Abstract
Objective
Although beneficial effects of potassium intake on blood pressure (BP) are well established, little is known about genetic factors that underlie interindividual variability in BP response to dietary potassium. In a previous study, we reported the first evidence for significant heritabilities for BP response in a dietary intervention study in rural Chinese. In this report, we extend our genetic studies to examine associations with polymorphisms in genes in vascular endothelial pathways.
Methods
We genotyped study participants for 23 SNPs in EDN1, NOS3, and SELE. We tested 17 of these SNPs for associations with BP response to potassium supplementation in 1,843 participants. Association tests used population based (GEE) and family based (QTDT) methods, as well as tests for gene by gene interaction (GMDR, GEE).
Results
Single SNP analysis identified significant associations for several SNPs in EDN1 with multiple measures of BP response to potassium supplementation. The cumulative effects of the minor EDN1 alleles that showed significant associations were to reduce measures of BP response by 0.5 to 0.9 mm Hg. We found significant evidence for effects of gene by gene interactions between EDN1 and SELE, even in the absence of individual associations with SELE variants.
Conclusions
Our results implicate variability in EDN1 and SELE as genetic factors that influence BP response to potassium intake. While such epidemiological studies do not allow direct determination of physiologic mechanisms, our findings of joint effects identify EDN1 and SELE as targets for functional studies to determine their interactions in BP response to potassium intake.
Keywords: potassium, blood pressure response, dietary intervention, hypertension, genetic association, gene by gene interaction
INTRODUCTION
Hypertension (HT) is the leading risk factor for premature death, which causes 7.6 million or 13.5% of total mortality worldwide [1]. HT is caused by interactions among numerous genetic and environmental factors. HT treatment using medication and/or life style modification is highly effective in reducing end organ damage [2–7]. Life style modifications for management of blood pressure (BP) include weight control, exercise, lower sodium and alcohol intake, and higher potassium intake [8]. Increased potassium intake through fruits and vegetables or dietary supplements is considered an inexpensive and safe way to lower BP, especially in moderately hypertensive individuals where medication is not appropriate [9, 10]. Beneficial effects of potassium intake on BP control is well documented by many epidemiological and animal studies, as well as numerous clinical trials [9–17]. However, BP response to potassium intake varies widely among individuals. In general, potassium appears to be more effective in older individuals, females, and African Americans [9, 11, 14]. In addition, increased BP response to potassium intake is associated with elevated BP [9] and increased consumption of dietary sodium [16]. Potassium can exert its beneficial effect on BP through different physiological mechanisms, including effects on vascular endothelial pathways such as stimulation of nitric oxide production, a potent vasodilator. Potassium also protects the integrity of vascular endothelium that is vital in maintaining normal BP, reducing the formation of oxygen free radicals to limit vulnerability to damaging factors such as platelet aggregation and leukocyte adherence [14, 18].
In this report, we investigate the influence of genetic variation on BP response to potassium supplementation in the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) study, a controlled feeding study of rural Chinese villagers [19]. The GenSalt study design involved BP measurements at baseline on their typical diet, and after controlled dietary interventions (one week each of a low sodium diet, high sodium diet, and high sodium diet with potassium supplementation). Lymphocyte DNA samples were collected from GenSalt participants for genotyping variants in genes from various physiological pathways involved in blood pressure homeostasis, including central vascular endothelial pathways. We previously reported the first evidence of genetic effects on BP response to potassium, including significant heritabilities for systolic BP (SBP) (h2=0.24), diastolic BP (DBP) (h2=0.21), and mean arterial pressure (MAP) (h2=0.25) in GenSalt participants [20]. In this study we further examine genetic factors that influence BP response to potassium intake, focusing on three genes from vascular endothelial pathways that control BP including endothelin 1 (EDN1) that encodes a potent endothelium derived vasoconstrictor, nitric oxide synthase 3 (NOS3) in the vascular endothelial nitric oxide signaling pathway, and E Selectin (SELE) that encodes a major adhesion molecule on vascular endothelial surfaces. We genotyped GenSalt participants for single nucleotide polymorphisms (SNPs) at these loci to identify associations with BP response to potassium intake, and to examine interactions among these genes that share endothelial pathways.
