Abstract
Each of the thirteen identified Fanconi anemia (FA) genes is required for resistance to DNA interstrand crosslinking agents, such as mitomycin C, cisplatin, and melphalan. While these agents are an excellent tool for understanding the function of FA proteins in DNA repair, it is uncertain whether a defect in the removal of DNA interstrand crosslinks (ICLs) is the basis for the pathophysiology of FA. For example, DNA interstrand crosslinking agents induce other types of DNA damage, in addition to ICLs. Further, other DNA damaging agents, such as ionizing or ultraviolet radiation, activate the FA pathway, leading to monoubiquitination of FANCD2 and FANCI. Also, FA patients display congenital abnormalities, hematologic deficiencies, and a predisposition to cancer in the absence of an environmental source of ICLs that is external to cells. Here we consider potential sources of endogenous DNA damage, or endogenous stresses, to which FA proteins may respond. These include ICLs formed by products of lipid peroxidation, and other forms of oxidative DNA damage. FA proteins may also potentially respond to telomere shortening or replication stress. Defining these endogenous sources of DNA damage or stresses is critical for understanding the pathogenesis of deficiencies for FA proteins. We propose that FA proteins are centrally involved in the response to replication stress, including replication stress arising from oxidative DNA damage.
Keywords: DNA interstrand crosslinks, endogenous stress, oxidative stress, lipid peroxidation, critically short telomeres, replication stress
1. Introduction
At present, thirteen Fanconi anemia (FA) complementation groups and the corresponding genes have been identified (reviewed in [1,2]). Importantly, the shared clinical and cellular phenotypes of patients from each complementation group suggest that the encoded proteins may function in a biochemical pathway. This pathway is the FA-BRCA pathway, including the basic FA pathway that consists of the FA core complex (composed of FANC- A, B, C, E, F, G, L, and M) and its substrates for monoubiquitination, FANCD2 and FANCI [3-10]. FANCD2 monoubiquitination is induced by various types of DNA damage, including exposure to DNA interstrand crosslinking agents, ionizing radiation (IR), and ultraviolet radiation [3]. Replication stress, generated by treatment of cells with aphidicolin (APH) or hydroxyurea (HU), also induces FANCD2 monoubiquitination [11,12].
The FANCD1/BRCA2, FANCN/PALB2, and FANCJ/BRIP1 proteins are also components of the FA-BRCA pathway. Biallelic mutation of the corresponding genes leads to FA, while monoallelic mutation is associated with breast cancer susceptibility in the general population [13-16]. FANCD1/BRCA2, FANCN/PALB2, and FANCJ/BRIP1 are not required for FANCD2 monoubiquitination [17-19], and may therefore function downstream of the basic FA pathway.
Among the phenotypes that are characteristic of cells derived from FA patients, irrespective of the complementation group, is hypersensitivity to DNA interstrand crosslinks (ICLs) induced by agents such as mitomycin C (MMC), cisplatin, melphalan, and psoralen/UV-A [3,20,21]. ICLs block strand separation necessary for passage of the replication fork and for transcription, and are thereby highly deleterious to cells [22,23]. It should be noted, however, that DNA interstrand crosslinking agents also induce other types of DNA damage, including alkylation of bases and DNA intrastrand crosslinks [22,24] (Fig. 1). In fact, alkylation typically predominates over ICLs [25].
Figure 1. DNA adducts formed by interstrand crosslinking agents.

Bifunctional compounds can form monoadducts that affect a single nucleotide. Or these compounds can form adducts affect two nucleotides in the same strand or two paired strands to generate DNA intrastrand and interstrand crosslinks, respectively.
FA cells also display chromosome abnormalities, such as chromosome breakage and the formation of radial chromosomes, in the presence of diepoxybutane or MMC [26]. In fact, such chromosome instability is used in the diagnosis of Fanconi anemia ([27] and “Fanconi Anemia and its Diagnosis” Auerbach, this issue). Additionally, FA cells also accumulate in G2-M of the cell cycle in response to treatment with DNA interstrand crosslinking agents ([21,28,29] and “Genotype-Phenotype Correlations in Fanconi Anemia” Neveling et al., this issue). Together, these cellular phenotypes suggest that FA proteins may function in cellular responses to DNA damage.
