Abstract
Objectives
To characterize the lipid profile of individuals with newly diagnosed type 1 diabetes mellitus using LC-MS-based lipidomics and the accurate mass and time (AMT) tag approach.
Design and methods
Lipids were extracted from plasma and sera of 10 subjects from the Diabetes Antibody Standardization Program (years 2000–2005) and 10 non-diabetic subjects and analyzed by capillary liquid chromatography coupled with a hybrid ion-trap-Fourier transform ion cyclotron resonance mass spectrometer. Lipids were identified and quantified using the AMT tag approach.
Results
Five hundred fifty-nine lipid features differentiated (q < 0.05) diabetic from healthy individuals in a partial least-squares analysis, characterizing of individuals with recently diagnosed type 1 diabetes mellitus.
Conclusions
A lipid profile associated with newly diagnosed type 1 diabetes may aid in further characterization of biochemical pathways involved in lipid regulation or mobilization.
Keywords: lipidomics, AMT tag approach, capillary liquid chromatography, mass spectrometry, type 1 diabetes, Diabetes Antibody Standardization Program
Introduction
Type 1 diabetes mellitus (T1DM) is a chronic, autoimmune disease affecting over 1 million individuals in the United States alone and is characterized by insulin deficiency as a result of pancreatic beta-cell death. At clinical diagnosis, only about 10% of beta-cell mass remains [1] and their protection (at and prior to hyperglycemia onset) and regeneration are active areas of research interest. Currently, the best approach for predicting who may be at risk for developing T1DM before onset of clinical symptoms is by measurement of autoantibodies to islet cell antigens [2–4].
We previously conducted a global proteomics analysis of plasma and serum samples from the Diabetes Antibody Standardization Program (DASP) in an attempt to identify novel protein biomarkers of T1DM. The DASP is a collaborative agreement between the U.S. Centers for Disease Control and Prevention’s (CDC) National Diabetes Laboratory and the Immunology of Diabetes Society, and has as its goal the periodic evaluation (since 2000) of assays for islet cell autoantibodies by selected laboratories. In our proteomics study, we reported the identification of five candidate protein markers of recently diagnosed T1DM in a DASP sample subset [5]. In particular, zinc alpha-2-glycoprotein (ZAG) was strongly upregulated in individuals with T1DM. ZAG is a member of the immunoglobulin superfamily [6] and displays lipid mobilization activity [7, 8]. Patient samples collected as part of the DASP study correspond to recently diagnosed individuals (within 14 days of starting insulin treatment); thus, increased levels of ZAG in patient relative to control samples may be an indication of a system-wide mobilization of lipids for energy production [9–11]. We therefore hypothesized that perturbations may be present in the components of the blood lipidome of individuals with newly diagnosed T1DM.
To evaluate this hypothesis, we performed lipidomics analyses using the same DASP samples in an attempt to identify perturbations in the lipids of individuals with recently diagnosed T1DM, as well as to potentially identify a lipid profile predictive or diagnostic of the disease, and which may also reflect beta-cell lipotoxins. We used capillary liquid chromatography (LC) coupled with Fourier transform ion cyclotron resonance (FTICR) mass spectrometry (MS) and the accurate mass and time (AMT) tag approach [12] to identify and quantify lipids present in healthy and diabetic individuals. The AMT tag approach relies on initial, low-throughput shotgun LC-MS/MS analyses to populate a database of identified molecules. The identified species, or AMT tags, are annotated with their associated calculated monoisotopic mass and LC normalized elution time (NET) information, among other parameters. The AMT tag strategy is based on the uniqueness of the measured molecular mass and LC retention time for a specific lipid and makes use of the fact that the probability will be low that “new” species detected in additional analyses of the same or similar biological sample will be observed at the same mass and LC retention time as a previously assigned species. Subsequent high-throughput analyses using LC-MS with high-mass measurement accuracy allows molecules to be identified by matching their measured monoisotopic masses and NETs to those of the entries in the AMT tag database within user-defined mass and NET tolerances. By performing the subsequent analyses with high-mass measurement accuracy instrumentation and avoiding additional MS/MS experiments for lipid identification, the dynamic range of detection is greatly expanded and allows for detection of low abundance species that would otherwise not be observed by MS/MS. Our preliminary lipid AMT tag database [13] contains over 250 lipids that were identified from human plasma, erythrocytes and lymphocytes. While the coverage, in terms of number of identified species, of the human blood lipidome represented by our lipid AMT tag database is modest, it provides the means to define the lipid profiles among comparative samples in preliminary, proof-of-principle studies. Herein, we report the identification of perturbations in the lipid profile of individuals with recently diagnosed T1DM, providing evidence of altered lipid metabolism in patients enrolled in the DASP.
Methods and Materials
Human plasma and serum samples
The DASP is conducted in accordance with the Human Subjects policies and regulations of the CDC. Similarly, this work was approved by the Institutional Review Board of the Pacific Northwest National Laboratory. Human plasma and serum samples from the DASP (years 2000–2005) were received frozen on dry ice; these samples (Table 1) correspond to healthy control individuals (n = 10) and patients recently diagnosed with type 1 diabetes (n = 10). The patient samples were collected from donors under the age of 30 within 14 days of starting exogenous insulin treatment [14] before insulin antibodies (as opposed to insulin autoantibodies) were induced. The control individuals were self-reporting healthy blood donors.
Table 1.
Control and patient demographic data.
| Status | Race | Gender | Age |
|---|---|---|---|
| Control | Caucasian | Male | 18 |
| Control | Caucasian | Female | 18 |
| Control | Caucasian | Female | 19 |
| Control | Black | Female | 20 |
| Control | Hispanic | Female | 20 |
| Control | Caucasian | Male | 21 |
| Control | Caucasian | Female | 21 |
| Control | Black | Female | 22 |
| Control | Caucasian | Female | 23 |
| Control | Caucasian | Male | 26 |
| Patient | Caucasian | Female | 10 |
| Patient | Caucasian | Female | 12 |
| Patient | Caucasian | Male | 12 |
| Patient | Caucasian | Male | 12 |
| Patient | Caucasian | Male | 13 |
| Patient | Caucasian | Male | 16 |
| Patient | Caucasian | Female | 16 |
| Patient | Hispanic | Female | 18 |
| Patient | Caucasian | Female | 20 |
| Patient | Caucasian | Female | 29 |
Age Gender Race Status
Serum glucose, total cholesterol, high-density lipoprotein (HDL)-cholesterol, low-density lipoprotein (LDL)-cholesterol, and triglycerides were determined (Table 2) by enzymatic assays at the Northwest Lipid Metabolism and Diabetes Research Laboratory at the University of Washington (Seattle, WA), using appropriate Roche reagents (e.g. hexokinase for glucose) and a Roche Double Modular P Analytics automated analyzer (Roche Diagnostics, Indianapolis, IN). The Roche methods are standardized to the CDC Reference Methods.
