate anatomical display of organs such as the heart, kidney, prostate, liver, etc., would be a beneficial aid to health practioners in diagnosing ailments or assessing the health of that organ. Although there are a wide range of medical imaging modalities that could be utilized, the use of ultrasound is advantageous for the following reasons: only safe nonionizing sound waves are used in the scanning process; the portability of the hardware; cost is inexpensive when compared to other medical imaging modalities; and real-time or near real-time imaging is attainable.
Although there are many advantages of using ultrasound as an imaging modality, the images produced are hampered by a phenomena known as speckle. Speckle is visible in all ultrasound images as a dense granular noise1 or “snake-like” objects that appear throughout the image. Unfortunately, the presence of speckle does adversely effect image contrast, edge detection, and segmentation.
An explanation on the cause of speckle is offered by Goodman in [1]. Goodman mathematically models speckle as a sum of a large number of complex phasors, a.k.a. a complex random walk. These complex phasors can have either constructive or destructive relationship with each other. Thus, bright and dark points can be observed in close proximity. The relationship and relevance of acoustic speckle to laser speckle is given by Abbott and Thurstone in [2]. More relevant and detailed descriptions of ultrasound image formation and the statistics of ultrasound speckle can be found in [3], [4].
The experimentation performed in [3] provides evidence that the intensity or prelogarithm compressed envelop detection amplitudes J (n, m) can be modeled as the multiplicative model given in (1)
| (1) |
where the multiplicative noise η×(n, m) is samplewise independent and uncorrelated to the ideal image pixel value I(n, m) and P(n, m) is the point spread function (PSF) of the ultrasound imaging system. The multiplicative model could be made equivalent to the additive model
| (2) |
if the additive noise term η+(n, m) is made dependent upon the ideal image I(n, m) or other additive noise values.
The intuition behind the proposed despeckling method is that speckle as seen in the intensity image or as a logarithmically compressed intensity image, commonly called B-mode image, is a stochastic process where outliers inevitably occur. The despeckling of the B-mode image that is proposed iteratively removes outliers. The essence of our despeckling method has been proposed in [5] to remove highly dense impulse noise. The ultimate goal of this paper is to enhance ultrasound slices for the purpose of providing optimal contrast enhancement, which will subsequently be used to segment substantial anatomic features via a gradient based edge detection method.
A smoothing of speckle and preservation of edges are in a general sense divergent. This problem is not only directly relevant to enhancing ultrasound images, but also relevant to enhancing synthetic aperture radar (SAR), optical laser, and other imaging modalities. A wide variety of methods have been proposed to address speckle removal or reduction for a variety of applications. When addressing speckle as unwanted noise, adaptive filtering methods like the Nagao [6], Lee [7], Frost [8], Kuan [9], [10], adaptive weighted median filter (AWMF) [11], speckle reducing anisotropic diffusion [12], and other [13]-[28] methods have been proposed.
The comparison in this paper is unlike the one published in [29], which was based upon subjective interpretation by expert radiologists. The results reported in [29] found that adaptive local statistics filters like the Lee [7], the Frost et al. [8], and the Kuan et al. filters [10] were robust in providing a visually enhanced image to make an accurate diagnosis of the amount, composition, and texture of plaque in the carotid artery.
Our application differs from the one in [29] in that we are interested in autonomous or semi-autonomous segmentation. We will rely on several performance metrics to quantify improvements to contrast, structural similarity, and segmentation. The general goal being to determine which despeckling method best removes speckle while producing the best contrast and preservation of edges. Best contrast is meant in the sense of decreasing the variance in a homogeneous region while distinct regions are well defined. Although the proposed despeckling method has been favorably evaluated in [30], the evaluation offered in Section IV is based upon three objective metrics. A generalization of the Fisher discriminant [31] and the structural similarity (SSIM) index [32] are used to quantify how well various algorithms achieve this general goal. Additionally, an evaluation of a despeckling method’s ability to promote robust segmentation of actual B-mode ultrasound cine data of a mouse heart captured in vivo is performed where a shape metric is used to evaluate the results.
II. Theoretical Background
If it is allowed to assume that the complex-valued IQ data is modeled as a sum of complex phasors as Goodman suggests in [1]
for some positive integer K, then it can be shown that the amplitudes of the IQ data |IQ(n, m)| in a constant reflectivity region are Rayleigh or more generally Rician distributed when K is sufficiently large and with the added assumptions that the amplitudes ak(n, m) and phases φk(n, m) are independent.2 Logarithmically transformed random variables that are Rayleigh or Rician distributed has been shown to followa Fisher-Tippett distribution [33].
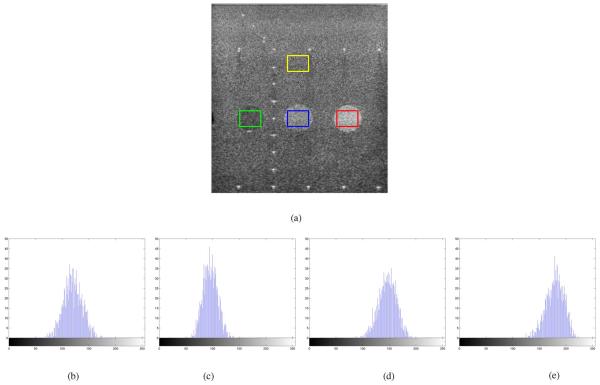
Fig. 1(a) shows a eight bit full scale contrast stretched B mode ultrasound image of a phantom consisting of three disks representing regions with various return echo intensities. This B mode image was acquired from a Siemens Sequoia system. The location of the linear array transducer operating at 3 Mhz is at the top of the image. In Fig. 1-(e) the histograms of the pixel values enclosed in the yellow, green, blue, and red boxes in Fig. 1(a) are shown, respectively. These histograms highlight that the pixel values in a homogeneous region are the result of some stochastic process.
Fig. 1.
B-mode ultrasound image of a phantom and histograms of pixel values enclosed in the (b) yellow, (c) green, (d) blue, and (e) red boxes of Fig. 1(a). (a) Ultrasound phantom image, (b) yellow box, (c) green box, (d) blue box, and (e) red box.
The following subsections will outline some methods that were tested in Section IV. The algorithms outlined in the following subsection are not meant to be a comprehensive list or even a list of the most robust despeckling methods. Rather, each algorithm was chosen because it represents a unique approach to achieve despeckling. Moreover, most recent methods have compared their results with at least one or more of the following methods.
A. Wiener Filter
Suppose the detected image is given as the additive model in (2) where the PSF P is known or estimated. Applying the Wiener filter as described in [34] to the detected image J results in a low-pass filtered approximation of the ideal image I. The Wiener filtered image is determined as
where , likewise for J,I,η+ and ,,, respectively,
and is the variance of the noise.
B. Lee Filter
Lee in [7] proposed methods to contrast enhance an image and to restore an image corrupted by noise. Lee’s noisy image models in [7] are given in (1) and (2) where the PSF is ignored. Lee’s paper proposes an adaptive filter to aggressively smooth, via local averaging, in homogeneous regions while regions that contain significant image features are to be left unmolested.