While comparative studies have suggested that FA cells display a greater sensitivity to DNA interstrand crosslinking agents [20,21], there is evidence that FA cells may also be sensitive to other stresses. For example, there is conflicting evidence which suggests that FA cells may be hypersensitive to agents that induce oxidative DNA damage [21,30,31]. Further, deficiency of FA proteins is associated with sensitivity to formaldehyde, which crosslinks protein to DNA [32]. And it has been reported that some FA cells may show sensitivity to DNA monoalkylating agents [20]. Together, these results call into question whether FA proteins respond primarily, or exclusively, to ICLs.
Some human diseases, such as Xeroderma pigmentosum (XP), are caused, in part, by defective repair of lesions that clearly originate from environmental sources of DNA damage. XP patients display abnormal skin pigmentation and a greatly increased incidence of skin cancer that is associated with exposure to solar radiation [33,34]. In contrast, it is not clear that the phenotypes of Fanconi anemia patients derive from exposure to exogenous agents that induce ICLs. For example, a striking array of tissues and organs are potentially affected by congenital abnormalities ([35] and “Fanconi Anemia and its Diagnosis” Auerbach, this issue). Further, FA patients display an increased incidence of a wide variety of cancers, including leukemia and tumors of the head/neck, urogenital tract, digestive tract, lung, and brain [36]. An environmental agent that would effect this diverse spectrum of organs is not readily apparent. Thus, FA proteins may instead have a role in responding to sources of DNA damage or replication stress that are intrinsic to the cell. Identifying the roles of FA proteins in responding to endogenous stresses is critical as a foundation for understanding the pathophysiology of FA.
In this review, we will consider endogenous stresses which may activate the FA pathway and for which FA proteins may mediate a cellular response that results in increased cell survival or proliferation. Among these stresses, lipid peroxidation generates compounds that can generate ICLs (reviewed in [37,38]). Since FA cells are hypersensitive to ICLs, endogenous DNA interstrand crosslinking agents are obviously a relevant source of DNA damage. Oxidation of bases and sugar moieties is a major source of endogenous DNA damage [39] and is another stress to which FA cells may be sensitive. We will consider evidence for a function of FA proteins in responding to oxidative stress and to oxidative DNA damage. Additionally, telomeres shorten with proliferation and critically short telomeres are recognized as DNA double-strand breaks [40,41]. We will review evidence for increased telomere shortening in FA. Finally, FANCD2 is recruited to stalled replication forks [12,42] and it has been reported that FANCD1/BRCA2 is required to stabilize stalled replication forks [43]. We will consider evidence as to whether FA proteins have a critical function at the stalled replication fork. An overview of these potential sources of endogenous DNA damage is shown in Fig. 2.
Figure 2. Outline of potential sources of endogenous DNA damage that could activate the FA-BRCA pathway.
(A) Reactive oxygen species (ROS) can induce lipid peroxidation, which can generate bifunctional compounds that induce ICLs in DNA. (B) ROS can directly oxidize bases in DNA. (C) Telomere dysfunction, through disruption of telomere packaging or through shortening of telomeres to a critical length, can lead to recruitment of DNA damage response proteins (DDR) to telomeres. (D) Endogenous DNA damage, replication errors, or chromatin structures that are difficult to replicate, can induce replication stress and/or fork collapse.
2. Reactive oxygen species and the generation of ICLs by products of lipid peroxidation
2.1. Intracellular reactive oxygen species, oxidative stress, and antioxidants
Reactive oxygen species (ROS) refer to a variety of molecules and free radicals derived from molecular oxygen, which can be reduced to generate relatively stable intermediates. The biologically relevant ROS, such as hydrogen peroxide (H2O2), superoxide (O2), and the hydroxyl radical (HO), are generated from the precursor superoxide anion [44]. Superoxide can be rapidly converted to H2O2 by superoxide dismutases (SODs; [45]). In the presence of transitional metals, H2O2 can generate hydroxyl radicals. In addition to ROS, the cell also contains reactive nitrogen species (RNS), such as nitric oxide (●NO). Nitric oxide is a short-lived molecule with a free electron that regulates many physiological functions by itself, some of which are associated with development [46,47].
