Abstract
An outstanding biological question is why tissue regeneration in mammals is limited, whereas urodele amphibians and teleost fish regenerate major structures, largely by cell cycle reentry. Upon inactivation of Rb, proliferation of postmitotic urodele skeletal muscle is induced, whereas in mammalian muscle this mechanism does not exist. We postulated that a tumor suppressor present in mammals but absent in regenerative vertebrates, the Ink4a product ARF (alternative reading frame), is a regeneration suppressor. Concomitant inactivation of Arf and Rb led to mammalian muscle cell cycle reentry, loss of differentiation properties, and upregulation of cytokinetic machinery. Single postmitotic myocytes were isolated by laser micro-dissection-catapulting and transient suppression of Arf and Rb yielded myoblast colonies that retained the ability to differentiate and fuse into myofibers upon transplantation in vivo. These results show that differentiation of mammalian cells is reversed by inactivation of Arf and Rb, and support the hypothesis that Arf evolved at the expense of regeneration.
Introduction
Tissue regeneration in humans is extremely limited, which constitutes a major challenge to the repair of damaged organ and tissue function. Humans and other mammals do not regenerate large portions of lost muscles or other mesenchymal structures after traumatic injury or surgical excision. By contrast, some vertebrates such as the urodele amphibians and the teleost fish have a remarkable capacity to regenerate entire limbs, the lens of the eye, and portions of the heart (Poss et al. 2002; Brockes and Kumar 2008; Tanaka and Weidinger 2008). Although classically defined resident stem cells clearly play a role in tissue regeneration, their relatively low frequency in a given tissue may be insufficient to account for the massive regeneration observed in some lower vertebrates. In zebrafish, heart regeneration results from dedifferentiation and subsequent proliferation of cardiomyocytes (Poss et al. 2002). Substantial evidence from studies of newts and axolotls supports a similar regenerative mechanism, in which postmitotic limb tissues including muscles lose their differentiation markers, re-enter the cell cycle, proliferate and then recapitulate differentiation in the blastema. (Hay and Fischman 1961; Lentz 1969; Kintner and Brockes 1984; Lo et al. 1993; Gardiner and Bryant 1996; Echeverri et al. 2001). Recent observations strongly suggest that dedifferentiated cells of the limb remain lineage-committed during this process (Kragl et al. 2009). In marked contrast, there is no evidence that dedifferentiation occurs as a natural part of tissue regeneration in mammals. This raises the possibility that a mechanism of regeneration involving reversal of differentiation of mesenchymal tissues, such as muscle, may have been lost or suppressed during evolution of higher vertebrates that, if elucidated, could significantly impact regenerative medicine.
Muscle differentiation in mammals occurs by a stepwise progression. This process entails morphological and functional changes driven by the expression of a series of muscle regulatory factors (MRFs), which induce expression of differentiation-specific genes such as creatine kinase and myosin heavy chain (MHC) (Molkentin and Olson 1996). In particular, myogenin heralds a transition from proliferative myoblast to committed post-mitotic muscle cell (Walsh and Perlman 1997; Charge and Rudnicki 2004). Of critical importance to this transition is the expression of the retinoblastoma protein (Rb) (Gu et al. 1993; Lassar and Munsterberg 1994; Novitch et al. 1996; Huh et al. 2004). The role of Rb in differentiation is multi-faceted, including not only the orchestration of mitotic arrest and prevention of cell cycle reentry, but also inhibition of apoptosis, and enforcement of stable tissue-specific gene expression (Burkhart and Sage 2008). Since the differentiated state requires continuous active control (Blau et al. 1985; Blau and Baltimore 1991), (Yamanaka and Blau, 2010) ongoing expression of Rb or possibly redundant pocket proteins would be predicted to be necessary for the maintenance of the specialized muscle cell phenotype.
Attempts to reverse differentiation and postmitotic arrest in mammalian skeletal muscle cells by either acute suppression or permanent elimination of Rb have produced conflicting results. In newt muscle cells, cell cycle reentry and DNA synthesis occur when Rb is inactivated by phosphorylation, (Tanaka et al. 1997). Similarly, the inactivation of Rb by viral oncoproteins in immortalized mammalian myoblast cell lines, such as C2C12, readily results in BrdU incorporation and S-phase reentry in nuclei of differentiated myotubes (Gu et al. 1993; Crescenzi et al. 1995), in agreement with more recent studies using siRNA to suppress Rb (Blais et al. 2007). In marked contrast, in similar experiments using primary muscle cells isolated directly from mammalian muscle tissues, Rb reduction or elimination by Cre-mediated excision, failed to result in significant S-phase reentry (Sacco et al. 2003; Camarda et al. 2004; Huh et al. 2004). These data suggest that Rb loss in primary differentiated skeletal muscle cells is not sufficient to induce reversal of the post-mitotic state in mammals, in stark contrast to the situation in urodeles.
We reasoned that components of the mammalian cell cycle machinery known to be absent in lower organisms could have evolved at the expense of regeneration. A prime candidate is the Ink4a locus, which encodes the structurally and functionally unrelated products, p16 and ARF (Alternative Reading Frame). Both of these proteins are potent tumor suppressors that are frequently inactivated in human and mouse cancers. p16 specifically inhibits cdk4 and is thought to function upstream of Rb, while ARF binds MDM2 which results in p53 stabilization, in addition to having p53 independent functions (Sherr et al. 2005). Notably, ARF responds to oncogenic stimuli, including the inactivation of Rb, by inducing p53-dependent growth arrest or apoptosis (Sharpless and DePinho 1999; Sherr et al. 2005). While the Rb and p53 pathways are evolutionarily ancient, their regulation by the Ink4a locus is a relatively new phenomenon. Homologs of p16 exist in fish (Kazianis et al. 1999; Gilley and Fried 2001). But the earliest identified ARF ancestor is in chickens, with no candidates in databases of lower organisms (Gilley and Fried 2001; Kim et al. 2003; Brookes et al. 2004). Thus, the absence of ARF relatives could underlie certain fundamental differences in growth control in lower vertebrates.
Here we test the hypothesis that reversal of differentiation of mammalian skeletal muscle cells can be induced by inactivation of Rb in conjunction with ARF. Specifically, we postulated that upon loss of Rb, growth arrest and differentiation are maintained by induction of expression of the tumor suppressor ARF. Our data show that transient suppression of both Rb and ARF results in the ability of skeletal muscle cells to lose their differentiated properties, cycle, and then redifferentiate in a manner that mimics urodele cells. We used both genetic and biochemical approaches in primary cells and monitored single cell behavior by time lapse microscopy, which enabled a rigorous analysis of the intrinsic control of muscle cell proliferation and modulation of the differentiated phenotype. The use of photoactivated laser microdissection (PALM) and laser pressure catapulting (LPC) to isolate single, morphologically intact, adherent, primary differentiated cells (myocytes) allowed the unambiguous identification of clones from individual cells. Based on our findings we propose a molecular mechanism whereby evolutionary loss of ARF explains differences between urodele and mammalian skeletal muscle regeneration. The results suggest a means of replicating in mammals the robust regenerative response typical of urodeles, demonstrating that the transient induction of dedifferentiation could serve as an adjunct to classical tissue specific stem cells.
Results
Suppression of Rb by siRNA induces S-phase reentry in C2C12 myotubes
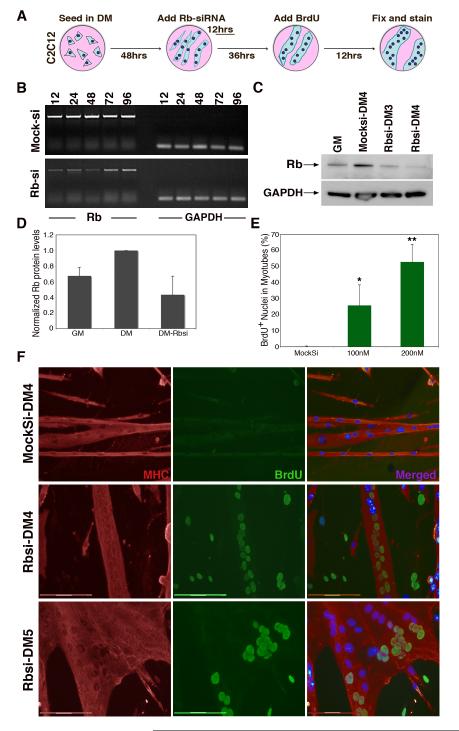
The myoblast cell line C2C12, is a model system for studying muscle differentiation in vitro. In differentiation medium (DM), confluent C2C12 myoblasts exit the cell cycle and fuse with one another to form multinucleated muscle cells (myotubes), which express muscle proteins and are contractile. We developed a protocol to transiently express siRNAs in 66.8% of myotubes using the siImporter reagent (Figure S1 A-B). To determine the duration and efficiency of siRNA treatment, C2C12 myotubes were treated with siRNAs against Rb (Rbsi) (Fig 1A-B). Semi-quantitative RT-PCR and western analysis confirmed transient suppression of Rb transcript and protein levels (Fig. 1B-D). Treatment with Rbsi resulted in a decrease of Rb protein levels (Fig. 1C) to approximately 50% ± 22 % of the levels in control myotubes (Fig. 1D). These data show that using siRNAs, Rb can be substantially reduced for a period of 48 hours.
Figure 1. Suppression of Rb is sufficient for cell-cycle reentry in C2C12 myotubes.
(A) Schematic representation of the treatment of C2C12 myotubes. (B) sqRT-PCR of Rb expression timecourse in hours following treatment with mocksi or Rbsi. GAPDH expression shown as RNA loading control. (C) Western blot of protein expression levels of Rb (100 kDa) in C2C12 myoblasts (GM), myotubes (DM4), and myotubes at DM3 and DM4, 24hrs and 48hrs respectively after treatment with Rb-siRNA. GAPDH (35 kDa) as a loading control. (D) Histogram representing the levels of Rb in GM, in DM4 and in DM4 treated with Rb-siRNA for 48hrs. Samples are normalized to Rb levels of DM4 myotubes. (E) Histogram represents BrdU incorporation in myonuclei of myotubes in day 4 of differentiation (DM4), at least 36hrs following Rbsi treatment. A minimum of 500 nuclei were counted from random fields for each trial. (F) Immunofluorescence images from mock treated DM4 C2C12 myotubes and 200nM Rbsi-treated myotubes in DM4 and DM5. Myotubes were labeled with primary antibody for MHC (red) and BrdU (green), as well as with Hoechst 33258 dye (blue). Bar, 150μm. Growth medium (GM); Myotubes cultured in differentiation medium for 4 or 5 days (DM4 or DM5 respectively). Error bars indicate the mean ± SE of at least three independent experiments, P value was determined with a t test (*P<0.05, **P<0.01).
