Abstract
RAGE (Receptor for Advanced Glycation End-products) and its ligands are overexpressed in multiple cancers. RAGE has been implicated in tumorigenesis and metastasis, but little is known of the mechanisms involved. In this study we define a specific functional role for an alternate splice variant termed RAGEv1 that encodes a soluble endogenous form of the receptor that inhibits tumorigenesis. RAGEv1 was downregulated in lung, prostate, and brain tumors relative to control matched tissue. Overexpressing RAGEv1 in tumor cells altered RAGE-ligand stimulation of several novel classes of genes that are critical in tumorigenesis and metastasis. Additionally, RAGEv1 inhibited tumor formation, cell invasion, and angiogenesis induced by RAGE-ligand signaling. Analysis of signal transduction pathways underlying these effects revealed marked suppression of JNK pathway signaling and JNK inhibition suppressed signaling through the RAGE pathway. Tumors expressing RAGEv1 were significantly smaller than wild-type tumors and displayed prominently reduced activation of JNK. Our results identify RAGEv1 as a novel suppressor function the study of which may offer new cancer therapeutic directions.
Keywords: RAGE, tumorigenesis, JNK, alternative splicing, angiogenesis
Introduction
Tumorigenesis is a multi-step process involving the alteration of a number of key cellular properties including uncontrolled proliferation, evasion of cell death (apoptosis and necrosis), vascularization (angiogenesis) and subsequent invasion and migration of tumor cells into the surrounding tissues (1). Understanding the molecular processes underlying these cellular phenotypic changes is critical in order to develop novel therapies. Central to mediating these changes is the interaction between cell surface receptors and their cognate ligands, which through intracellular signaling induce alterations in gene expression. In this context, recent studies have identified that the Receptor for Advanced Glycation End-products (RAGE) and its ligands may play an important role in cancer (2-5). RAGE is a multi-ligand receptor that has been implicated in the pathogenesis of numerous disease states including diabetes, vascular and inflammatory diseases (3;6;7). RAGE-ligand interaction triggers activation of a diverse array of signaling pathways that lead to processes integrally linked to the tumorigenic sequelae including cellular migration, invasion, proliferation and survival (2;4). These pathways include the mitogen activated protein kinases (MAPK), Rho GTPases (Rac-1 and Cdc42) as well as activation of nuclear factor κB (NF-κB), which in-turn result in the expression and activation of proinflammatory cytokines (2;8-13). Notably, in mice, blocking RAGE-ligand induced signaling led to a striking reduction in tumor development and suppressed the motility and invasiveness of tumor cells (2). Furthermore, studies have indicated that RAGE and its ligands are expressed in human tumors, and often the extent of tumor invasiveness and metastatic potential was correlated with the degree of RAGE/lignad up-regulation (9;14-22).
In recent years, the discovery that endogenous soluble isoforms exist for the RAGE gene suggest a potential innate mechanism to counter-act the adverse effects of RAGE ligands (23-25). In particular, endogenous soluble RAGE (esRAGE or RAGE splice variant 1; RAGEv1) is produced by alternative splicing, resulting in a product lacking the transmembrane domain and cytoplasmic domain of RAGE and is readily secreted from cells (23;24). Intriguingly, soluble forms of RAGE have been detected in human plasma, and these levels correlate with the presence and/or extent of RAGE-mediated diseases (26-29). In cancer, specifically, a number of studies have reported that levels of soluble RAGE were lower in subjects afflicted with breast or lung cancer compared to controls (30;31). Yet, no experiments to date have elucidated a functional role of soluble RAGE isoforms detected in human subjects. Therefore, our goal here was to establish the mechanistic effects of RAGEv1 on RAGE ligand-mediated tumorigenesis, and to test if RAGEv1 might suppress tumor-provoking signaling pathways. Here, we therefore report on a novel mechanism by which the soluble splice variant of RAGE (RAGEv1) inhibits tumorigenesis, and may represent a novel therapeutic target in the treatment of cancer.
Materials and Methods
Cell Culture, Antibodies and Reagents
Rat C6 glioma cells were obtained from ATCC and maintained in DMEM supplemented with 10% FBS (Invitrogen). Human primary aortic endothelial cells (ECs) were purchased from Lonza and maintained in EGM2 media (Lonza, Basel, Switzerland). The RAGEv1 rabbit polyclonal antibody was raised against the purified peptide sequence (Ac- CGEGFDKVREADSPQHM-amide) and affinity purified by QCB. This antibody recognizes the unique C-terminus region of RAGEv1, but not full-length RAGE.
Plasmid Engineering and RAGEv1 Stable Clone Generation
RAGEv1 (Genbank no.AY755620) was cloned as previously described (24). The RAGE-ligand, S100A12 was cloned in frame with the 6xHis-tag (C-terminus tag) in the vector pET101 (Invitrogen) from lung cDNA, using the primers (5′-CTCCATGACAAAACTTGAAGAGC-3′) and (5′-TTCTTTGTGGGTGTGGTAATG-3′). C6 cells were stably transfected with pcDNA3.1-RAGEv1 or empty pcDNA3.1 (mock transfected) constructs and screened for RAGEv1 expression by western blot of cell lysates and conditioned using anti-RAGE IgG, anti-RAGEv1 IgG. RAGEv1 expression was further verified by measurement of conditioned media using the sRAGE Quantikine ELISA (R&D Systems).
RAGEv1 in vitro binding assay
Binding of RAGEv1 to RAGE-ligand was tested using pull-down assays with the Pierce His Protein Interaction Pull-Down Kit (Pierce) according to the manufacturer's instructions using His-tagged s100A12 (bait) and cultured media from RAGEv1 (prey).
Cancer PathwayFinder PCR Array
RAGEv1 and mock C6 cells were incubated with/without 10 μg/ml S100B for 2 h and total RNA extracted using Trizol (Invitrogen). Gene expression of 84 genes representative of the six biological pathways involved in tumorigenesis (including 5 housekeeping genes for normalization; Rplp1, Hprt1, Rpl13a, Ldha and Actb) was assessed using the Rat Cancer PathwayFinder™ RT2 Profiler™ PCR Array (SABiosceinces) according to the manufacturer's instructions and analyzed on an MX3005P Real-time PCR System (Stratagene). Only genes demonstrating a 1.5-fold or greater change were considered for further analysis. PCR array data was validated using a combination of Taqman QPCR and western blot analysis as described in the supplementary methods.