Aerobic organisms utilize O2 for metabolism, which inevitably generates ROS. The intracellular ROS are usually derived from oxidative phoshorylation, P450 metabolism, peroxisomes, and inflammatory cell activation (Fig. 3) (for review see references [48,49]). O2− and H2O2 are generated by a variety of oxidoreductases through a leak of electrons to O2 from the mitochondrial electron transport chain at Complex III [50-52], as well as redox cycling of quinones and numerous other autooxidation reactions [53]. ●NO is synthesized in cells through a 5-electron oxidation of l-arginine by members of a family of heme containing enzymes called nitric oxide synthases [54]. ROS are enzymatically produced as signaling molecules within several major organelles (Fig. 3).
Figure 3. Major organelles and enzymes known to generate ROS.
The activity of the respiratory chain in the mitochondria is responsible for most ROS produced in aerobiosis. On the other hand, the metabolic pathway that drives the degradation of long chain fatty acids in the peroxisome is also an important ROS source. The activity of ROS produced by cytochrome p450 or NADPH oxidases may be restricted to the area where they are located, and not all ROS are sufficiently stable to traverse a cell and damage DNA in the nucleus. 5-lipoxygenase can be found associated with membranes or with the nuclear envelope. Peroxisomal xanthine oxidase is an enzyme that produces ROS from molecular oxygen.
ROS are highly reactive and readily cause oxidative modifications to biomolecules like DNA, proteins, and lipids. Because of a short half-life and limited diffusion distance, most ROS cause damage locally near the site of production. Both the sugar and the base moieties are susceptible to oxidation, causing base modifications such as 8-oxoguanine (8-oxoG), single-strand breakage, and cross-linking to proteins [55,56]. In addition, adjacent single-strand breaks in opposite strands may be converted to DNA double-strand breaks (DSBs) upon replication. DSBs are lethal unless repaired [57], whereas base damage may be mutagenic, cytotoxic or both. Protein can be damaged by oxidative modifications of amino acids, as well as by ROS-mediated peptide cleavage. This results in malfunction of many enzymes, including cytochrome c oxidase, glutathione oxidase, and catalase, which in turn reduces the capacity of the cell to eliminate ROS. Lipid peroxidation, in which ROS attack lipid membranes in a chain reaction manner, produces a series of organic radicals, and results in disruption of membranes and their function. Recent studies show that chronic low levels of ROS induce DNA or protein damage that contribute importantly to cancer and aging [48,58,59].
Cells utilize enzymatic and non-enzymatic antioxidants to counteract ROS. The balance between the production and degradation of ROS maintains cellular homeostasis. Important cellular antioxidant enzymes include superoxide dismutases (SODs), which act to scavenge the superoxide radical by converting it into H2O2 and oxygen [60], and the peroxidases and catalases, which convert H2O2 into H2O [61]. There are two major non-enzymatic antioxidants synthesized by the cell: glutathione and thioredoxin [62]. The balance between the reduced form (GSH) and the oxidized form (GSSG) of glutathione is an indicator of the redox state within the cell. On the other hand, thioredoxins are small proteins, which contain an active site with a redox-active disulfide and thus serve to maintain a reduced intracellular redox state in mammalian cells by the reduction of protein thiols [63].
Under physiological conditions, ROS are maintained at proper levels by a balance between its production and degradation. The balance between the generation and elimination of ROS can readily change if any step in ROS production or scavenging is disturbed. Therefore, an increase in ROS generation, a decrease in antioxidant capacity, or both will lead to oxidative stress. Oxidative stress resulting from ROS imbalance could be detrimental to cells due to oxidative damage to lipids, proteins, and DNA. Important oxidized products are lipid hydroperoxides [64], carbonylated proteins [65], and DNA with oxidized bases such as 8-oxoG [66]. The consequence of oxidative damage to these macromolecules can contribute to a cascade of events that leads to apoptosis or uncontrolled proliferation by modifying many intracellular signaling pathways including protein phosphatases, protein kinases, and transcription factors. However, exact mechanisms by which redox status induces cells to proliferate or to die are still under intensive investigation.
