Abstract
Vitamin K epoxide reductase (VKOR) generates vitamin K hydroquinone to sustain γ-carboxylation of many blood coagulation factors. Here, we report the 3.6Å crystal structure of a bacterial homolog of VKOR from Synechococcus sp. The structure shows VKOR in complex with its naturally fused redox partner, a thioredoxin-like domain, and corresponds to an arrested state of electron transfer. The catalytic core of VKOR is a four transmembrane helix bundle that surrounds a quinone, connected through an additional transmembrane segment with the periplasmic thioredoxin-like domain. We propose a pathway for how VKOR uses electrons from newly synthesized proteins to reduce a quinone, a mechanism confirmed by in vitro reconstitution of vitamin K-dependent disulfide bridge formation. Our results have implications for the mechanism of the mammalian VKOR and explain how mutations can cause resistance to the VKOR inhibitor warfarin, the most commonly used oral anticoagulant.
Mammalian VKOR catalyzes a decisive step in the vitamin K cycle (Fig. S1a) that is required to sustain blood coagulation (reviewed in ref. 1,2). The cycle occurs in the endoplasmic reticulum (ER) membrane and begins with the γ-carboxylation of select glutamic acids in coagulation factors, a modification required for Ca2+-dependent activation at sites of injury (reviewed in ref. 3,4). The γ-carboxylase requires vitamin K hydroquinone, which is oxidized to vitamin K epoxide. VKOR is necessary to regenerate the hydroquinone; VKOR reduces the epoxide in two steps, first to the quinone and then to the hydroquinone. The reducing equivalents have been proposed to come from protein disulfide isomerase (PDI), an ER-luminal enzyme with several thioredoxin (Trx)-like domains; PDI would in turn be reduced during disulfide bridge formation of newly synthesized proteins 5,6.
Both reaction steps catalyzed by VKOR are inhibited by warfarin 7 (Fig. S1b), a drug that is widely used to treat thromboembolic diseases, including deep vein thrombosis, pulmonary embolism, stroke, and myocardial infarction. One challenge of warfarin therapy is its narrow therapeutic window; overdose can cause severe hemorrhage that is often lethal. Genetic variation among patients causes significant differences in the required warfarin doses and has served as a model case for pharmacogenetic drug treatment 8. Many warfarin resistance mutations map to VKORC1, the gene encoding VKOR 9. However, how exactly warfarin inhibits VKOR is unclear.
Homologs of VKOR have been identified in a variety of organisms, including some plants, bacteria, and archaea 10–12 (Fig. S2). All homologs contain an active site CXXC motif, which in mammalian VKOR switches between reduced and disulfide-bonded states during the reaction cycle, as well as an additional pair of cysteines and a conserved serine/threonine. Bacterial VKOR homologs catalyze disulfide bridge formation in secreted proteins by cooperating with a periplasmic, Trx-like redox partner 11,12. In some species, a single polypeptide chain contains both VKOR and a Trx-like domain.
We have determined the crystal structure of such a natural fusion protein derived from Synechococcus sp., in which VKOR and its Trx-like partner are caught in an intermediate stage of the redox reaction. The structure of this VKOR-complex not only sheds light on the redox pathway, but also has significant implications for the function of the mammalian counterpart and the mechanism by which VKOR mutations cause warfarin resistance. Because VKOR homologs are found in a number of medically important bacteria, including M. tuberculosis 12, our structure could also aid in the development of new antibiotics.
Structure of VKOR with its redox partner
The VKOR homolog from Synechococcus sp., naturally fused to a Trx-like domain, was expressed in E. coli and purified in the detergent n-dodecyl-β-D-maltoside (DDM). The purified protein contained bound endogenous quinones, primarily ubiquinone (mass spectrometry data not shown), and showed a characteristic UV absorption at 400nm 13. Because the wild type protein did not yield high-quality crystals, we tested mutants, in which cysteines were changed to alanines or serines to stabilize an interdomain disulfide bridge 14. The VKOR domain mutant C56S gave crystals that belonged to the P61 space group (Table S1). Experimental phases with a maximum resolution of 3.6Å were obtained by the multiple isomorphous replacement method and were dramatically improved by solvent flattening due to the 80% solvent content. The membrane helices were well resolved in the resulting electron density map (Fig. S3), likely because they are packed side by side (Fig. S4). A model could be built with confidence, and the register was confirmed by the positions of aromatic residues and by specific binding sites of heavy atoms (Fig. S5a). In addition, the positions of all methionines were confirmed by crystallizing seleno-methionine substituted protein (Fig. S5b). The Trx-like domain was also crystallized by itself; these crystals diffracted to 1.7Å (Table S1). The phases were obtained by the single anomalous diffraction method using a mercury derivative and a model was built automatically (Fig. S6). The structure of the Trx-like domain docked well into the map of the full-length protein. The model of the complex of VKOR and its redox partner was refined to Rwork and Rfree factors of 24.6% and 30.5%, respectively. The model lacks the 15 N-terminal and 5 C-terminal residues of the full-length protein and has uncertainties in the region encompassing amino acids 53 to 55.