In vitro tumorigenic assays
In vitro angiogenesis assays were performed by seeding ECs on top of Matrigel (BD Biosciences) and incubated with 50% EGM-2 media : 50% conditioned media from RAGEv1 and mock cells incubated with/without s100B. After 2 h, tube formation was assessed and images taken from 4 independent fields. Tumor cell adhesion assays were performed by incubating cells with/without 10 μg/ml S100B for 24 h were seeded into tissue culture plates for 2 h to adhere. Attached cells were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet and OD measured at 590nm. Cellular apoptosis was assessed using Annexin V / propidium iodide staining. Cells stimulated with/without 10 μg/ml S100B for 24 h, were stained with the ApoTarget Annexin-V FITC Apoptosis Kit and analyzed by flow cytometry. Invasion assays were performed using the BD BioCoat™ BD Matrigel™ Invasion Chambers (BD Biosciences) by seeding cells into the upper chamber, after 24 h, invaded cells were assessed by staining with 0.1% crystal violet solution. In vitro tumor growth was performed using the Cell Transformation Detection Assay (Millipore) by plating cells in a layer of 0.4% agarose in DMEM. After 14 days, colonies were visualized using Cell Stain solution (Millipore).
In vivo tumor growth
All animal studies were performed with the approval of the Institutional Animal Care and Use Committee of Columbia University and conform to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. Weanling female SCID mice were subcutaneously injected in the right flank with 1×105 of mock or RAGEv1 expressing C6 cells suspended in 0.1 ml PBS. After 31 days, tumors were measured with calipers and the volume of the tumor calculated using the following formula: V = π×h(h2 + 3a2)/6, where h = height of the tumor section; a = (length + width of the tumor)/4; and V = volume of the tumor. Excised tumors sections were snap frozen for protein analysis.
Cell signaling assays
Activation of cell signaling proteins were performed by western blot using antibodies to phospho/total MEK1/2, SAPK/JNK, p38 and AKT (Cell Signaling). Inhibition of these pathways was performed with chemical for SAPK/JNK (SP600125, 10μM), MEK1/2 (U0126, 10μM), p38 (SB203580, 10μM) and AKT (Triciribine, 10μM) or control (0.1% DMSO), using stimulated cells (10 μg/ml S100B for 2h) and QCR analysis performed for Apaf1, Pdgfb and Tnf performed as above. Invasion assays were performed by incubating cells with either 2.5μM SP600125 or 0.1% DMSO as vehicle, and invasion performed with ligand stimulation as described above.
Statistical analyses
In all experiments, unless otherwise indicated, data are reported as mean ± SEM in at least 3 replicates per group. Data were analyzed by post-hoc comparisons using 2-tailed t test and a P value less than or equal to 0.05 was considered significant.
Results
RAGEv1 is down-regulated in various human cancers
Previous studies have monitored the presence of RAGEv1 in human plasma and the relationship with various inflammatory disease states. To analyze the expression profile of RAGEv1 directly in human tumor tissue samples, we studied normal and tumor matched tissue retrieved from the same subject, and examined the expression of RAGEv1 by western blot analysis (Supplementary Fig. S1). Interestingly, these experiments suggested that levels of RAGEv1 tended to be lower in tumor versus normal matched adjacent tissue. These data suggest for the first time that RAGEv1 levels may be down-regulated in the tumor, but not in adjacent non-malignant tissue, and thus we sought to test the hypothesis that RAGEv1 might act as a molecular decoy to block tumorigenesis through preventing the cellular effects of RAGE ligands.
The ectopic expression of RAGEv1 by tumor cells
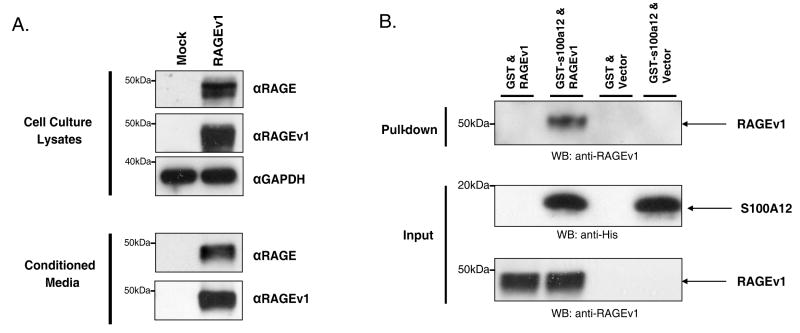
To test these concepts, we firstly generated rat C6 glioma cells which stably expressed RAGEv1. C6 rat glioma cells are an established model system for both in vitro and in vivo analysis of tumorigenesis, and we have previously seen RAGE-ligand dependent responses in this cell line (s100B / CML) (13;32). Western blot analysis confirmed the expression of RAGEv1 in transfected cells compared to control empty vector transfected (mock) cells, by using antibodies to the unique C-terminus sequence of RAGEv1 and antibodies to the extracellular domain of human RAGE (Fig. 1A). Furthermore, western blot analysis of cell culture media confirmed that RAGEv1 protein was actively secreted by the C6 cells (Fig. 1A). Measurement of RAGEv1 in cell culture media using an sRAGE ELISA demonstrated levels of ∼1500-2000 pg/ml, which is in the physiological range detected in human plasma (7). To confirm that RAGEv1 binds RAGE ligands, pull-down experiments were performed with recombinant RAGE ligand (GST-s100A12), using the conditioned cell culture media (CM) containing RAGEv1. Western blot analysis of pull-down elutant showed a clear interaction between RAGEv1 and s100A12, but not with GST alone (Fig. 1B).
Figure 1.
RAGEv1 is secreted by tumor cells and binds RAGE-ligand. (A) Western blot analysis was performed to assess the secretion of RAGEv1 from C6 glioma cells stably transfected with empty vector control (mock) or RAGEv1. Total cell lysate or conditioned media were analyzed for RAGEv1 expression with antibodies to human RAGE or the specific RAGEv1 C-terminus epitope. GAPDH was used as a loading control for total cell lysate. (B) Binding of RAGEv1 to RAGE-ligands was assessed by incubation of GST-tag or GST-tagged s100A12 with conditioned media from mock and RAGEv1 transfected cells. Pulldown assays were performed to assess RAGEv1 binding and western blot analysis performed with antibodies against RAGEv1 on the elutant from pulldown reactions.