Table 2. Glucose and lipid values for control and patient individuals.
Values are mean ± standard error. n.s., not significant.
| Control (n = 10) | Patient (n = 10) | Significance | |
|---|---|---|---|
| Glucose (mg/dL) | 83 ± 7 | 200±29 | P < 0.001 |
| Total Cholesterol (mg/dL) | 140 ± 10 | 150±24 | n.s. |
| HDL cholesterol | 24 ± 1 | 27±5 | n.s. |
| LDL cholesterol | 93 ± 14 | 90±17 | n.s. |
| Triglycerides | 112 ± 19 | 102±29 | n.s. |
Lipid extraction
Plasma and serum lipids were extracted in triplicate via the addition of 100 μL cold (−20ºC) chloroform/methanol (2:1, v/v) to 20 μL plasma [13], and the mixture was vortexed for 10 s. The sample was then allowed to stand at 4ºC for 15 min, followed by vortexing for 10 s. Protein was separated from the two liquid phases by centrifugation at 13,400 g for 10 min. The lower chloroform phase was removed by pipetting and placed into a sterile, siliconized eppendorf tube, while the protein interlayer and upper aqueous phase were discarded. The chloroform phase was then dried in vacuo and stored at −80ºC until analysis. Prior to analysis, dried lipid extracts were reconstituted in 60 μL methanol and centrifuged at 13,400g for 5 min to remove any particulates. Our previous studies have indicated no significant difference in the solubility of triglycerides, cholesterol esters, or related lipids containing longer chain fatty acids when using methanol, the Bligh and Dyer solvent (chloroform/methanol/water, 1/2/0.8, v/v/v), or a commonly used solvent for shotgun (infusion) lipidomics (chloroform/methanol/water, 1/2.2/0.12, v/v/v) for reconstitution of dried plasma lipid extracts.
Reversed-phase capillary LC-FTICR analyses
An automated LC system with two 150 μm x 65 cm capillary columns was used, as previously described [13]. All samples in this study were analyzed on the same capillary column in random order. The mobile phases were (A) 10 mM ammonium acetate in 50:50 water/methanol (v/v) and (B) 10 mM ammonium acetate in 50:50 methanol/acetonitrile (v/v). The LC system was equilibrated at 6,000 psi with mobile phase A prior to injecting 1 μL of sample. Exponential gradient elution was initiated 3 min after sample loading with an initial column flow of ~1 μL/min. After 90 min of gradient separation, the mobile phase mixer was purged with 3 mL of mobile phase B, followed by a 5 min column wash. Finally, the mobile phase mixer was purged with 10 mL of mobile phase A, which represented the end of one separation cycle. While gradient elution is performed on one column, the other column is equilibrated with mobile phase A.
The capillary LC system was coupled to a hybrid linear ion-trap-Fourier transform ion cyclotron resonance (FTICR) mass spectrometer (LTQ-FT, ThermoFisher, San Jose, CA). The capillary temperature and electrospray voltage were 200ºC and +2.2 kV, respectively. The FT was used as the mass analyzer over the m/z range 100–1000, with a duty cycle of ~1.0 s and mass resolution of 100,000.
Processing of quantitative LC-FTICR datasets
LC-FTICR datasets, defined as the data obtained from a single LC-FTICR analysis, were processed using the PRISM Data Analysis system [15], a series of software tools freely available at http://ncrr.pnl.gov/software/ and developed in-house. The first step involved deisotoping of the raw MS data to give the monoisotopic mass, charge state, and intensity of the major peaks in each mass spectrum using Decon2LS [16]. The data were next examined in a 2-D fashion using MultiAlign to identify groups of mass spectral peaks that were observed in sequential spectra using an algorithm that computes a Euclidean distance in n-dimensional space for combinations of peaks. Each group, generally ascribed to one detected species and referred to as a “feature”, has a median monoisotopic mass, central normalized elution time (NET), and abundance estimate computed by summing the intensities of the MS peaks that comprise the entire LC-FTICR feature. LC-FTICR features were then chromatographically aligned across all 60 datasets using the LCMSWARP algorithm [17] in MultiAlign, and the lipid identities of detected features were determined by comparing their measured monoisotopic masses and NETs to calculated monoisotopic masses and observed NETs for lipids in an AMT tag database [13] within search tolerances of ±3 ppm and ±0.03 NET for monoisotopic mass and elution time, respectively.
Statistical analysis of processed LC-MS data
Following chromatographic alignment and database matching, the abundances of all detected features (both AMT tag database matched and unmatched) were loaded into DAnTE [18] for statistical analysis. Feature abundances were transformed to log2 scale then subjected to central tendency normalization [19]. Comparative data analysis was then performed on lipid features that were observed in a minimum of two out of three technical replicates in at least eight of the ten individuals per sample type (control and patient) a so-called minimum observation filter. It is important to note that, for most lipid features within a sample type, more observations than the required minimum were present (e.g. a given lipid feature was detected in at least 2 out of 3 technical replicates for 95.2% and 92.6% of control and patient samples, respectively). Statistically significant differences between the lipid profiles of patient and healthy control individuals were determined using ANOVA. Partial least-squares (PLS) [20] was also performed using the data matrices containing either AMT tag database matched features alone or all features (both database matched and unmatched) that met the minimum observation threshold described above, in order to identify lipid profiles characteristic of T1DM.
Results
Summary of data
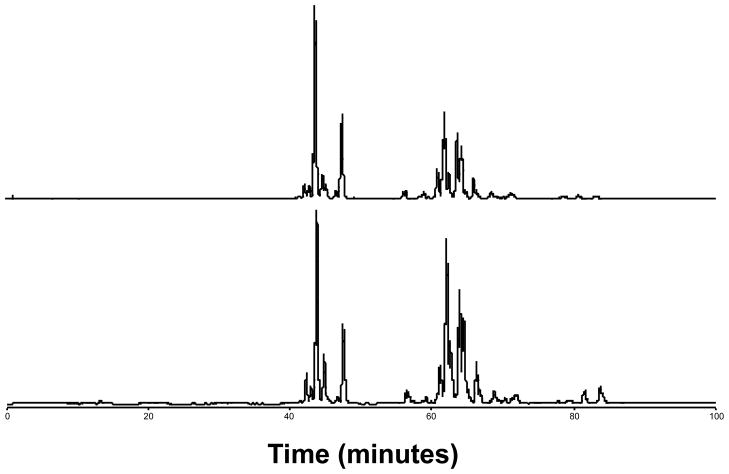
Representative LC-FTICR chromatograms (Figure 1) of healthy control and patient lipid extracts showed that the respective lipid profiles were generally similar. However, lipid peak intensities in some regions do appear to differ between the two individuals, although it is difficult to ascertain whether these differences are due to biological variation or the presence of T1DM. Therefore, the data from all control and patient lipid extracts were processed using the AMT tag approach in order to identify those lipids whose differential abundance could be ascribed to T1DM.