2.2. Endogenous ICLs can result from oxidative stress
Lipid peroxidation, resulting from oxidative stress, can induce the formation of ICLs (reviewed in [37,38]). Endogenous ICLs are rare, in comparison to other types of DNA damage, so it has been difficult to study them. It should be kept in mind, however, that ICLs interfere with DNA replication, as well as transcription, and are thereby among the most toxic lesions in the cell [22,37]. Evidence for endogenous ICLs comes from their formation in vitro or by inference from a mutational spectrum that includes large rearrangements or deletions [38,67].
Malondialdehyde is a known lipid peroxidation product that generates ICLs both in vitro and in vivo [67]. Malondialdehyde also forms monoadducts, which are more prevalent than ICLs, and which can be mutagenic [67,68]. Crotonaldehyde, acrolein, and trans-4-hydroxy-2-nonenal are other products of lipid peroxidation that can induce ICLs [38,69]. In addition, cigarette smoke and automobile exhaust are environmental sources of crotonaldehyde and acrolein. Nitric oxide is a potential endogenous source of ICLs that is generated independently of lipid peroxidation [70].
The sensitivity of FA cells to endogenous ICLs could be due either to defects in detoxifying ROS or in repairing ICLs. Some FA proteins interact with enzymes involved in detoxifying ROS (see next section), while other FA proteins, such as FANCD1/BRCA2, have obvious roles in DNA repair. It would therefore be interesting to determine whether different FA complementation groups have distinct sensitivities to endogenous ICLs that result from ROS.
3. Fanconi anemia and oxidative DNA damage
3.1 Evidence suggesting a relationship between FA proteins and oxidative stress/oxidative DNA damage
Many diseases of clinical interest, including those associated with aging, involve oxidative stress in their underlying etiology. There is ample evidence that FA cells are abnormally sensitive to oxidative stress. Joenje et al. [71] first reported that cultured FA cells are vulnerable to oxygen-induced chromosomal aberrations. The observation was later confirmed by two other groups that showed FA fibroblasts and primary bone marrow cells grow better under hypoxic conditions than in ambient air [72,73]. Over the last decade, FA oxidant hypersensitivity has been documented in many studies using primary and immortalized cell lines derived from FA patients, as well as FA knockout mouse models ([31,73-78] and “Genotype-Phenotype Correlations in Fanconi Anemia” Neveling et al., this issue).
Direct evidence implicating the connection between FA and oxidative stress comes from two lines of studies using mice deficient for the FA complementation group C gene (Fancc-/-). Hadjur et al. [78] reported defective hematopoiesis in mice with combined deficiencies of the anti-oxidative enzyme, Cu/Zn superoxide dismutase (SOD), and FANCC. Although the Fancc-/-;Sod-/- mice do not display development defects or increased spontaneous chromosomal aberrations typical of FA, the animals have a phenotype of BM hypocellularity, which is not detected in single mutant mice. Furthermore, hepatocytes from Fancc-/-;Sod-/- mice exhibit a zonal pattern of microvesicular steatosis. The Haneline group [31] demonstrated that BM hematopoietic progenitors and murine embryonic fibroblasts isolated from Fancc-/- mice exhibit hypersensitivity to oxidative stress generated by H2O2. Pretreatment with the antioxidants selenomethionine or N-acetylcysteine preferentially enhances the survival of Fancc-/- cells. Furthermore, the authors found that H2O2 induces over-activation of the serine-threonine kinase apoptosis signal-regulating kinase 1 (ASK1), which has been shown to be an important kinase involved in oxidant-induced apoptosis; [79]) in Fancc-/- cells. This was correlated with elevated H2O2-induced apoptosis in these mutant cells. All of the observations indicate that the altered redox state of the hematopoietic progenitors in these mice was responsible for an impairment of cell proliferation or survival.