METHODS
Study population
GenSalt study participants were comprised of Han Chinese families from six rural villages in Northern China. Families were recruited through 18–60 year old probands who were either prehypertensive or had stage-1 HT (SBP 130–160 mm Hg and/or DBP of 85–100 mm Hg), but were never treated with hypertension medication. Parents, spouses, siblings, and offspring were invited to participate in the study. Family members were excluded if they had stage-2 HT, a history of CVD or diabetes, pregnancy, heavy alcohol consumption, or on low sodium diet or anti-hypertensive treatment. A total of 3,142 individuals in 631 families participated in this study. Of the 1,906 eligible family members (parents were excluded), a total of 1,843 subjects participated in the dietary intervention (97% participation rate). The study was performed over the period from October, 2003 to July, 2005. In addition to BP measures, a large number of demographic, anthropomorphic, and medical variables were measured in GenSalt subjects. More information about subject recruitment and measurements are available elsewhere [19]. All of the GenSalt subjects signed an informed consent, and Institutional Review Board approvals for this study were obtained from all participating institutions.
Dietary intervention and study measurements
Probands and their siblings, spouses, and offsprings (but not parents) participated in the dietary intervention (1,843 individuals). Since BP response to potassium intake tends to be higher in individuals with high sodium intake, all study participants were on a high sodium diet (18 g of salt or 308 mmol of sodium per day) for 7 days, followed by 7 days of potassium supplementation (60 mmol of potassium per day) while keeping the high sodium diet.
All unsalted meals were cooked and served by the study staff, and prepackaged salt was added at the time of consumption. Dietary compliance by the participants was confirmed by measurements of 24-hour urinary excretion of sodium and potassium. For the 7 day potassium supplementation period, average measures of potassium in 24-hour urinary excretions were 77.3 mmol as compared to 36.8 mmol at baseline before dietary interventions. BP measurements were taken in the sitting position using a random-zero sphygmomanometer. Each average BP value was calculated from 9 BP measurements from 3 clinical visits at baseline, and in the last 3 days of each dietary intervention. BP response to potassium supplementation was expressed as absolute differences between BP values from the high potassium/sodium intervention and the preceding high sodium intervention. More detailed descriptions of the diets and BP measurements are available in previous reports from the GenSalt study [19, 20].
SNP selection and genotyping
We selected 23 SNPs in genes for Endothelin 1(EDN1), nitric oxide synthase 3 (NOS3), and E selectin (SELE) including tag SNPs that represent the common Chinese haplotypes identified by Tagger software based on linkage disequilibrium patterns determined by the International HapMap Project [21]. In addition, we included potentially functional SNPs that introduce amino acid changes (missense substitutions) or that have shown significant associations in previous studies. SNP genotyping was performed using the high throughput SNPlex platform (Applied Biosystems) according to the manufacturer's protocol [22]. Six of the SNPs were excluded due to low call rate (<80%, one SNP), and/or low minor allele frequency (MAF <0.05, 5 SNPs), and/or severe deviation from Hardy-Weinberg Equilibrium (HWE) (p<0.001, one SNP), yielding 17 SNPs for further analysis. Table 1 presents more detailed information for these SNPs concerning their allelic identities, gene and genome locations, HWE p-values, call rates, and allele frequencies.
Table 1.
SNPs in genes involved in vascular endothelial pathways (EDN1, NOS3, SELE) tested for associations with BP response to potassium supplementation.
| Ch | Gene | SNP | Position | Region | HWpvala | Call rate | MAFb | Alleles- Maj/Min |
|---|---|---|---|---|---|---|---|---|
| 6 | EDN1 | rs1476046 | 12401206 | intron | 0.570 | 0.87 | 0.28 | G/A |
| 6 | EDN1 | rs1630736 | 12403972 | intron | 1.000 | 0.81 | 0.45 | T/C |
| 6 | EDN1 | rs1800541 | 12397204 | promoter | 0.990 | 0.88 | 0.20 | T/G |
| 6 | EDN1 | rs1800543 | 12402122 | intron | 0.920 | 0.85 | 0.28 | T/C |
| 6 | EDN1 | rs2859338 | 12406969 | intergenic | 0.700 | 0.88 | 0.07 | G/A |
| 6 | EDN1 | rs3087459 | 12397624 | promoter | 0.475 | 0.88 | 0.21 | A/C |
| 6 | EDN1 | rs5370 | 12404240 | exon | 0.788 | 0.84 | 0.27 | G/T |