RAGEv1 expression impacts on pro- tumorigenic gene expression profiles
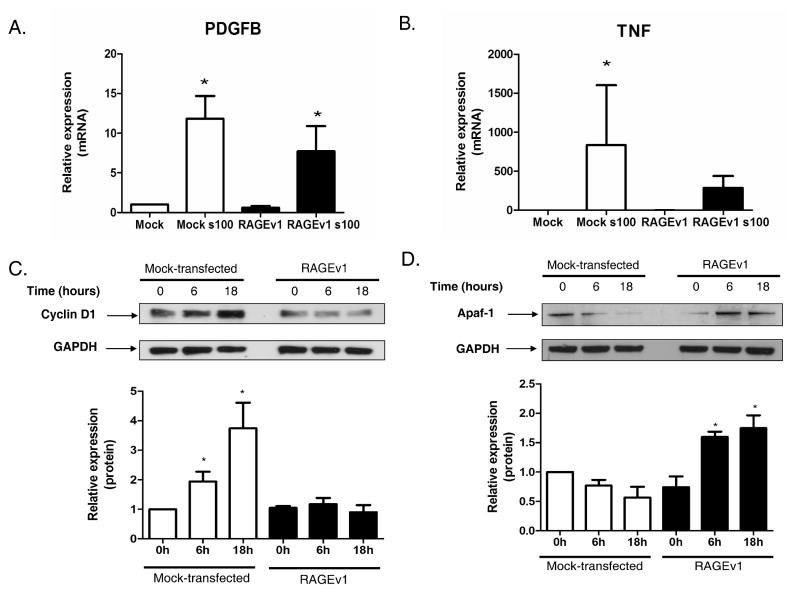
Next, we tested the impact of RAGEv1 on RAGE /ligand-activated tumorigenic pathways. We hypothesized that RAGE activation regulates the expression of genes that promote tumor cell properties, and that RAGEv1 might block these effects. To test this hypothesis, we performed QPCR array analysis on the control and RAGEv1 transfected cells, and assessed the impact of RAGE -ligand stimulation using the Cancer PathwayFinder array. This array consists of 84 genes representing six major biological pathways involved in tumorigenesis. We identified several classes of genes that were altered by RAGEv1 expression including angiogenesis, adhesion, apoptosis, invasion/metastasis, cell cycle control and signaling (Supplementary Table 1). Of the 84 cancer pathway focused genes, 19 demonstrated at least a 1.5-fold difference in gene expression between ligand-stimulated cells expressing either RAGEv1- or mock-transfected cells. Interestingly, genes implicated in angiogenesis represented the major class altered both in number and expression level, which include Fgf1, Ifna1, Pdgfb, Thbs1, Tnf and Vegfc. As shown in Fig. 2 (and Supplementary Fig. S2), we validated several of these genes for changes in expression by Taqman QPCR and western blot analysis. Taqman QPCR analysis confirmed that RAGEv1 impaired RAGE-ligand induced expression of Pdgfb (Fig. 2A) and Tnf (Fig. 2B), and increased expression of the apoptotic gene, Apaf-1 (Supplementary Fig. S2A),. Western blot analysis demonstrated that RAGEv1 blocked RAGE-ligand (s100B) induced expression of cyclin D1 (Fig. 2C) and increased expression of Apaf-1 (Fig. 2D). Similar changes in protein expression (cyclin D1) were seen for other RAGE-ligands (CML-HSA) (data not shown). Together these data indicate that expression of RAGEv1 inhibits expression of genes critically involved in tumorigenesis through blocking RAGE-ligand interaction.
Figure 2.
RAGEv1 inhibits RAGE-ligand induced changes in tumorigenic gene expression. (A-B) Relative gene expression of PDGFB and TNF were determined by QPCR on mock and RAGEv1 expressing cells, stimulated with RAGE-ligand (s100B), gene expression normalized to GAPDH levels. (C-D) Western blot analysis of cyclin D1 and Apaf1 performed on cell lysates from mock and RAGEv1 cells, stimulated with RAGE-ligand (s100B), normalized to GAPDH levels. Data are means ± SEM from three independent experiments and significant differences (P ≤ 0.05) indicated by an asterisk.
RAGEv1 impacts on the mechanisms of tumorigenesis in vitro
As RAGE has been reported to have a role in promoting cancer cell survival, migration and invasion, we hypothesized that RAGEv1 may impair these functions. To test this, we analyzed in vitro whether RAGEv1 impacts on RAGE-mediated tumor cell angiogenesis, adhesion, apoptosis and invasion. We first evaluated angiogenesis, as the gene expression data suggested pathways involved in this process were most affected by RAGEv1 blockade of RAGE signaling. Assays were performed by culturing endothelial cells on Matrigel in conditioned medium (CM) from RAGEv1-transfected or mock-transfected tumor cells. We observed markedly reduced tube formation of ECs exposed to CM from RAGEv1 cells versus mock (Fig. 3A). Consistent with these findings, CM from RAGE ligand-stimulated mock cells induced a two-fold increase in tube formation compared to unstimulated mock CM (Fig. 3A), whereas CM from RAGEv1 failed to stimulate tube formation (Fig. 3A). We next analyzed the effect of RAGEv1 expression on cellular adhesion. As shown in Fig. 3B, RAGEv1-expressing cells adhered more to the substratum than mock cells. RAGE ligand stimulation of did not affect tumor cell adhesion in either mock or RAGEv1 cells (Fig. 3B). To determine the effect of RAGEv1 on tumor cell survival, we next studied cellular apoptosis. Using the Annexin-V/PI FACS assay, compared to control cells, RAGEv1 expressing cells displayed more apoptotic cells (4% vs 8%, Fig. 3C). However, RAGE ligand stimulation did not appear to alter the percentage of apoptotic cells in either control or RAGEv1- expressing cells. To confirm that RAGEv1 expression affects tumor cell survival, we performed cell viability and caspase 3/7 activation assays. We observed a significant decrease in cellular viability of RAGEv1- expressing cells compared to controls, independent of ligand stimulation (Supplementary Fig. S3A). This was further confirmed by an increase in caspase 3/7 activation in RAGEv1-expressing cells compared to control (Supplementary Fig. S3B). These data suggest that by blocking RAGE activation, RAGEv1 prevents tumor cell evasion of apoptosis and cell death.
Figure 3.