Figure 1. Representative LC-FTICR base peak ion chromatograms.
Shown are chromatograms of lipid extracts from control (top) and patient (bottom) individuals.
Chromatographic alignment of LC-FTICR datasets produced a data matrix containing 24,323 individual features that were observed in as few as 1 dataset to as many as 60. It is important to note that the data matrix included both lipid features and features resulting from chemical noise. In general, the abundances of lipid features among technical replicates for each individual were very similar, based on Pearson correlation coefficient (Table 3). These values are a reflection of the day-to-day precision of our method, since the average time between the first and third technical replicate was ~16 hr, and the entire time required for the analysis of all samples was 10 days. However, Replicate B for Patient 10 appeared to be an outlier. Inspection of the raw data corresponding to this sample revealed that the intensity of the total ion chromatogram (TIC) was lower by an order of magnitude than the average intensities of the TICs from the other samples (data not shown), likely due to a pipetting error during sample preparation or an injection error during sample analysis; otherwise, the chromatography appeared normal for this replicate. The lower TIC intensity for this sample resulted in fewer identified features: 2503 versus an average of 4441 for the other 59 samples. Indeed, the data was found to be an outlier with a p < 0.01 when using Grubb’s test for outliers, based on the total number of lipid features identified in each LC-FTICR dataset. Thus, data from this replicate were excluded from further downstream processing and analysis to avoid compromising the final results.
Table 3.
Pearson correlation coefficients between replicate analyses from each individual.
| Rep A/Rep B | Rep A/Rep C | Rep B/Rep C | mean | |
|---|---|---|---|---|
| Control 1 | 0.85 | 0.90 | 0.93 | 0.89 |
| Control 2 | 0.92 | 0.95 | 0.93 | 0.93 |
| Control 3 | 0.90 | 0.90 | 0.93 | 0.91 |
| Control 4 | 0.93 | 0.94 | 0.95 | 0.94 |
| Control 5 | 0.91 | 0.95 | 0.92 | 0.93 |
| Control 6 | 0.90 | 0.93 | 0.94 | 0.92 |
| Control 7 | 0.94 | 0.92 | 0.94 | 0.93 |
| Control 8 | 0.92 | 0.94 | 0.95 | 0.94 |
| Control 9 | 0.90 | 0.82 | 0.87 | 0.86 |
| Control 10 | 0.95 | 0.95 | 0.97 | 0.96 |
| Patient 1 | 0.84 | 0.88 | 0.79 | 0.84 |
| Patient 2 | 0.96 | 0.96 | 0.95 | 0.96 |
| Patient 3 | 0.89 | 0.96 | 0.89 | 0.91 |
| Patient 4 | 0.96 | 0.96 | 0.95 | 0.96 |
| Patient 5 | 0.86 | 0.92 | 0.92 | 0.90 |
| Patient 6 | 0.90 | 0.94 | 0.90 | 0.91 |
| Patient 7 | 0.92 | 0.93 | 0.92 | 0.92 |
| Patient 8 | 0.91 | 0.95 | 0.91 | 0.92 |
| Patient 9 | 0.86 | 0.91 | 0.83 | 0.87 |
| Patient 10 | 0.60 | 0.91 | 0.61 | 0.71 |
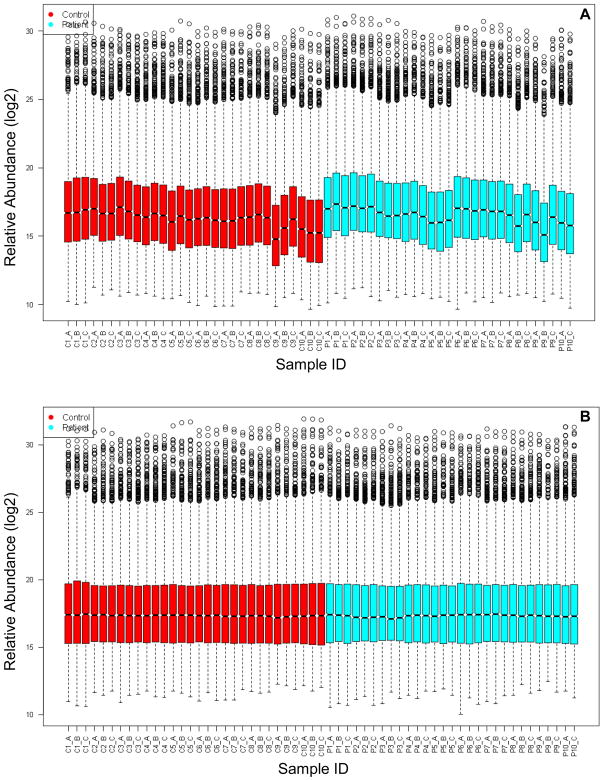
Central tendency normalization was used to correct for systematic biases in the data caused by changes in LC-MS performance over time (Figure 2), and subsequent application of the minimum observation filter resulted in a final data matrix containing 2014 lipid features (both AMT tag database matched and unmatched). The minimum observation filter served to retain as many reproducibly observed lipid features as reasonable while filtering out those lipid features due to chemical noise. ANOVA was then performed to identify those lipid features (both AMT tag database matched and unmatched) that were significantly different between control and diabetic samples. This analysis revealed 559 candidate lipid markers of T1DM with q < 0.05 (Supplemental Table 1), of which 55 matched entries in the lipid AMT tag database (Table 4). The q value is a false discovery rate (FDR)-based measure of significance for genome-wide studies [21] and is essentially an adjusted p value. Thus a q value less than 0.05 indicates a FDR below 5%.
Figure 2. Box plots of detected lipid feature abundances.
Relative abundances of detected lipid features (log2 scale) are shown for 59 samples analyzed by LC-FTICR. Each box in the plot describes the abundance distribution (log2 scale) of an average of 4441 individual features based on five-number summaries: the smallest observation, lower quartile, median, upper quartile, and largest observation. Replicate B for Patient 10 is not shown and was removed as an outlier prior to normalization. (A) Before normalization. (B) After central tendency normalization. C: control; P: patient.