| 7 | NOS3 | rs10277237 | 150120991 | intergenic | 0.695 | 0.87 | 0.25 | A/G |
| 7 | NOS3 | rs1800781 | 150130091 | intron | 0.683 | 0.87 | 0.08 | G/A |
| 7 | NOS3 | rs3918227 | 150138593 | intron | 0.819 | 0.82 | 0.06 | C/A |
| 7 | NOS3 | rs743507 | 150145135 | intron | 0.736 | 0.84 | 0.26 | A/G |
| 7 | NOS3 | rs7830 | 150147218 | intron | 0.069 | 0.83 | 0.40 | C/A |
| 1 | SELE | rs3917406 | 166433935 | intron | 0.890 | 0.82 | 0.48 | C/T |
| 1 | SELE | rs3917419 | 166431476 | intron | 0.973 | 0.86 | 0.15 | C/T |
| 1 | SELE | rs4786 | 166423789 | utr | 0.569 | 0.87 | 0.44 | G/A |
| 1 | SELE | rs5368 | 166428603 | exon | 0.611 | 0.84 | 0.28 | C/T |
| 1 | SELE | rs932307 | 166434362 | intron | 0.446 | 0.86 | 0.48 | A/G |
HWpval, p-values for tests of Hardy-Weinberg equilibrium
MAF, minor allele frequency
Statistical analysis
Plink and PedCheck programs were used to assess the Mendelian consistency of the SNP genotype data [23, 24]. Programs from the Affected-Sib-Pair Interval Mapping and Exclusion package (ASPEX) and the Graphical Representation of Relationships (GRR) package were used to check for potential misreported relationships within pedigrees [25, 26]. Haploview and the Pedstat procedure in QTDT were used for SNP descriptive statistics [27, 28]. BP response variables were adjusted for covariates (age, age2, age3, gender, generation (in a multi-generation family), BP examination room temperature, and field center), and standardized to ensure a mean value of 0 and a standard deviation of 1. For family-based analysis, we used the quantitative transmission disequilibrium test (QTDT version 2.6), we used the orthogonal model which incorporates the variance component method, and a 1000 Monte-Carlo permutation test to adjust for multiple testing [29]. For population-based analysis, we used the Generalized Estimation Equation (GEE) method [30] that accounts for familial correlation. GEE analysis was performed using SAS 9.1 (proc genmod) and exchangeable working correlation matrix with an additive genetic model (coding each SNP as 0 for homozygotes for the major allele, 1 for heterozygotes, and 2 for homozygotes for the minor allele. False Discovery Rate (FDR) was used to correct for multiple testing in GEE analysis [31]. We used both family-based statistical methods (QTDT) which are more robust to hidden population substructure and population-based methods (GEE) that provide increased power by including all subjects rather than only informative families. The application of these complementary approaches provided increased confidence in the validity of the association results. For interactions among genes, we used the Generalized Multifactor Dimensionality Reduction program (GMDR version 0.7) to determine joint effects of each pair of SNPs. GMDR uses cross-validation and permutation tests to eliminate false positive results. We used GMDR for an exhaustive search for all possible two locus models with ten-fold, leave-one-out cross validation. The best model identified by GMDR for each BP measure was verified using GEE to account for familial correlation. For GEE verification, we created a new variable for the interaction by classifying SNP genotype combinations according to BP response (coded 0 for genotype combinations with low response, and 1 for genotype combinations with high response). We calculated mean values for blood pressure traits (SBP, DBP, MAP) for genotypic classes using GEE residuals that were adjusted for familial correlations.
RESULTS
We tested variants in three genes (EDN1, NOS3, SELE) involved in key vascular endothelial pathways for associations with BP response to potassium supplementation in rural Chinese participants in the GenSalt dietary intervention study [19] (Table 1). We genotyped 1,843 individuals (mean age of 38.7 years, 53% male) that participated in the dietary intervention consisting of seven days each of low sodium diet, high sodium diet, and the high sodium diet with potassium supplementation. Table 2 shows results for associations between each SNP and BP response (SBP, DBP, MAP) to potassium supplementation using both GEE and QTDT. GEE identified significant associations for four SNPs in EDN1 (rs1476046, rs1630736, rs1800541, rs3087459) with multiple BP measurements. Of these SNPs, rs1800541 and rs3087459 are in almost complete LD (R2 = 0.98). The FDR for all significant p-values for GEE was less than 0.09, which means that less than one of these p-values may be a false positive. QTDT analysis verified associations for the two correlated SNPs in EDN1 with DBP and MAP.
Table 2.