Since FA cells exhibit hypersensitivity to oxidative stress, FA proteins have been proposed to play roles in the maintenance of redox homeostasis during oxidant metabolism. Indeed, an extensive body of evidence suggests that FA proteins play an important role in oxidative stress signaling in a variety of cell types. Three FA proteins, FANCA, FANCC, and FANCG, as parts of the FA protein complex [80], are found to associate with a variety of cellular factors that primarily function in redox-related processes (Table 1). For example, the FANCC protein interacts with NADPH cytochrome P450 reductase and glutathione S-transferase P1-1 [76,81], two enzymes involved in either triggering or detoxifying reactive intermediates including ROS. Fancc-/- mice deficient for the anti-oxidative enzyme Cu/Zn superoxide dismutase demonstrate defective hematopoiesis [78]. Another FA protein, FANCG, interacts with cytochrome P450 2E1, a member of the P450 superfamily that is associated with the production of reactive oxygen intermediates, and the mitochondrial anti-oxidant enzyme peroxiredoxin [75,82], suggesting a possible role for FANCG in protection against oxidative DNA damage. Finally, it has also been shown that loss of Fancc function might result in an altered redox regulation and hyperactivation of ASK1, a serine-threonine kinase that plays an important role in redox apoptotic signaling [31].
Table 1. Interaction of FA proteins with cellular redox-signaling factors.
Cells defend against oxidative DNA damage through several repair mechanisms: base excision repair, nucleotide excision repair (NER), mismatch repair (MMR), translesion synthesis (TLS), homologous recombination (HR), and non-homologous end-joining (NHEJ) (Reviewed in [83]). FA proteins have been implicated in TLS, MMR and DNA recombinational repair [84-89]. Translesion bypass for oxidized bases is well documented. For example, the translesion polymerases pol η and ι are both able to insert a C opposite 8-oxoG and proficiently extend from this base pair [90,91]. MMR is also involved in repair of 8-oxoG:A mismatches [92,93]. It is well documented that HR and NHEJ all contribute to repair of oxidative DNA damage [83,94]. It is thus speculated that the FA pathway is integrated with these mechanisms in repairing oxidative DNA damage.
The most important clinical features of FA are hematological. FA patients often develop pancytopenia during the first few years of life. Bone marrow (BM) failure is the major cause of morbidity and mortality of FA, and more than 80% of FA patients die from BM failure [95,96]. In addition, FA patients have increased susceptibility to developing myelodysplasia or acute myeloblastic leukemia [80,95,97]. Increasing evidence indicates progressive BM failure in FA patients results from excessive apoptosis and subsequent failure of the hematopoietic stem cell and progenitor (HSC/P) compartment.
Whether oxidative stress is a pathological factor in FA disease progression remains an issue of debate. Several studies indicate that FA hematopoietic cells, including HSC/P, are hypersensitive to oxidative stress [31,73-78,98]. A number of hypotheses regarding the effect of oxidative stress in FA have been suggested, including the proposal that ROS could damage DNA and that the inability of FA HSC/P to repair such damage would result in exacerbated genomic instability, leading to apoptosis and malignant transformation.
BM hematopoietic cells from FA patients and knockout mice are hypersensitive to oxidative stress-induced DNA damage and growth arrest. This suggests FA proteins may function specifically in response to oxidative stress. While certain FA proteins interact with some cellular redox signaling factors (Table 1), whether these interactions are relevant to cellular anti-oxidant defense or repair of oxidative DNA damage is not clear. We recently tested the hypothesis that FA proteins may functionally interact with the DNA-damaging checkpoint signals in response to oxidative stress. Our results suggest that murine FANCA protein may coordinate the regulation of the oxidative DNA damage response with the tumor suppressor p53 [99]. This notion is supported by the observation that hypersensitivity of cells derived from Fanca-/- mice to H2O2 is correlated with persistent p53 hyperactivation. Further, the H2O2-induced cell cycle checkpoint and DNA damage response in primary Fanca-/- cells depends on p53.