To determine if S-phase reentry in C2C12 myotubes occurred after transient suppression of Rb, we labeled Mocksi and Rbsi-treated myotubes with BrdU, as indicated in the scheme in Fig. 1A. BrdU-labeled myonuclei were observed inside MHC+ myotubes only after Rbsi treatment. S-phase reentry was dependent on the dose of Rbsi used and doubled from 25.7% to 52.8% as the siRNA concentration was increased from 100 to 200nM (Fig. 1E). The 200nm concentration was used in subsequent experiments. The percentage of myotubes that contained any BrdU positive nuclei was analyzed in a separate set of experiments and found to be 25% (Fig. S1 H), which is explicable since myotubes with Brdu positive nuclei often had large clusters of nuclei that underwent DNA synthesis, likely reflecting the successful transfection of a fraction of the cells in the culture (Fig. S1 A). Fig. 1F shows representative images of BrdU incorporation together with MHC immunostaining to confirm differentiation. We found a marked change in myotube morphology 72 hours after transfection of Rbsi (Fig. 1D bottom panels) from a compact elongated structure with characteristic linear nuclear alignment, to an amorphous structure, with nuclei aggregated in clusters, many of which were BrdU-positive. These data confirm that transient suppression of Rb is sufficient to induce S-phase reentry in differentiated C2C12 myotube nuclei, and show that the extent of this effect depends on the concentration of Rbsi used.
Both Rb and p19ARF must be suppressed for S-phase reentry in primary myotubes
The majority of the data that suggest that suppressing Rb is sufficient for cell cycle reentry in mature mammalian myotubes comes from experiments using the C2C12 cell line (Gu et al. 1993; Velloso et al. 2001; Blais et al. 2007). Although useful for studying muscle cell differentiation and fusion (Pajcini et al. 2008), we reasoned that C2C12 cells, like other cell lines, may have acquired mutations essential to their immortalization. Therefore, we considered it critical to use primary cells to assess the role of Rb in maintaining the post-mitotic state. We harvested primary myoblasts as previously described (Rando and Blau 1994), from Rosa26-CreERT2 Rblox/lox mice (Viatour et al. 2008) crossed to mice carrying a Cre-responsive β-galactosidase reporter allele (Soriano 1999). In these mice, Cre expression and Rb excision is dependent on tamoxifen (TAM) induction (Figure S1 C-D). In primary myotube cultures, a single 24hr treatment with 1μM TAM was sufficient to reduce Rb expression (Figure S1 E). Both transcript and protein levels dropped substantially by 96hrs following TAM treatment (Figure S1 F and Fig. 2C).
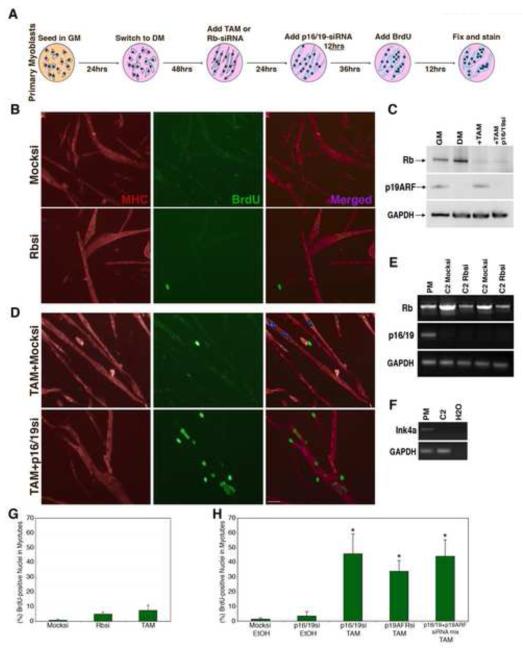
Figure 2. Suppression of Rb and p16/19 is necessary for cell-cycle reentry in primary myotubes.
(A) Schematic representation of the treatment of primary myotubes with either tamoxifen (TAM) and/or siRNA. (B) Immunofluorescence images from mock si-Glo treated primary myotubes and Rbsi treated myotubes. Myotubes were labeled with primary antibody for MHC (red) and BrdU (green), as well as with Hoechst 33258 dye (blue). Bar, 50μm. (C) Western blot of primary myotube protein levels; Rb (100 kDa) and p19ARF (20 kDa) in GM and DM5, after TAM treatment or TAM and p16/19si treatment. GAPDH (35 kDa) as loading control. (D) Immunofluorescence images indicating BrdU incorporation in TAM and mock siGlo treated primary myotubes compared to TAM and p16/19si-RNA treated primary myotubes. (E) sqRT-PCR showing Rb and Ink4a (p16/19) expression in primary myotubes as well as in two different C2C12 myotube populations treated with Mocksi or Rbsi. (F) sq-PCR amplification using primers for the shared exon 2-3 region of the ink4a locus, from genomic DNA prepared from primary myoblasts and C2C12 myoblasts. (G) Histogram represents BrdU incorporation in primary myotube nuclei at DM5, following suppression of Rb with either siRNA or TAM. (H) Histogram represents BrdU incorporation in primary myotube nuclei following treatment with TAM and siRNAs against ink4a gene products. A minimum of 500 nuclei were counted from random fields for each trial in F and G. Error bars indicate the mean ± SE of at least three independent experiments. (*P<0.01).
We analyzed S-phase reentry in primary myotubes after loss of Rb expression according to the scheme in Fig. 2A. In contrast to C2C12 myotubes, in primary myotubes regardless of whether Rb was knocked-down by Rbsi (Fig. 2B) or excised by Cre expression (Fig. 2D top panels), in over 1500 nuclei scored from random fields of MHC+ myotubes, BrdU-labeling of myonuclei was relatively rare (Fig. 2G). In the representative images in Fig. 2B and 2D, BrdU+ nuclei are detectable, but these are generally not within MHC+ myotubes. Thus, loss of Rb in primary myotubes is not sufficient to induce cell cycle reentry.
We observed that acute suppression of Rb in primary myotubes led to upregulation of p19ARF mRNA (Figure S1 F) and protein (Fig. 2C, third lane). These results are in agreement with previous reports showing that disruption of Rb function by mutation or by hyperphosphorylation in response to mitogenic signals leads to the activation of ARF (DeGregori et al. 1997; Sage et al. 2003). ARF, in turn, serves to block inappropriate cycling. Induction of ARF was accompanied by a mild increase in baseline levels of apoptosis, but the majority of myotubes remained robust and viable. Apoptosis was rarely observed in si-RNA treated cells in which p16/p19 was reduced (Figure S1 I).
In contrast to primary cells, p19ARF was not detected in C2C12 myotubes in response to Rb suppression (Fig. 2E). We determined by genomic PCR analysis that the lack of p19ARF expression in C2C12 cells was due to a deletion in the Ink4a locus (Fig. 2F), a characteristic of murine cell lines immortalized by continuous passage in culture (Sherr and DePinho 2000). These observations suggested that the disparate cell cycle reentry responses of primary and immortalized muscle cells to loss of Rb might be related to the status of ARF. To test whether the suppression of the Ink4a gene products in Rb-deficient primary myotubes would permit reentry, we designed siRNAs that target the shared exon 2 region of Ink4a mRNA (p16/19si), the ARF-specific exon 1β (p19ARFsi), and the p16-specific exon 1α. Primary myotubes were first treated with TAM or Rbsi, then 24hrs later transfected either with p16si, p16/19si, p19ARFsi alone or with both p16/19si and p19ARFsi together (see scheme in Fig. 2A). Gene knockdown in myotubes was verified by western analysis (Fig. 2C), and by RT-PCR (Fig. S1 G). When labeled with BrdU following TAM and p16/19si treatment, differentiated myonuclei incorporated BrdU in MHC-positive myotubes (Fig. 2D). Quantification of the BrdU labeling indicated that 45.7 ± 7.2% of myonuclei enter S-phase in TAM and p16/19si treated myotubes, a marked increase over baseline values observed in myotubes treated only with TAM or p16/19si (Fig. 2 G-H). Different combinations of siRNAs that exclusively induced knockdown of p19ARF or that targeted both Ink4a gene products in TAM treated cells yielded similar results (Fig. 2H). Notably knockdown of p16 alone after TAM treatment did not increase BrdU incorporation above background levels (data not shown), suggesting that ARF was primarily responsible for the observed Ink4a effects. Therefore, for subsequent experiments we used the p16/19si (exon 2) because it produced the strongest suppression of p19ARF. Thus robust S-phase reentry in differentiated primary mammalian myotubes occurs only after combined suppression of both Rb and p19ARF.
Upregulation of mitotic and cytokinetic components in Rb and p16/19-deficient myonuclei
To determine if S-phase reentry in primary myonuclei marked the initiation of the mitotic process, we analyzed control or TAM and p16/19si treated nuclei for the induction of expression of a panel of mitotic and cytokinetic proteins. AuroraB and survivin are components of the chromosome passenger complex (CPC), which controls spindle structure, kinetochore attachment and chromosome segregation (Ruchaud et al. 2007). Anillin is important in the organization of the cleavage furrow during cytokinesis (Hickson and O’Farrell 2008). In addition, we characterized expression of cyclins D1 and E as well as Emi1/FBXO5, which regulates progression through early mitosis by preventing premature activation of APC (Reimann et al. 2001). We analyzed mitotic progression by investigating the localization of Eg5, a motor protein in the kinesin-like family involved in spindle dynamics during mitosis (Sawin and Mitchison 1995)
Anillin and each of the CPC components described above are expressed in primary proliferating myoblasts, but their mRNA and protein levels drop precipitously once myotubes form (Figure S2 A-B). When Rb was excised in differentiated primary multinucleated myotubes after TAM-treatment, anillin, AuroraB and survivin mRNA levels all rose, despite high levels of p19ARF expression (Figure S2 A). However, protein levels of AuroraB and survivin were comparable to those of growing myoblasts only after concomitant suppression of Rb and p19ARF (Figure S2 B). To verify that upregulation of CPC components occurred specifically in myonuclei that had entered S-phase, primary myoblasts were labeled with BrdU for 12hrs then stained for BrdU and survivin. Dividing primary myoblasts were positive for both BrdU and survivin, with the latter marking the cleavage furrow (data not shown). Differentiated, control-treated myotubes did not have myonuclei that expressed survivin, but those treated with TAM and p16/19si exhibited clustered BrdU+ myonuclei, and in these same myonuclei survivin expression was upregulated (Figure S2 C bottom panels). Although survivin was not localized in organized cleavage furrows, quantification of the immunostaining results indicated that nearly 80% of the myonuclei that had re-entered S-phase also upregulated survivin (Figure S2 D).