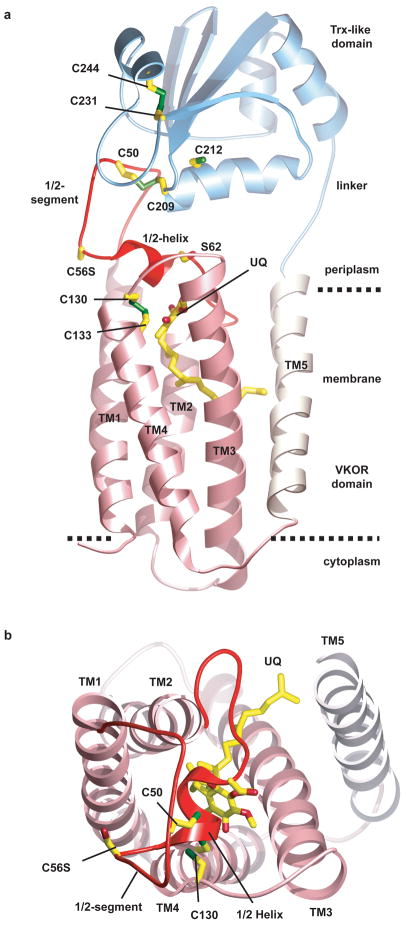
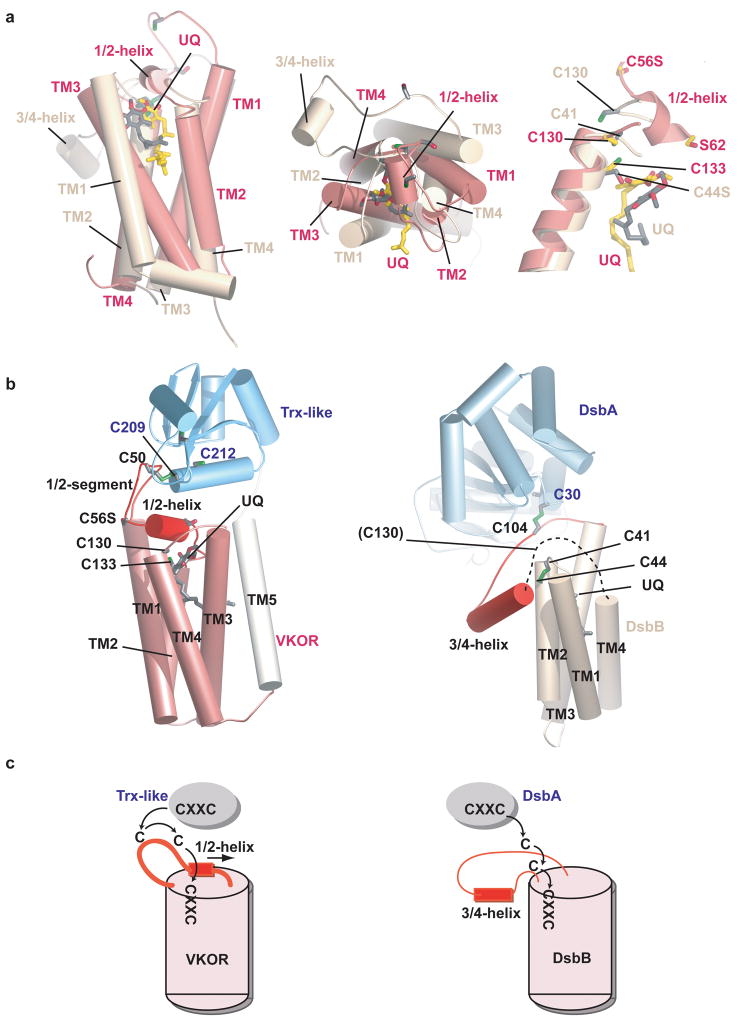
The protein consists of the membrane-embedded domain containing VKOR, connected via a linker segment to the extracellular Trx-like domain (Fig. 1a; see also Movie S1). The membrane-embedded part contains five transmembrane helices (TM) (Figs. 1a, b). TMs 1-4 are homologous to human VKOR (Fig. S2; ~24% identical amino acids). Consistent with in vivo experiments 15, both termini and a loop between TMs 2 and 3 are located in the cytosol. The helices form a four-helix bundle with a quinone in its interior. The quinone is close to the periplasmic side of the membrane, in proximity to the CXXC active site motif in TM4 (formed by Cys130 and Cys133). TM5 passes through the membrane to connect with the fused periplasmic Trx-like domain. TM5 is located on the outside of the four-helix bundle, contacting TM3 (Fig. 1b) and the loop between TM1 and 2.
Figure 1. Architecture of Synechococcus VKOR in complex with its redox partner.
a, Overall structure of the protein, consisting of VKOR and the thioredoxin (Trx)-like domain. TMs 1-4 that are homologous to the mammalian VKOR are colored in pink. The loop between TMs 1 and 2, containing the 1/2-segment and 1/2-helix, is shown in red. Ubiquinone (UQ) is shown in stick presentation. Sulfur atoms in cysteines are indicated in green. The dotted lines indicate the membrane boundaries. b, View of VKOR from the periplasm. The Trx-like domain was removed for clarity.
The longest loop between TMs is on the periplasmic side of the VKOR homolog, between TM1 and TM2. This 1/2-loop contains an extended N-terminal part (called the “1/2-segment”), a short helix (called the “1/2-helix”), and a C-terminal segment that contacts TM5 (Fig. 1a). The 1/2-segment contains a pair of cysteines (Cys50 and Cys56), which are present in all VKORs. The 1/2-helix begins after Cys56 and ends with the conserved serine/threonine residue (Ser62). In most species, there are five residues between the second cysteine and the conserved serine/threonine (Fig. S2), suggesting that the 1/2-helix is a common feature. The 1/2-helix forms a lid on the four-helix bundle, shielding the quinone from the periplasmic space. The helix is amphipathic and likely lies on top of the lipid surface, with the well-conserved Trp64 as a major anchor point.
Consistent with the introduced mutation Cys56Ser, an inter-domain disulfide bridge with strong density at 3.5σ is observed between Cys50 in the VKOR domain and Cys209 in the CXXC motif of the Trx-like domain. Formation of the inter-domain disulfide bridge is facilitated by the insertion of the 1/2-segment of VKOR into the active site groove of the Trx-like domain (Fig. S7). This groove is formed by several loops between the secondary structure elements, has a hydrophobic surface, and contains the active site CXXC motif. The groove likely also interacts with newly synthesized protein substrates, allowing disulfide exchanges. The Trx-like domain also contains two other cysteines (Cys231 and Cys244), which are disulfide bonded and likely play a structural role.