RAGEv1 inhibits mechanisms of tumorigenesis in vitro. (A) RAGEv1 blocks RAGE-ligand induced angiogenesis. Matrigel tube formation assay of ECs exposed to conditioned media from mock and RAGEv1 transfected tumor cells exposed to vehicle or RAGE-ligand (s100B). Tube number was counted in three random fields from three independent experiments. (B) RAGEv1 expression increases cellular adhesion. Serum starved cells were incubated with vehicle or RAGE-ligand (s100B) for 24 h, seeded into plates, cultivated for 2 h and bound cells fixed, stained with crystal violet and quantified by absorbance at 570nm. (C) RAGEv1 expression increases cellular apoptosis. Serum starved cells were incubated with vehicle or RAGE-ligand (s100B) for 24 h and apoptosis assessed by staining for annexin V-FITC (x-axis) and /PI (y-axis) and flow cytometry. (D) RAGEv1 blocks RAGE-ligand induced tumor cell invasion. Matrigel invasion assays were performed with mock and RAGEv1 transfected cells in response to vehicle or RAGE-ligand (s100B). Data are means ± SEM from three independent experiments. Significant differences (P ≤ 0.05) between groups are indicated by an asterisk.
Ultimately in the tumorigenic process, cells invade the surrounding tissue and metastasize (33). Therefore we tested whether RAGEv1 blocks RAGE ligand-induced cellular invasion. RAGE ligand stimulation of control cells induced a ∼2.5-fold increase in cell invasion (Fig. 3D), whereas in contrast, RAGEv1 expression blocked cell invasion (Fig. 3D). Previous studies demonstrated that RAGE activation leads to an increased invasive phenotype though activation of matrix metalloproteinases (MMPs) (2). To investigate this, we analyzed both protein levels and activity of MMP-2/9 in control and RAGEv1 expressing cells. MMP-9 protein levels were upregulated in control cells in response to RAGE ligand stimulation (Supplementary Fig. S3C), whereas in RAGEv1-expressing cells, MMP-9 was not detectable. Furthermore, activity levels of MMP-9 by gelatin zymography in control cell CM revealed increased MMP-9 activation in response to RAGE-ligand, whereas active MMP-9 was not detected in RAGEv1-expressing cells (Supplementary Fig. S3D). MMP-2 (antigen or activity) was not detected in either control or RAGEv1-expressing cells (data not shown). Together these data demonstrate that RAGEv1 acts to block RAGE ligand-induced tumor invasion.
RAGEv1 suppresses tumorigenic signaling
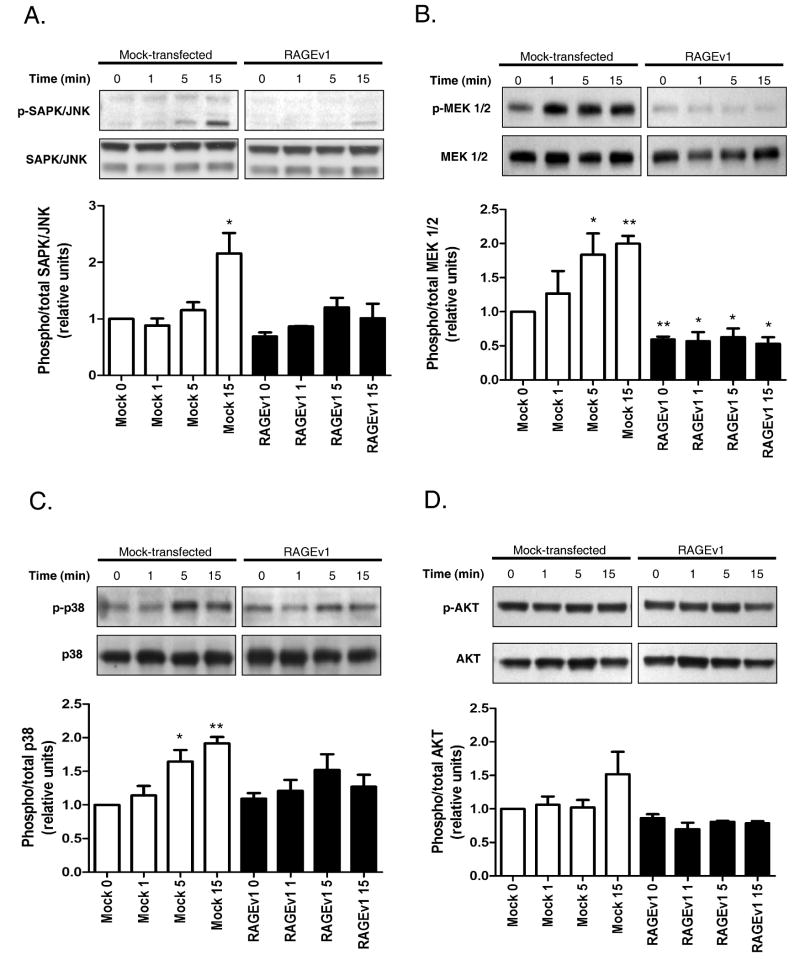
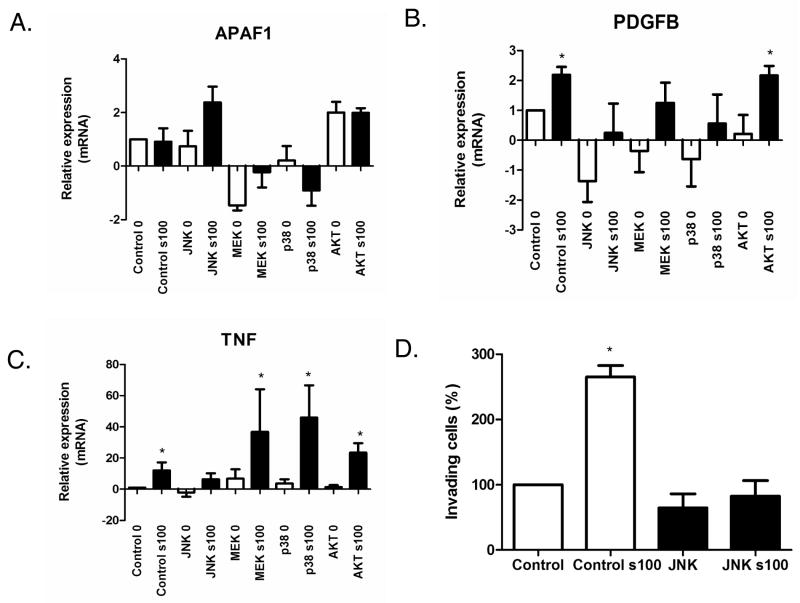
Previous studies have suggested that RAGE signals through these MAPK pathways to regulate cellular processes (2;8;8;9). To explore the specific signaling mechanisms modulated by RAGEv1, we examined the MAPK signaling pathway, including MEK1/2, p38 and SAPK/JNK. RAGE-ligand stimulation (s100B) resulted in ∼2 fold activation of MEK 1/2, p38 and SAPK/JNK in control cells (Fig. 4A-C). In contrast, in cells expressing RAGEv1, activation of MEK 1/2, p38 and SAPK/JNK was markedly reduced (Fig. 4A-C). Interestingly, AKT was not activated by RAGE ligand (s100B), nor did activity levels differ between control and RAGEv1-expressing cells (Fig. 4D). Experiments with mock / RAGEv1 expressing cells using the CML-HSA RAGE-ligand (13), revealed similar changes in MEK 1/2, p38, SAPK/JNK and AKT (data not shown). To evaluate the specific signaling pathway(s) responsible for the tumorigenic changes observed in these cells, we tested the effect of various inhibitors. Control cells were incubated with inhibitors of MEK1/2 (U0126), p38 (SB203580), SAPK/JNK (SP600125) and AKT (Triciribine), followed by