Analysis of the expression of cyclins D and E1, showed that these gene products are expressed in proliferating myoblasts and downregulated in differentiated myotubes. Upon treatment with either TAM or with TAM and p16/19si together, there was a rise in the expression of these cyclins as well as Emi1/FBXO5 (Sup Fig. 2E). These data indicate that mature myonuclei in myotubes progress through S-phase. In order to determine the extent of mitotic progression, we analyzed Eg5 localization in myotubes treated with TAM and p16/19si. Our results indicate that despite upregulation of Eg5, bi-polar spindles did not form in Rb and p16/19 deficient myotubes, although monopolar clustering of Eg5 was frequently observed (Sup Fig. 2F). Full karyokinesis or cytokinesis was not observed in myotubes.
Taken together, our data suggest that primary multinucleated myotubes deficient in both Rb and ARF synthesize DNA and components of the mitotic machinery, but fail to assemble these proteins in a manner required for nuclear division. While the incorporation of BrdU and expression of Ki67 mark the occurrence of S-phase, Aurora B is maximally expressed in G2-M. Moreover, survivin is only synthesized in G2-M in accordance with the cell cycle dependent CDE/CHR boxes in its promoter and post-translational stabilization that occurs at this cell cycle phase (Kobayashi et al. 1999; Li and Altieri 1999). However, the diffuse pattern of survivin immunostaining (Fig. S2 C) indicates that the nuclei in dedifferentiating mytotubes did not enter prophase, during which survivin staining becomes punctate (Caldas et al. 2005). Furthermore Eg5, although upregulated at the protein level, does not form the typical peri-centrosomal clustering signifying the onset of prophase (Sawin and Mitchison 1995). Thus, our data indicate that suppression of Rb and ARF in intact myotubes results in progression through S-phase to G2, but the nuclei cannot complete M-phase and are arrested at the onset of mitosis.
Dedifferentiation accompanies S-phase reentry in primary muscle cells
Postmitotic differentiated muscle cells express phenotypic markers such as myosin heavy chain (MHC), and a characteristic elongated morphology that results, in part, from the arrangement of the microtubule network. During differentiation the microtubule cytoskeleton shifts from the radial, centrosomal arrangement typical of myoblasts and most mononucleated mitotic cells, to a non-centrosomal array which extends along the longitudinal axis of the multinucleated myotube (Bartolini and Gundersen 2006). We investigated the role of the Rb and Ink4a gene products in sustaining expression of the differentiated phenotype by analyzing morphology and expression of markers of differentiation. Immunostaining demonstrated that MHC is not expressed in proliferating myoblasts or during early differentiation, but is readily detectable in all nascent myotubes by day 4 in differentiation media (Fig. 3A top panels). Treatment with tamoxifen after differentiation did not significantly alter myotube morphology and did not result in a detectable change in MHC levels by immunostaining (Fig. 3A bottom right panel). However, combined deletion of Rb and suppression of ink4a caused a marked decrease in MHC expression in cultures that had previously been differentiated (Fig. 3A bottom panels). Although intact, multinucleated and viable myotubes were present at days 6 and 7 of differentiation, cells that were rendered deficient in Rb and ink4a products lost structural integrity and began to collapse by day 6. By day 7, primary myotube morphology and MHC levels had severely deteriorated when compared to myotubes treated only with TAM and mock siRNAs (Fig. 3A bottom panels).
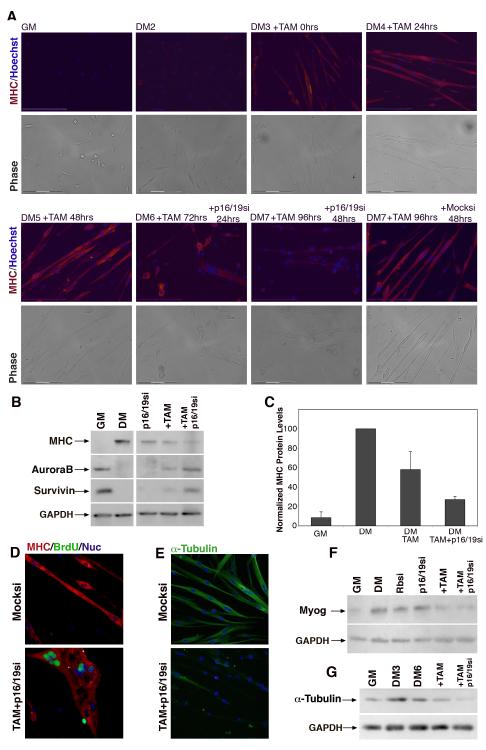
Figure 3. Dedifferentiation of mature myotubes.
(A) Immunofluorescence images of primary myoblasts and myotubes cultured for indicated times in DM and treated with TAM and either non-specific siRNA (Mocksi) or p16/19si at the indicated time points. Myotubes were labeled with primary antibodies for MHC (red), and Hoechst 33258 (blue). Bottom panels: phase images of the same image fields. Bar, 150μm. (B) Western blot analysis of primary myoblasts in GM and DM5 showing expression of MHC (220 kDa), AuroraB (38 kDa) and Survivin (20 kDa) as well as expression of these same proteins in myotubes after treatment for at least 48hrs with siRNA against p16/19, with TAM or both TAM and p16/19si. (C) MHC protein levels normalized to differentiated myotube cultures. Primary myotubes were treated with TAM or TAM and p16/19si. Growth medium (GM), Differentiation medium (DM). Error bars indicate the mean ± SE of at least three independent experiments. (D) Representative immunofluorescence images of DM6 primary myotubes treated with EtOH and non-specific siRNA or TAM and p16/19si for at least 48hrs. Myotubes were labeled with primary antibodies for MHC (red), BrdU (green), and Hoechst 33258 (blue). (E) Representative immunofluorescence images of primary DM6 myotubes after Mocksi or TAM and p16/19si treatment. Myotubes were labeled with primary antibodies for α-tubulin (green) and Hoechst 33258 (blue). (F) Western blot analysis of Myogenin (36 kDa) and (G) α-tubulin (50 kDa) in primary myoblasts and myotubes treated as indicated with siRNA and/or TAM. In each of the blots, GAPDH (35 kDa) is the loading control. Growth medium (GM); Myotubes were cultured in differentiation medium for 3 or 6 days (DM3 or DM6 respectively).
The combined suppression of Rb and p16/19 resulted in a decrease in levels of differentiation-specific muscle proteins in addition to MHC. Rb loss alone in primary myotubes treated with TAM led to a moderate reduction in some myogenic protein levels, such as MHC and myogenin (Fig. 3B, 3C and 3F), and a moderate increase in the mitotic proteins AuroraB and survivin (Fig. 3B). However, suppression of both Rb and p19ARF was more profound and accompanied by substantially reduced levels of myogenin and M-CK in addition to MHC (Fig. 3F and Figure S3 D upper band). In agreement with findings in primary myotubes in which both Rb and p19/ARF proteins were absent, Western blot analyses of C2C12 myotubes, which already lack p19ARF, exhibited decreased MHC levels when treated only with Rbsi (Figure S3 A). Expression levels of MRF4, also declined. Semi-quantitative RT-PCR analyses revealed a decrease in RNA levels that paralleled these protein data (Figure S3 C).
Immunofluorescence (IF) analysis of myotubes revealed several morphological differences in TAM and p16/19si myotubes when compared to Mocksi myotubes. In addition to incorporating BrdU by day 6 of differentiation, treated myotubes were significantly less compact than controls, never exhibited striations, and appeared to have large gaps in their MHC network (Fig. 3D). While mock-treated primary myotubes retained their elongated morphology and nuclear organization, BrdU+ myotubes collapsed into amorphous multinucleated syncytial structures, highlighted by the clustering of myonuclei. Although not all of the myonuclei entered S-phase, loss of structural integrity of the myotube is likely due to the lack of maintenance of myotube nuclear protein domains. Morphological changes and loss of MHC expression were also correlated with BrdU+ staining in C2C12 myotubes, however, only loss of Rb was required (Figure S3 B).
To further investigate the deterioration in myotube morphology we analyzed the expression and arrangement of α-tubulin in mature myotubes. In mock-treated myotubes α-tubulin is expressed at high levels and microtubules are arranged longitudinally, while in TAM and p16/19si-treated myotubes, α-tubulin levels are lower by IF (Fig. 3E) and western analysis (Fig. 3G and Figure S3 E-F). To dynamically visualize structural deterioration, myotubes were imaged by time-lapse microscopy following treatment with TAM and Mocksi (Video 1) or TAM and p16/19si (Video 2). A time-lapse comparison shows that complete morphological collapse of myotube structure only takes place after loss of both the Rb as well as Ink4a gene products. Despite extensive structural differences, the Rb and ink4a deficient primary myotubes do not die or detach faster than Mocksi-treated myotubes, and retain their motility and membrane activity, such as filopodia and lamellapodia.
Terminally differentiated myocytes are capable of proliferation after suppression of Rb and p16/19
The extent of muscle differentiation can be characterized based on the sequential expression of muscle regulatory transcription factors (MRFs). MyoD and Myf5 are early and characteristic of cycling myoblasts, whereas late transcription factors include myogenin and MRF4, which are expressed in terminally differentiated myocytes and myotubes (Shen et al. 2003; Charge and Rudnicki 2004). Based on this pattern of transcription, we developed a system to study cycling and dedifferentiation in prospectively isolated populations derived from individual differentiated muscle cells. We infected low passage Rosa26-CreERT2 Rblox/lox primary myoblasts with a retroviral vector in which GFP expression is under the control of the myogenin promoter (pLE-myog3R-GFP). To verify the fidelity of myogenin and GFP co-expression, pLE-myog3R-GFP myoblasts were sparsely seeded in growth medium or differentiation medium for 72 hours and stained for myogenin and GFP. IF analysis clearly showed that GFP and myogenin expression were upregulated in the differentiating (DM3) myocyte population (Fig. 4A). Quantification of IF data indicated that only 0.9% of the cells are GFP-positive in growth conditions, likely representing spontaneous differentiation. In differentiated cultures, GFP expression correlated with myogenin immunoreactivity, and 27 ± 2.0% of the cells had detectable GFP levels (Fig. 4Bi). To verify the fidelity of myogenin expression in GFP-positive cells, we scored elongated GFP-positive myocytes for myogenin. Our data indicate that 98.4 ± 1.8% of cells were GFP+ myogenin+ (Fig. 4Bii). High-throughput analysis of GFP expression in myoblasts and myocytes was also performed by fluorescence activated cell sorting (FACS), which showed that 35% of the cells expressed GFP after 3 days in differentiation medium, while only 1.8% of cells in growth medium were GFP+ (Fig. 4D). Thus pLE-myog3R-GFP infection of primary myoblasts allows for reliable identification of myogenin-expressing cells.