Table 4. Identities of significant features matching the lipid AMT tag database.
Those features matching the lipid AMT tag database and identified as significantly different (ANOVA, q < 0.05) are reported.
| Lipid Identity | Monoisotopic mass | NET | q value | Control abundance (mean ± std error) | Patient abundance (mean ± std error) |
|---|---|---|---|---|---|
| LPC 14:0 | 467.3019 | 0.33 | 0.041 | 22.9 ± 0.3 | 23.7 ± 0.1 |
| LPC 18:0 | 523.3646 | 0.42 | < 0.001 | 30.5 ± 0.1 | 29.8 ± 0.1 |
| LPC 18:2 | 519.3335 | 0.36 | 0.009 | 27.8 ± 0.3 | 28.9 ± 0.1 |
| LPC 18:3 | 517.3173 | 0.29 | 0.002 | 21.2 ± 0.4 | 23.0 ± 0.2 |
| LPC 20:3 | 545.3484 | 0.40 | < 0.001 | 26.0 ± 0.1 | 25.2 ± 0.1 |
| LPC 20:5 | 541.3162 | 0.36 | 0.037 | 25.2 ± 0.4 | 26.2 ± 0.2 |
| LPC 22:0 | 579.4273 | 0.49 | 0.006 | 18.5 ± 0.2 | 18.0 ± 0.1 |
| LPC 22:6 | 567.3331 | 0.37 | 0.010 | 22.6 ± 0.6 | 24.7 ± 0.1 |
| PC 30:0 | 705.5343 | 0.57 | < 0.001 | 21.6 ± 0.2 | 23.0 ± 0.1 |
| PC 32:0 | 733.5653 | 0.62 | 0.048 | 25.7 ± 0.1 | 26.0 ± 0.1 |
| PC 32:1 | 731.5490 | 0.57 | < 0.001 | 24.6 ± 0.2 | 25.8 ± 0.1 |
| PC 32:2 | 729.5335 | 0.55 | 0.004 | 22.4 ± 0.3 | 23.4 ± 0.1 |
| PC 34:0 | 761.5949 | 0.63 | 0.043 | 24.1 ± 0.1 | 23.7 ± 0.1 |
| PC 34:2 | 757.5650 | 0.53 | 0.001 | 20.8 ± 0.2 | 21.5 ± 0.1 |
| PC 36:4 | 781.5643 | 0.53 | 0.003 | 22.1 ± 0.2 | 22.7 ± 0.1 |
| PC 36:5 | 779.5480 | 0.57 | 0.015 | 23.9 ± 0.1 | 24.5 ± 0.1 |
| PC 38:3 | 811.6093 | 0.63 | 0.021 | 27.2 ± 0.1 | 26.7 ± 0.2 |
| PC 38:4 | 809.5947 | 0.61 | 0.015 | 28.6 ± 0.2 | 27.8 ± 0.2 |
| PC 38:5 | 807.5791 | 0.58 | 0.022 | 27.2 ± 0.2 | 26.3 ± 0.2 |
| PC 40:4 | 837.6270 | 0.63 | 0.005 | 23.1 ± 0.2 | 21.8 ± 0.3 |
| PC 40:5 | 835.6117 | 0.61 | 0.003 | 24.0 ± 0.2 | 22.5 ± 0.4 |
| PC (16:0e/18:2;16:0p/18:1) | 743.5849 | 0.61 | < 0.001 | 23.1 ± 0.1 | 23.8 ± 0.1 |
| PC (16:0p/20:1;16:0e/20:2) | 771.6158 | 0.64 | 0.032 | 23.0 ± 0.1 | 22.5 ± 0.1 |
| PC (16:0p/22:2; 16:0e/22:3) | 797.6315 | 0.64 | 0.017 | 21.2 ± 0.2 | 20.4 ± 0.2 |
| PC (16:0p/22:4;16:0e/22:5) | 793.6004 | 0.61 | 0.028 | 25.6 ± 0.2 | 24.8 ± 0.3 |
| PC (18:0p/22:4;18:0e/22:5) | 821.6322 | 0.63 | 0.006 | 22.7 ± 0.2 | 21.4 ± 0.3 |
| SM (d16:1/24:0) | 786.6639 | 0.66 | 0.003 | 26.9 ± 0.0 | 26.7 ± 0.1 |
| SM (d18:1/18:0) | 730.5999 | 0.61 | 0.004 | 26.8 ± 0.1 | 26.3 ± 0.1 |
| SM (d18:1/18:1) | 728.5856 | 0.58 | < 0.001 | 26.1 ± 0.1 | 25.3 ± 0.1 |
| SM (d18:1/18:2) | 726.5701 | 0.56 | < 0.001 | 23.4 ± 0.1 | 21.9 ± 0.1 |
| SM (d18:1/20:0) | 758.6317 | 0.64 | 0.015 | 26.2 ± 0.1 | 26.0 ± 0.1 |
| SM (d18:1/20:1) | 756.6165 | 0.61 | 0.003 | 24.9 ± 0.1 | 24.3 ± 0.1 |
| SM (d18:1/24:1) | 812.6788 | 0.68 | 0.030 | 28.1 ± 0.0 | 27.8 ± 0.1 |
| SM (d18:1/24:2) | 810.6621 | 0.65 | 0.001 | 27.6 ± 0.1 | 27.0 ± 0.1 |
| SM (d20:1/22:3) | 808.6475 | 0.62 | < 0.001 | 24.3 ± 0.1 | 23.1 ± 0.2 |
| Oxidized PC (1-16:0/2-O-Hydroxy-7:2) / (16:0/7:1) | 665.4292 | 0.40 | 0.012 | 21.0 ± 0.1 | 21.6 ± 0.1 |
| Oxidized PC (16:0/Hydroxy-18:2) | 773.5588 | 0.52 | 0.001 | 27.7 ± 0.2 | 28.6 ± 0.1 |
| Oxidized PC (1-16:0/2-Hydroxy-18:3) | 831.5654 | 0.48 | 0.012 | 18.8 ± 0.2 | 19.9 ± 0.3 |
| Oxidized PC (16:0/Hydroperoxy-18:3) | 787.5387 | 0.48 | 0.010 | 23.3 ± 0.2 | 24.3 ± 0.3 |