RAGE ligand stimulation. Analysis of gene expression by QPCR revealed that in the presence of the SAPK/JNK inhibitor SP600125, RAGE ligand stimulation induced Apaf1 and blocked Pdgfb and Tnf expression, compared to control stimulated cells (Fig. 5A-C). Pre-treatment of cells with U0126, SB203580 or Triciribine had little or no effect on RAGE ligand dependent expression of Apaf1, Pdgfb and Tnf (Fig. 5A-C). To test the functional role of JNK inhibition in RAGE-ligand mediated tumorigenesis, we analyzed its effects on RAGE ligand- driven cellular invasion. Stimulation of control cells induced a ∼3-fold increase in cell invasion (Fig. 5D), whereas treatment of cells with the JNK inhibitor blocked RAGE ligand stimulated cellular invasion (Fig. 5D).
Figure 4.
RAGEv1 inhibits MAPK signaling (A-D) Cells (mock and RAGEv1) were stimulated with RAGE-ligand for the indicated times, lysed and subjected to western blot with antibodies for phospho-status and total of SAPK/JNK, MEK 1/2, p38 and AKT for the indicated times. Activity levels are expressed as a ratio of phospho/total levels. Data are means ± s.e.m from three independent experiments. Significant differences (P ≤ 0.05) between groups are indicated by an asterisk.
Figure 5.
RAGEv1 inhibits tumorigenesis through blocking a JNK-dependent signaling mechanism. (A-C) Cells were incubated with control (DMSO) or inhibitors for SAPK/JNK (SP600125), MEK1/2 (U0126), p38 (SB203580) and AKT (Triciribine) for 1h before stimulation with RAGE-ligand (s100B). RNA was then extracted and QPCR performed for APAF1, PDGFB and TNF, normalized to GAPDH levels. (D) JNK inhibition blocks RAGE-ligand induced tumor cell invasion. Matrigel invasion assays were performed with mock transfected cells, treated with control (DMSO) or SAPK/JNK (SP600125) and in response to vehicle or RAGE-ligand (s100B). Data are means ± s.e.m from three independent experiments. Significant differences (P ≤ 0.05) between groups are indicated by an asterisk.
RAGEv1 suppresses tumorigenesis
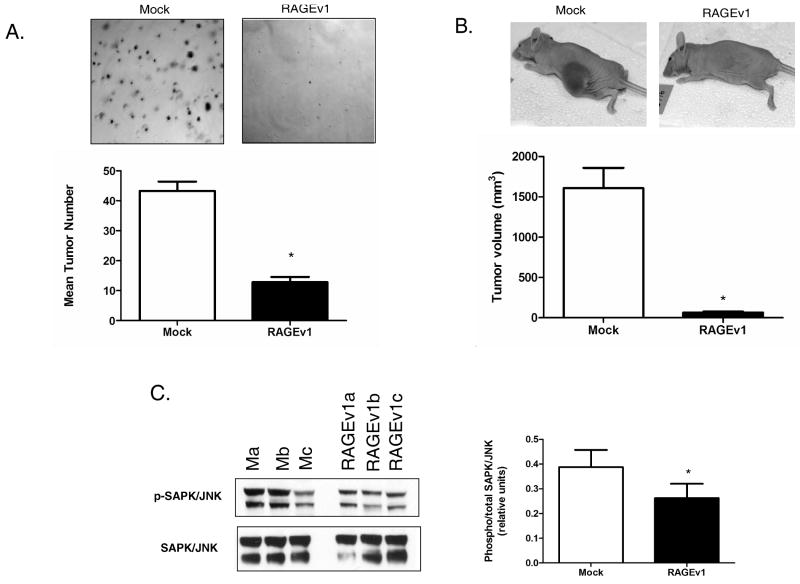
Finally, we investigated whether RAGEv1 expression affected tumor cell growth in vitro and in vivo, and if these effects are linked to SAPK/JNK signaling. To determine whether RAGEv1 modulates tumor formation, we performed the soft agar assay. This is perhaps the most consistent in vitro method to replicate the in vivo tumorigenic environment. Control and RAGEv1 cells were assayed for anchorage-independent growth for 14 days. Compared to mock cells, RAGEv1 expression drastically reduced the ability of tumor cells to form colonies in soft agar (Fig. 6A). Athymic nude mice were injected with mock or RAGEv1 cells and tumor growth was measured after 31 days. Compared to control cells, there was a ∼25-fold lower volume in tumors developed from RAGEv1-expressing cells (Fig. 6B). In parallel with these effects, western blotting revealed that expression of RAGEv1 significantly reduced SAPK/JNK activation by ∼2 fold compared to control tumors (Fig. 6C).
Figure 6.
RAGEv1 inhibits tumorigenesis in vitro and in vivo. (E) RAGEv1 inhibits tumorigenic growth in vitro. Soft agar assay were performed with mock and RAGEv1 transfected cells cultured for 14 days and stained with crystal violet. (A) Athymic nude mice were injected with either mock or RAGEv1 expressing tumor cells and tumor growth measured after 31 days. (B) Protein was extracted ex vivo from mock (Ma-c) and RAGEv1 (RAGEv1a-c) tumors and subjected to western blot for phospho and total levels of SAPK/JNK. Significant differences (P ≤ 0.05) between groups are indicated by an asterisk.