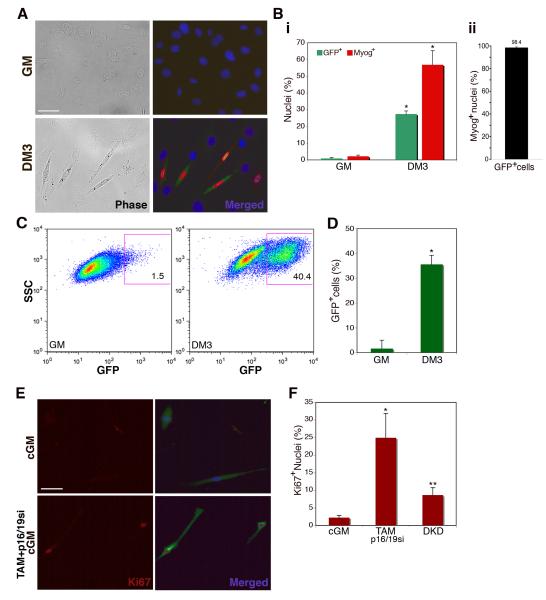
Figure 4. Myogenin-expressing myocytes enter S-phase only after loss of Rb and ink4a gene products.
(A) Immunofluorescence images of myoblasts (GM) and myocytes (DM3) infected with pLE-myog3R-GFP. Cells were labeled with primary antibodies for GFP (green), myogenin (red) and Hoechst 33258 (blue). Bar 50μm. (B.) (i) Histogram represents percentage of GFP-positive (green bars) and myogenin-positive (red bars) cells in growth medium (GM) or differentiation medium (DM3). A minimum of 1000 nuclei were counted from random fields for each trial. Error bars indicate the mean ± SE of at least three independent experiments. (*P<0.005). (ii) Histogram represents percentage of GFP-positive cells that also express detectable myogenin by immunostaining. Individual cells were evaluated for expression of each marker. A minimum of 250 cells were counted from random fields. Error bars indicate the mean ± SE of three independent experiments. (C) Representative FACS plots of myoblasts (GM) and myocytes (DM3) infected with retroviral pLE-myog3R-GFP construct. Gated population indicates GFP-positive myocyte population employed in subsequent experiments. (D) Histogram representation of GFP expression in three independent experimental FACS profiles on myoblasts (GM) and myocytes (DM3) (*P<0.001). (E) Immunofluorescence images of GFP-positive FACS-sorted myocytes cultured in conditioned GM only (cGM) or in cGM with TAM and p16/19si-RNA. Cells were labeled for Ki67 (red) and GFP (green) as well as Hoechst 33258 (blue). Bar 50μm. (F) Histogram represents percent of Ki67-positive nuclei in GFP-positive FACS sorted population, in cGM, treated with TAM and p16/19si, or treated with Rbsi and p16/19si-RNA in tandem (DKD). Growth medium (GM); A minimum of 100 nuclei were counted from random fields for each trial. Error bars indicate the mean ± SE of three independent experiments. (*P<0.01, **P<0.005).
Myoblasts seeded at low density express myogenin and undergo terminal differentiation in the absence of fusion (Shen et al. 2003). We took advantage of this in vitro property of muscle cells to investigate whether the dedifferentiation and cell cycle reentry observed in differentiated multinucleated myotubes could lead to proliferation after suppression of Rb and p19ARF in differentiated mononucleated myocytes. In order to follow the fate of single cells, the following experiments were performed using individual pLE-myog3R-GFP myocytes. First, sparsely seeded primary muscle cells, maintained in DM for 72hrs, were sorted on the basis of GFP expression as depicted by the gated population in Fig. 4C. To determine whether the differentiated muscle cells were capable of proliferation, the FACS sorted population was cultured in conditioned growth medium (cGM) for up to 48hrs and then stained for GFP and Ki67 (Fig. 4E top panels). Only 2.3% of GFP+ cells had Ki67 positive nuclei. In contrast, GFP+ cells treated with TAM 24hrs prior to FACS sorting, and with p16/19si 12hrs after FACS sorting (Fig. 4E bottom panels) exhibited Ki67 nuclear staining in 25% of the GFP+ population 48hrs after culture in cGM (Fig. 4F).
As an alternative method to suppress Rb and p16/19, siRNAs against Rb and p16/19 were used to transiently reduce both transcripts. Analyses to determine the ideal dosage and method of siRNA application showed that most efficient knockdown occurred after tandem treatment of primary muscle cells with Rbsi, a 12hr recovery period, followed by treatment with p16/19si. DKD treatment of FACS-sorted GFP+ cells, resulted in 8.6% Ki67+ nuclei (Fig. 4F). That the frequency of DNA synthesis was lower in DKD treated cells than in those treated with TAM and p16/19si was expected given the lower efficiency of knockdown compared to TAM treatment for Rb suppression. The data from these two types of experiments show at the single cell level, that differentiated myogenin-expressing myocytes, like myotubes, efficiently enter S-phase only after suppression of both Rb and Ink4a gene expression.
We reasoned that if myocytes could divide, they would give rise to clones. However, to rule out cell migration and definitively show that a postmitotic myocyte divided, single-cell resolution and clonal analysis was critical. Accordingly, to assess the proliferative potential of myocytes, we first FACS-purified cells twice in order to isolate individual differentiated GFP+ myocytes. Individual myocytes were then sorted directly into microarrays of hydrogel wells (Lutolf et al. 2009) (Fig. S4 A), while a subset was stained for co-expression of GFP and myogenin (Fig. S4 B). After treatment with Mocksi or DKD, proliferative myocytes were scored as the percentage of microwells that had a minimum of 8 cells at 96hrs after treatment. 6.8% of DKD treated myocytes gave rise to clones in contrast to only 0.6% of Mocksi treated cells (Figure S4 C). Taken together these data support the conclusion that myogenin-positive myocytes acquire proliferation potential after suppression of Rb and p16/19 expression.
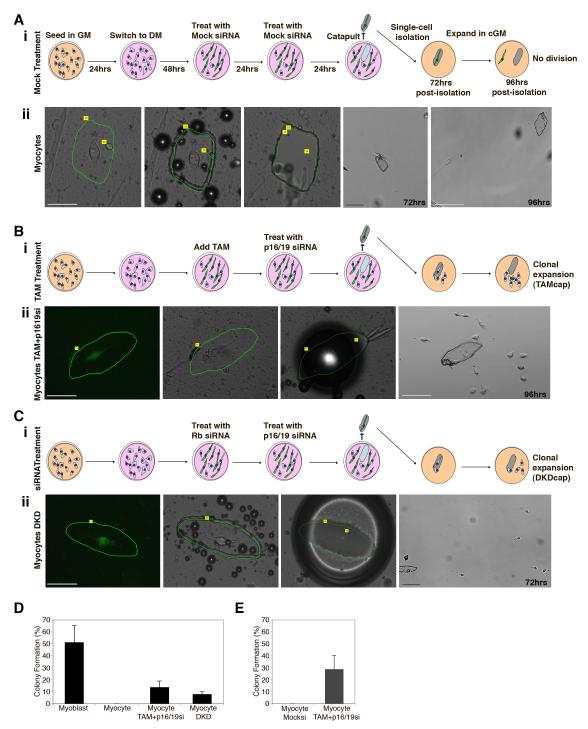
Clonal expansion and proliferation of individually purified myocytes after laser microdissection and laser pressure catapulting (LPC)
To determine definitively if differentiated myocytes dedifferentiate and proliferate, it is essential to monitor individual myocytes before and after loss of Rb and ARF. For this purpose we pioneered the use of photoactivated laser microdissection (PALM) and laser pressure catapulting (LPC) with primary cells. LPC is extensively used to obtain precise regions of fixed tissues for DNA, RNA and protein analyses. However, the use of LPC for studies of living cells has only been reported in methodological descriptions using robust cell lines in protocols by the developers of PALM (Stich et al. 2003; Schutze et al. 2007). We optimized LPC in order to ensure cell viability and isolate single cells from mass cultures of primary myoblasts and myocytes. Using the PALM LPC system, cells are selected for isolation by the operator, who encircles the cell of interest with a unique software-simulated shape; then the laser is directed to cut the membrane precisely along the operator-drawn lines. The cut membrane is subsequently catapulted by laser pressure and captured by a robotic arm carrying an inverted media-containing cap. During this procedure, the captured cell remains attached to the membrane on which it grew, so that cell adhesion and morphology are not disrupted. This technology allows a single cell to be isolated intact on the surface on which it is growing. Using LPC, the identification of a clone derived from a captured cell is unambiguous as it is plated singly in a well. In this way there is no question regarding the origin of the cells that proliferate.
Laser microsdissection and pressure catapulting analysis was performed with myocytes. For this purpose primary myoblasts expressing pLE-myog3R-GFP+ were sparsely seeded and differentiated to become GFP+ myocytes, and then treated with either mock siRNA (Fig. 5A i) or for suppression of Rb and ARF (Fig. 5B i and 5C i.) Individual myocytes were selected based on their differentiated phenotype including elongated morphology evident by phase microscopy (Fig. 5A i left panel) and bright GFP expression (Fig. 5B ii and 5C ii left panels). Laser microdissection was used to mark and cut the membrane surrounding prospectively identified single myocytes (green lines Fig. 5B ii and Fig. 5C ii). Marking and recording membrane shapes allowed for tracking of each membrane and the isolated cell on its surface. The LPC burst locations that lead to catapulting are indicated by the blue dots in the images in Fig. 5B ii, which also serve as a means of identifying the membrane. Typical images obtained during the steps of the PALM and LPC isolation process are shown in Fig. 5B ii and 5C ii. Myocyte morphology before and after membrane ablation did not significantly change. Note in the example shown that 72hrs after capture the mock treated myocyte is still associated with the membrane (Fig. 5Aii, panel 4), and by 96hrs it has left the membrane but still exists as a single adherent cell, although it was cultured in conditioned growth medium since the time of capture (Fig. 5Aii, panel 5). Similarly, in individual myocytes treated for reduction of Rb and ARF, morphology remained intact during the cutting and catapulting (Fig. 5B ii and 5C ii, left 3 panels). However, in contrast to mock-treated myocytes, reduction of Rb (TAM or siRNA) and ARF (siRNA) led to cell division and colony formation in the immediate vicinity of the membrane (Fig. 5B ii, fourth panel from left, Fig. 5C ii, fourth panel from left). Additional examples of single GFP+ myocyte laser capture are shown in Figure S4 D-E.