Significance values for associations of individual SNPs with BP response to potassium supplementation using population based (GEE) and family based (QTDT) statistical methods.
| GEE | QTDT | ||||||
|---|---|---|---|---|---|---|---|
| Gene | SNP | SBP | DBP | MAP | SBP | DBP | MAP |
| EDN1 | rs1476046 | 0.191 | 0.042* | 0.037* | 0.909 | 0.144 | 0.178 |
| EDN1 | rs1630736 | 0.037* | 0.390 | 0.136 | 0.447 | 0.357 | 0.304 |
| EDN1 | rs1800541 | 0.030* | 0.013* | 0.010* | 0.159 | 0.006** | 0.010* |
| EDN1 | rs1800543 | 0.235 | 0.092 | 0.058 | 0.598 | 0.190 | 0.161 |
| EDN1 | rs2859338 | 0.840 | 0.656 | 0.824 | 0.708 | 0.771 | 0.648 |
| EDN1 | rs3087459 | 0.064 | 0.029* | 0.025* | 0.161 | 0.021* | 0.023* |
| EDN1 | rs5370 | 0.217 | 0.093 | 0.056 | 0.692 | 0.225 | 0.212 |
| NOS3 | rs10277237 | 0.879 | 0.666 | 0.451 | 0.897 | 0.830 | 0.959 |
| NOS3 | rs1800781 | 0.859 | 0.178 | 0.275 | 0.544 | 0.785 | 0.957 |
| NOS3 | rs3918227 | 0.681 | 0.909 | 0.809 | 0.586 | 0.992 | 0.808 |
| NOS3 | rs743507 | 0.829 | 0.113 | 0.196 | 0.774 | 0.191 | 0.382 |
| NOS3 | rs7830 | 0.165 | 0.898 | 0.275 | 0.391 | 0.171 | 0.501 |
| SELE | rs3917406 | 0.402 | 0.500 | 0.408 | 0.898 | 0.665 | 0.836 |
| SELE | rs3917419 | 0.823 | 0.799 | 0.883 | 0.530 | 0.572 | 0.416 |
| SELE | rs4786 | 0.359 | 0.305 | 0.239 | 0.756 | 0.946 | 0.995 |
| SELE | rs5368 | 0.500 | 0.567 | 0.515 | 0.781 | 0.900 | 0.945 |
| SELE | rs932307 | 0.178 | 0.088 | 0.071 | 0.805 | 0.554 | 0.604 |
SBP: Systolic blood pressure, DBP: diastolic blood pressure, MAP: Mean arterial pressure, GEE: Generalized Estimation Equation.
P<0.05
P<0.01
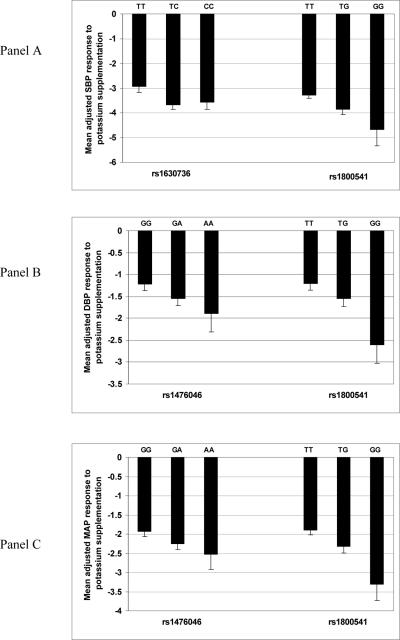
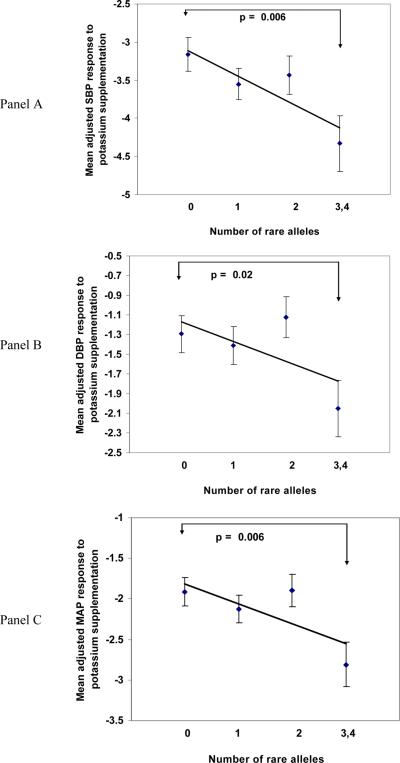
Figure 1 shows mean BP response for genotypes for EDN1 SNPs that showed significant associations with SBP (Panel A), DBP (Panel B), and MAP (Panel C). For these comparisons, we excluded genotypes for EDN1 rs3087459 due to almost complete LD with EDN1 rs1800541 (R2=0.98). With one exception (SBP for rs1630736), homozygotes for the minor allele had the strongest BP responses to potassium supplementation, and heterozygotes had intermediate responses. Figure 2 shows the cumulative effect of associated EDN1 SNPs on mean BP response in groups that carry increasing numbers of minor alleles. For SBP (Panel A), the combined effect of minor alleles (rs1630736 and rs1800541) was to decrease mean values of SBP by approximately 0.9 mm Hg (p=0.006). For DBP (Panel B) and MAP (Panel C), the combined effect of the minor alleles (rs1476046, rs1800541) was to lower DBP (p=0.02) and MAP (p=0.006) by approximately 0.5 and 0.7 mm Hg respectively.