Contrast enhancement is promoted at each pixel by a linear rescaling of the local mean summed with the multiplication of a gain applied to the difference of the pixel value with the local mean to determine a new pixel value. Formally, let J(n, m) be the original noisy pixel value, then the new pixel value is determined as
| (3) |
where μ is the local mean. The function g(·) is a linear rescaling of the mean that is g(μ) = aμ+b where the parameters were determined to allow the new pixel value to utilize the full eight bit dynamic range. As pointed out in [7], if 0 ≤ k < 1, then (3) determines a smoothing filter, i.e., a low-pass filter. When the gain k is greater than one, then (3) attempts to “sharpen” image features that is enhance the edges.
The gain k is adaptively chosen as a function of the local statistics. The gain k is determined as
| (4) |
where σ2 is the local variance in the same window about J(n, m) that determined μ and is the global noise variance. When , the gain parameter is less than but approximately one, in which case the filter in (3) performs like an identity filter that is . If the local variance σ2 is greater than but nearly equal to the global noise variance , then and the filter specified by (3) serves as a low-pass filter. Since the global noise variance can only be greater than or equal to zero, the gain parameter k can never exceed one. Thus, in homogeneous regions k should be set to zero and (3) provides local smoothing. When J(n, m) is determined to be an edge pixel, then k should be set to one and the pixel is left undeteriorated.
When the image is determined or assumed to be degraded by multiplicative noise as in (1), then Lee in [7] approximates the image as an additive noise model of the form
| (5) |
where are chosen so that mean square error between the J(n, m) of the multiplicative model from (1) and of the additive model in (5) is minimized. Since the image polluted by multiplicative noise is recast as an image with additive noise, the adaptive filter employed in the additive case, given in (3), can be used, though the gain parameter k is redefined. Given an a priori determination of the mean and variance of the stationary noise, μη and respectively, the adaptive gain parameter is determined as
where μI is the mean of I(n, m) determined as
within some fixed window and μ is the local mean of J(n, m) within the same window. The variable Q is determined as
where σ2 is the local variance of J(n, m) within some window.
C. Speckle Reduction by Using Anisotropic Diffusion
The speckle reducing anisotropic diffusion (SRAD) method of Yu and Acton in [12] adaptively applies an anisotropic diffusion method. Inspired by the time dependant heat diffusion equation, Perona and Malik in [35] published a method to perform anisotropic diffusion on noisy images. In [35], the diffusion equation of an image with continuous variables over time J(x, y, t) is given as
| (6) |
where div is the divergence operator, ∇ is the gradient operator, ∇2 is the Laplacian operator, v · w represents the dot product of two vectors, and J(x, y, 0) = J(x, y). When c(x, y, t) is a constant, then (6) defines isotropic diffusion. For anisotropic diffusion, the function known as the conduction coefficient (a.k.a. coefficient of variation, diffusion coefficient) c(·) is given as
where K is some constant and ∥ · ∥ is the norm.
Yu and Acton in [12] proposed a anisotropic diffusion method where the coefficient of variation is determined as a function of the ratio of the local variance to the local squared mean. In other words, SRAD utilizes the instantaneous coefficient of variation (ICOV) as a variable in their conduction coefficient function. The ICOV of SRAD determines whether a pixel should be smoothed or left intact. The SRAD algorithm iteratively processes a nonzero valued image I(x, y) := I(x, y; 0) according to
| (7) |
and
| (8) |
where is the outward normal vector to B(Ω), the border of ω. Equations (7) and (8) are known as the SRAD partial differential equations. The diffusion coefficient c(·) is defined as the quotient
| (9) |
or as the exponential function
| (10) |
In (9) and (10), if q(x, y; t) ≈ q0(t), then c(q(x, y; t)) ≈ 1 and smoothing with respect to (7) is enacted. If q(x, y; t)≫ q0(t), then the diffusion coefficient is very small and smoothing in a local region around (x, y) is averted. When at time t, if (x, y) resides in a homogeneous region, then smoothing can be promoted by allowing q(x, y; t) ≈ q0(t). When (x, y) lie on an edge or in the vicinity of an edge, then defining q(x, y; t)≫ q0(t) would prohibit deterioration of the edge. Yu and Acton defined q0(t), the coefficient of variation in fully developed speckle, as the ratio of the noise standard deviation to the noise mean
| (11) |
and the ICOV is defined as
The standard deviation and the mean of the noise in (11) are determined by calculating the mean and standard deviation within a homogeneous region
Lastly, it should be noted that although the intuitive ideas of SRAD is outlined using continuous variables , our implementation of SRAD used in the evaluation given in Section IV is the discrete form which is detailed in [12].
III. Materials and Methods
Since the publication of the Lee filter [7], numerous despeckling techniques [8]-[13], [16], [19] have been proposed that utilized the same basic adaptive filtering concept. The general concept of these filters is to adaptively determine some degree of local smoothing that should be performed. The differences of the various despeckling methods are the various methods used to choose how to smooth and/or the diverse criteria used to determine the degree of smoothing. The parameter k in (3) determines the degree or amount of smoothing and is generally derived from some function of the local statistics. This parameter usually can be any continuous value between zero and one. Ideally, the smoothing should occur when the pixel is within a homogeneous region and the local mean μ should be the mean determined from this homogeneous region. If the pixel value lies on an edge due to some significant anatomical feature of interest, then the original pixel value should be preserved.
The propose despeckling method is aimed at removing outliers. Removal of outliers for denoising images is a method suggested by Abreu et al. in [5] to remove highly dense impulse noise. The general concept of the proposed despeckling method is outlier pixel values are smoothed aggressively. Otherwise, the current pixel value is preserved. Thus, the adaptive parameter k of (3) is binary (either k = 0 or k = 1) in the proposed despeckling method. Iteratively filtering in this manner the variance in each homogeneous region or class (a collection of similar regions) is reduced while the mean values from differing regions or classes are maintained.
The proposed despeckling method relies on how outliers are defined. Although there are numerous definitions of an outlier that merit investigation, the proposed despeckling method considers the pixel value that is being scrutinized an outlier if it is a local extremum. As prescribed in [5] the outliers are replaced by a value which does not rely on the outlier value. The impulse removal method in [5] replaces the outliers with the rank order mean, which is determined from ordering the pixel values in some odd size window (the outlier value is not included in this window) and the average of the two middle values of this ordering, i.e., the median value from an even number of windowed samples, is used to replace the outlier.
Although the rank order mean is worth considering in future improvements, we prefer to adhere to computational efficiency (no ordering of pixel values is needed) and replace each outliers with the local mean from some surrounding neighborhood where the outlier value is not a member of . The choice of the neighborhood is important. A large neighborhood fully contained in a homogeneous region will produce a local mean that is close to the mean of the homogeneous region. Yet, a neighborhood that is too large will extend past the homogeneous region and yield a misleading value. If a small neighborhood is used, then more iterations will be needed to achieve the same result as with a larger neighborhood. A small neighborhood would be beneficial in preserving fine structure(s) in an ultrasound image.
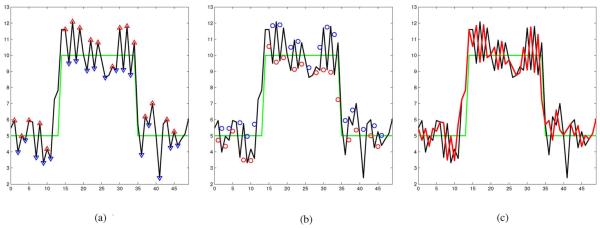
Each iteration of the propose method produces a sequence with locally reduced variance. The local extrema of the new sequence are consider outliers and the process is iterated. The steps of our proposed iterative method applied to a 1-D signal or a 2-D image J are described in the following. For the sake of clarity in explanation, the steps of the proposed despeckling algorithm is illustrated in 1-D in Fig. 2.