| Oxidized PC (1-18:0/2-C7-oxo) | 649.4338 | 0.43 | 0.004 | 23.2 ± 0.2 | 24.1 ± 0.2 |
| Oxidized PC (18:0/Hydroxy-18:3) | 799.5748 | 0.52 | 0.032 | 25.3 ± 0.1 | 26.1 ± 0.3 |
| Oxidized PC (1-20:0/2-C7-oxo) | 677.4649 | 0.45 | 0.004 | 21.8 ± 0.2 | 22.8 ± 0.2 |
| LPE 16:0 | 453.2869 | 0.37 | 0.001 | 20.9 ± 0.3 | 22.2 ± 0.1 |
| LPE 18:2 | 477.2871 | 0.36 | 0.032 | 20.7 ± 0.5 | 22.0 ± 0.1 |
| PE 36:2 | 743.5502 | 0.57 | < 0.001 | 22.8 ± 0.2 | 23.9 ± 0.1 |
| PE 38:5 | 765.5346 | 0.57 | 0.017 | 18.9 ± 0.4 | 17.6 ± 0.3 |
| TAG 46:1 | 793.7166 | 0.78 | 0.020 | 23.2 ± 0.2 | 23.8 ± 0.2 |
| TAG 50:4 | 843.7342 | 0.76 | 0.033 | 22.3 ± 0.1 | 21.7 ± 0.2 |
| TAG 54:4 | 899.7925 | 0.82 | 0.011 | 26.4 ± 0.2 | 25.6 ± 0.2 |
| TAG 54:5 | 897.7778 | 0.79 | 0.006 | 24.8 ± 0.2 | 23.9 ± 0.2 |
| TAG 54:6 | 895.7642 | 0.77 | 0.032 | 22.6 ± 0.3 | 21.6 ± 0.3 |
| TAG 56:2 | 931.8585 | 0.90 | 0.022 | 17.2 ± 0.2 | 18.1 ± 0.3 |
| TAG 56:7 | 921.7789 | 0.78 | 0.039 | 21.1 ± 0.3 | 19.7 ± 0.5 |
| CE 20:3 | 691.6269 | 0.79 | 0.030 | 20.6 ± 0.1 | 20.0 ± 0.2 |
| CE 20:5 | 687.5970 | 0.74 | 0.038 | 19.0 ± 0.2 | 19.8 ± 0.3 |
Mean ± standard errors of integrated LC-MS peak areas (log2 transformed) in bold or italics indicate increase or decrease in T1DM, respectively, relative to controls.
LPC: lysophosphatidylcholine; PC: phosphatidylcholine; SM: sphingomyelin; LPE: lysophosphatidylethanolamine; PE: phosphatidylethanolamine; TAG: triacylglycerol; CE: cholesterol ester; NET: normalized elution time.
Partial Least-Squares
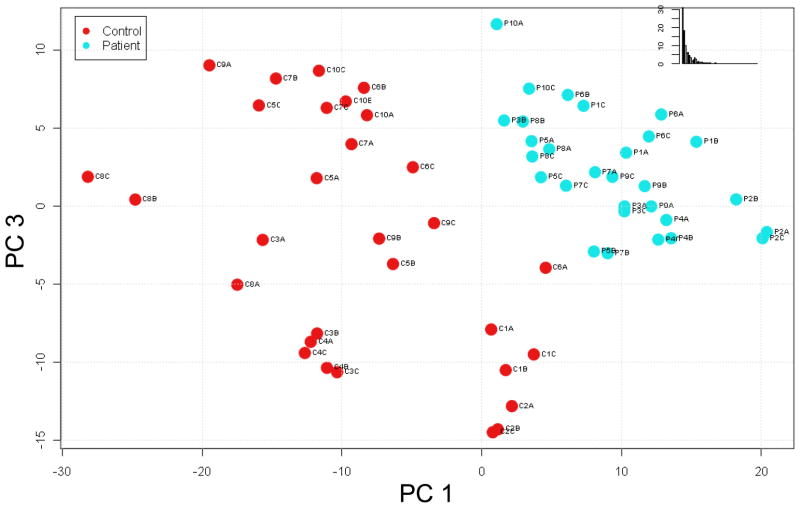
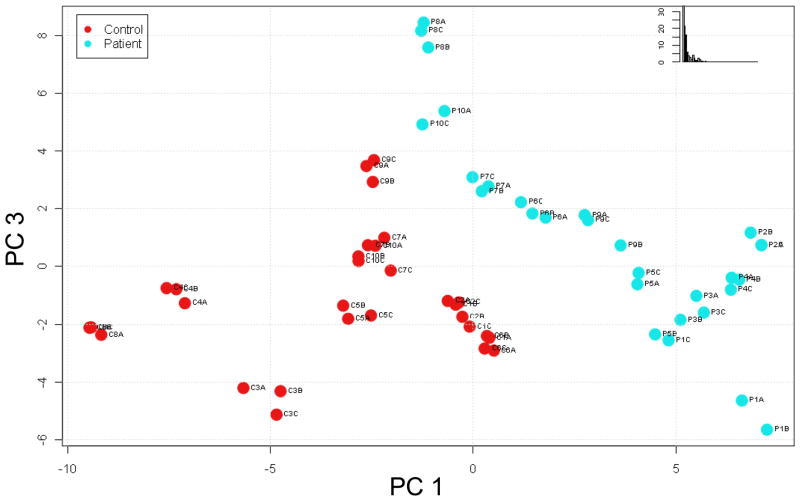
We further assessed trends within the data using partial least-squares (PLS) to identify lipid profiles specific for T1DM. PLS is a chemometrics method for identifying the fundamental relations in a matrix of predictors in which there are more variables than observations, as in the case of omics data. For example, there are typically thousands to tens of thousands of features (variables) detected in an omics experiment compared to the usual tens or hundreds of samples (observations) analyzed. The resulting PLS scores plots are used to visualize any natural clustering of observations, in this case samples, within the data matrix. PLS analysis of the 559 lipid features (both AMT tag database matched and unmatched) that differed significantly (q < 0.05) between control and diabetic samples resulted in segregation of control samples away from patient samples (Figure 3). This observation implies that the significantly different features (both AMT tag database matched and unmatched) may be useful as a profile for diagnostic purposes since, on average, the significantly different lipid features were detected in 96% of the datasets examined. We further performed PLS analysis of the 55 significantly different (q < 0.05) lipid features that matched entries in the lipid AMT tag database. This analysis indicated that these identified lipids might also be used as a profile characteristic of T1DM, since the patient samples were reasonably segregated from the control samples (Figure 4).
Figure 3. Partial least squares (PLS) Score plot based on significantly different lipid features.