Figure 5. Laser microdissection and PALM LPC; single-cell isolation and clonal expansion of dedifferentiated myocytes.
(A. i) Schematic representations of the culture conditions, treatment with Mock siRNA, and isolation by laser microdissection and catapulting of myogenin-GFP+ myocytes. Diagram also shows the fate of isolated cells 72 and 96hrs after isolation. (A.ii) Representative images of mock-treated myocyte (DM4) (panels left to right) prior to microdissection, immediately after microdissection, after LPC isolation, 72hrs post-isolation and 96hrs post-isolation. Bar 50μm, 100μm. (B. i) Schematic representations of the culture conditions, treatment with TAM and p16/19 siRNA, and isolation by laser microdissection and catapulting of myogenin-GFP+ myocytes. Diagram also shows the fate of isolated cells 72 and 96hrs after isolation. (B.ii) Representative image of TAM and p1619si-RNA treated myocyte (DM4); First panel- native GFP expression marks myogenin expression prior to microdissection. Second panel- the same cell during microdissection, Third panel-after LPC isolation, Fourth panel- 96hrs post-isolation and visualization of expansion. Bar 50μm, 100μm. (C. i) Schematic representations of the culture conditions, treatment with Rb and p16/19 siRNA , and isolation by laser microdissection and catapulting of myogenin-GFP+ myocytes. Diagram also shows the fate of isolated cells 72 and 96hrs after isolation. (C.ii) Representative image of DKD treated myocyte; First panel- GFP expression marks myogenin expression prior to microdissection. Second panel- the same cell after microdissection. Third panel- after LPC. Fourth panel- 72hrs post-isolation with visualization of expansion. Bar 50μm, 200μm. (D) Histogram represents percentage of colony formation after PALM LPC cell isolation. Error bars indicate the mean ± SE of at least five independent experiments, in which at least 50 membranes were captured for each myocyte trial, and at least 20 myoblast membranes were captured to verify cell survival and capture efficiency. (E) Histogram represents percentage of colony formation after PALM LPC cell isolation following FACS isolation of GFP+ myocyte population as indicated by the scheme in Figure S5 A. Error bars indicate the mean ± SE of at least four independent experiments, in which at least 50 membranes were captured for each myocyte trial. Primary muscle cells were differentiated for 4 days (DM4) at the time of LPC isolation.
In five independent PALM LPC isolation experiments, an analysis of a total of 250 membranes verified that without reduction of Rb and ARF, not a single captured myogenin-GFP+ myocyte divided to produce a colony. In contrast, deletion of Rb and suppression of ARF produced 34 colonies, a frequency of 13.8% (Fig. 5D), and transient suppression of both Rb and ARF (DKD) resulted in a colony frequency of 8.0%.
To increase the efficiency of identification of single GFP+ myocytes, and to thereby avoid desiccation we increased viability by first using FACS to enrich for myocytes expressing myogenin-GFP, thus facilitating identification of single myocytes (Fig. S5 A). Images of the captured control or TAM and p16/19si treated myocytes are shown (Fig. S5 B and C). Again, control treated myocytes failed to produce colonies in conditioned growth medium (Fig. S5 B), while myocytes in which Rb and Ink4a genes were silenced under conditions of greater survival (Fig. S5 C), exhibited a high frequency of 28.8% colony formation (Fig. 5E).
Redifferentiation of captured myocyte colonies after exposure to low serum
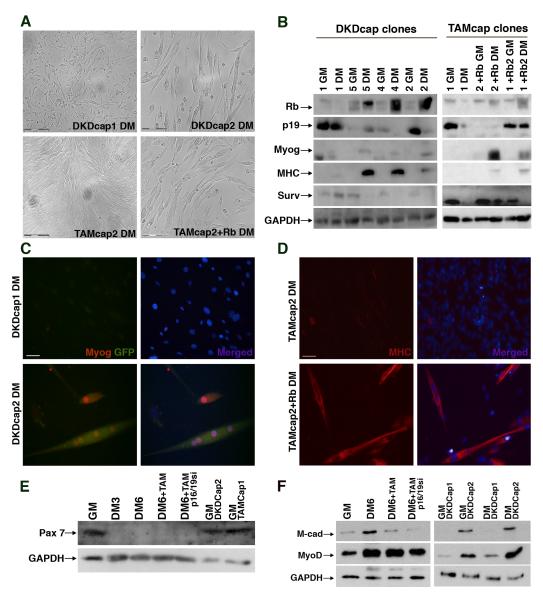
Dedifferentiated muscle cells in axolotls have been shown to be capable of proliferation and contribution to regenerating muscle, although single cell tracking using genetic marking has not been possible to date. After LPC capture, TAM and p16/19si-treated myocytes proliferated rapidly in GM, and most of the cells lost myogenin expression after 72hrs, evidenced by lack of GFP (Fig. S6 B). To determine if such dedifferentiated, actively dividing mammalian myocytes were capable of redifferentiation, we exposed these cells to differentiation conditions. However, they continued to proliferate and never fused (Fig. S6 B lower panels), by comparison with control LPC captured myoblasts, which had started to fuse by three days in DM (Fig. S6 A). This lack of differentiation is expected because the TAM treated cells had permanently lost Rb expression. In contrast, Rb suppression in DKD captured myocytes was transient. Four colonies derived from single DKD-treated captured myocytes were expanded, exposed to DM for four days and then assayed for muscle markers, either by microscopy or biochemically. The DKD colonies spanned the spectrum of differentiation potential, as shown in the top panels of Fig. 6A and Fig. 6C. One colony (DKDcap1) continued to proliferate despite being in DM while another (DKDcap2) differentiated and fused. Heterogeneity in the behavior of captured cells after transient knockdown may be due to clonal variability following extensive proliferation.
Figure 6. Dedifferentiated myocytes are capable of expansion and redifferentation into mature myotubes.
(A) Phase contrast images of Rbsi and p16/19si double-knockdown captured (DKDcap) myocyte colonies and TAM with p16/19si captured myocyte (TAMcap) colonies at DM4, prior to protein harvest for expression analysis. Bar 150μm (B) Western blot analysis of captured colonies in GM and DM arranged from left to right according to their differentiated morphologies in DM4; protein levels of Rb (100 kDa), p19ARF (20 kDa), myogenin (36 kDa), MHC (220 kDa) and Survivin (20 kDa) as well as GAPDH (35 kDa) as a loading control. (C) Representative images of two DKDcap myocyte colonies in DM4, labeled for GFP (green) and myogenin (red) as well as Hoechst 33258 (blue). Bar 25μm. (D) Representative images of TAMcap myocyte colony in DM4 labeled for MHC (red) and Hoechst 33258 (blue), a portion of which (lower panels) was infected with retrovirus re-introducing Rb expression. Bar 50μm. (E) Western blot analysis of Pax-7 protein (57 kDa) levels in muscle cells in GM, DM at indicated time points and with indicated treatments, and in the DKDcap dedifferentiated clones. (F) Western blot analysis of M-cadherin protein (88 kDa) and MyoD (34 kDa) levels in primary muscle cells under growth conditions (GM), differentiated conditions (DM6) with indicated treatments, and in the isolated dedifferentiated clones (DKDcap1 and DKDcap2) in GM and DM4 (DM).
Analysis of captured DKD colonies supports a function for Rb in successful redifferentiation, since we found that DKDcap1 cells lost Rb expression, while the colonies that readily fused and differentiated (DKDcap4 and DKDcap2) expressed high levels of Rb in DM (Fig. 6B). p19ARF expression in DKDcap1 was high, the cells failed to upregulate GFP and endogenous Myogenin remained undetectable in DM (Fig. 6C). In contrast, in the colonies that differentiated well, downregulation of p19ARF was observed in DM. Expression of GFP, Myogenin and MHC provided confirmation of differentiation potential of clones DKDcap4 and DKDcap2 (Fig. 6B and C). The relative changes in expression patterns of Rb, Ink4a and myogenic proteins by the DKDcap2 colony and by captured myoblasts were similar (Fig. S6 C-D).
We reasoned that if Rb was re-introduced into the TAM and p16/19si captured cells, redifferentiation and fusion should occur. Thus, we infected a subset of captured colonies with a pMIG retrovirus vector expressing the human Rb cDNA (Sage et al. 2000). We monitored the TAMcap myoblast colonies after infection with pMIG-Rb or control pMIG retrovirus and determined that differentiation and fusion occurred in those colonies that re-expressed Rb (Fig. 6A lower panels). Analysis of Rb protein levels showed the expected increase in Rb protein that paralleled the upregulation of the myogenic proteins, myogenin and MHC (Fig. 6B). In addition, Survivin levels decreased in the TAMcap1 colony following pMIG-Rb infection (Fig. 6B, lane 6). However, ARF levels remained variable among clones. The myogenic potential of TAMcap colonies infected with pMIG-Rb was also verified by immunofluorescence for MHC (Fig. 6D). Together, these data demonstrate that dedifferentiated myocytes are capable of successful redifferentiation in vitro after capture and expansion, and that this process is dependent upon expression of Rb.