Discussion
The generation of soluble receptor isoforms represents an important mechanism to regulate aberrant receptor signaling in biological systems and a potential therapeutic strategy for cancer. Here, we have identified for the first time a functional role for the endogenous soluble RAGEv1 isoform as a molecular decoy for RAGE, and as an inhibitor of key cellular properties that facilitate tumorigenesis. Firstly, RAGEv1 expression in tumor cells inhibited RAGE-induced gene expression profiles favoring tumor cell mediated angiogenesis, invasion, cell cycle progression and evasion of cell death. Secondly, functional studies in vitro demonstrated that RAGEv1 strongly inhibited RAGE ligand induced angiogenesis, tumor cell invasion / metastasis and JNK activation. Finally and importantly, in vitro and in vivo studies firmly establish that expression of RAGEv1 strongly suppresses tumor growth. We conclude that RAGEv1 acts as a molecular decoy for RAGE to block the tumorigenic process and therefore strategies to increase RAGEv1 levels could be useful in the treatment of cancer.
A growing body of evidence suggests that the inhibition of RAGE has the potential to be a specific and effective anti-cancer therapeutic strategy. In particular, RAGE and its diverse ligands have been shown to be over-expressed in numerous tumorigenic states and blocking their expression/interaction inhibits tumor growth and metastasis (2;4). In this regard, these means of inhibition have included the use of RAGE blocking antibodies, dominant-negative receptor constructs and the recombinant soluble ligand-binding extracellular domain (sRAGE) (2;4;13). However, the occurrence of an endogenous soluble receptor system (RAGEv1) suggests the possibility of a natural means to block RAGE signaling. Furthermore, serum levels of RAGEv1 have been shown to be inversely correlated with various inflammatory disease states, suggesting the role of RAGEv1 as a potentially useful biomarker (26-28). These findings indicate that RAGEv1 may affect the clinical course of RAGE-driven pathologies, including various tumorigenic states, by blocking RAGE:ligand interaction. However, to-date no studies have investigated the molecular and cellular significance of RAGEv1 in vascular and tumor biology. Here, we tested the hypothesis that RAGEv1 acts as a molecular decoy for RAGE, blocking tumor- promoting signaling and in turn, inhibiting the tumorigenic processes both in vitro and in vivo.
To explore the molecular mechanisms through which RAGEv1 may affect RAGE-induced tumorigenesis, we analyzed gene expression profiles in tumor cells. Our data revealed that compared to control cells, RAGEv1 altered expression of genes associated with invasion/metastasis, apoptosis and cell cycle control. However, intriguingly the genes most dramatically altered were associated with angiogenesis. In vitro cellular assays revealed RAGEv1 strikingly inhibited RAGE ligand induction of angiogenesis and cellular invasion. However, although RAGEv1 induced tumor cell adhesion and cell death, we were unable to demonstrate any further effects of exogenous RAGE ligand stimulation on these processes, therefore suggesting a role for endogenous ligand generation by these cells. These results suggest that RAGEv1 inhibits multiple RAGE ligand-mediated processes leading to tumor formation and progression. The surprising finding that RAGEv1 blocks tumor cell expression of genes influencing angiogenesis implicates RAGE/RAGEv1 in multiple facets of tumor development. The recent finding that RAGE influences tumor development through inflammatory cell recruitment (5) and our angiogenesis data in this study, suggest that the RAGE/RAGEv1 system can influence not only tumor cells, but also cells of the tumor microenvironment. Furthermore, previous studies have demonstrated in human tumor tissue a correlation between RAGE expression levels and tumor vessel density (34;35). This therefore further implicates an essential role for RAGEv1 in countering the angiogenic effects of RAGE in the process of tumor angiogenesis.
To identify the signaling pathways eliciting these RAGEv1 mediated effects, we studied various components of the MAPK pathway. Work from both our group and others have definitely shown that RAGE predominantly acts as a signal transduction receptor for its ligands, and interruption of RAGE signaling, abrogates cellular dysfunction and disease pathogenesis (2;8-13). Furthermore, numerous studies have demonstrated a central role of MAPK signaling in mediating RAGE-ligand effects (9;36). In agreement with these data, we observed that RAGEv1 blocked RAGE-ligand activation of the major MAPK pathways including MEK, p38 and JNK. Importantly to note, neither RAGE-ligand stimulation nor RAGEv1 affected AKT signaling in these cells. Further investigation of the specific MAPK pathway responsible for driving pro-tumorigenic gene expression, revealed JNK signaling to predominantly play a role. The JNK pathway has been shown to be crucially involved in cancer development through affecting a wide range of cellular processes including proliferation, survival and invasion (37;38). Moreover, our results demonstrate that JNK activation is responsible for the increased tumor cell invasion seen through activation by RAGE ligand. Together, these data suggest that RAGEv1 acts to block tumorigenesis through inhibiting JNK signaling induced by RAGE ligand interaction.
To examine whether RAGEv1 may play a role in tumor formation, we used an in vitro tumor formation assay and an in vivo ectopic murine tumor model. In our experiments, RAGEv1 significantly inhibited tumor growth, which was consistent with previous observations using approaches to block RAGE-ligand signaling (2). In agreement with the in vitro signaling data, JNK activation was downregulated in tumors expressing RAGEv1, suggesting that JNK may be involved in RAGE ligand mediated tumorigenesis in vivo. However, due to the highly significant inhibition of tumorigenic growth in the RAGEv1 expressing cells, the very small size of the resulting tumors rendered it difficult to perform extensive histological analyses, such as quantification of blood vessel density.