Fig. 2.
Illustration of one iteration of the proposed despeckling method. (a) Ideal (green) and noisy (black) signals. The △s are local maximums and the ▽s are local minimums of the noisy signal. (b) △s and ▽s in Fig. 2(a) are replaced with the local means (shown as ○s and ○s, respectively). (c) Resulting (red) signal after one iteration.
- The algorithm is initialized by the original speckled image
- White Gaussian noise with a small variance is initially and periodically added to the the current image
This step ensures that the filter acts on a local region of the current signal or image . -
An iteration i begins by determining the set of locations of local maxima and local minima of . The locations of these extrema are defined by the set
where m = n ± 1 for 1-D signals and for 2-D images.An illustration using a signal of fifty samples is shown in Fig. 2. Fig. 2(a) show the ideal noise free signal in green, the noisy signal (the ideal signal with Gaussian noise added) in black. The red upward pointed (△) and blue downward pointed (▽) triangles denote the local peaks and valleys of the noisy signal, respectively. These local extrema are considered outliers.
-
Without using the local extrema values, the algorithm replaces each extremum with the local mean taken from neighboring samples. For all (n, m) ∈ E
where is some local neighborhood of , is the cardinality of set , and . In the 1-D illustration shown in Fig. 2 the local maximums shown as △s and the local minimums shown as ▽s in Fig. 2(a) are replaced with corresponding local averages. The local averages are depicted as either red or blue circles (○ or ○) in Fig. 2(b). That is each △ value in Fig. 2(a) is replace with the corresponding ○ value in Fig. 2(b) and each ▽ value in Fig. 2(a) is replace with the corresponding ○ value depicted in Fig. 2(b).This produce the reduced local variance red signal shown in Fig. 2(c).
- If convergence is attained, that is
for some predefined ∈ > 0 and the total number of iteration is not met, then the process is iterated from either step 2 or step 3, depending upon whether more zero mean small variance noise should be added. If the total number of iterations is met, then the filtering process is terminated.
By removing outliers at each iteration, this method reduces the local variance of the signal/image. In effect, this method produces a converging sequence of signals or images by squeezing or compressing the stochastically distributed pixel values to some limiting value. Since the proposed filtering method described is able to compress the distribution of pixel values, the proposed method is fittingly named the squeeze box filter (SBF).
IV. Results
A. Using a Field ii Simulated Image
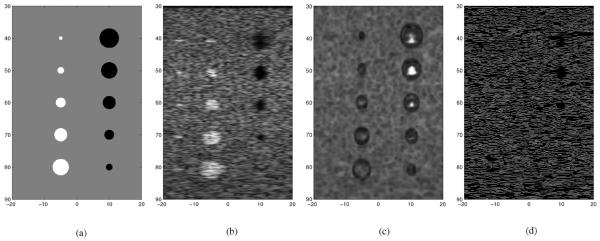
To evaluate the despeckling performance of various filters against the proposed SBF despeckling method, a template image was created and is denoted as T. An eight bit full scale contrast stretched template image is used as the ideal noise free images and is shown in Fig. 3(a). The template T consists of three classes. The background class is shaded in gray and the pixel values in this class are set at one. In the template T, a class consisting of five disks of various diameter vertically aligned on the left is shaded in white and the pixel values in this class are set at five. The pixel values of the vertically aligned class of five black disk of various diameters on the right of template T are set at zero.
Fig. 3.
Two and three class templates and ultrasound simulations used in the comparison: (a) three class template, (b) Field II simulation, (c) SSIM map, and (d) canny edge map of Fig. 3(b).
The Mathworks MatLab® Field II simulation [36], [37] version 3.16 is used to simulate a B-mode ultrasound image of template T from a linear array. The Field II simulated B-mode ultrasound image of template T is shown in Fig. 3(b). The indices of the images in Fig. 3 represents distances in units of millimeters (mm). The simulation is generated with the linear array transducer at the top of the image. Thus, the row indices indicate axial distance away from the transducer and the column indices indicate lateral distance. The five small bright white dots along the left side at axial distances 40, 50, 60, 70, and 80 of Fig. 3(b) were placed to simulate the ultrasound system’s PSF at various distance from the transducer. These simulated PSFs are not included in any of the classes. The other relevant parameters of the Field II simulation of an ultrasound system attached to a linear array are listed in Table I.
TABLE I.
Field II Parameters Used to Simulate the Ultrasound Image Shown in Fig. 3(B)
| Transducer center frequency | 3.5 MHz |
| Sampling frequency | 100 MHz |
| Speed of sound | 1540 m/s |
| Wavelength | 0.44 mm |
| Element width | 0.44 mm |
| Element height | 5 mm |
| Kerf | 0.05 mm |
| Focal point | 70 mm |
| Number of physical elements | 192 |
| Number of active elements | 64 |
The image in Fig. 3(c) is the eight bit full scale contrast SSIM index map of the Field II simulated image in Fig. 3(b) against the template in Fig. 3(a). The SSIM index provides a metric based upon local measures of luminance and contrast. The SSIM index compares the local structural similarity of an image against an ideal image, which is free of noise and distortion. The SSIM index varies between −1 and 1, where −1 indicates the worst local similarity to the ideal image and 1 indicates the best local similarity to the ideal image. The image in Fig. 3(c) shows the SSIM at each pixel of the Field II simulated image in Fig. 3(b) against the template image in Fig. 3(a). In Fig. 3(c), whiter pixels would indicate better structural similarity and darker pixels would indicate poorer structural similarity. The SSIM index map in Fig. 3(c) shows that there are only three regions in the Field II simulated image that exhibit good structural similarity. The three regions of good structural similarity are located within the three larger dark disks in Fig. 3(b).
Fig. 3(c) shows the Canny edge map of the Field II simulated image in Fig. 3(b). This edge map does not exhibit any edge gradient that are relevant to the dark and white disks. Thus, the Field II simulated image in Fig. 3(b) accurately portrays most actual ultrasound images.
Evaluation of despeckling methods’ improvements to an ultrasound image requires a meaningful quantifiable measure. This measure should indicate when different homogeneous regions are properly defined. Additionally, this metric should account for differences in pixel values from the mean of a class. A class is taken to be a collection of homogeneous regions. This measure in essence will determine how well an algorithm despeckles the simulated ultrasound image while keeping the distinct classes well separated.
The method we propose to quantify the improvements made to an ultrasound image is as follow:
A despeckling algorithm is applied to the simulated image J in Fig. 3(b) to produce the eight bits full scale contrast stretched processed image, denoted as Ialg.
The means and variances in each predefined class of Ialg are computed.
-
The ratio of the average squared differences of the interclass means to the sum of the intraclass variances are calculated and this quantity is denoted as Q(Ialg). Explicitly, the quantity to evaluate the performance of a despeckling algorithm to preserve distinct classes and promote smoothing within homogeneous regions of each class is given as
(12) whereand |Ck| denotes the number of pixels in class Ck.