Figure 1.
Mean adjusted SBP (a), DBP (b), and MAP (c) response (mm Hg) to potassium supplementation according to genotypes for EDN1 SNPs that showed significant associations with each measure. BP response variables were adjusted for age, age2, age3, gender, generation, BP examination room temperature, and field center.
Figure 2.
Cumulative effects of the minor alleles for EDN1 SNPs that showed significant associations with BP response to potassium supplementation. Mean values of adjusted BP response for groups carrying 0, 1, 2, and 3 or 4 rare alleles (545, 596, 448, and 218 individuals, respectively) are shown for SBP (Panel A), DBP (Panel B), and MAP (Panel C). The best fitting trend line and p value from t-tests between those with 0 risk allele and those with 3 or 4 risk alleles are shown (R2 =0.76 for SBP, R2=0.40 for DBP, and R2=0.54 for MAP). BP response variables were adjusted for age, age2, age3, gender, generation, BP examination room temperature, and field center.
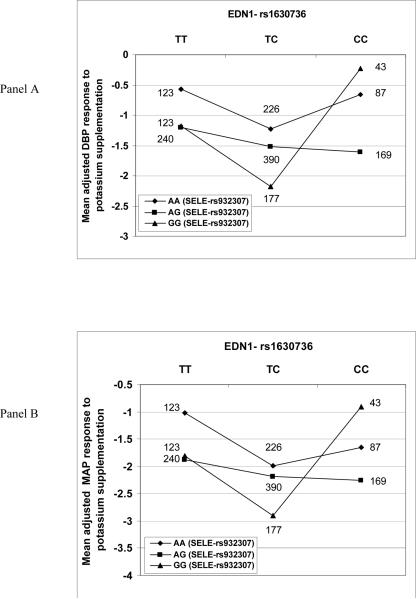
Because these genes are all involved in vascular endothelial pathways that control BP, we used GMDR to identify interactions among the variants at the three loci (EDN1, NOS3, SELE) that influence response to potassium supplementation. GMDR found significant joint effects between EDN1 rs1630736 and SELE rs932307 for both DBP and MAP. We used GEE analysis with the best GMDR model to confirm the interaction for both DBP (p = 0.006) and MAP (p = 0.003). Figure 3 shows the joint effect of these EDN1 and SELE variants on DBP (Panel A) and MAP (Panel B) in response to potassium supplementation. In single gene analysis, EDN1 rs1630736 showed significant associations with SBP (p=0.04), but not DBP and MAP (Table 2). This lack of association with DBP and MAP may be explained by interaction with SELE genotypes. In combination with particular SELE rs932307 genotypes (AG and GG), EDN1 TT and TC genotypes showed a trend for decreasing DBP and MAP response (Figure 3). However, in combination with the SELE GG genotype, EDN1 CC homozygotes reversed that trend with a larger response than the TT and TC genotypes.
Figure 3.
Mean adjusted values (mm Hg) for DBP response (Panel A) and MAP response (Panel B) to potassium supplementation as a function of interaction between genotypes for EDN1 rs1630736 and SELE rs932307. Number of individuals for each EDN1/SELE genotype group are shown. BP response variables were adjusted for age, age2, age3, gender, generation, BP examination room temperature, and field center.
DISCUSSION
The GenSalt dietary intervention study in rural Chinese first reported evidence for genetic factors that underlie individual variation in BP response to potassium intake, with significant heritabilities for SBP, DBP, and MAP. In this study, we present the first examination of the effects of variants in key genes in vascular endothelial pathways on response to potassium including endothelin I (EDN1), nitric oxide synthase 3 (NOS3), and E selectin (SELE). The GenSalt target population and study design provided several advantages for this genetic association study. For example, the study participants were recruited from a relatively homogenous population (Han Chinese) thus limiting statistical confounding due to hidden population substructure or admixture in association tests. In addition, the GenSalt participants are rural villagers with similar lifestyles (farmers) and seasonal diets.