Intriguingly, to support these findings, our studies of human tumor versus unaffected tissue suggest that RAGEv1 expression levels are decreased in tumor tissue. These findings suggest that at some point in the tumorigenic process, shut-down of RAGEv1 expression may facilitate tumor growth and invasion. Whether RAGEv1 is downregulated in the process preceding the tumor state, or in the course of tumorigenesis is a question that future studies need to address. Certainly, however, findings in human tumor versus adjacent normal tissue indicate that RAGEv1 levels are mutable. Recent data from multiple human cohort studies suggest that not only are RAGEv1 levels inversely correlated with inflammatory disease states, but therapies that target these pathologies lead to increased production of RAGEv1 (39;40). These data provide further evidence that not only are RAGEv1 levels modulatable, but they may be changed as a consequence of altering the inflammatory state during the course of disease pathology. Understanding the mechanism(s) regulating expression of cell surface RAGE and the production and/or release of RAGEv1, including the potential impact of ligand-RAGE interaction itself in these processes, will be important to investigate in future experimentation. Furthermore, what role RAGE-ligand signaling plays in their regulation will be interesting to investigate. This therefore raises the tantalizing prospect that RAGEv1 may not only act as a biomarker for inflammatory disease states, but targeted strategies to increase RAGEv1 levels may prove to be a useful therapy in the treatment of cancer by countering the effects of RAGE-ligand signaling.
In conclusion, our results suggest for the first time that RAGEv1 is likely not only a potential tumor biomarker, but that it may possess innate functions. Here, we show that RAGEv1 modulates tumor cell properties by affecting not only tumor cell survival, migration and invasion, but by regulating molecular pathways that influence the microenvironment to support tumor growth such as angiogenesis. We therefore propose that strategies to either suppress RAGE-ligand signaling in malignant tissue and/or to boost endogenous production and release of RAGEv1 in the tumor bed might represent highly potent strategies in the control of tumor growth and metastasis.
Supplementary Material
Acknowledgments
This work was supported by grants from the United States Public Health Service and the Juvenile Diabetes Research Foundation. BIH is a recipient of a Career Development Award from the Juvenile Diabetes Research Foundation International. AMS is a recipient of Scholar Award from the Juvenile Diabetes Research Foundation International. AZK is a recipient of a Postdoctoral Fellowship from the Juvenile Diabetes Research Foundation International.
This publication was also made possible by Grant Number UL1 RR024156 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000 Jan 7;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, et al. Blockage of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–60. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 3.Sparvero LJ, safu-Adjei D, Kang R, Tang D, Amin N, Im J, et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J Transl Med. 2009;7:17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logsdon CD, Fuentes MK, Huang EH, Arumugam T. RAGE and RAGE ligands in cancer. Curr Mol Med. 2007 Dec;7(8):777–89. doi: 10.2174/156652407783220697. [DOI] [PubMed] [Google Scholar]
- 5.Gebhardt C, Riehl A, Durchdewald M, Nemeth J, Furstenberger G, Muller-Decker K, et al. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med. 2008 Feb 18;205(2):275–85. doi: 10.1084/jem.20070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan SF, Ramasamy R, Schmidt AM. Mechanisms of disease: advanced glycation end-products and their receptor in inflammation and diabetes complications. Nat Clin Pract Endocrinol Metab. 2008 May;4(5):285–93. doi: 10.1038/ncpendmet0786. [DOI] [PubMed] [Google Scholar]
- 7.Kalea AZ, Schmidt AM, Hudson BI. RAGE: a novel biological and genetic marker for vascular disease. Clin Sci (Lond) 2009 Apr;116(8):621–37. doi: 10.1042/CS20080494. [DOI] [PubMed] [Google Scholar]
- 8.Lander HM, Tauras JM, Ogiste JS, Hori O, Moss RA, Schmidt AM. Activation of the receptor for advanced glycation end products triggers a p21(ras)-dependent mitogen-activated protein kinase pathway regulated by oxidant stress. J Biol Chem. 1997;272:17810–4. doi: 10.1074/jbc.272.28.17810. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann MA, Drury S, Fu CF, Qu W, Taguchi A, Lu Y, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for s100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 10.Kislinger T, Tanji N, Wendt T, Qu W, Lu Y, Ferran LJ, et al. Receptor for Advanced Glycation End Products Mediates Inflammation and Enhanced Expression of Tissue Factor in Vasculature of Diabetic Apolipoprotein E-Null Mice. Arterioscler Thromb Vasc Biol. 2001 Jun 1;21(6):905. doi: 10.1161/01.atv.21.6.905. [DOI] [PubMed] [Google Scholar]
- 11.Huttunen HJ, Fages C, Rauvala H. Receptor for Advanced Glycation End Products (RAGE)-mediated Neurite Outgrowth and Activation of NF-kappa B Require the Cytoplasmic Domain of the Receptor but Different Downstream Signaling Pathways. J Biol Chem. 1999 Jul 9;274(28):19919–24. doi: 10.1074/jbc.274.28.19919. [DOI] [PubMed] [Google Scholar]
- 12.Kislinger T, Fu C, Huber C, Qu W, Taguchi A, Yan SD, et al. N(epsilon)-(carboxymethyl)lysine adducts of proteins are ligands for receptor for advanced glycation end products that activate cell signaling pathways and modulate gene expression. J Biol Chem. 1999;274:31740–9. doi: 10.1074/jbc.274.44.31740. [DOI] [PubMed] [Google Scholar]
- 13.Hudson BI, Kalea AZ, Arriero MD, Harja E, Boulanger E, D'Agati V, et al. Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42. J Biol Chem. 2008 Oct 15;283:34457–68. doi: 10.1074/jbc.M801465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YCP, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267(21):14998–5004. [PubMed] [Google Scholar]