-
To avoid sensitivity to resolution, the quantity we will use to measure the improvements to the image J due to algorithm alg is
(13) We refer to this relative performance metric as the ultrasound despeckling assessment index (USDSAI).
If the classes are well separated, then the numerator in (12) will be large. If the segmentation or restoration algorithm produces small intraclass variances, then the denominator of (12) will be small, resulting in the USDSAI quantity to be large. A large value would indicate that alg produces the desirable restoration result. If is greater than one, then the algorithm being tested improved the ultrasound image J. Larger values indicate better performance when comparing various algorithms. If is less than one, then we consider the algorithm being tested as detrimental to the ultrasound image J. If is equal to one, then the contrast of an image provide by the algorithm being tested is equivalent to the contrast of the original unprocessed image.
To elucidate the significance of the USDSAI quantity defined in (13), we examine the three extreme cases. In the first case, the simulated image J is not constant and the resulting image of some algorithm Ialg is constant. Clearly, Q(Ialg) = (0/0), which we define as zero. Thus, the quantity is equal to zero. We consider algorithms that yield small values, less than one, as indicative of poor performance.
In the second case, the simulated image J is neither a constant nor a scaled version of the template image. The resulting image of some algorithm is the template image, that is Ialg = T. In this case
when the average of the squared differences of the interclass means represented as K is not equal to zero. We will consider this quantity as indicative of an ideal improvement to the image J and the algorithm being tested performed ideally.
In the third and final case, the simulated image J is composed of classes C1, C2, C3, …, CN and the pixel values of the resulting image Ialg is some constant except for a single point within each class C1, C2, C3, …, CN, without loss of generality, say the pixel values are all zero except at differing points in each class have values K1|R1|, K2|R2|, K3|R3|, … KN|RN|, where Ki > 0 and |Ri| is the number of pixels in class Ci. In other words, the algorithm being tested sets the pixels in each class to the same constant except one pixel, which differs from the rest. In this case, the sum of the interclass mean differences squared is
| (14) |
The inequality of (14) is attained from the safe assumption that Kk > 0 for all k. The variance for each class Ck where k = 1, 2, 3, …, N is
The sum of the intraclass variances is
Let Cmin = min{|Ck| where k = 1, 2, 3, …, N}, so that
| (15) |
Using the inequalities from (14) and (15), the quantity Q(Ialg) defined in (12) is bounded above by
| (16) |
In this present case, the algorithm being evaluated produces an image composed of a scaled Kronecker delta function in each class. When Cmin is very large, the original unprocessed image contains large classes of homogeneous regions and the metric Q(Ialg) returns a small number. Thus, the value would be smaller than .
To present an objective comparison, we perform a mesh search algorithm varying the parameters of each algorithm so that the USDSAI value was maximized. It should be noted that the mesh search optimization method is problematic in attaining optimal performance, since the search can get stuck on a local values and never attain global optimality.3
The USDSAI values along with the mean SSIM (MSSIM) of the various tested filter (AWMF, wavelet soft thresholding, Lee, SRAD, Wiener, and SBF) are given in Table II. The USDSAI and MSSIM of the unprocessed simulated image is listed in Table II for reference. The USDSAI and MSSIM values and given in Table II are slightly greater than the USDSAI of the unprocessing image , indicating that the contrast improvements and the structural similarity improvements against the ideal template image due to these two filters are modest.
TABLE II.
USDSAI Values and MSSIM Index for the Various Algorithms Tested
| USDSAI | MSSIM | ||
|---|---|---|---|
| FILTERS | Unprocessed | 1 | 0.2279 |
| AWMF | 1.2559 | 0.2537 | |
| Wavelet | 1.3723 | 0.2410 | |
| Lee | 3.4641 | 0.3036 | |
| SRAD | 3.3494 | 0.8080 | |
| Wiener | 3.5586 | 0.0160 | |
| SBF | 4.0291 (SD=0.0771) | 0.8432 (SD=0.0041) | |
The USDSAIs , , and indicate significant contrast enhancement by the Lee, SRAD, and Wiener filters, respectively, are possible. The images despeckled by the Lee, SRAD, and Wiener filters are shown in Fig. 4(a), (b),, and (c), respectively. The unprocessed pixel values enclosed in the green box of the SRAD filtered image shown in Fig. 4(b) are used to initialized the ICOV of the SRAD algorithm. The eight bit full scale contrast stretched SSIM index map of the Lee, SRAD, and Wiener filters images are shown in Fig. 4(e), (f), and (g), respectively. It is interesting and should be noted that the MSSIM of the Wiener filter is remarkably decreased when compared to the MSSIM of the unprocessed image. This is due to the poor local SSIM in the class composed of the five white disks and the background class. The SSIM indices of theWiener filtered image shown in Fig. 4(g) are displayed as black in these regions, which indicate low local values. But the SSIM index in the dark disks of the Wiener filtered image exhibit excellent structural similarity, which are shown as white in the SSIM index map of Fig. 4(g).
Fig. 4.
Resulting images from Lee, SRAD, Wiener, and SBF filters and SSIM map. (a) Lee, (b) SRAD where the green box shows the region used to initialized the ICOV, (c) Wiener, (d) SBF, (e) Lee SSIM map, (f) SRAD SSIM map, (g) Wiener SSIM map, and (h) SBF SSIM map.
Since the SBF require periodic addition of noise, it is stochastic in nature. The SBF algorithms was also optimized using a mesh search over a wide range of parameters. The SBF algorithm was evaluated using the optimal parameters, which include size of the neighborhood to compute the local mean and total number of iterations, with adding Gaussian distributed noise of standard deviation equal to 0.1 at the zeroth and 50th iterations and 100 total iterations. The SBF algorithm was applied ten consecutive times to the Field II simulated image shown in Fig. 3(b). After each application of SBF, the USDSAI and MSSIM index were computed. The SBF USDSAI and MSSIM index listed in Table II are the mean USDSAI and the mean MSSIM index, respectively, from the ten application of SBF with the optimized parameters. The standard deviation of each index is listed within parentheses in Table II. Since the mean USDSAI and mean MSSIM index are greater than the USDSAI and MSSIM indices from the other despeckling method, it is possible to achieve better despeckling results with the SBF method than with the other tested methods. Additionally, since the standard deviation is small, the SBF method is capable of providing consistently robust despeckling results.
B. In Vivo Mouse Heart
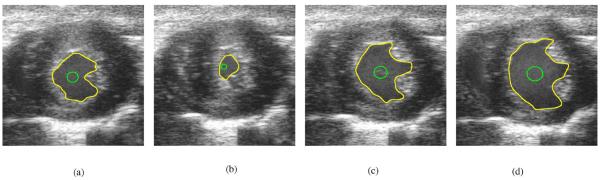
The ability to capture the left ventricle (LV) of the heart is of great importance to cardiac research and a potentially valuable tool to clinically assess cardiac health. We performed an experiment on short axis cardiac cine ultrasound data of a mouse heart. The data was acquired in vivo using a specifically designed small animal echocardiograph (ECG) ultrasound imaging system manufactured by VisualSonics® Inc. (Toronto, Ontario, Canada). The VisualSonics® ultrasound system used a signal element mechanically sweeping transducer, which operated at 30 Mhz.4 The VisualSonics system is able to attain 1000 frames per second capture rate. Mouse heart rate is roughly ten cardiac cycles per second. Thus, the VisualSonics® ultrasound system is able to provide image samples well over the Nyquist rate. Moreover the system is connected to an ECG system so that image acquisition can be begin and finish at End Diastole (where the heart is most relaxed). This system produces roughly 100 frame cine data over one mouse cardiac cycle. Figs. 5 and 6 show the B-mode short axis slices of the LV of a mouse heart at various times during the cardiac cycle that are produced by the VisualSonics® system. Fig. 5 is the zeroth frame of the cine data and shows the mouse heart near End Diastole. Fig. 6(a) shows frame 30 of the cine data, which is the mouse heart at a midway point between End Diastole and End Systole (where the heart is most compressed). Frame 60 is shown in Fig. 6(b). This image shows the short axis cross section of the mouse heart at End Systole. Frame 80 is shown in Fig. 6(c), which is a cross section of the mouse heart near a midpoint betweeen End Systole and End Diastole. Fig. 6(d) shows the mouse heart at End Diastole again.