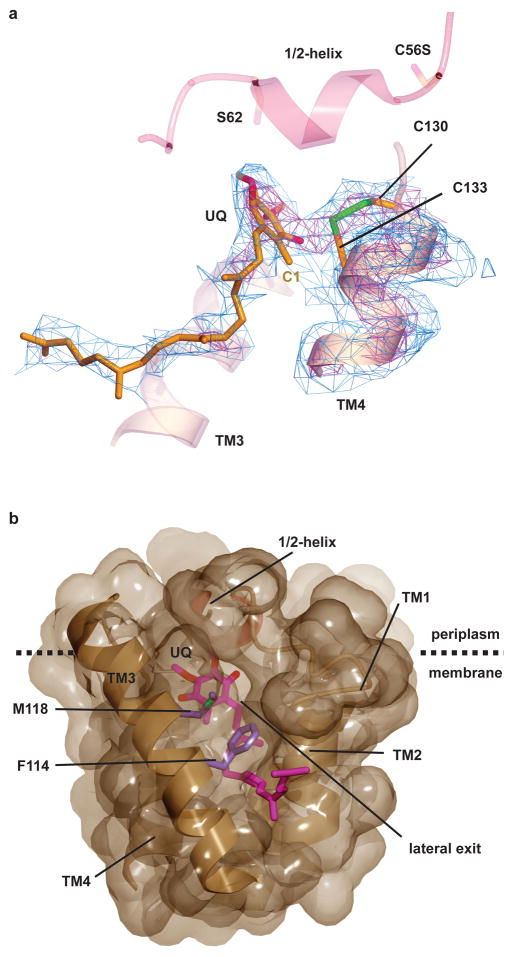
The plane of the quinone ring is clearly defined (Fig. 2a; see also Fig. S8 and Movie S2), tilted by 70° with respect to the membrane surface. Much of the isoprenyl tail of the ubiquinone can also be seen, intercalated into the V-shaped cleft between TMs 2 and 3 (Fig. 2b). The location of the isoprenyl tail establishes the positions of the substituents on the quinone ring (Fig. 2a; for chemical structure, see Fig. S1b). Strong, continuous density is observed in the ~2Å gap between the C1 position of ubiquinone and Cys133 in the CXXC motif of VKOR, suggesting that they are linked by a covalent bond. Cys130 and Cys133 are located at the side of the quinone ring and there is strong density between the two sulfur atoms. It thus appears that the structure corresponds to a situation in which electrons are shared between the disulfide bridge in the CXXC motif and the quinone ring.
Figure 2. The active site of VKOR.
a, The active site of VKOR, including the cysteines of the CXXC motif in TM4 and the ubiquinone molecule (UQ). The experimental electron density map is contoured at 1σ (blue) and 3.5σ (red). Note the strong electron density between Cys130 and Cys133, indicative of a disulfide bridge (in green), as well as between Cys133 and the C1 position of the quinone. TM3 and the 1/2-helix are shown without electron density for orientation. b, Surface representation of the binding pocket for the quinone (in red), viewed from the side. TM5 was removed for clarity. Note the cleft between TMs 2 and 3, blocked only by the side chains shown in purple, which could provide a lateral exit for the quinone into lipid. The lower part of the membrane is not shown.
Implications for the mammalian VKOR
Although a three-TM model has been proposed for human VKOR 16, our structure and multiple sequence alignments predict that the core of all VKORs, including those in mammals, consist of four TMs 10 (Fig. S2). This topology model places all enzymatically important residues on or close to the extracellular/luminal side of the membrane. These residues include the active site CXXC motif, the conserved serine/threonine in the 1/2-helix, and the two cysteines in the 1/2-segment. This topology would allow transfer of reducing equivalents from the Trx-like redox partner via the loop cysteines to the quinone in VKOR (see below).
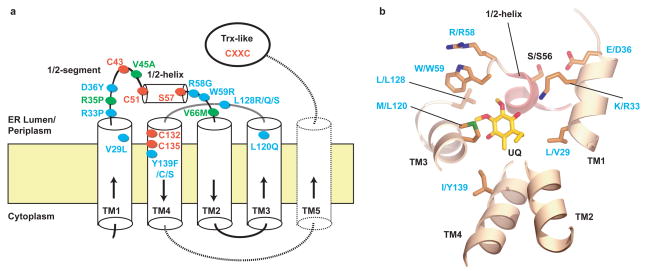
The structure allows us to understand how mutations in the mammalian VKOR can cause warfarin resistance, particularly because warfarin-resistance mutations selected in a bacterial VKOR homolog map to the same residues 15. There are eleven confirmed mutations in mammalian VKORs 9,17, eight of which map close to the binding site of the quinone (Figs. 3a, b). These include mutations of Val29, Arg33, and Asp36 in TM1 (numbering for the human VKOR), Arg58 and Trp59 immediately following the 1/2-helix, Leu120 and Leu128 in TM3, as well as Tyr139 in TM4, a residue that has been implicated in warfarin binding 2,18. The striking clustering of the mutations indicates that warfarin binds to the same site as the quinone seen in our structure, or at least close to it (Fig. 3b). The binding pocket is large enough to accommodate either the naphthoquinone vitamin K or warfarin, but not both, indicating that warfarin replaces vitamin K to cause inhibition. Some of the mutations around the quinone-binding site (Tyr139Phe 2,18,19, Leu120Gln, Leu128Gln/Ser/Arg 9,17,18, and Val29Leu) may lower the affinity for the drug. The Arg58Gly, Trp59Arg, and Asp36Tyr mutations might disturb the 1/2-helix as a lid for the binding pocket; Trp59Arg could displace the helix from the membrane and Asp36Tyr in TM1 might disrupt the interaction of the residue with Ser56 in the 1/2-helix. Mutations Arg35Pro, Val45Ala, and Val66Met 9,17,20 map to the 1/2-loop and could affect the active site of VKOR indirectly.