Muscle stem cells and proliferating primary myoblasts express the marker Pax 7 (Charge and Rudnicki 2004). We tested whether muscle cells with reduced Rb and ink4a dedifferentiated to a point such that expression of Pax 7 was reactivated. Primary myoblasts expressed Pax7 when they were proliferating under growth conditions, but once in differentiation media for at least 72hrs, Pax7 protein levels decreased and remained undetectable by western analysis in mature myotubes (Fig 6E lanes 2 and 3). After reduction of Rb and ARF, dedifferentiating myotubes did not re-express Pax7 (Fig. 6E lanes 4 and 5), however myocytes from the PALM captured myogenin-positive population re-expressed Pax7 once they established colonies in conditioned growth media (Fig 6E lanes 6 and 7). We also tested expression of MyoD and M-cadherin, which have characteristic expression patterns during muscle differentiation. Reduction of Rb and Arf caused a decrease of MyoD and M-cadherin protein levels in myotubes (Fig. 6F left panel and Fig. S6 E). In PALM generated clones, expression of MyoD and M-cadherin, like morphological differentiation was correlated with expression of Rb (Fig. 6F right panel). Only clones that expressed Rb (DKDCap2) expressed relatively high levels of MyoD and M-cadherin, both of which increased somewhat during differentiation (Fig. 6F right panel, lanes 2 and 4). By contrast, the clone that did not express Rb (DKDCap1), possibly due to mutation or deletion during clonal selection, did not express detectable levels of M-cadherin and expressed only low levels of MyoD in growth and differentiation medium (Fig. 6F right panel, lanes 1 and 3).
Taken together these results strongly support a role for Rb and Ink4a in the maintenance of myogenic differentiation, as their suppression in myogenin-expressing post-mitotic myocytes leads to division, expansion, reactivation of Pax 7, as well as MyoD and m-cadherin expression patterns consistent with myoblasts.
Captured and expanded myocytes are capable of contribution to muscle in vivo
Finally, we tested whether PALM LPC isolated, expanded myocytes could contribute to existing muscle in vivo. 1.5×105 DKD derived dedifferentiated myocytes (DKDcap1 and DKDcap2) were injected into the tibialis anterior (TA) of NOD/SCID mice. Myogenin-GFP expression was used to track the injected cells, as this marker was reliably detected in vitro upon differentiation (Fig. 6C). Ten days after injection, DKDcap1 cells, which exhibited no fusion in vitro, proliferated excessively in vivo and caused severe disruption of the muscle laminin network at the site of injection (Figure S7 top panels). On the other hand, DKDcap2 cells readily fused to the existing muscle fibers and caused no disruption of the laminin network (Figure S7 bottom panels).
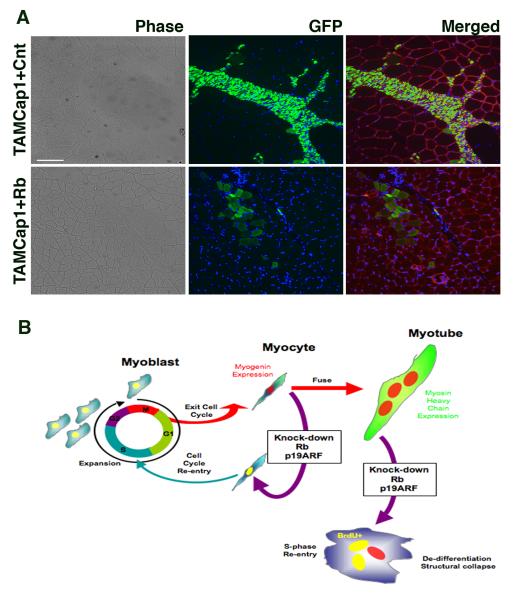
We also analyzed in vivo contribution of colonies in which Rb was deleted and ARF suppressed (TAMcap). Infection with pMIG-Rb restored Rb expression and pLE-GFP retrovirus was used to enhance visualization by expressing GFP constitutively and at higher levels than those produced by the myogenin reporter. Control TAMcap1 cells, which received control pMIG and pLE-GFP infections, proliferated excessively in vivo (Fig. 7A top panels). By contrast, TAMcap1 cells with re-introduced Rb expression, efficiently fused to existing muscle fibers, brightly labeling them with GFP, without any apparent proliferation and without disruption of the existing laminin network (Fig. 7A bottom panels). We conclude that if Rb expression is either transiently suppressed or adequately restored dedifferentiated myocytes are capable of redifferentiation and incorporation by fusion to existing muscle in vivo.
Figure 7. Dedifferentiated myocytes are capable of fusing to muscle in vivo.
(A) Representative cross-sections of tibialis anterior 10 days post-injection of 2.5 ×105 cells from TAMcap1 and TAMcap1+Rb clonally expanded myocytes (see Fig. 6). Incorporation of dedifferentiated myocytes into pre-existing fibers can be visualized in merged fields by GFP+ staining (green) of laminin-bound fibers (red), nuclei (blue); to enhance visualization, cells were infected with a constitutive-eGFP expressing retroviral vector prior to injection. (n=4, three of which had GFP+ fibers detected) Bar 50μm. (B) Schematic representation of the events following suppression of Rb and p19ARF in primary differentiated myocytes and multi-nucleated myotubes.
Discussion
The molecular basis for the extraordinary disparity between mammals and certain lower vertebrates, such as newts, axolotls and zebrafish, in their capacity to regenerate injured or amputated tissues remains a major unresolved biological question. Mammalian regeneration of a muscle occurs only if that muscle is replaced after mincing or grafting of small muscles, or when chemical agents spare stem cells. Experiments of this type indicate that significant architectural remodeling can occur in lieu of scarring but that the extent of regeneration is limited by the amount of replaced or surviving muscle (Carlson 2003). Although endogenous muscle stem cells can account for some degree of regeneration (Collins and Partridge 2005; Sacco et al. 2008), they do not seem to suffice in extreme circumstances. Indeed, to date there is no evidence that the extensive regeneration of entire muscles seen in urodeles can be achieved by any known mammalian mechanism when a significant mass of tissue is removed and not replaced. This regenerative potential is likely limited in part by a combination of excessive demand for cell proliferation at the site of injury in order to replace lost tissue, and an inhibition of architectural remodeling by fibrosis. A possible basis for the failure of regeneration in mammals, which has fascinated scientists for centuries, is a capacity, which regenerative vertebrates have retained and mammals have lost: dedifferentiation.
Does dedifferentiation endow regenerative vertebrates with capabilities that mammals lack? This question underscores the need for novel insights into the molecular mechanisms of dedifferentiation in order to discover what is missing in mammals. In urodeles, compelling evidence from studies of skeletal muscle suggests that dedifferentiation is a major mode of tissue regeneration (Hay and Fischman 1961; Lentz 1969; Kintner and Brockes 1984; Lo et al. 1993; Tanaka et al. 1997; Echeverri et al. 2001) Dedifferentiation involves two processes, which are separable and independent: muscle cell fragmentation into individual mononuclear cells, and cell cycle reentry followed by proliferation (Velloso et al. 2000; Brockes and Kumar 2008). In mammalian myotubes produced using the immortalized cell line, C2C12, overexpression of transcriptional factors present in the blastema such as msx1 (Odelberg et al. 2000) or twist (Hjiantoniou et al. 2008), or exposure to small molecules that disrupt the cytoskeleton, have been reported to result in myotube fragmentation (Perez et al. 2002). In the dedifferentiation studies reported here, fragmentation of muscle cells was not observed, which is in good agreement with reports showing that the fragmentation process in urodeles is independent of cell cycle reentry (Velloso et al. 2000). In addition, since the trigger and mechanism for muscle fragmentation and cellularization in urodeles remains unknown, it is currently not possible to determine if a similar pathway exists in mammals. Understanding how fragmentation occurs and delineating the dedifferentiation mechanisms are complementary but distinct goals in muscle regeneration biology.
Molecular regulation of dedifferentiation by Rb and ARF
Our decision to suppress Rb in experiments directed at elucidating the mechanisms underlying muscle dedifferentiation was based on evidence in urodeles that Rb coordinates muscle cell cycle entry in response to damage (Tanaka et al. 1997). In mammals as in urodeles, cell cycle regulation by Rb is dynamic and controlled primarily by its phosphorylation state (Lipinski and Jacks 1999; Burkhart and Sage 2008). There is also a wealth of data firmly establishing the tumor suppressor Rb as a necessary player in the orchestration of mammalian muscle cell differentiation, including evidence for a dual role in both muscle cell cycle progression and exit (Gu et al. 1993; Zacksenhaus et al. 1996; Puri et al. 2001; Blais et al. 2007). Indeed, Rb null mice die before birth and lack differentiated muscles (Zacksenhaus et al. 1996; Takahashi et al. 2003). Furthermore, Rb has also been shown to act not only as a cell cycle regulator, but to impact differentiation and tissue specific gene expression directly by binding histone deacetylase 1 (HDAC1) and promoting activation of muscle genes such as MyoD (Puri et al. 2001). Once differentiation occurs, this state is stably maintained, at least in mammals. This stability is underscored by the inability to reverse differentiation simply by inactivating Rb in primary differentiated mammalian muscle cells (Camarda et al. 2004; Huh et al. 2004). Indeed, studies reporting otherwise have been confounded by the use of immortalized cell lines such as C2C12 (Gu et al. 1993; Blais et al. 2007), which we show here have a deletion in the gene and do not express the ink4a products. Loss or suppression of Rb leads only to moderate dedifferentiation, as demonstrated by the reduced accumulation of myogenin and MHC (Fig 3). In fact, as shown in this report and by others previously (Camarda et al. 2004; Huh et al. 2004), it is remarkable how little phenotypic change occurs in primary differentiated skeletal muscle cells when Rb is suppressed.
The minimal impact of Rb absence alone on muscle dedifferentiation suggested that maintenance of mammalian differentiation is ensured by a separate mechanism. Whereas the basal Rb and p53 pathways are functional in lower vertebrates, we reasoned that a modulator that is absent in regeneration competent vertebrates would be a good candidate for a regeneration suppressor in mammals. We therefore focused on the ink4a locus and ARF in particular. Unlike Rb, inactivation of ARF alone in knockout mice has no apparent effect on differentiation (Serrano et al. 1996; Kamijo et al. 1997). In agreement with these reports, we found that suppressing ARF alone had no effect on muscle differentiation or dedifferentiation. Loss of Rb led to elevated expression and protein levels of ARF. Concomitant inactivation of Rb and ARF caused extensive loss of differentiation. Previously differentiated myotubes exhibited robust DNA synthesis and expression of mitotic proteins upon acute loss of Rb and p19ARF, suggesting that these two are nodal points for intrinsic control of muscle cell cycle reentry. The profound loss of architectural integrity and downregulation of myogenin, MRF-4, MHC and M-CK, further suggests that Rb and ARF together are potent stabilizers of the differentiated state. Notably, alternative approaches to induce cycling by altering growth factor signaling and regeneration in mammalian cardiac muscle cells (Bersell et al. 2009) produce a very moderate effect when compared to regenerating urodele muscle. We speculate that ARF may inhibit robust cycling and regeneration in this setting as well.