Fig. 5.
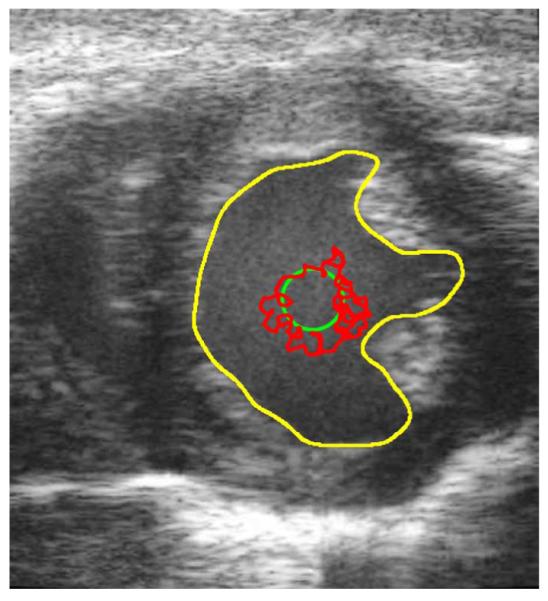
Unprocessed in vivo B-mode cardiac ultrasound image at End Diastole. The yellow contour is the manually defined contour. The green contour is the initial contour. The red contour is the result of 150 iteration of a BFAC.
Fig. 6.
Unprocessed in vivo B-mode cardiac ultrasound images at various times during the cardiac cycle. The yellow and green contours represent the desired and initial contours, respectively, (a) Frame 30, (b) Frame 60 (End Systole), (c) Frame 80, (d) Frame 100 (End Diastole).
The yellow contours in Figs. 5 and 6, and later in the images of Figs. 7 and 8 are the manually defined contours of the endocardial border of the LV. These yellow contours are used as the gold standard to evaluate the segmentation results produced from despeckles the B-mode images, then applying a balloon force active contour (BFAC) [38] segmentation algorithm to segment the endocardial border of the LV of each frame. The green contour is the initial contour. The green initial contour is a circle whose center is determined as the mean of the manually defined contour of each frame. The radius of the initial circular contour of each frame is one quarter the average distance of all the manually defined contour points to the center.
Fig. 7.
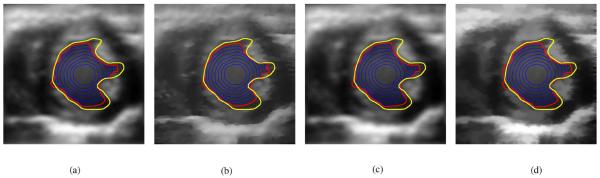
Performance of each despeckling algorithm was optimized to give the best final BFAC (red) contour in frame 0. The blue contours are the BFAC contours at every 50th iteration. The yellow contour is the desired manually defined contour. (a) Frame 0: Lee, (b) Frame 0: SRAD, (c) Frame 0: Wiener, and (d) Frame 0: SBF.
Fig. 8.
BFAC segmentation after despeckling with the various filter. Each blue contours are the BFACs at every 50th iterations. The final BFAC contours are shown in red. (a) Frame 30: Lee, (b) Frame 60: Lee, (c) Frame 80: Lee, (d) Frame 100: Lee, (e) Frame 30: SRAD, (f) Frame 60: SRAD, (g) Frame 80: SRAD, (h) Frame 100: SRAD, (i) Frame 30: Wiener, (j) Frame 60: Wiener, (k) Frame 80: Wiener, (l) Frame 100: Wiener, (m) Frame 30: SBF, (n) Frame 60: SBF, (o) Frame 80: SBF, and (p) Frame 100: SBF.
The BFAC is used to autonomously segment the endocardial border after each despeckling filter has been applied. The red chaotic contour in Fig. 5 is the final BFAC segmentation after 150 iterations were applied to the unprocessed frame 0 image. The chaotic contour is due to significant gradients caused by speckle in a region that is occupied by blood. An accurate segmentation of the LV would require the speckle within the LV to be smoothed. Yet, the endocardial border of the LV should remain intact. The BFAC parameters used are fixed throughout this experiment and are listed in Table III.
TABLE III.
Parameters Used in the BFAC
| Elasticity | 0.05 | External Force Weight | 20 |
| Rigidity | 0 | Pressure Force Weight | 0.15 |
| Viscosity | 0.75 | Iterations | 400 |
The average directed Hausdorff distance [39] is used to provide a quantitative assessment of the segmentation results. The directed Hausdorff distance of a point on the red contour to the yellow contour is the minimum Euclidean distance from the point on the red contour to some point on the yellow manually defined contour. The average directed Hausdorff distances of all points on each final BFAC red contour are used to quantify the robustness of the despeckling-segmentation results. A smaller average directed Hausdorff distance equate to a contour that is closer to the manually defined yellow contour of each frame.
Since the SBF is a stochastic method to despeckle ultrasound images, the SBF despeckling followed by BFAC segmentation is performed ten times on each test frame. The average and standard deviation (SD) of the average directed Hausdorff distances from these ten applications of despeckling with SBF followed by BFAC segmentation are given in the last two rows of Table IV.
TABLE IV.
Hausdorrf Distances From Balloon Force Active Contour Segmentation After Despeckling With Various Algorithms
| Frame Number | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | Mean | |
| Lee | 7.4902 | 8.5134 | 7.8473 | 6.4972 | 6.4997 | 7.7184 | 7.0269 | 6.7038 | 6.9753 | 7.7989 | 11.0135 | 7.6441 |
| SRAD | 6.2154 | 7.2243 | 9.9661 | 9.6284 | 8.3389 | 10.9107 | 18.6815 | 9.2456 | 5.6883 | 24.7845 | 12.8406 | 11.2295 |
| Wiener | 7.6064 | 8.262 | 7.7059 | 6.8277 | 6.7774 | 8.1183 | 7.4165 | 7.583 | 7.8816 | 8.7228 | 10.5028 | 7.9459 |
| SBF (SD) |
6.6654 (0.0598) |
6.8411 (0.0706) |
7.6376 (0.5716) |
6.5657 (0.0673) |
4.9165 (0.1105) |
5.0478 (0.094) |
5.5436 (0.246) |
7.5547 (0.3727) |
6.3962 (0.1485) |
9.0679 (0.1266) |
10.1526 (0.2651) |
6.9445 |
The Lee, SRAD, Wiener, and SBF filters were tested. Since the performance of each despeckling method is dependent upon the choice of various parameters, the optimal choice of parameters of each despeckling method was determined through trial and error so that the BFAC segmentation of frame 0 was optimal with respect to minimizing the average directed Hausdorff distance of the final red contour to the frame 0 manually defined yellow contour. The results of the trial and error optimization are shown in Fig. 7, where the red contours are the results of the segmentation. The blue contours in Fig. 7 show the evolution of the BFAC, in that each blue contour represents the BFAC at every 50th iteration. That is the most inner contour is the BFAC at iteration 50. Each blue contour outward represents the BFAC after an additional 50 iterations. The optimal parameters of each despeckling algorithm determine by trial and error in frame 0 were used to despeckle the remaining test frames of the ultrasound cine data.