Figure 3. Mutations causing warfarin resistance in mammalian VKOR.
a, Membrane topology of VKORs. TMs 1-4 form the essential four-helix bundle found in all VKORs. Dotted lines indicate TM5 and the linker to the Trx-like domain found in Synechococcus sp. The conserved cysteines and serine/threonine are shown as red ovals. Warfarin resistance mutations in mammalian VKOR located close to the bound quinone or elsewhere are indicated as blue or green ovals, respectively. b, Mutations indicated in blue in a are mapped into the Synechococcus structure, based on sequence alignment (Fig. S2). Some TM segments are removed for clarity. Labels refer to the residue in Synechococcus VKOR, followed by the residue and its position in human VKOR. The positions V29, R33, and D36 are in a helical extension of TM1, but their location is somewhat uncertain because of alignment ambiguities.
Our structure supports the idea that the ER luminal redox partner of the mammalian VKOR is a Trx-like protein. Although there is a whole family of such proteins in the ER lumen, PDI is proposed to be the partner of VKOR 5,6. In this case, the first Trx-like domain of the enzyme (a-domain) would likely be responsible for the interaction, as it contains an active site CXXC motif and is the only thioredoxin fold of PDI that can be superimposed onto our structure without causing significant parts of the molecule to clash with the membrane (Fig. S9). Because the Trx-like protein in the ER lumen ultimately donates its disulfide bridge to newly synthesized proteins, it is possible that VKOR or an uncharacterized VKOR-like protein 9 may contribute more generally to disulfide bond formation, even when uncoupled from its better characterized role in γ-carboxylation.
Electron transfer pathway
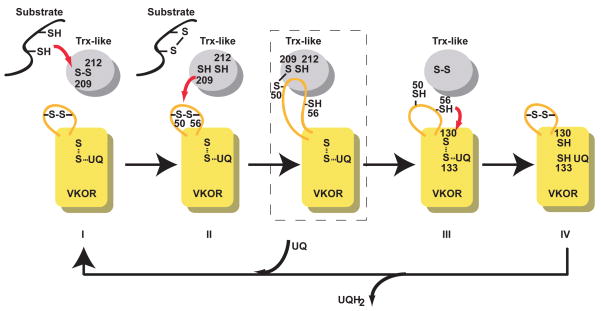
Our structure corresponds to an arrested intermediate during the transfer of electrons from the Trx-like domain to the VKOR. The disulfide bridge between Cys209 of the Trx-like domain and Cys50 of VKOR would normally be transient, as it would be resolved by the formation of a disulfide bond between Cys50 and Cys56 (confirmed by mass spectrometry of the wild type protein). In our structure, this step is prevented by mutation of Cys56.
We propose the following pathway of electron transfer, which would be applicable to all VKORs (Fig. 4). First, a newly synthesized protein reduces the CXXC motif of the Trx-like protein in the periplasm/ER lumen. Second, the Trx-like protein reduces the disulfide bridge between Cys50 and Cys56 in the 1/2-loop of VKOR, with a mixed disulfide bridge as an intermediate. Third, the loop cysteines reduce the CXXC motif of VKOR. Finally, the disulfide in this CXXC motif is reformed by reduction of the quinone. The net effect would be the movement of reducing equivalents from cysteines in newly synthesized proteins to the quinone.
Figure 4. Electron transfer pathway.
The scheme shows the flow of electrons from a newly synthesized protein (substrate) in the periplasm of bacteria or the ER lumen of eukaryotes to a quinone (UQ). The different states are indicated by Roman numerals. The boxed state corresponds to an intermediate state represented by the crystal structure. A similar intermediate was detected in vivo 15.
One implication of the structure is that a major conformational change is required to allow electrons to flow from the Trx-like domain to the quinone. Specifically, the amphipathic 1/2-helix must move out of the way, perhaps by sliding parallel to the plane of the membrane, to allow Cys56 in the loop to gain access to Cys130 of the CXXC motif. In the resting state, the 1/2-helix would shield the active site, preventing it from non-selectively oxidizing proteins in the periplasm.
Our model assumes that during the reaction cycle, a covalent bond is formed between the quinone and Cys133, consistent with the observed continuous electron density between them and with a previously proposed catalytic mechanism 21. Cys133 is located on the side of the quinone ring, a geometry that likely facilitates electron transfer to the aromatic systems and has been observed for the soluble FAD-containing thiol oxidases Ero1p 22 and Erv2p 23. Interestingly, the quinone appears to interact with Cys133 at an early stage of the redox process, when Cys133 is still disulfide bonded with Cys130 (Fig. 4, boxed intermediate, as represented by our structure). We propose that once the quinone is fully reduced, it would no longer be covalently linked to Cys133 and could then exit the binding pocket into the lipid phase through the cleft between TM2 and TM3 (Fig. 2b). The quinone would thus be exchanged during each round of the redox cycle.