In statistical tests of individual SNPs, we found significant associations for EDN1 variants with multiple measures of BP response to potassium supplementation using both family-based (QTDT) and population-based (GEE) methods (Table 2). Two of the associated SNPS (rs1800541, and rs3087459) were in strong LD (R2=0.98), and located together in the EDN1 promoter region with potential regulatory effects on gene expression (Table 1). The rs1800541 variant previously has shown associations with hypertension in haplotype analysis of British rheumatoid arthritis patients [32], and with asthma in 2 different European populations [33]. We also found significant associations for rs1476046 and rs1630736 that are located in EDN1 introns. While the evidence is compelling for the influence of EDN1 variation on BP response to potassium, we are not able to provide mechanistic or physiological explanations. However, these results support a potential role in potassium response for EDN1 that is expressed in vascular endothelium as well as other tissues (smooth muscle, fibroblasts, myocytes, macrophages, and neurons). EDN1 encodes preproendothelin that is processed to form endothelin 1, a potent vasoconstricter that also mediates water and salt homeostasis via G-protein coupled receptors targeted by HT drug therapies [34–36].
We found significant evidence for interaction of SNPs in SELE (rs932307) and EDN1 (rs1630736) for DBP (p = 0.006) and MAP (p = 0.003) in response to potassium supplementation. Interestingly, we did not find main effects of SELE rs932307 on these BP measures in the absence of interaction with EDN1 rs1630736 (Table 2). These results underscore the importance of gene by gene interactions in complex traits, particularly among genes that share physiologic and metabolic pathways. This statistical evidence of interaction identifies SELE and EDN1 as good candidates for future studies to define their functional interactions on BP response to potassium. SELE is expressed in endothelial cells, producing E selectin that mediates cell adhesion of circulating monocytes to vascular endothelial surfaces, leading to atherosclerosis [37]. Rare SELE mutations were previously found to be associated with BP [38–40] and atherosclerosis [37] in several studies. Elevated plasma levels of soluble E selectin have been found associated with HT, perhaps as a result of endothelial damage or SELE involvement in endothelial dysfunction. A tentative connection between potassium supplementation and SELE is provided by studies in rats that found high potassium diets reduced endothelial injuries with a concomitant reduction in leukocyte adherence to the vascular wall [41]. Previous studies have identified effects of gene by gene interaction on BP response, including evidence for interaction between Gly460Trp in the alpha-adducin gene (ADD1) and the insertion/deletion polymorphism in the gene for angiotensin converting enzyme (ACE) on BP response to a sodium load [42].
Conclusion
We have identified associations of variation in genes in vascular endothelial pathways with BP response to potassium supplementation in a dietary intervention study in rural Chinese. We found significant associations of promoter and intronic SNPs in EDN1 with multiple measures of BP response, as well as evidence for significant effects of gene by gene interaction between variants in EDN1 and SELE in the absence of main effects of SELE variants. One of the goals of genetic studies is to provide diagnostic genetic markers to identify individuals with specific types of response to medication, as well as dietary and lifestyle treatments. This early report of genetic associations indicates that BP response to potassium may ultimately become a target for development of such genetic tests. However, these results will require replication in other cohorts to determine generalizability to other ethnicities and to populations with diets that are lower in sodium than these rural Chinese villagers. While individual responses due to genetic factors may be modest, from a public health perspective, only a 2 mm Hg reduction in SBP leads to a 10% decrease in stroke attributable mortality and 7% decrease in ischemic heart disease attributable mortality [43].
CONDENSED ABSTRACT.
We tested polymorphisms in genes in vascular endothelial pathways for associations with blood pressure response to potassium in a dietary intervention study in rural Chinese. We used family and population based methods to test for associations with 17 SNPs in EDN1, NOS3, and SELE. We identified significant associations for several EDN1 SNPs with multiple measures of BP response. Cumulative effects of the minor alleles for associated SNPs were reductions of 0.5–0.9 mm Hg in BP response. We also found significant evidence for gene by gene interactions between EDN1 and SELE, even in the absence of individual associations with SELE variants.