Our findings support the hypothesis that tumor suppression mediated by the ink4a locus arose at the expense of regeneration. Both p16ink4a and p19ARF have been recently shown to contribute to the decline in regenerative potential of multiple tissues during aging by affecting stem cell self-renewal (Sharpless and DePinho 2007; Levi and Morrison 2008). Our study suggests that the ink4a locus has an additional negative impact on tissue regeneration, i.e., suppression of cell cycle reentry and dedifferentiation. The remarkable combined effect of acute Rb and ARF loss strongly suggests (i) that continuous expression of Rb itself has an important function in maintaining the differentiated state and (ii) that the maintenance of the differentiated state in mammals depends on complementary activities of Rb and ARF. These findings are explicable in view of the known need for continuous regulation of differentiation (Blau and Baltimore 1991); (Yamanaka and Blau, 2010) as well as the documented functions of Rb and ARF in preventing inappropriate cycling as tumor suppressors.
Single cell analyses of reversal of differentiation
Bulk cultures do not allow a definitive assessment that a given cell has divided. The lack of transgenic animals, cellular complexity, and rapid developmental changes observed in the blastema has hindered analysis of dedifferentiation at the single-cell level in urodele regeneration until recently (Sobkow et al. 2006). In mammalian muscle culture systems in which S-phase reentry was observed, the persistence of cells at earlier stages of differentiation cannot be ruled out (Gu et al. 1993; Schneider et al. 1994; Blais et al. 2007). In addition, continuous timelapse monitoring (Duckmanton et al. 2005) and single cell analysis are essential, since reports of division of differentiated muscle cells could be the result of cell migration. To overcome these problems we employed (i) dynamic single-cell tracking of myocytes isolated in microwells by time lapse microscopy and (ii) isolation of single myocytes by PALM laser capture microscopy. In particular, the laser microsdissection and catapulting technology allows for unambiguous documentation of the division of individual differentiated cells, as they are each selected and isolated on the basis of both their morphology and their expression of a genetic marker of differentiation, myogenin. These single cell studies clearly demonstrated that cell cycle entry and expansion of individual differentiated postmitotic myocytes occurs after Rb and p19ARF loss. By contrast, untreated myocytes, crawled off of membranes, but never divided. We further demonstrated the regenerative potential of dedifferentiated myocytes by inducing redifferentiation. A subset of captured colonies produced by transient inactivation of Rb and p19ARF were exposed to differentiation medium in culture and fused to form myotubes. Additionally, in myocytes that had irreversibly lost Rb expression due to Cre-mediated excision, reintroduction of Rb by retroviral delivery not only induced myotube formation and muscle gene expression in vitro, but also resulted in fusion and regeneration of myofibers in vivo with typical architecture and no evidence of the tumorigenic characteristics of cells that did not receive Rb. These findings suggest that transient inactivation of the two tumor suppressors could yield dedifferentiated cells with extensive regenerative potential as depicted in the diagram in Fig 7B.
We capitalized on evolutionary differences to genetically modify the mammalian cell-cycle regulatory pathways to more closely mimic those found in lower vertebrates. Our results reveal that it is possible to derive regenerative cells from differentiated, post-mitotic muscle in addition to classically defined stem cells. Skeletal muscle cells can alternate between a differentiated, post-mitotic state and a proliferative, regenerative state, retaining the essential characteristics of their cell type of origin during the regenerative cycle. Our experiments implicate ARF in the suppression of regeneration in mammalian cells by impeding dedifferentiation. We postulate that a combination of transient interventions to inactivate the Rb pathway while suppressing ARF may be fruitfully employed to maximize a mammalian regenerative response.
Experimental Procedures
Mice and primary myoblast preparation
Rosa26-CreERT2 Rblox/lox mice (Viatour et al. 2008) were crossed to mice carrying a Cre-responsive β-galactosidase reporter allele (Ventura et al. 2007). The hind leg muscles of 6-8 weeks old offspring were prepared for primary myoblast harvest as described (Rando and Blau 1994).
Cell culture
C2C12 and primary mouse myoblasts were cultured as described in (Pajcini et al. 2008) and Supplementary Materials.
Cloning and vector construction
pLE-myog3R-GFP retroviral vector was constructed by subcloning the myogenin promoter elements driving GFP expression from peGFPN1-hmyg (a gift from Daniel Kemp-Novartis) into the pLE-GFP vector. See Supplementary Materials for details of the construct preparation and virus production.
siRNA silencing and semi-quantitative RT-PCR
RNA-interference was carried out using small-interfering RNA duplexes designed then screened for specific and effective knockdown of target genes. Duplexes were designed for p16/19 sense sequence AGGUGAUGAUGAUGGGCAAUU and p19ARF sense sequence GCUCUGGCUUUCGUGAACAUG or ordered directly as ON-TARGETplus siRNA Rb1 (J-047474-06) from Thermo/Dharmacon. For control transfections non-targeting siRNA#1 (D-001810-01-05) and siGlo-Green were purchased from Thermo/Dharmacon. Transfections of siRNA duplexes, resuspended in siRNA buffer (Dharmacon) were carried out after differentiation of myocytes or after fusion of myotubes at 48-72hrs in DM with siImporter transfection reagent (Millipore) as per manufacturer’s instructions. Transfection mix was added to cells after supplemented to differentiation media for 12 hrs. RNA was harvested from C2C12 or primary cells in GM or DM by RNeasy mini kit (Qiagen) and 200ng of total RNA was used in semi-quantitative RTPCR analysis with Superscript III One-Step RT-PCR (invitrogen). Primers designed for genes tested and details of the PCR reactions may be found in Suplementary Materials.
BrdU analysis
5-bromo-2′-deoxy-uridine (BrdU) labeling and detection kit (Roche) was used according to the manufacturer’s instructions. BrdU labeling reagent was added to the cells with fresh DM media for 12hrs.
Western analysis
Cells were lysed at room temperature in lysis buffer (50mM Tris pH 7.5, 10mM MgCl2, 0.3 M NaCl, 2% IGEPAL). When necessary, membranes were stripped by incubating at 50oC for 45min and then 1hr at RT in stripping buffer (100mM 2-mercaptoethanol, 2% SDS, 62mM Tris, ph 7).
See Supplementary Materials for details of antibody use.
Immunofluorescence
C2C12 myotubes and primary myocytes or myotubes seeded densely for fusion or sparsely for differentiation as described above were fixed and permeabilized as per manufacturer’s instructions when co-staining with BrdU or with 1.5% paraformaldehyde 15 min at room temperature (RT) then permeabilized with 0.3% Triton-PBS 10 min at RT. See Supplementary Materials for details of antibody use.
Cells were imaged with Zeiss Axioplan2 using 40x water immersion objective, Zeiss Axiovert 200M, or Zeiss Observer Z1 using NeoFluar 10x or LD Plan NeoFluar 20x objectives while ORCA-ER C4742-95; Hamamatsu Photonics, or Axiocam MRm cameras were used to capture images. Openlab 5.0.2, Volocity 3.6.1 (Improvision), and PALM Robo V4 (Zeiss) were the software used for image acquisition. Images were composed and edited in Photoshop CS(Adobe). Background was reduced using contrast adjustments and color balance was performed to enhance colors. All modifications were applied to the entire image.
FACS sorting
Cells were harvested from culture dishes after 0.05% trypsin treatment, centrifuged and resuspended in FACS buffer (PBS + 2%GS+2mM EDTA), and kept on ice until analysis. Cells were analyzed and sorted using a FACSVantage SE (BD Biosciences), with the DIVA analysis software. Dead cells were gated out by staining with PI (1μg/ml), and cells were sorted for GFP expression at low pressure to preserve cell viability. Double sort was carried out in order to obtain a purity of 99% viable cells. In the second sort, cells were sorted directly in GM or DM as indicated and then seeded in different platforms (microwells or PALM duplex dishes).
PALM LPC
Primary myoblasts were sparsely seeded for differentiation in 50mm or 35mm laminin (Roche) coated Duplexdishes (Zeiss). See Supplemental Materials for details. Laser ablation was carried out after stage calibration, laser focus and optical focus calibration as per manufacturer’s instructions, in a Zeiss Observer Z1 inverted microscope outfitted with PALM Microbeam (Zeiss). Ablated membranes were catapulted by LPC bursts into Roboarm SingleTube Capture II receptacles (500μL eppendorf tube cap) with 80μL of media. Membrane/myocyte capture was verified after capture by direct observation of the captured receptacle. Total volume of the receptacle was transferred into 12 or 24-well collagen coated plates, where captured myocytes were cultured in conditioned growth media (cGM), which was harvested from actively dividing myoblasts and 0.2μm filtered.
Immunocytochemistry
10 days after injection, TA muscles were dissected and immersed in PBS/0.5% EM-grade PFA (Polysciences) for 2 h at RT followed by overnight immersion in PBS with 20% sucrose at 4°C. Section staining and image analysis was performed as described in (Pajcini et al. 2008).
Timelapse microscopy
Primary myotubes were imaged using Zeiss Axiovert 200M equipped with timelapse apparatus CTI-Controller 3700 Digital; Tempcontrol 37-2 digital; scanning stage Incubator XL 100/135 (PECON). Frames were captured every 10min for a total of 50hrs, encompassing days 5 and 6 during primary myotube fusion and maturation. Images were acquired and analyzed using Volocity 3.6.1 (Improvision).
Highlights.
Inactivation of Rb and ARF induces cell cycle reentry of mammalian muscle.
Single postmitotic myocytes give rise to clones after transient loss of Rb and ARF
Myocyte-derived clones differentiate and fuse to mature muscle fibers in vivo
Regeneration by reversal of differentiation is possible in mammalian cells
PALM LPC analysis to isolate and follow the fate of individual primary cells
Supplementary Material
Acknowledgements
We would like to thank Alessandra Sacco for TA injections and numerous helpful discussion on this manuscript; Daniel Kemp (Novartis) for providing us with the peGFPN1-hmyg plasmid from which the myogenin promoter elements were subcloned; Rainer Gangnus and Renate Burgemeister from Carl Zeiss MicroImaging GmbH for invaluable help in PALM LPC methodology; Justine Seidenfeld for her early work on the project; Karen Havenstrite and Penney Gilbert for microwell array construction; and Rose Tran for sectioning. Warren Pear for use of laboratory facilities.