Electron transfer in vitro
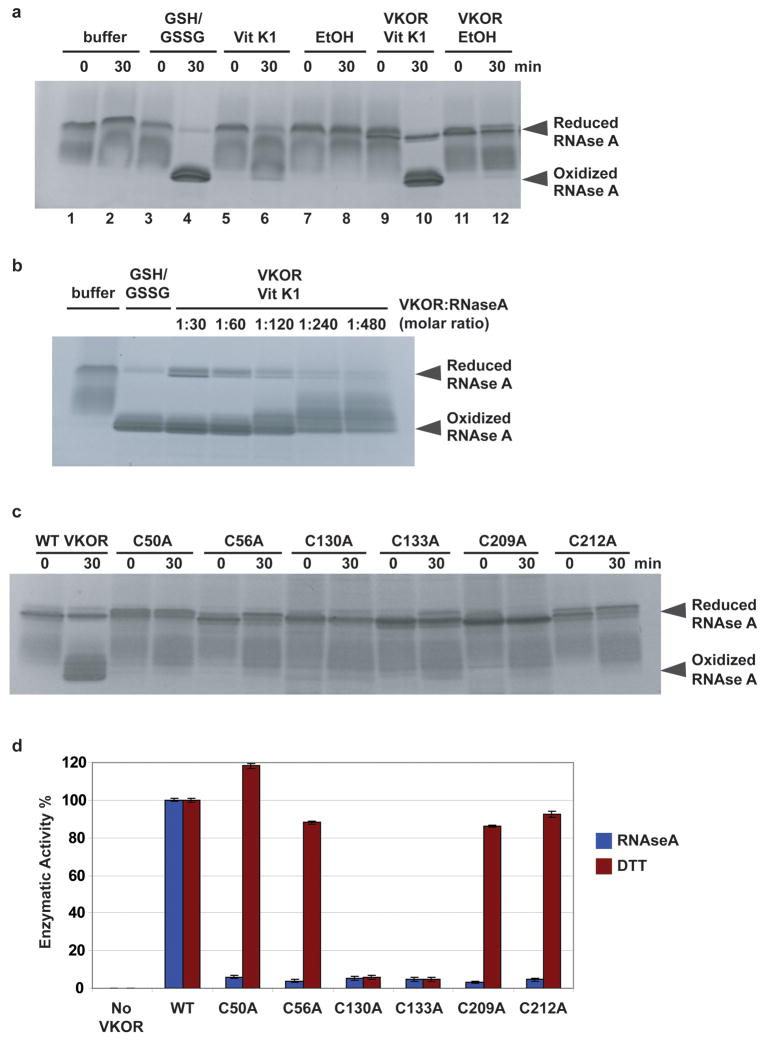
To verify the proposed electron transfer pathway, we established a reconstituted system for vitamin K-dependent oxidative protein folding. The purified Synechococcus sp. protein, containing both the VKOR and Trx-like domains, was incubated with reduced RNAse A, and disulfide bridge formation was assayed by protection from a cysteine-modifying reagent. The cysteines in RNAse A were oxidized only when vitamin K was added (Fig. 5a, lane 10 versus 12). Other quinones, such as ubiquinone Q1 and menaquinone (vitamin K2), were also active (data not shown). Dilution experiments show that a single VKOR molecule can catalyze the oxidation of ~500 cysteines (Fig. 5b). Mutation of any of the cysteines in the electron transfer pathway (Cys209 and Cys212 in the Trx-like domain and Cys50, Cys56, Cys130, and Cys 133 in VKOR; Fig. 4) abolish disulfide bridge formation (Fig. 5c). The activity of the bacterial VKOR homolog could also be demonstrated when the reduction of vitamin K was followed with a fluorometric assay. Again, with reduced RNAse A as substrate, none of the cysteine mutants was active (Fig. 5d). However, with dithiothreitol (DTT) as reductant, only the active site mutants Cys130Ala and Cys133Ala were inactive. Thus, DTT can directly reduce the active site, bypassing the other cysteines. These results argue that under physiological conditions, the Trx-like domain reduces the loop cysteines of VKOR, which in turn reduce the active site cysteines. Warfarin only inhibited the reaction at high concentrations (data not shown), but ferulenol, a close analog, was almost as potent as with the mammalian enzyme (half-maximum inhibition 0.2 μM versus 0.1 μM) (2; 24; Fig. S10). These data again emphasize the similarities between the bacterial and mammalian enzymes. It should be noted, however, that the mammalian VKOR catalyzes both the reduction of vitamin K epoxide to the quinone, and from the quinone to the hydroquinone, employing the same redox-active cysteines 25 (Fig. 4). The bacterial VKOR homologs might perform only the quinone to hydroquinone conversion. In fact, we have not been able to detect reduction of vitamin K epoxide by the enzyme from Synechococcus sp. (data not shown), and quinone epoxides have not been described in bacteria.
Figure 5. In vitro reconstitution of vitamin K-dependent oxidative folding.
a, Purified Synechococcus protein, containing the VKOR and Trx-like domains, was incubated with reduced, denatured RNAse A and vitamin K1 (Vit K1) (lanes 9, 10). The reaction was quenched with AMS, which modifies free sulfhydryl groups. Samples were analyzed by SDS-PAGE and Coomassie staining. Controls were performed without Vit K1 (in ethanol, EtOH) or VKOR, as indicated. RNAse A was also oxidized with glutathione (GSH/GSSG) (lanes 3,4). b, As in a, but with VKOR serially diluted. c, As in a, but with wild type (WT) or mutants in cysteines in the predicted electron transfer pathway (Fig. 4). d, As in c, except reduction of Vit K1 was followed in a fluorometer, using reduced RNAse or dithiothreitol (DTT) as reductant. Error bars represent standard error of mean instrumental fluorescence variation.