The experiment was performed on frames 0, 10, 20, …, 100 of the in vivo short axis cardiac B-mode cine data. The average directed Hausdorff distance of all tested methods on the tested frames are given in Table IV. The segmentation results of each despeckling method for frames 30, 60, 80, and 100 are shown in Fig. 8.
It should be noted than all tested despeckling algorithm perform well with respect to segmentation of the LV endocardial border. A visual inspection of Fig. 8 shows that the segmentation of the SRAD processed image is the worst. Although SRAD performed well in frame 80, which is shown in Fig. 8(g), the BFAC segmentation results in frame 30, 60, and 100, which are shown in Fig. 8(e), (f), and (h), respectively, poorly reflect the manual segmented yellow contours of each respective frame. The BFAC red contours in these SRAD processed frames consist of erroneous loops and extend well beyond the endocardial border. These errors are most evident in the End Systole frame 60 shown in Fig. 8(f). These errors are reflected in Table IV where the Hausdorff distance of the final BFAC contour of the SRAD despeckled frame 60 is 18.6815. This is the largest Hausdorff distance of all resulting BFAC.
It should also be noticed that all four BFAC segmentations of frame 100 failed to capture the left endocardial border of the LV. This is due to image drop out, weak contrast of edges that are axially parallel to the transducer. This problem is inherent to all ultrasound imaging system. The solution of this problem is beyond the scope of this paper. It is suspected that the segmentation errors caused by the image drop out in frame 100 is the cause of significantly large Hausdorff distances in the second rightmost column of Table IV.
The quantitative results of Table IV shows that SBF despeckling provided a better BFAC segmentation in six out the eleven frames. The SBF despeckling method promoted a better BFAC segmentation more often than when despeckling with the other three methods. The mean average directed Hausdorff distance, listed in the rightmost column of Table IV, is smaller when despeckling with SBF. Further, except in the frame 20 evaluation, the small standard deviation of the average directed Hausdorff distances from contours produced by ten trials of SBF despeckling followed by BFAC segmentation signify that the SBF despeckling promotes consistently accurate BFAC segmentation. When evaluating the segmentation results using frame 20, the Hausdorff distances of the Wiener and Lee filtered images produced contours that are within one standard deviation of the average directed Hausdorff distance produced by the SBF processed image. Thus, it is reasonable to concede that the Lee, Wiener, and SBF despeckling algorithms provided equivalently accurate BFAC segmentation results in frame 20.
V. Discussion
The evaluation in this paper provides evidence that despeckling ultrasound images to promote robust contrast enhancement is possible with the adaptive Lee filter, the linear Wiener method, the anisotropic SRAD method, and the stochastically driven SBF method. The evaluation was based upon the USDSAI and SSIM (or rather MSSSIM) indices produced by each method’s ability to despeckle a Field II simulated image. Although the optimized Wiener filter was able to achieve a USDSAI close to the Lee and SRAD USDSAI performance, the MSSIM index was decreased when compared to the MSSIM index of the unprocessed Field II simulated image. Surprisingly, the SSIM map of the Wiener despeckled simulated image exhibited excellent local similarity in the class composed of the five dark disks, while the remainder of the Wiener processed image exhibited poor local structural similarity. Both SRAD and SBF were able to attain excellent contrast enhancement (high USDSAI) and high MSSIM index. The SRAD and SBF SSIM maps showed poor performance in the class consisting of the five dark disks of varying size. An interesting method that is left for future work would be to despeckle using a hybrid of SBF (or SRAD) with Wiener filtering. This method might consist of replacing a speckle polluted image pixel with a pixel from a Wiener, SBF, or SRAD filtered version of that image.
When testing on actual ultrasound B-mode images, the BFAC was used to determine if the degree of despeckling provide by the Lee, SRAD, Wiener, and SBF methods were sufficient enough to provide a robust segmentation of a mouse LV. Although the BFAC parameters given in Table III were fixed throughout the experiment performed in Section IV.B, it may be possible to conjointly tune the parameters of a despeckling method with the parameters of the BFAC (or other segmentation method) to provide better performance than reported in Section IV.
Great efforts were made in optimizing the tested algorithms. It is foreseeable that optimization of a despeckling algorithm would be dependent of transducer geometry, operating frequency, focal point(s), distribution of pixel values due to speckle, subject being scanned, etc. Automatic selection of optimal despeckling algorithm’s parameters would provide a useful tool for research and clinical applications.
The evaluations provide in this paper are far from being comprehensive and all inclusive. More algorithms need to be tested and for more applications to include contrast enhancement and automatic segmentation. The evaluation should include metric that are based upon visual information, information fidelity, perception, and other novel concepts.
VI. Conclusion
The removal or reduction of speckle while preserving or enhancing edge information of an ultrasound image is an challenging task. An evaluation of various despeckling algorithms is presented. In general, the previously published despeckling methods scrutinize every pixel values and attempt to determine the amount or degree of local or global smoothing to apply. In contrast, an iterative despeckling method, SBF, that at each iteration smoothes only outlying pixel values, is proposed. Outliers are described as local extrema. Without using the outlier values, the SBF method replaces these outliers with the local mean. Each iteration of the proposed method compresses the image pixel values so that the differences in interclass means are preserved or possibly enhanced while the intraclass variance is decreased. The superior contrast enhancement and structural similarity performances of the SBF method are established by experimentation using a Field II simulated ultrasound image and evaluated with the USDSAI and SSIM (or MSSIM) performance metrics.
An evaluation using actual B-mode in vivo cardiac cine data was performed. This evaluation consist of applying a BFAC segmentation to B-mode images despeckled by the Lee, SRAD, Wiener, and SBF methods. Robust segmentation was determined by the lowest average directed Hausdorff distance. Although all tested despeckling algorithms provided robust segmentation of a mouse LV throughout the cardiac cycle, the quantitative results showed that the SBF method consistently and more often provided contours that better resembled the manually defined contours.
Acknowledgments
This work was supported in part by NIH NIBIB grant EB001826, in part by U.S. Army CDMRP grant W81XWH-04-1-0240, and in part by NIH NCRR RR022582 grant. The associate editor coordinating the review of this manuscript and approving it for publication was Dr. Margaret Cheney.
Biographies

Peter C. Tay (S’01–M’03) received the B.S. and M.A. degrees in mathematics and the Ph.D. degree in electrical and computer engineering, all from the University of Oklahoma, Norman, OK.