We gratefully acknowledge grants 2T32 HD007249 of The Developmental and Neonatal Biology Program and NIH training grants 5T32 AI07328 and 5T32 HD007249 to KVP; NIH grants AG009521, AG020961, HL096113, MDA grant 4320, LLS grant TR6025-09, JDRF grant 34-2008-623, CIRM grant RT1-01001 and the Baxter Foundation to HMB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. Journal of cell science. 2006;119(Pt 20):4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138(2):257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. The Journal of cell biology. 2007;179(7):1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Baltimore D. Differentiation requires continuous regulation. The Journal of cell biology. 1991;112(5):781–783. doi: 10.1083/jcb.112.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC, Webster C. Plasticity of the differentiated state. Science (New York, NY. 1985;230(4727):758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annual review of cell and developmental biology. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- Brookes S, Rowe J, Gutierrez Del Arroyo A, Bond J, Peters G. Contribution of p16(INK4a) to replicative senescence of human fibroblasts. Experimental cell research. 2004;298(2):549–559. doi: 10.1016/j.yexcr.2004.04.035. [DOI] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8(9):671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas H, Jiang Y, Holloway MP, Fangusaro J, Mahotka C, Conway EM, Altura RA. Survivin splice variants regulate the balance between proliferation and cell death. Oncogene. 2005;24(12):1994–2007. doi: 10.1038/sj.onc.1208350. [DOI] [PubMed] [Google Scholar]
- Camarda G, Siepi F, Pajalunga D, Bernardini C, Rossi R, Montecucco A, Meccia E, Crescenzi M. A pRb-independent mechanism preserves the postmitotic state in terminally differentiated skeletal muscle cells. The Journal of cell biology. 2004;167(3):417–423. doi: 10.1083/jcb.200408164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BM. Muscle regeneration in amphibians and mammals: passing the torch. Dev Dyn. 2003;226(2):167–181. doi: 10.1002/dvdy.10223. [DOI] [PubMed] [Google Scholar]
- Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiological reviews. 2004;84(1):209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- Collins CA, Partridge TA. Self-renewal of the adult skeletal muscle satellite cell. Cell cycle (Georgetown, Tex. 2005;4(10):1338–1341. doi: 10.4161/cc.4.10.2114. [DOI] [PubMed] [Google Scholar]
- Crescenzi M, Soddu S, Sacchi A, Tato F. Adenovirus infection induces reentry into the cell cycle of terminally differentiated skeletal muscle cells. Annals of the New York Academy of Sciences. 1995;752:9–18. doi: 10.1111/j.1749-6632.1995.tb17402.x. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(14):7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckmanton A, Kumar A, Chang YT, Brockes JP. A single-cell analysis of myogenic dedifferentiation induced by small molecules. Chemistry & biology. 2005;12(10):1117–1126. doi: 10.1016/j.chembiol.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Clarke JD, Tanaka EM. In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Developmental biology. 2001;236(1):151–164. doi: 10.1006/dbio.2001.0312. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Bryant SV. Molecular mechanisms in the control of limb regeneration: the role of homeobox genes. The International journal of developmental biology. 1996;40(4):797–805. [PubMed] [Google Scholar]
- Gilley J, Fried M. One INK4 gene and no ARF at the Fugu equivalent of the human INK4A/ARF/INK4B tumour suppressor locus. Oncogene. 2001;20(50):7447–7452. doi: 10.1038/sj.onc.1204933. [DOI] [PubMed] [Google Scholar]
- Gu W, Schneider JW, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72(3):309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- Hay ED, Fischman DA. Origin of the blastema in regenerating limbs of the newt Triturus viridescens. An autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Developmental biology. 1961;3:26–59. doi: 10.1016/0012-1606(61)90009-4. [DOI] [PubMed] [Google Scholar]
- Hickson GR, O’Farrell PH. Anillin: a pivotal organizer of the cytokinetic machinery. Biochemical Society transactions. 2008;36(Pt 3):439–441. doi: 10.1042/BST0360439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjiantoniou E, Anayasa M, Nicolaou P, Bantounas I, Saito M, Iseki S, Uney JB, Phylactou LA. Twist induces reversal of myotube formation. Differentiation; research in biological diversity. 2008;76(2):182–192. doi: 10.1111/j.1432-0436.2007.00195.x. [DOI] [PubMed] [Google Scholar]
- Huh MS, Parker MH, Scime A, Parks R, Rudnicki MA. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. The Journal of cell biology. 2004;166(6):865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91(5):649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kazianis S, Morizot DC, Coletta LD, Johnston DA, Woolcock B, Vielkind JR, Nairn RS. Comparative structure and characterization of a CDKN2 gene in a Xiphophorus fish melanoma model. Oncogene. 1999;18(36):5088–5099. doi: 10.1038/sj.onc.1202884. [DOI] [PubMed] [Google Scholar]
- Kim SH, Mitchell M, Fujii H, Llanos S, Peters G. Absence of p16INK4a and truncation of ARF tumor suppressors in chickens. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(1):211–216. doi: 10.1073/pnas.0135557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies identify blastemal cells derived from dedifferentiating limb regeneration. Nature. 1984;308(5954):67–69. doi: 10.1038/308067a0. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Hatano M, Otaki M, Ogasawara T, Tokuhisa T. Expression of a murine homologue of the inhibitor of apoptosis protein is related to cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(4):1457–1462. doi: 10.1073/pnas.96.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460(7251):60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Lassar A, Munsterberg A. Wiring diagrams: regulatory circuits and the control of skeletal myogenesis. Current opinion in cell biology. 1994;6(3):432–442. doi: 10.1016/0955-0674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Lentz TL. Cytological studies of muscle dedifferentiation and differentiation during limb regeneration of the newt Triturus. The American journal of anatomy. 1969;124(4):447–479. doi: 10.1002/aja.1001240404. [DOI] [PubMed] [Google Scholar]
- Levi BP, Morrison SJ. Stem cells use distinct self-renewal programs at different ages. Cold Spring Harbor symposia on quantitative biology. 2008;73:539–553. doi: 10.1101/sqb.2008.73.049. [DOI] [PubMed] [Google Scholar]
- Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. The Biochemical journal. 1999;344(Pt 2):305–311. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Jacks T. The retinoblastoma gene family in differentiation and development. Oncogene. 1999;18(55):7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(15):7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf MP, Doyonnas R, Havenstrite K, Koleckar K, Blau HM. Perturbation of single hematopoietic stem cell fates in artificial niches. Integrative Biology. 2009;1(1):59–69. doi: 10.1039/b815718a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Defining the regulatory networks for muscle development. Current opinion in genetics & development. 1996;6(4):445–453. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Mulligan GJ, Jacks T, Lassar AB. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. The Journal of cell biology. 1996;135(2):441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odelberg SJ, Kollhoff A, Keating MT. Dedifferentiation of mammalian myotubes induced by msx1. Cell. 2000;103(7):1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- Pajcini KV, Pomerantz JH, Alkan O, Doyonnas R, Blau HM. Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. The Journal of cell biology. 2008;180(5):1005–1019. doi: 10.1083/jcb.200707191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez OD, Chang YT, Rosania G, Sutherlin D, Schultz PG. Inhibition and reversal of myogenic differentiation by purine-based microtubule assembly inhibitors. Chemistry & biology. 2002;9(4):475–483. doi: 10.1016/s1074-5521(02)00131-x. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science (New York, NY. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, Giordano A, Kedes L, Wang JY, Sartorelli V. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Molecular cell. 2001;8(4):885–897. doi: 10.1016/s1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. The Journal of cell biology. 1994;125(6):1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105(5):645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nature reviews. 2007;8(10):798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456(7221):502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Siepi F, Crescenzi M. HPV E7 expression in skeletal muscle cells distinguishes initiation of the postmitotic state from its maintenance. Oncogene. 2003;22(26):4027–4034. doi: 10.1038/sj.onc.1206353. [DOI] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424(6945):223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes & development. 2000;14(23):3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(10):4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JW, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science (New York, NY. 1994;264(5164):1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Current opinion in genetics & development. 1999;9(1):22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- - How stem cells age and why this makes us grow old. Nature reviews. 2007;8(9):703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Shen X, Collier JM, Hlaing M, Zhang L, Delshad EH, Bristow J, Bernstein HS. Genome-wide examination of myoblast cell cycle withdrawal during differentiation. Dev Dyn. 2003;226(1):128–138. doi: 10.1002/dvdy.10200. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Bertwistle D, W DENB, Kuo ML, Sugimoto M, Tago K, Williams RT, Zindy F, Roussel MF. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harbor symposia on quantitative biology. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, DePinho RA. Cellular senescence: mitotic clock or culture shock? Cell. 2000;102(4):407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- Sobkow L, Epperlein HH, Herklotz S, Straube WL, Tanaka EM. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Developmental biology. 2006;290(2):386–397. doi: 10.1016/j.ydbio.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Takahashi C, Bronson RT, Socolovsky M, Contreras B, Lee KY, Jacks T, Noda M, Kucherlapati R, Ewen ME. Rb and N-ras function together to control differentiation in the mouse. Molecular and cellular biology. 2003;23(15):5256–5268. doi: 10.1128/MCB.23.15.5256-5268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Gann AA, Gates PB, Brockes JP. Newt myotubes reenter the cell cycle by phosphorylation of the retinoblastoma protein. The Journal of cell biology. 1997;136(1):155–165. doi: 10.1083/jcb.136.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Weidinger G. Micromanaging regeneration. Genes & development. 2008;22(6):700–705. doi: 10.1101/gad.1660508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velloso CP, Kumar A, Tanaka EM, Brockes JP. Generation of mononucleate cells from post-mitotic myotubes proceeds in the absence of cell cycle progression. Differentiation; research in biological diversity. 2000;66(4-5):239–246. doi: 10.1046/j.1432-0436.2000.660410.x. [DOI] [PubMed] [Google Scholar]
- Velloso CP, Simon A, Brockes JP. Mammalian postmitotic nuclei reenter the cell cycle after serum stimulation in newt/mouse hybrid myotubes. Curr Biol. 2001;11(11):855–858. doi: 10.1016/s0960-9822(01)00234-2. [DOI] [PubMed] [Google Scholar]
- Viatour P, Somervaille TC, Venkatasubrahmanyam S, Kogan S, McLaughlin ME, Weissman IL, Butte AJ, Passegue E, Sage J. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell stem cell. 2008;3(4):416–428. doi: 10.1016/j.stem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Current opinion in genetics & development. 1997;7(5):597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes & development. 1996;10(23):3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.