Five hundred sixty lipid features (both AMT tag database matched and unmatched) determined to be significantly different (q < 0.05) by ANOVA were used in a PLS analysis in an attempt to identify natural clustering of the samples. PC 1: principal component 1; PC 3: principal component 3; Inset: % variability in data captured by the principal components.
Figure 4. Partial least squares (PLS) Score plot based on significantly different lipids.
Sixty-three lipids determined to be significantly different (q < 0.05) by ANOVA and matching entries in the lipid AMT tag database were used in a PLS analysis in an attempt to identify natural clustering of the samples. PC 1: principal component 1; PC 3: principal component 3; Inset: % variability in data captured by the principal components.
Discussion
In a previous proteomics study, we reported that ZAG, a member of the immunoglobulin superfamily [6] that displays lipid mobilization activity [7, 8], was strongly upregulated in individuals with T1DM [5]. Because patients enrolled in the DASP provide samples within 14 days of diagnosis, increased levels of ZAG in patients relative to controls may be an indication of a system-wide mobilization of lipids for energy production via the induction of lipolysis in adipose tissue [9–11]. We hypothesized that perturbations may be present in individual molecular species of the blood lipidome of individuals with newly diagnosed T1DM. In general, dyslipidemia is well-established in diabetes mellitus, particularly in individuals with type 2 diabetes. In individuals with T1DM, especially those with poorly controlled blood glucose, triacylglycerol-rich lipoproteins (e.g. chylomicrons; very low density lipoprotein, VLDL; and LDL) are often elevated because of (1) increased VLDL production related to increased circulating glucose and free fatty acids due to lack of insulin, and (2) a reduction in the activity of chylomicron- and VLDL-catabolizing lipoprotein lipase, an insulin-regulated enzyme [22]. In contrast, T1DM patients who have well-controlled blood glucose levels and no renal damage have serum lipid and lipoprotein levels similar to those of healthy controls [22]. Indeed, the type 1 diabetic individuals in the DASP sample subset in our study had a normal lipid panel relative to controls (Table 2). Many studies have characterized the dyslipidemia associated with newly diagnosed T1DM in terms of the affected lipoproteins and in terms of general lipid class [23, 24]; however, very few, have considered the perturbations present on the level of individual lipid molecular species [25].
We applied LC-MS and the AMT tag approach in a preliminary lipidomics analysis of a DASP sample subset. While the chromatograms corresponding to healthy controls and recently diagnosed T1DM individuals are fairly similar, downstream analysis of the lipidomics data revealed several molecular species whose abundances were significantly altered between the two conditions. Further, application of PLS resulted in the segregation of control individuals from those with T1DM when using those lipid features that were identified as statistically significant (Figure 3). These data support that there is a lipid profile characteristic of newly diagnosed T1DM and that this profile may be useful in the diagnosis of the disease and exploration of potential beta-cell lipotoxins.
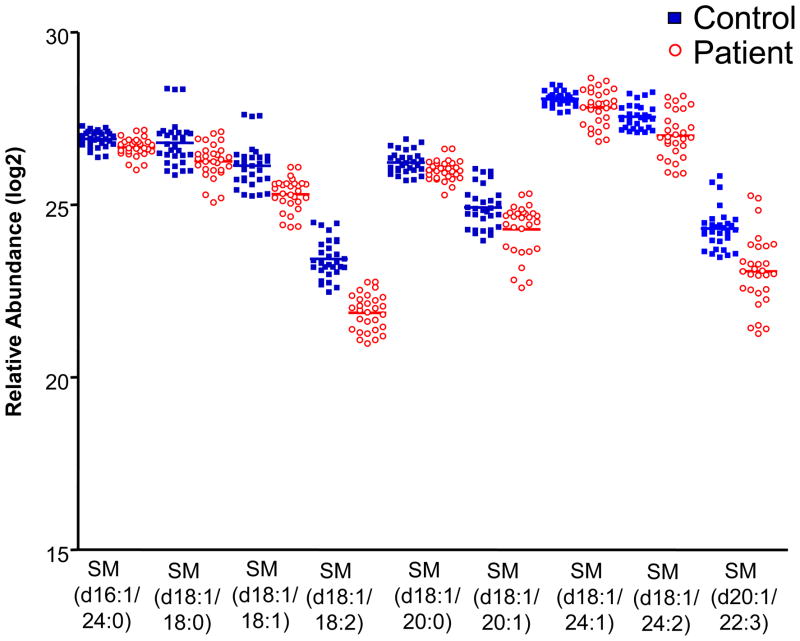
The lipid species that were most different between healthy controls and patients with T1DM belong to phosphatidylcholine-containing classes, such as lysoglycerophosphatidylcholine (LPC), glycerophosphatidylcholine (PC), and sphingomyelin (SM) (Table 4). Nine SM species were significantly decreased in individuals with T1DM relative to controls (Figure 5). Previous studies have also reported decreased plasma and serum SM levels in patients with T1DM relative to controls. Watała and Jóźwiak reported decreased levels of SM in both plasma and isolated LDL and HDL fractions from patients with T1DM relative to age- and sex-matched controls, based on thin layer chromatography [26]. Decreased levels of SM have also been reported by others studying the phospholipid composition of HDL and apolipoprotein B-containing lipoprotein particles[27], [28]. Similarly, Orešič and colleagues reported a decrease in SM molecular species in type 1 diabetic patients relative to controls [25]. In that paper, LC-MS-based lipidomics was applied in the study of serum samples from children enrolled in the Type 1 Diabetes Prediction and Prevention (DIPP) study. The authors consistently detected 8 molecular species of SM, with 4 species (d18:1/16:0, d18:1/23:1, d18:1/24:0, and d18:1/25:1) detected in significantly (Wilcoxon rank-sum test, p < 0.05) lower abundance in type 1 diabetic children before and after the appearance of autoantibodies, relative to healthy controls [25]. In contrast, we found d16:1/24:0, d18:1/18:0, d18:1/18:1, d18:1/18:2, d18:1/20:0, d18:1/20:1, d18:1/24:1, d18:1/24:2, and d20:1/22:3 SM to be significantly decreased in patients relative to controls. In general, there was not a high degree of overlap in the SM molecular species identified between our study and that of Orešič and colleagues; however, this is not unexpected, as the ionization of molecules by ESI and their subsequent detection is a stochastic process that depends, among other things, on the efficiency of the front-end LC separation. The discrepancy in identified SM species may also be due to dietary differences among the individuals participating in the DASP and DIPP. Importantly, the overall trend of decreased SM in individuals with T1DM remains consistent between the two studies and is supported by other previous work [27, 28].