Pathways of disulfide formation
It is revealing to compare the VKOR pathway with the other known system of disulfide bridge formation in bacteria. Instead of VKOR, many bacteria possess DsbB, a sequence-unrelated membrane protein that shuttles electrons from the periplasmic, Trx-like DsbA protein to a quinone 26–28. Like VKOR, DsbB contains four TM segments and a quinone 14,29–31. DsbB also has an active site CXXC motif, a long loop containing two shuttle cysteines, and an amphipathic helix (3/4-helix) within this loop. A comparison of the structures shows that the TM segments of DsbB form a four-helix bundle that is similar to that of VKOR (Fig. 6a; left and middle panels). In both cases, the helices are arranged in a clock-wise manner when viewed from the periplasm, and encircle the quinone. However, the shuttle cysteines in DsbB are located in a loop between TMs 3 and 4, rather than between TMs 1 and 2, and the CXXC motif is present on the periplasmic side of TM2, rather than in TM4. In both enzymes, an amphipathic helix, the 1/2-helix in VKOR and the 3/4-helix in DsbB, sits at the membrane surface and may serve to anchor a loop cysteine close to the active site CXXC motif. The quinone is located at about the same position in VKOR and DsbB 30 (Fig. 6a; right panel). We postulate that in wild type DsbB, Cys44 would interact with the quinone, similarly to Cys133 in VKOR, even when the CXXC motif is oxidized. As with VKOR, the hydroquinone generated during each redox cycle might loose its interaction with the cysteine and be laterally released into lipid, although an alternative model in which the quinone is permanently bound has also been proposed 32.
Figure 6. Comparison of VKOR with DsbB.
a, Superposition of the four-helix bundles of VKOR (pink) and DsbB (yellow) 30, based on the helices containing the CXXC motifs. The left two panels show views from the side and periplasm. The right panel shows a close-up view of the active sites. b, Overall architecture of VKOR with its Trx-like redox partner (left panel) and of DsbB with its redox partner DsbA 14. The dotted line in the right panel indicates a segment disordered in the X-ray structure. c, Electron transfer pathways for VKOR and DsbB. Note that for VKOR the loop cysteine cannot access the active site CXXC motif unless the 1/2-helix is displaced. By contrast, in DsbB the 3/4-helix is peripheral and electron transfer can occur without major conformational change; the loop cysteines could not interact if the helix were positioned as in VKOR.
The structural similarities indicate that the electron transfer mediated by VKOR and DsbB occurs by a common general pathway. However, while the amphipathic 1/2-helix of VKOR follows the loop cysteines (Fig. 6b, 6c, left panel) and must relocate to allow electron transfer from the Trx-like domain to the quinone, DsbB does not require major conformational changes. DsbB’s amphipathic 3/4-helix separates the two loop cysteines, which are only able to interact due to the unusual, peripheral location of the helix with respect to the four-helix bundle (Fig. 6b, 6c, right panel). The reactive cysteines are all lined up (Cys30 in DsbA, Cys104, Cys41, and Cys44 in DsbB), allowing unimpeded electron transfer from the Trx-like domain to the quinone 30,31.
The two pathways of disulfide bridge formation in bacteria also share similarities with those in eukaryotes. Although the oxidases Ero1p and Erv2p are present in the ER lumen, rather than integrated into the membrane, they also contain four helices in association with a cofactor (FAD), a loop with shuttling cysteines, and an active site CXXC motif 22,23. Thus, disulfide bridge formation appears to generally employ similarly structured components for electron transfer, with VKOR representing the newest class of sulfhydryl oxidases.
Methods summary
The codon-optimized VKOR gene from Synechococcus sp. was expressed with a His tag in E. coli and the protein was purified by Ni- affinity and gel-filtration chromatography in 0.05% DDM. Crystals were obtained by the hanging drop diffusion method and frozen in liquid nitrogen. To obtain heavy atom derivatives, the crystals were soaked with mercury, platinum, or lead compounds. Phases were obtained by the MIR method and improved by solvent flattening. The Trx-like domain (amino acids 186 to 283) was expressed in E. coli, purified, and crystallized. SAD phases were calculated using heavy atom sites found by the direct method, and the model was built and refined automatically. This structure was docked into the density map of the full-length protein. The final model of the full-length protein was manually built and refined to Rwork and Rfree factors of 24.6% and 30.5%, respectively. The structure factors and coordinates were deposited in the protein data bank (accession code 3KP9 for the full-length protein and accession code 3KP8 for the Trx-like domain).
Disulfide bridge formation in RNAse A was assayed by combining purified VKOR protein (0.5 μM) with reduced, denatured RNAse A (15 μM) and vitamin K 1 quinone (100 μM); the reaction was quenched by addition of 5 × non-reducing SDS buffer supplemented with 50 mM AMS (4-Acetamido-4′-maleimidylstilbene 2,2′-disulfonic acid). Reduction of vitamin K (50 μM) was assayed fluorometrically by excitation at 250 nm and following the emission at 430 nm. The reaction was initiated by addition of either reduced RNAse A (15 μM) or DTT (1 mM). A detailed outline of the experimental procedures is given in Methods.