ACKNOWLEDGEMENTS
This report is from the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) study that is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (U01HL072507). Upsher-Smith Laboratories, Inc. has provided Klor-Con M20 potassium tablets for the GenSalt study.
Support: NIH grant U01HL072507, Upsher-Smith Laboratories, Inc. provided potassium tablets
Footnotes
Conflict of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lawes CM, Vander Hoorn S, Rodgers A. International Society of Hypertension. Global burden of blood-pressure-related disease, 2001. Lancet. 2008;371:1513–1518. doi: 10.1016/S0140-6736(08)60655-8. [DOI] [PubMed] [Google Scholar]
- 2.Baker DW. Prevention of heart failure. J Card Fail. 2002;8:333–346. doi: 10.1054/jcaf.2002.0805333. [DOI] [PubMed] [Google Scholar]
- 3.Rigaud AS, Seux ML, Staessen JA, Birkenhager WH, Forette F. Cerebral complications of hypertension. J Hum Hypertens. 2000;14:605–616. doi: 10.1038/sj.jhh.1001118. [DOI] [PubMed] [Google Scholar]
- 4.Ritz E. Total cardiovascular risk management. Am J Cardiol. 2007;100:53J–60J. doi: 10.1016/j.amjcard.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Messerli FH, Williams B, Ritz E. Essential hypertension. Lancet. 2007;370:591–603. doi: 10.1016/S0140-6736(07)61299-9. [DOI] [PubMed] [Google Scholar]
- 6.Mancia G. Blood pressure reduction and cardiovascular outcomes: past, present, and future. Am J Cardiol. 2007;100:3J–9J. doi: 10.1016/j.amjcard.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Wong TY, Mitchell P. Hypertensive retinopathy. N Engl J Med. 2004;351:2310–2317. doi: 10.1056/NEJMra032865. [DOI] [PubMed] [Google Scholar]
- 8.Whelton PK, He J, Appel LJ, Cutler JA, Havas S, Kotchen TA, et al. Primary prevention of hypertension: clinical and public health advisory from The National High Blood Pressure Education Program. JAMA. 2002;288:1882–1888. doi: 10.1001/jama.288.15.1882. [DOI] [PubMed] [Google Scholar]
- 9.Braschi A, Naismith DJ. The effect of a dietary supplement of potassium chloride or potassium citrate on blood pressure in predominantly normotensive volunteers. Br J Nutr. 2008;99:1284–1292. doi: 10.1017/S0007114507864853. [DOI] [PubMed] [Google Scholar]
- 10.Gu D, He J, Wu X, Duan X, Whelton PK. Effect of potassium supplementation on blood pressure in Chinese: a randomized, placebo-controlled trial. J Hypertens. 2001;19:1325–1331. doi: 10.1097/00004872-200107000-00019. [DOI] [PubMed] [Google Scholar]
- 11.Naismith DJ, Braschi A. The effect of low-dose potassium supplementation on blood pressure in apparently healthy volunteers. Br J Nutr. 2003;90:53–60. doi: 10.1079/bjn2003861. [DOI] [PubMed] [Google Scholar]
- 12.He FJ, MacGregor GA. Fortnightly review: Beneficial effects of potassium. BMJ. 2001;323:497–501. doi: 10.1136/bmj.323.7311.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He FJ, Markandu ND, Coltart R, Barron J, MacGregor GA. Effect of short-term supplementation of potassium chloride and potassium citrate on blood pressure in hypertensives. Hypertension. 2005;45:571–574. doi: 10.1161/01.HYP.0000158264.36590.19. [DOI] [PubMed] [Google Scholar]
- 14.Coca SG, Perazella MA, Buller GK. The cardiovascular implications of hypokalemia. Am J Kidney Dis. 2005;45:233–247. doi: 10.1053/j.ajkd.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 15.Adrogue HJ, Madias NE. Sodium and potassium in the pathogenesis of hypertension. N Engl J Med. 2007;356:1966–1978. doi: 10.1056/NEJMra064486. [DOI] [PubMed] [Google Scholar]
- 16.Krishna GG. Effect of potassium intake on blood pressure. J Am Soc Nephrol. 1990;1:43–52. doi: 10.1681/ASN.V1143. [DOI] [PubMed] [Google Scholar]
- 17.Whelton PK, He J, Cutler JA, Brancati FL, Appel LJ, Follmann D, Klag MJ. Effects of oral potassium on blood pressure. Meta-analysis of randomized controlled clinical trials. JAMA. 1997;277:1624–1632. doi: 10.1001/jama.1997.03540440058033. [DOI] [PubMed] [Google Scholar]
- 18.Haddy FJ, Vanhoutte PM, Feletou M. Role of potassium in regulating blood flow and blood pressure. Am J Physiol Regul Integr Comp Physiol. 2006;290:R546–52. doi: 10.1152/ajpregu.00491.2005. [DOI] [PubMed] [Google Scholar]