He is currently an Assistant Professor in the Department of Engineering and Technology, Western Carolina University, Cullowhee, NC. He was an Oak Ridge Institute for Science and Education Research Fellow at the National Weather Service (NWS) and subsequently a Software Engineer for the NWS in Norman, OK. Prior to his current appointment he was a Postdoctoral Research Associate in the Departments of Biomedical Engineering and ECE at the University of Virginia, Charlottesville, VA. His research interests include biomedical imaging and signal/image/video processing for medical applications.

Christopher D. Garson (S’04) received the B.S. degree in biomedical engineering from Duke University, Durham, NC, and the M.S. degree the University of Virginia, Charlottesville.
He is currently self-employed as an independent software developer; projects include webcam-based gaze estimation and the development of interactive video content for mobile phones in American and Chinese markets.

Scott T. Acton(S’89–M’93–SM’99) graduated from Oakton High School, Vienna, Virginia, in 1984. He received the B.S. degree in electrical engineering from Virginia Tech, Blacksburg in 1988 as a Virginia Scholar, and received the M.S. degree in electrical and computer engineering and the Ph.D. degree in electrical and computer engineering from the University of Texas at Austin in 1990 and 1993, respectively, where he was a student of Prof.
Al Bovik. He has worked in industry for AT&T, Oakton, VA, the MITRE Corporation, McLean, VA and Motorola, Inc., Phoenix, AZ and in academia for Oklahoma State University, Stillwater. Currently, he is Professor of Electrical and Computer Engineering and Biomedical Engineering at the University of Virginia (UVA). At UVA, he was named the Outstanding New Teacher in 2002, Faculty Fellow in 2003, and Walter N. Munster Chair for Intelligence Enhancement in 2003.
Dr. Acton is an active participant in the IEEE, currently serving as Associate Editor for the IEEE transactions on image processing and, formerly, as Associate Editor the IEEE signal processing letters. He was the 2004 Technical Program Chair and the 2006 General Chair for the Asilomar Conference on Signals, Systems and Computers. His research interests include anisotropic diffusion, active models, basketball, biomedical segmentation problems and biomedical tracking problems. From 2007–2008, he was on sabbatical in Santa Fe, NM.

John A. Hossack (S’90–M’92–SM’02) received the B. Eng. and Ph.D. degrees from the University of Strathclyde, Glascow, U.K.
He is currently a Professor in the Department of Biomedical Engineering and Department of Electrical and Computer Engineering, University of Virginia, Charlottesville. Previously, he was a postdoctoral researcher in the E. L. Ginzton Laboratory at Stanford University and then a Design Engineer, and later Technical Fellow, at Acuson Corp in Mountain View, CA. He is active Vice President, Publications for the Ultrasonics, Ferroelectrics and Frequency Control Society at IEEE and is an Associate Editor for the transactions on ultrasonics, ferroelectrics and frequency control. He was Technical Program Chair at the 2005 IEEE Ultrasonics Symposium. His current research interests relate to ultrasonic transducer design (including intravascular devices), mouse heart ultrasound imaging, cardiac ultrasound image quantification, and the use of ultrasound for image guiding and effecting focal drug/gene delivery in the context of management of arterial disease.
Footnotes
Some algorithms utilize the features of speckle and do not consider speckle as noise. The use of the term noise in the context of this paper is based upon the visual observation that speckle may cloud visual or automated detection of edges and significant borders of the object being imaged.
Independence is in the sense that prob(ak(n, m), al(n, m)) = prob(ak(n, m)) prob(al(n, m)), similarly for prob(φk(n, m), φl(n, m)) and prob(ak(n, m), φk(n, m)).
The best method to obtain global optimality of each algorithm is beyond the scope of this paper.
The location of the transducer is at the bottom of all the mouse heart images that are shown in this paper.
Contributor Information
Peter C. Tay, Department of Engineering and Technology, Western Carolina University, Cullowhee, NC 28723 USA (ptay@email.wcu.edu).
Christopher D. Garson, Independent software developer and was a student with the Department of Biomedical Engineering, University of Virginia, Charlottesville, VA 22904 USA (cdg6d@virginia.edu).
Scott T. Acton, Department of Electrical and Computer Engineering and also the Department of Biomedical Engineering, University of Virginia, Charlottesville, VA 22904 USA acton@virginia.edu.
John A. Hossack, Department of Electrical and Computer Engineering and also the Department of Biomedical Engineering, University of Virginia, Charlottesville, VA 22904 USA hossack@virginia.edu.
References
- [1].Goodman JW. Statistical properties of laser speckle patterns. In: Dainty JC, editor. Laser Speckle and Related Phenomena. Springer-Verlag; Berlin, Germany: 1984. pp. 9–75. [Google Scholar]
- [2].Abbott JG, Thurstone FL. Acoustic speckle: Theory and experimental analysis. Ultrason. Imaging. 1979;1:303–324. doi: 10.1177/016173467900100402. [DOI] [PubMed] [Google Scholar]
- [3].Wagner RF, Smith SW, Sandrik JM, Lopez H. Statistics of speckle in ultrasound B-scans. IEEE Trans. Sonics Ultrason. 1983 May;SU–30(3):156–163. [Google Scholar]
- [4].Wagner RF, Insana MF, Smith SW. Fundamental correlation lengths of coherent speckle in medical ultrasonic images. IEEE Trans. Ultrason., Ferroelectr., Freq. Control. 1988 Jan;35(1):34–44. doi: 10.1109/58.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Abreu E, Lightstone M, Mitra SK, Arakawa K. A new efficient approach for the removal of impulse noise from highly corrupted images. IEEE Trans. Image Process. 1996 Jun;5(6):1012–1025. doi: 10.1109/83.503916. [DOI] [PubMed] [Google Scholar]
- [6].Nagao M, Matsuyama T. Edge preserving smoothing. Comput. Graph. Image Process. 1979 Apr.9(4):394–407. [Google Scholar]
- [7].Lee JS. Digital image enhancement and noise filtering by use of local statistics. IEEE Trans. Pattern Anal. Mach. Intell. 1980 Mar.PAMI-2(2):165–168. doi: 10.1109/tpami.1980.4766994. [DOI] [PubMed] [Google Scholar]
- [8].Frost VS, Stiles JA, Shanmugan KS, Holtzman JC. A model for radar images and its application to adaptive digital filtering of multiplicative noise. IEEE Trans. Pattern Anal. Mach. Intell. 1982 Mar;PAMI-4(2):157–166. doi: 10.1109/tpami.1982.4767223. [DOI] [PubMed] [Google Scholar]