Figure 5. Vertical scatter plots of significantly different SM species.
Ten SM species determined to be significantly different (q < 0.05) by ANOVA are plotted.
We also identified perturbations in other glycerophosphatidylcholine lipids, including LPC, PC, and ether PC. Orešič and colleagues [25] reported decreased levels of ether PC in individuals with T1DM relative to controls; we identified 4 ether PC molecular species that were significantly decreased in recently diagnosed T1DM. In contrast, no solid conclusion could be drawn regarding LPC and PC. For LPC, half of the identified species were either significantly increased or decreased in T1DM. Similarly, we identified 6 molecular species of PC as significantly decreased in T1DM relative to controls, while 7 molecular species were identified as significantly increased in T1DM.
Consistent with our observations, other studies have reported perturbations in the glycerophosphatidylcholine lipid (lecithin) content of serum or isolated lipoprotein fractions, although these reports do not always agree in terms of increases or decreases in this lipid class in T1DM. For example, Ziegler et al. [29] reported a depletion of glycerophosphatidylcholine lipids in serum and apolipoprotein B-containing lipoprotein fractions in individuals with T1DM who otherwise presented with a normal lipid profile relative to controls. Similarly, Bagdade and Subbaiah [28] reported decreased SM, PC, and LPC in the HDL-containing fraction of plasma in women with T1DM relative to controls. These patients also presented with normal triglycerides, total and HDL-cholesterol, and lipoprotein phospholipids in whole plasma. However, in a separate study, the same authors reported significantly (p > 0.025) increased SM and PC in the HDL fraction of plasma in men with T1DM relative to controls [28]. Rabini and colleagues also reported increased concentrations of LPC in plasma of T1DM individuals [30]. These contradictions suggest that, aside from dietary differences and biological variation between healthy controls and patients, reported differences in glycerophosphatidylcholine lipid content of individuals may be due to differences in general phospholipid composition of individual lipoprotein particles as a result of T1DM. It is conceivable that these species originate from different lipoprotein particles, resulting in perceived increases or decreases in abundance depending on the composition of the parent particles. The work by Watała and Jóźwiak supports this hypothesis [26]. They analyzed various lipid classes in total plasma lipid extracts from control and T1DM patients, as well as in the LDL and HDL fractions. While SM was decreased in patients in all fractions (see above), LPC was decreased in patients in total plasma (p < 0.001) but increased in patients in the HDL fraction (p <0.002). In contrast, glycerophosphatidylethanolamine lipids (PE) were increased in patients in total plasma (p < 0.05) but were unchanged in the LDL and HDL fractions (similar to Watała and Jóźwiak, we identified 2 LPE and 1 PE molecular species as significantly increased in T1DM versus control). Glycerophosphatidylserine and glycerophosphatidylinositol lipids were decreased in patients in the LDL fraction (p < 0.001) but were unchanged in total plasma and not detected in the HDL fraction. Thus, lipidomic analyses of total lipid extracts should ideally be complemented by lipidomic analyses of isolated lipoprotein fractions in order to identify perturbations in the lipid compositions of the lipoproteins themselves. Ziegler and colleagues suggested that subtle abnormalities in the composition of atherogenic apo-B-containing lipoproteins may be a factor in the increased rate of atherosclerosis in diabetic patients, despite the existence of an otherwise normal lipid profile [29]. Bagdade and Subbaiah propose that such abnormalities may compromise reverse cholesterol transport in diabetic patients and promote atherosclerosis [28]. Along these lines, we identified 7 molecular species of oxidized PC that were significantly increased in T1DM. Oxidized phospholipids are implicated in many inflammatory diseases such as atherosclerosis [31–33], and their observed increase in individuals with recently diagnosed T1DM may be an indication of oxidative stress in these individuals. Finally, we identified 5 molecular species of triacylglyecerol (TAG) that were significantly decreased in T1DM, possibly indicating a recruitment of TAGs for energy production.
In conclusion, application of the AMT tag approach facilitated the identification of a lipid profile comprised of a number of significant features (both identified and unidentified; Supplemental Table 1) characteristic of recently diagnosed T1DM. While we have identified a portion of the species significantly altered in recently diagnosed T1DM (Table 3), additional work using targeted MS/MS to identify the remaining species may yield further insights into the classes of lipids that become perturbed. These classes and molecular species may eventually be targets for therapeutic intervention if they are found to mediate beta-cell toxicity. Lastly, we propose that future studies of the lipidome of individuals with T1DM should include analyses of both total lipid extracts and lipids extracted from isolated lipoprotein fractions.
Supplementary Material
Acknowledgments
The authors would like to thank the DASP Standardization Committee and the members of the diabetes research community who have contributed DASP samples, as well as Drs. Alicia J. Jenkins and Katrina M. Waters of the University of Melbourne and Pacific Northwest National Laboratory (PNNL), respectively, for helpful discussions. This work was supported by NIH grant DK070146 to R.D.S.; T.A. is supported by a post-doctoral fellowship from the Canadian Diabetes Association. Work was performed in the EMSL, the Environmental Molecular Sciences Laboratory, a national scientific user facility located at PNNL and sponsored by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research. PNNL is operated by Battelle for the DOE under Contract No. DE-AC06-76RLO-1830.
Footnotes
DISCLAIMER
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention and the Agency for Toxic Substances and Disease Registry.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental table of identified lipids (AMT tag database matched) and unidentified lipid features differentiating healthy control from diabetic individuals; associated q value from ANOVA, mass, NET, database identity, average abundance values, and standard errors of the means.