- 15.Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of RAGE and amphoterin in the developing nervous system. J Biol Chem. 1995;270(43):25752–61. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- 16.Hermani A, De SB, Medunjanin S, Tessier PA, Mayer D. S100A8 and S100A9 activate MAP kinase and NF-kappaB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp Cell Res. 2006 Jan 15;312(2):184–97. doi: 10.1016/j.yexcr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Harpio R, Einarsson R. S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma. Clin Biochem. 2004 Jul;37(7):512–8. doi: 10.1016/j.clinbiochem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 18.van Heijst JW, Niessen HW, Hoekman K, Schalkwijk CG. Advanced glycation end products in human cancer tissues: detection of Nepsilon-(carboxymethyl)lysine and argpyrimidine. Ann N Y Acad Sci. 2005 Jun;1043:725–33. doi: 10.1196/annals.1333.084. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh HL, Schafer BW, Sasaki N, Heizmann CW. Expression analysis of S100 proteins and RAGE in human tumors using tissue microarrays. Biochem Biophys Res Commun. 2003 Jul 25;307(2):375–81. doi: 10.1016/s0006-291x(03)01190-2. [DOI] [PubMed] [Google Scholar]
- 20.Kuniyasu H, Oue N, Wakikawa A, Shigeishi H, Matsutani N, Kuraoka K, et al. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J Pathol. 2002 Feb;196(2):163–70. doi: 10.1002/path.1031. [DOI] [PubMed] [Google Scholar]
- 21.Sasahira T, Akama Y, Fujii K, Kuniyasu H. Expression of receptor for advanced glycation end products and HMGB1/amphoterin in colorectal adenomas. Virchows Arch. 2005 Apr;446(4):411–5. doi: 10.1007/s00428-005-1210-x. [DOI] [PubMed] [Google Scholar]
- 22.Ishiguro H, Nakaigawa N, Miyoshi Y, Fujinami K, Kubota Y, Uemura H. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate. 2005 Jun 15;64(1):92–100. doi: 10.1002/pros.20219. [DOI] [PubMed] [Google Scholar]
- 23.Yonekura H, Yamamoto Y, Sakurai S, Petrova RG, Abedin MJ, Li H, et al. Novel splice variants of the receptor for advanced glycation end-products expressed in human vascular endothelial cells and pericytes, and their putative roles in diabetes-induced vascular injury. Biochem J. 2003 Mar 15;370(Pt 3):1097–109. doi: 10.1042/BJ20021371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson BI, Carter AM, Harja E, Kalea AZ, Arriero M, Yang H, et al. Identification, classification, and expression of RAGE gene splice variants. FASEB J. 2007 Dec 18; doi: 10.1096/fj.07-9909com. [DOI] [PubMed] [Google Scholar]
- 25.Galichet A, Weibel M, Heizmann CW. Calcium-regulated intramembrane proteolysis of the RAGE receptor. Biochem Biophys Res Commun. 2008 May 23;370(1):1–5. doi: 10.1016/j.bbrc.2008.02.163. [DOI] [PubMed] [Google Scholar]
- 26.Katakami N, Matsuhisa M, Kaneto H, Matsuoka TA, Sakamoto K, Nakatani Y, et al. Decreased endogenous secretory advanced glycation end product receptor in type 1 diabetic patients: its possible association with diabetic vascular complications. Diabetes Care. 2005 Nov;28(11):2716–21. doi: 10.2337/diacare.28.11.2716. [DOI] [PubMed] [Google Scholar]
- 27.Choi KM, Yoo HJ, Kim HY, Lee KW, Seo JA, Kim SG, et al. Association between endogenous secretory RAGE, inflammatory markers and arterial stiffness. Int J Cardiol. 2008 Jan 9; doi: 10.1016/j.ijcard.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 28.Koyama H, Shoji T, Fukumoto S, Shinohara K, Shoji T, Emoto M, et al. Low circulating endogenous secretory receptor for AGEs predicts cardiovascular mortality in patients with end-stage renal disease. Arterioscler Thromb Vasc Biol. 2007 Jan;27(1):147–53. doi: 10.1161/01.ATV.0000251502.88818.4b. [DOI] [PubMed] [Google Scholar]
- 29.Lindsey JB, de Lemos JA, Cipollone F, Ayers CR, Rohatgi A, Morrow DA, et al. Association between circulating soluble receptor for advanced glycation end products and atherosclerosis: observations from the Dallas Heart Study. Diabetes Care. 2009 Jul;32(7):1218–20. doi: 10.2337/dc09-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tesarova P, Kalousova M, Jachymova M, Mestek O, Petruzelka L, Zima T. Receptor for advanced glycation end products (RAGE)--soluble form (sRAGE) and gene polymorphisms in patients with breast cancer. Cancer Invest. 2007 Dec;25(8):720–5. doi: 10.1080/07357900701560521. [DOI] [PubMed] [Google Scholar]
- 31.Jing R, Cui M, Wang J, Wang H. Receptor for advanced glycation end products (RAGE) soluble form (sRAGE): a new biomarker for lung cancer. Neoplasma. 2010;57(1):55–61. doi: 10.4149/neo_2010_01_055. [DOI] [PubMed] [Google Scholar]
- 32.Grobben B, De Deyn PP, Slegers H. Rat C6 glioma as experimental model system for the study of glioblastoma growth and invasion. Cell Tissue Res. 2002 Dec;310(3):257–70. doi: 10.1007/s00441-002-0651-7. [DOI] [PubMed] [Google Scholar]
- 33.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008 Dec 25;359(26):2814–23. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasahira T, Kirita T, Bhawal UK, Ikeda M, Nagasawa A, Yamamoto K, et al. The expression of receptor for advanced glycation end products is associated with angiogenesis in human oral squamous cell carcinoma. Virchows Arch. 2007 Mar;450(3):287–95. doi: 10.1007/s00428-006-0359-2. [DOI] [PubMed] [Google Scholar]
- 35.Tsuji A, Wakisaka N, Kondo S, Murono S, Furukawa M, Yoshizaki T. Induction of receptor for advanced glycation end products by EBV latent membrane protein 1 and its correlation with angiogenesis and cervical lymph node metastasis in nasopharyngeal carcinoma. Clin Cancer Res. 2008 Sep 1;14(17):5368–75. doi: 10.1158/1078-0432.CCR-08-0198. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka N, Yonekura H, Yamagishi S, Fujimori H, Yamamoto Y, Yamamoto H. The receptor for advanced glycation end products is induced by the glycation products themselves and tumor necrosis factor-a through nuclear factor-kB and by 17B-estradiol through SP-1 in human vascular endothelial cells. J Biol Chem. 2000;275:25781–90. doi: 10.1074/jbc.M001235200. [DOI] [PubMed] [Google Scholar]
- 37.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009 Aug;9(8):537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 38.Salh B. c-Jun N-terminal kinases as potential therapeutic targets. Expert Opin Ther Targets. 2007 Oct;11(10):1339–53. doi: 10.1517/14728222.11.10.1339. [DOI] [PubMed] [Google Scholar]
- 39.Santilli F, Bucciarelli L, Noto D, Cefalu AB, Davi V, Ferrante E, et al. Decreased plasma soluble RAGE in patients with hypercholesterolemia: effects of statins. Free Radic Biol Med. 2007 Nov 1;43(9):1255–62. doi: 10.1016/j.freeradbiomed.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 40.Tan KC, Chow WS, Tso AW, Xu A, Tse HF, Hoo RL, et al. Thiazolidinedione increases serum soluble receptor for advanced glycation end-products in type 2 diabetes. Diabetologia. 2007 Sep;50(9):1819–25. doi: 10.1007/s00125-007-0759-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.