Online Methods
Purification, crystallization, and structure determination
The gene coding for the full-length VKOR protein from Synechococcus sp. was codon optimized (Genscript) and subcloned into the PET20b vector (Novagen). Cysteines in the protein were systematically mutagenized using Quikchange (Stratagene). The protein with the C56S mutation was expressed in a C43 E. coli strain and the membrane fraction was solubilized with 1% dodecyl-β-D-maltopyranoside (DDM). The protein was purified by Ni-affinity and gel filtration chromatography in a buffer containing 20mM Tris/HCl pH7.5, 0.3M NaCl, 10% glycerol, 0.05% DDM. The protein was concentrated to ~10 mg/ml and dialyzed against 20 mM Tris/HCl pH 7.5, 100 mM NaCl, 0.05% DDM. Crystals were grown by the hanging drop diffusion method at 22°C with 15% PEG1000, 0.2M magnesium chloride at pH 6.5–7.5 in the reservoir. The crystals were cryoprotected in 20% glycerol and frozen in liquid nitrogen. The data were collected at the ID24 beamline of Argonne National Laboratory and reduced by HKL2000 33. The crystals were soaked for 1 to 6 hours in 1mM ethylmercuricthiosalicylic acid, 1mM methylmercury chloride, 1mM p-chloromercuribenzoic acid, 1mM K2PtCl4, or 20mM acetoxytrimethyllead. Phases were obtained by MIR using a Patterson-based phasing method installed in SOLVE 34 and improved by solvent flattening using DM 35 and PARROT 36. The combined MIR phases have a figure of merit of 0.46 before density modification. Crystals soaked in ethylmercuricthiosalicylic acid diffracted to 3.6Å and were used for refinement. The Trx-like domain (amino acids 186 to 283) was expressed using the same vector and strain and purified by Ni-affinity and gel filtration chromatography. The protein was crystallized in 0.6M sodium potassium tartrate, 10% PEG MME 5000, pH 7.8 and cryoprotected with 30% PEG400. SAD phases were calculated in SHELX C/D/E 37 using heavy atom sites found by the direct method, and the model was built and refined automatically in ARP/WARP 38. This structure was docked into the density map of the full-length protein. The final models were built in O 39 and Coot 40, and refined with TLS parameters 41 in REFMAC 42. The model of the full-length protein has 3 residues (1.4%) in the disallowed region of the Ramachandran plot. All residues in the model of the Trx-like domain are in the allowed region of the Ramachandran plot.
In vitro reconstitution of electron transfer
Vitamin K-dependent oxidative folding was monitored by adapting a method used for Ero1 43. Purified VKOR protein (0.5 μM) was mixed with vitamin K quinone (100 μM) in a buffer containing 100 mM Tris/acetate pH 7.5, 65 mM NaCl, 2 mM EDTA, and 0.005% digitonin (or 0.05% DDM). The reaction was started by addition of reduced, denatured RNAse A (15 μM). Where indicated, the reaction buffer was supplemented with 1 mM reduced glutathione (GSH) and 0.2 mM oxidized glutathione (GSSG). Vitamin K was added in ethanol and its concentration was determined by the absorbance at 248 nm (E = 18,900 M−1cm−1). The reaction was quenched by addition of 5x non-reducing SDS buffer supplemented with 50 mM AMS (4-Acetamido-4′-maleimidylstilbene 2,2′-disulfonic acid, Molecular Probes) for 30 min at 25° C. Samples were analyzed by SDS-PAGE and visualized by Coomassie blue staining.
Reduced, denatured RNAse A was prepared by resuspending 5 mg RNAse A (Sigma, type XII) in 1 ml 100 mM Tris/acetate pH 8, 2 mM EDTA, 6 M guanidinium hydrochloride, and 140 mM DTT overnight at 4° C. Immediately prior to use, DTT and guanidinium hydrochloride were removed by gel filtration using a PD-10 gravity column (GE Healthcare). The concentration of reduced, denatured RNAse A was determined by the absorbance at 280 nm (E = 9.3 mM−1cm−1).
To follow the electron transfer reaction fluorometrically, purified VKOR protein (0.1 μM) was mixed with vitamin K 1 (300 μM) in a buffer containing 20 mM Tris/HCl pH 7.5, 0.1 M NaCl, 0.05% DDM. The reaction was initiated by adding reduced, denatured RNAse A (15 μM) or DTT (1 mM). Excitation was carried out at 250 nm and excitation was followed at 430 nm in a plate reader.
Supplementary Material
Acknowledgments
We thank C. Jao for help with mass spectroscopy, S. Harrison for insightful comments on the structure, B. Furie, B. C. Furie, and Y. Yu. for discussion, A. Osborne, B. van den Berg, J. Zimmer, and Y. Chen for critical reading of the manuscript, R. Zhang for help with the figures, the staff at Advanced Photon Source beamline ID-24C, and the SBGrid consortium at Harvard Medical School. S.S. is supported by an NIH Medical Scientist Training Program fellowship. J.B. is supported by grant GMO41883 from the National Institute of General Medical Sciences. W. L. is supported by a Charles King Trust fellowship and K99 grant 1K99HL097083 from National Heart, Lung, and Blood Institute (NIH). T.A.R. is an HHMI investigator.
Footnotes
Author Contributions
J.B., R.J.D., and D.B. conceived the project. W.L. purified and crystallized the proteins, and determined the structures. S.S. generated constructs and aided in crystallization. S.S. and W.L. performed biochemical analysis. W.L., S.S., and T.A.R. analyzed the data and wrote the paper.