- [9].Kuan DT, Sawchuk AA, Strand TC, Chavel P. Adaptive noise smoothing filter for images with signal-dependent noise. IEEE Trans. Pattern Anal. Mach. Intell. 1985 Mar;PAMI-7(2):165–177. doi: 10.1109/tpami.1985.4767641. [DOI] [PubMed] [Google Scholar]
- [10].Kuan DT, Sawchuk AA, Strand TC, Chavel P. Adaptive restoration of images with speckle. IEEE Trans. Acoust., Speech, Signal Process. 1987 Mar;ASSP-35(3):373–383. [Google Scholar]
- [11].Loupas T, McDicken WN, Allan PL. An adaptive weighted median filter for speckle suppression in medical ultrasonic images. IEEE Trans. Circuits Syst. 1989 Jan;36(1):129–135. [Google Scholar]
- [12].Yu Y, Acton ST. Speckle reducing anisotropic diffusion. IEEE Trans. Image Process. 2002 Nov;11(11):1260–1270. doi: 10.1109/TIP.2002.804276. [DOI] [PubMed] [Google Scholar]
- [13].Karaman M, Kutay MA, Bozdagi G. An adaptive speckle suppression filter for medical ultrasonic imaging. IEEE Trans. Med. Imag. 1995 Jun;14(2):283–292. doi: 10.1109/42.387710. [DOI] [PubMed] [Google Scholar]
- [14].Hao SGX, Gao X. A novel multiscale nonlinear thresholding method for ultrasonic speckle suppressing. IEEE Trans. Med. Imag. 1999 Sep;18(9):797–794. doi: 10.1109/42.802756. [DOI] [PubMed] [Google Scholar]
- [15].Achim A, Bezerianos A, Tsakalides P. Novel Bayesian multiscale method for speckle removal in medical ultrasound images. IEEE Trans. Med. Imag. 2001 Aug;20(8):772–783. doi: 10.1109/42.938245. [DOI] [PubMed] [Google Scholar]
- [16].Lopes A, Touzi R, Nezry E. Adaptive speckle filters and scene heterogeneity. IEEE Trans. Geosci. Remote Sens. 1990 Nov;28(6):992–1000. [Google Scholar]
- [17].Michailovich OV, Tannenbaum A. Despeckling of medical ultrasound images. IEEE Trans. Ultrason., Ferroelectr., Freq. Control. 2006 Jan;53(1):64–78. doi: 10.1109/tuffc.2006.1588392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dutt V, Greenleaf J. Adaptive speckle reduction filter for log-compressed B-scan images. IEEE Trans. Med. Imag. 1996 Dec;15(6):802–813. doi: 10.1109/42.544498. [DOI] [PubMed] [Google Scholar]
- [19].Tay PC, Acton ST, Hossack JA. A stochastic approach to ultrasound despeckling; Proc. IEEE Int. Symp. Biomedical Imaging; Arlington, Virginia. Apr. 6–9, 2006.pp. 907–910. [Google Scholar]
- [20].Aysal TC, Barner KE. Rayleigh-maximum-likelihood filtering for speckle reduction of ultrasound images. IEEE Trans. Med. Imag. 2007 May;26(5):712–727. doi: 10.1109/TMI.2007.895484. [DOI] [PubMed] [Google Scholar]
- [21].Gupta N, Swamy MNS, Plotkin E. Despeckling of medical ultrasound images using data and rate adaptive lossy compression. IEEE Trans. Med. Imag. 2005 Jun;24(6):743–754. doi: 10.1109/TMI.2005.847401. [DOI] [PubMed] [Google Scholar]
- [22].Rabbani H, Vafadust M, Abolmaesumi P, Gazor S. Speckle noise reduction of medical ultrasound images in complex wavelet domain using mixture priors. IEEE Trans. Biomed. Eng. 2008 Sep;55(9):2152–2160. doi: 10.1109/TBME.2008.923140. [DOI] [PubMed] [Google Scholar]
- [23].Xie J, Jiang Y, Tsui HT, Heng PA. Boundary enhancement and speckle reduction for ultrasound images via salient structure extraction. IEEE Trans. Biomed. Eng. 2006 Nov;53(11):2300–2309. doi: 10.1109/TBME.2006.878088. [DOI] [PubMed] [Google Scholar]
- [24].Yeoh WS, Zhang C. Constrained least squares filtering algorithm for ultrasound image deconvolution. IEEE Trans. Biomed. Eng. 2006 Oct;53(10):2001–2007. doi: 10.1109/TBME.2006.881781. [DOI] [PubMed] [Google Scholar]
- [25].Munteanu C, Morales FC, Ruiz-Alzola J. Speckle reduction through interactive evolution of a general order statistics filter for clinical ultrasound imaging. IEEE Trans. Biomed. Eng. 2008 Jan;55(1):365–369. doi: 10.1109/TBME.2007.897833. [DOI] [PubMed] [Google Scholar]
- [26].Abd-Elmoniem KZ, Youssef ABM, Kadah YM. Real-time speckle reduction and coherence enhancement in ultrasound imaging via nonlinear anisotropic diffusion. IEEE Trans. Biomed. Eng. 2002 Sep;49(9):997–1014. doi: 10.1109/TBME.2002.1028423. [DOI] [PubMed] [Google Scholar]
- [27].Sanchez JR, Oelze ML. An ultrasonic imaging speckle-suppression and contrast-enhancement technique by means of frequency compounding and coded excitation. IEEE Trans. Ultrason., Ferroelectr., Freq. Control. 2009 Jul;56(7):1327–1339. doi: 10.1109/TUFFC.2009.1189. [DOI] [PubMed] [Google Scholar]
- [28].Jirik R, Taxt T. High-resolution ultrasonic imaging using two-dimensional homomorphic filtering. IEEE Trans. Ultrason., Ferroelectr., Freq. Control. 2006 Aug;53(8):1440–1448. [PubMed] [Google Scholar]
- [29].Loizou CP, Pattichis C, Christodoulou CI, Istepanian RSH, Pantziaris M, Nicolaides A. Comparative evaluation of despeckle filtering in ultrasound imaging of the carotid artery. IEEE Trans. Ultrason., Ferroelectr., Freq. Control. 2005 Oct;52(10):1653–1669. doi: 10.1109/tuffc.2005.1561621. [DOI] [PubMed] [Google Scholar]
- [30].Sanches JM, Nascimento JC, Marques JS. Medical image noise reduction using the Sylvester-Lyapunov equation. IEEE Trans. Image Process. 2008 Sep;17(9):1522–1539. doi: 10.1109/TIP.2008.2001398. [DOI] [PubMed] [Google Scholar]
- [31].Fisher RA. The use of multiple measurements in taxonomic problems. Ann. Eugenics. 1936;7:179–188. [Google Scholar]
- [32].Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004 Apr;13(4):600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- [33].Arsenault HH, April G. Properties of speckle integrated with a finite aperture and logarithmically transformed. J. Opt. Soc. Amer. 1976 Nov;66(11):1160–1163. [Google Scholar]
- [34].Kailath T. Equations of Wiener-Hopf type in filtering theory and related applications. In: Masani P, editor. Norbert Wiener: Collected Works vol. III. MIT Press; Cambridge, MA: 1976. pp. 63–94. [Google Scholar]
- [35].Perona P, Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 1990 Jul;12(7):629–639. [Google Scholar]
- [36].Jensen JA, Svendsen NB. Calculation of pressure fields from arbitrarily shaped apodized and excited ultrasound transducers. IEEE Trans. Ultrason., Ferroelectr., Freq. Control. 1992 Mar;39(2):262–267. doi: 10.1109/58.139123. [DOI] [PubMed] [Google Scholar]
- [37].Jensen JA. Field: A program for simulating ultrasound systems. Med. Biol. Eng. Comput. 1996;34(1):351–353. [Google Scholar]
- [38].Xu C, Prince JL. Snakes, shapes, and gradient vector flow. IEEE Trans. Image Process. 1998 Mar;7(3):359–369. doi: 10.1109/83.661186. [DOI] [PubMed] [Google Scholar]
- [39].Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 1993 Sep;15(9):850–863. [Google Scholar]