The functional relationship between oxidative DNA damage-induced p53 signaling and the FA pathway has not been well defined. Early work from the Hoehn group showed that oxidative stress altered p53 expression and function in cultured FA cells [100-102]. Recently, studies from several other groups demonstrated that Fancd1-/-, Fancd2-/- and Fancc-/- mice have accelerated tumor development in a Trp53-deficient background [103-105], and that p53 knockdown in FANCD2-deficient zebrafish leads to increased apoptosis and developmental defects [106]. The redox imbalance in FA cells could alter p53 activity or expression, leading to an abnormal cellular response to ROS-generated DNA damage. In this context, additional investigations into the functional interaction between the p53 and FA pathways in the oxidative DNA damage response may provide new insights into the role of oxidative stress in FA pathogenesis and disease progression.
3.2 The role of inflammatory ROS in FA leukemogenesis
Inflammatory reactions, particularly chronic ones, can be a significant source of oxidative damage in hematopoietic cells. Leukocytes such as activated macrophages and neutrophils release a number of ROS, including hydrogen peroxide, superoxide, and hydroxyl radical, which can damage the DNA of nearby cells, particularly HSC/P cells in the BM [107-109]. There is compelling evidence that chronic inflammation increases the risk of human cancer [110]. In pathological conditions, unresolved inflammation provokes cell turnover, and coupled with ROS generated at sites of inflammation, leads to mutation of DNA and malignant transformation [111-113].
The cytokine TNF-α is a major mediator of inflammation and plays a key role in the pathogenesis of such inflammatory diseases as rheumatoid arthritis, Crohn's disease, and psoriasis, as demonstrated by the successful treatment of such conditions with antibodies to TNF-α or with a soluble TNF-α receptor fusion protein [114]. With respect to abnormal hematopoiesis, it is well recognized that TNF-α is involved in many disease situations, including anemia, myelodysplasia, and leukemia [115]. TNF-α exerts many of its biological effects through the activation of the MAPK stress signaling cascade, including JNK, p38MAPK, and ERK, as well as the NF-κB family of transcription factors [116]. One important feature is that inflammation generated by TNF-α induces overproduction of ROS, partially through prolonged activation of the MAPK signaling pathway [117].
Patients with FA have abnormally high levels of TNF-α in hematopoietic tissues [118-121]. The presence of this pro-inflammatory cytokine and increased oxidative stress, as a result of a persistent inflammatory response, may account for profound physiologic changes in these patients, including the development of BM failure and progression to leukemia. Many studies have shown a correlation between elevated circulating pro-inflammatory cytokines and anemia in patients with BM diseases, but direct evidence for the mechanistic link between inflammation and BM failure/leukemogenesis is lacking. Recent pioneering work from the laboratories of Wade Clapp and Laura Haneline [122] has demonstrated that Fancc-/- BM cells cultured ex vivo develop increased cytogenetic abnormalities and myeloid malignancies that are associated with an acquired resistance to TNF-α when transplanted into recipient mice. These results suggest that FA hematopoietic cells are prone to clonal hematopoiesis and malignancy. Studies from our own laboratory have suggested that excessive apoptosis of FA hematopoietic cells induced by TNF-α may contribute to the pathophysiology of BM failure and leukemia in FA children [123-125]. In addition, there is evidence that the production of ROS by TNF-α at inflammatory sites causes DNA damage [109,113]. Indeed, TNF-α-treated Fancc-/- HSC/P display high levels of oxidative DNA damage [123,125]. While the role of FA proteins in modulating the cellular response to inflammatory ROS-induced DNA damage remains to be elucidated, it is likely FA proteins can act to protect chromosomal DNA from ROS attack or facilitate the repair of oxidative DNA damage. Recently, the Grompe group reported that treatment of Fancd2-/-;Trp53+/- mice with the anti-oxidant compound Tempol delayed solid tumor development [126].
4. Telomeres and Fanconi anemia
4.1 Telomeres and activation of DNA damage responses
Telomeres are a specialized chromatin structure that cap the ends of linear chromosomes and prevent these ends from being recognized as DSBs (reviewed in [127,128]). The importance of telomeres is illustrated by the observation that telomere dysfunction can lead to the fusion of chromosomes. Chromosomes that result from such fusions are dicentric and can be broken by chromosome segregation during mitosis to generate an unstable chromosome [129]. Since FA is also characterized by chromosome instability [26], the question arises as to whether FA proteins may be involved in the maintenance of telomere length or the protection of telomeres against recognition as DSBs.