The structure factors and coordinates for the full-length protein and the Trx-like domain were deposited in the protein data bank under accession codes 3KP9 and 3KP8, respectively. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Oldenburg J, Marinova M, Muller-Reible C, Watzka M. The vitamin K cycle. Vitam Horm. 2008;78:35–62. doi: 10.1016/S0083-6729(07)00003-9. [DOI] [PubMed] [Google Scholar]
- 2.Tie JK, Stafford DW. Structure and function of vitamin K epoxide reductase. Vitam Horm. 2008;78:103–30. doi: 10.1016/S0083-6729(07)00006-4. [DOI] [PubMed] [Google Scholar]
- 3.Stenflo J, Suttie JW. Vitamin K-dependent formation of gamma-carboxyglutamic acid. Annu Rev Biochem. 1977;46:157–72. doi: 10.1146/annurev.bi.46.070177.001105. [DOI] [PubMed] [Google Scholar]
- 4.Furie B, Bouchard BA, Furie BC. Vitamin K-dependent biosynthesis of gamma-carboxyglutamic acid. Blood. 1999;93:1798–808. [PubMed] [Google Scholar]
- 5.Wajih N, Hutson SM, Wallin R. Disulfide-dependent protein folding is linked to operation of the vitamin K cycle in the endoplasmic reticulum. A protein disulfide isomerase-VKORC1 redox enzyme complex appears to be responsible for vitamin K1 2,3-epoxide reduction. J Biol Chem. 2007;282:2626–35. doi: 10.1074/jbc.M608954200. [DOI] [PubMed] [Google Scholar]
- 6.Soute BA, Groenen-van Dooren MM, Holmgren A, Lundstrom J, Vermeer C. Stimulation of the dithiol-dependent reductases in the vitamin K cycle by the thioredoxin system. Strong synergistic effects with protein disulphide-isomerase. Biochem J. 1992;281:255–9. doi: 10.1042/bj2810255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu PH, Huang TY, Williams J, Stafford DW. Purified vitamin K epoxide reductase alone is sufficient for conversion of vitamin K epoxide to vitamin K and vitamin K to vitamin KH2. Proc Natl Acad Sci U S A. 2006;103:19308–13. doi: 10.1073/pnas.0609401103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein TE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rost S, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–41. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 10.Goodstadt L, Ponting CP. Vitamin K epoxide reductase: homology, active site and catalytic mechanism. Trends Biochem Sci. 2004;29:289–92. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Singh AK, Bhattacharyya-Pakrasi M, Pakrasi HB. Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J Biol Chem. 2008;283:15762–70. doi: 10.1074/jbc.M800982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A. 2008;105:11933–8. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inaba K, et al. DsbB elicits a red-shift of bound ubiquinone during the catalysis of DsbA oxidation. J Biol Chem. 2004;279:6761–8. doi: 10.1074/jbc.M310765200. [DOI] [PubMed] [Google Scholar]
- 14.Inaba K, et al. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 15.Dutton RJ, et al. Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0912952107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tie JK, Nicchitta C, von Heijne G, Stafford DW. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J Biol Chem. 2005;280:16410–6. doi: 10.1074/jbc.M500765200. [DOI] [PubMed] [Google Scholar]
- 17.Rost S, et al. Novel mutations in the VKORC1 gene of wild rats and mice--a response to 50 years of selection pressure by warfarin? BMC Genet. 2009;10:4. doi: 10.1186/1471-2156-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelz HJ, et al. The genetic basis of resistance to anticoagulants in rodents. Genetics. 2005;170:1839–47. doi: 10.1534/genetics.104.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rost S, et al. Site-directed mutagenesis of coumarin-type anticoagulant-sensitive VKORC1: evidence that highly conserved amino acids define structural requirements for enzymatic activity and inhibition by warfarin. Thromb Haemost. 2005;94:780–6. doi: 10.1160/TH05-02-0082. [DOI] [PubMed] [Google Scholar]
- 20.D’Ambrosio RL, D’Andrea G, Cafolla A, Faillace F, Margaglione M. A new vitamin K epoxide reductase complex subunit-1 (VKORC1) mutation in a patient with decreased stability of CYP2C9 enzyme. J Thromb Haemost. 2007;5:191–3. doi: 10.1111/j.1538-7836.2006.02261.x. Epub 2006 Oct 12. [DOI] [PubMed] [Google Scholar]
- 21.Silverman RB. Chemical model studies for the mechanism of vitamin K epoxide reductase. Journal of American Chemical Society. 1981;103:5939–5941. [Google Scholar]
- 22.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in. Cell. 2004;117:601–10. doi: 10.1016/s0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 23.Gross E, Sevier CS, Vala A, Kaiser CA, Fass D. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase. Nature structural biology. 2002;9:61–7. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
- 24.Gebauer M. Synthesis and structure-activity relationships of novel warfarin derivatives. Bioorg Med Chem. 2007;15:2414–20. doi: 10.1016/j.bmc.2007.01.014. Epub 2007 Jan 17. [DOI] [PubMed] [Google Scholar]
- 25.Jin DY, Tie JK, Stafford DW. The conversion of vitamin K epoxide to vitamin K quinone and vitamin K quinone to vitamin K hydroquinone uses the same active site cysteines. Biochemistry. 2007;46:7279–83. doi: 10.1021/bi700527j. Epub 2007 May 25. [DOI] [PubMed] [Google Scholar]
- 26.Bardwell JC, et al. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci U S A. 1993;90:1038–42. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bader M, Muse W, Zander T, Bardwell J. Reconstitution of a protein disulfide catalytic system. J Biol Chem. 1998;273:10302–7. doi: 10.1074/jbc.273.17.10302. [DOI] [PubMed] [Google Scholar]
- 28.Kishigami S, Kanaya E, Kikuchi M, Ito K. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J Biol Chem. 1995;270:17072–4. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- 29.Malojcic G, Owen RL, Grimshaw JP, Glockshuber R. Preparation and structure of the charge-transfer intermediate of the transmembrane redox catalyst DsbB. FEBS Lett. 2008;582:3301–7. doi: 10.1016/j.febslet.2008.07.063. Epub 2008 Sep 5. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, et al. NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inaba K, et al. Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB. Embo J. 2009;28:779–91. doi: 10.1038/emboj.2009.21. Epub 2009 Feb 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regeimbal J, et al. Disulfide bond formation involves a quinhydrone-type charge-transfer complex. Proc Natl Acad Sci U S A. 2003;100:13779–84. doi: 10.1073/pnas.1935988100. Epub 2003 Nov 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, WM Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowtan KD, Main P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr D Biol Crystallogr. 1996;52:43–8. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- 36.Zhang KY, Cowtan K, Main P. Combining constraints for electron-density modification. Methods Enzymol. 1997;277:53–64. doi: 10.1016/s0076-6879(97)77006-x. [DOI] [PubMed] [Google Scholar]
- 37.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–22. doi: 10.1107/S0108767307043930. Epub 2007 Dec 21. [DOI] [PubMed] [Google Scholar]
- 38.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. Epub 2004 Nov 26. [DOI] [PubMed] [Google Scholar]
- 41.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–33. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–4. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.