- 19.GenSalt Collaborative Research Group GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu D, Rice T, Wang S, Yang W, Gu C, Chen CS, et al. Heritability of blood pressure responses to dietary sodium and potassium intake in a Chinese population. Hypertension. 2007;50:116–122. doi: 10.1161/HYPERTENSIONAHA.107.088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 22.Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, et al. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech. 2005;16:398–406. [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinds DR, M The ASPEX package:affected sib-pair exclusion mapping v1.88. 1999 [Google Scholar]
- 26.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Lou XY, Chen GB, Yan L, Ma JZ, Mangold JE, Zhu J, et al. A Combinatorial Approach to Detecting Gene-Gene and Gene-Environment Interactions in Family Studies. Am J Hum Genet. 2008 doi: 10.1016/j.ajhg.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanley JA, Negassa A, Edwardes MD, Forrester JE. Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 32.Panoulas VF, Douglas KM, Smith JP, Taffe P, Stavropoulos-Kalinoglou A, Toms TE, et al. Polymorphisms of the endothelin-1 gene associate with hypertension in patients with rheumatoid arthritis. Endothelium. 2008;15:203–212. doi: 10.1080/10623320802228708. [DOI] [PubMed] [Google Scholar]
- 33.Zhu G, Carlsen K, Carlsen KH, Lenney W, Silverman M, Whyte MK, et al. Polymorphisms in the endothelin-1 (EDN1) are associated with asthma in two populations. Genes Immun. 2008;9:23–29. doi: 10.1038/sj.gene.6364441. [DOI] [PubMed] [Google Scholar]
- 34.Hynynen MM, Khalil RA. The vascular endothelin system in hypertension--recent patents and discoveries. Recent Patents Cardiovasc Drug Discov. 2006;1:95–108. doi: 10.2174/157489006775244263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shreenivas S, Oparil S. The role of endothelin-1 in human hypertension. Clin Hemorheol Microcirc. 2007;37:157–178. [PubMed] [Google Scholar]
- 36.Dhaun N, Goddard J, Kohan DE, Pollock DM, Schiffrin EL, Webb DJ. Role of endothelin-1 in clinical hypertension: 20 years on. Hypertension. 2008;52:452–459. doi: 10.1161/HYPERTENSIONAHA.108.117366. [DOI] [PubMed] [Google Scholar]
- 37.Andreotti F, Porto I, Crea F, Maseri A. Inflammatory gene polymorphisms and ischaemic heart disease: review of population association studies. Heart. 2002;87:107–112. doi: 10.1136/heart.87.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marteau JB, Sass C, Pfister M, Lambert D, Noyer-Weidner M, Visvikis S. The Leu554Phe polymorphism in the E-selectin gene is associated with blood pressure in overweight people. J Hypertens. 2004;22:305–311. doi: 10.1097/00004872-200402000-00014. [DOI] [PubMed] [Google Scholar]
- 39.Sass C, Pallaud C, Zannad F, Visvikis S. Relationship between E-selectin L/F554 polymorphism and blood pressure in the Stanislas cohort. Hum Genet. 2000;107:58–61. doi: 10.1007/s004390000325. [DOI] [PubMed] [Google Scholar]
- 40.Chang YP, Liu X, Kim JD, Ikeda MA, Layton MR, Weder AB, et al. Multiple genes for essential-hypertension susceptibility on chromosome 1q. Am J Hum Genet. 2007;80:253–264. doi: 10.1086/510918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishimitsu T, Tobian L, Sugimoto K, Everson T. High potassium diets reduce vascular and plasma lipid peroxides in stroke-prone spontaneously hypertensive rats. Clin Exp Hypertens. 1996;18:659–673. doi: 10.3109/10641969609081773. [DOI] [PubMed] [Google Scholar]
- 42.Barlassina C, Schork NJ, Manunta P, Citterio L, Sciarrone M, Lanella G, et al. Synergistic effect of alpha-adducin and ACE genes causes blood pressure changes with body sodium and volume expansion. Kidney Int. 2000;57:1083–1090. doi: 10.1046/j.1523-1755.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 43.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R, Prospective Studies Collaboration Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]