References
- 1.Gepts W. Pathologic anatomy of the pancreas in juvenile diabetes mellitus. Diabetes. 1965;14:619–633. doi: 10.2337/diab.14.10.619. [DOI] [PubMed] [Google Scholar]
- 2.Bingley PJ, Bonifacio E, Williams AJ, Genovese S, Bottazzo GF, Gale EA. Prediction of IDDM in the general population: strategies based on combinations of autoantibody markers. Diabetes. 1997;46:1701–1710. doi: 10.2337/diab.46.11.1701. [DOI] [PubMed] [Google Scholar]
- 3.Seissler J, Hatziagelaki E, Scherbaum WA. Modern concepts for the prediction of type 1 diabetes. Exp Clin Endocrinol Diabetes. 2001;109 (Suppl 2):S304–316. doi: 10.1055/s-2001-18590. [DOI] [PubMed] [Google Scholar]
- 4.LaGasse JM, Brantley MS, Leech NJ, et al. Successful prospective prediction of type 1 diabetes in schoolchildren through multiple defined autoantibodies: an 8-year follow-up of the Washington State Diabetes Prediction Study. Diabetes Care. 2002;25:505–511. doi: 10.2337/diacare.25.3.505. [DOI] [PubMed] [Google Scholar]
- 5.Metz TO, Qian WJ, Jacobs JM, et al. Application of proteomics in the discovery of candidate protein biomarkers in a diabetes autoantibody standardization program sample subset. J Proteome Res. 2008;7:698–707. doi: 10.1021/pr700606w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uria JA, Fueyo A, Balbin M, Velasco G, Pendas AM, Lopez-Otin C. Alternative splicing gives rise to two novel long isoforms of Zn-alpha 2-glycoprotein, a member of the immunoglobulin superfamily. Gene. 1996;169:233–236. doi: 10.1016/0378-1119(95)00727-x. [DOI] [PubMed] [Google Scholar]
- 7.Todorov PT, McDevitt TM, Meyer DJ, Ueyama H, Ohkubo I, Tisdale MJ. Purification and characterization of a tumor lipid-mobilizing factor. Cancer Res. 1998;58:2353–2358. [PubMed] [Google Scholar]
- 8.Hirai K, Hussey HJ, Barber MD, Price SA, Tisdale MJ. Biological evaluation of a lipid-mobilizing factor isolated from the urine of cancer patients. Cancer Res. 1998;58:2359–2365. [PubMed] [Google Scholar]
- 9.Singer P, Gnauck G, Honigmann G, Thoelke H, Schliack The fatty acid pattern of triglycerides in liver, adipose tissue and serum of diabetics with hyperlipoproteinemia before and during clofibrate treatment. Acta Diabetol Lat. 1978;15:40–52. doi: 10.1007/BF02581006. [DOI] [PubMed] [Google Scholar]
- 10.Bing C, Bao Y, Jenkins J, et al. Zinc-alpha2-glycoprotein, a lipid mobilizing factor, is expressed in adipocytes and is up-regulated in mice with cancer cachexia. Proc Natl Acad Sci U S A. 2004;101:2500–2505. doi: 10.1073/pnas.0308647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell ST, Zimmerman TP, Domin BA, Tisdale MJ. Induction of lipolysis in vitro and loss of body fat in vivo by zinc-alpha2-glycoprotein. Biochim Biophys Acta. 2004;1636:59–68. doi: 10.1016/j.bbalip.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding J, Sorensen CM, Jaitly N, et al. Application of the accurate mass and time tag approach in studies of the human blood lipidome. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:243–252. doi: 10.1016/j.jchromb.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bingley PJ, Bonifacio E, Mueller PW. Diabetes Antibody Standardization Program: first assay proficiency evaluation. Diabetes. 2003;52:1128–1136. doi: 10.2337/diabetes.52.5.1128. [DOI] [PubMed] [Google Scholar]
- 15.Kiebel GR, Auberry KJ, Jaitly N, et al. PRISM: a data management system for high-throughput proteomics. Proteomics. 2006;6:1783–1790. doi: 10.1002/pmic.200500500. [DOI] [PubMed] [Google Scholar]
- 16.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics. 2009;10:87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaitly N, Monroe ME, Petyuk VA, Clauss TR, Adkins JN, Smith RD. Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal Chem. 2006;78:7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 18.Polpitiya AD, Qian WJ, Jaitly N, et al. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callister SJ, Barry RC, Adkins JN, et al. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J Proteome Res. 2006;5:277–286. doi: 10.1021/pr050300l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 21.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chahil TJ, Ginsberg HN. Diabetic dyslipidemia. Endocrinol Metab Clin North Am. 2006;35:491–510. vii–viii. doi: 10.1016/j.ecl.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Rubba P, Capaldo B, Falanga A, et al. Plasma lipoproteins and lipoprotein lipase in young diabetics with and without ketonuria. J Endocrinol Invest. 1985;8:433–436. doi: 10.1007/BF03348532. [DOI] [PubMed] [Google Scholar]
- 24.Loh KC, Thai AC, Lui KF, Ng WY. High prevalence of dyslipidaemia despite adequate glycaemic control in patients with diabetes. Ann Acad Med Singapore. 1996;25:228–232. [PubMed] [Google Scholar]
- 25.Oresic M, Simell S, Sysi-Aho M, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205:2975–2984. doi: 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watala C, Jozwiak Z. The phospholipid composition of erythrocyte ghosts and plasma lipoproteins in diabetes type 1 in children. Clin Chim Acta. 1990;188:211–219. doi: 10.1016/0009-8981(90)90202-4. [DOI] [PubMed] [Google Scholar]
- 27.Fievet C, Ziegler O, Parra HJ, Mejean L, Fruchart JC, Drouin P. Depletion in choline containing phospholipids of LpB particles in adequately controlled type I insulin-dependent diabetes mellitus. Diabete Metab. 1990;16:64–69. [PubMed] [Google Scholar]
- 28.Bagdade JD, Subbaiah PV. Abnormal high-density lipoprotein composition in women with insulin-dependent diabetes. J Lab Clin Med. 1989;113:235–240. [PubMed] [Google Scholar]
- 29.Ziegler O, Mejean L, Igau B, Fruchart JC, Drouin P, Fievet C. Accessibility of human apolipoprotein B-100 epitopes in insulin-dependent diabetes: relation with the surface lipid environment of atherogenic particles. Diabetes Metab. 1996;22:179–184. [PubMed] [Google Scholar]
- 30.Rabini RA, Galassi R, Fumelli P, et al. Reduced Na(+)-K(+)-ATPase activity and plasma lysophosphatidylcholine concentrations in diabetic patients. Diabetes. 1994;43:915–919. doi: 10.2337/diab.43.7.915. [DOI] [PubMed] [Google Scholar]
- 31.Leitinger N. The role of phospholipid oxidation products in inflammatory and autoimmune diseases: evidence from animal models and in humans. Subcell Biochem. 2008;49:325–350. doi: 10.1007/978-1-4020-8830-8_12. [DOI] [PubMed] [Google Scholar]
- 32.Tsimikas S, Witztum JL. The role of oxidized phospholipids in mediating lipoprotein(a) atherogenicity. Curr Opin Lipidol. 2008;19:369–377. doi: 10.1097/MOL.0b013e328308b622. [DOI] [PubMed] [Google Scholar]
- 33.Fruhwirth GO, Loidl A, Hermetter A. Oxidized phospholipids: from molecular properties to disease. Biochim Biophys Acta. 2007;1772:718–736. doi: 10.1016/j.bbadis.2007.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.