Telomeres shorten with each division [130]. When telomeres become critically short, senescence is induced by a checkpoint mechanism [40,41]. Presumably short telomeres can no longer be protected against recognition as a DSB. Consistent with this possibility, γ-H2AX foci, which are a marker for DSBs [131], are induced in fibroblasts with shortened telomeres [40,41]. Foci formation by other DNA damage response proteins, including 53BP1, MDC1, NBS1, phospho-ATM (S1981), phospho-Chk1 (S317), and phospho-Chk2 (T68), is also induced in senescent cells [40,41,132]. Importantly, ATM, along with p53, is required for the induction of senescence in cells with short telomeres [132]. Together, these results suggest that critically short telomeres could be a source of endogenous DNA damage in FA cells.
Cancer cells invariably have mechanisms that stabilize telomeres by maintaining their length. The major pathway is through the activity of telomerase, which is a ribonucleoprotein complex that can elongate telomeric DNA (reviewed in [128]). Another form of telomere maintenance is termed alternative lengthening of telomeres (ALT). The ALT pathway is utilized in about 10% of tumors, and may be found more frequently in tumors of mesenchymal origin [133]. ALT cells are characterized by ALT-associated PML-bodies (APBs), which contain telomeric DNA, PML bodies, and various DNA damage response proteins. APBs form in a cell cycle-dependent manner during late S phase-G2-mitosis [134], and RPA, RAD51, NBS1, BRCA1, and BLM are among the DNA damage response proteins that are present [135-138]. The role of many of these proteins in ALT telomere maintenance, if any, is not well characterized. DSBs, as determined by the formation of γ-H2AX foci, are also present at APBs [139]. This observation suggests that ALT cells, like senescent cells, have dysfunctional telomeres. In general, ALT cells do not express telomerase and instead maintain telomeres through inter-telomeric HR [140,141].
4.2 Telomeres, Fanconi anemia and FA proteins
There have been a number of studies suggesting that telomere maintenance may be disrupted in cells from FA patients. In multiple studies, telomeres in peripheral blood mononuclear cells (PBMCs) from FA patients were, as a group, significantly shorter than telomeres from age-matched controls [142-144]. In one study, an increase in undetectable telomeres in cells from FA patients was accompanied by a strong increase in fusions at chromosome ends, presumably due to telomere dysfunction. Telomere dysfunction in cells from FA patients does not appear to be due to defective recruitment of TRF2 to telomeres, however [145].
The telomere shortening observed in FA cells could be due to a defect in telomerase-dependent maintenance of telomere length in HSC/P. Alternatively, telomere shortening in FA cells could be an indirect consequence of increased proliferation of HSC/P in an attempt to maintain normal levels of blood cells [144]. In this case, the rate of telomere shortening with each cell division would not be affected by deficiencies for FA proteins. Indeed, a detailed study of various cell types, and of both mouse and human cells, strongly suggests that the rate of telomere shortening does not differ in FA and non-FA cells [146]. The observation that FANCD2 does not normally colocalize with telomeres in primary fibroblasts or tert-expressing cells is consistent with FA proteins not having a direct role in telomere maintenance [147].
FA proteins appear to have a function in ALT telomere maintenance, however. FANCD2 colocalizes with telomeres in various ALT cell lines [147,148], and depletion of FANCA or FANCD2 inhibits HR required for telomere maintenance in these cells [147]. Because ALT cells have an activated DNA damage response at dysfunctional telomeres [139], the function of the FA pathway at ALT telomeres may be related to its general role in DNA damage responses. Whether the FA pathway is activated by dysfunctional telomeres present in senescent cells is an important question that has not been answered.
5. Replication stress and FA
5.1. Evidence for the potential relationship of FA proteins to DNA replication
Monoubiquitination of FANCD2 is required for it to assemble into nuclear foci [149]. Immunolocalization studies have revealed that FANCD2 is monoubiquitinated and forms foci both spontaneously and in response to DNA damage induced by agents such as IR or MMC, or replication stress induced by APH or HU [3,4]. FANCD2 strongly colocalizes with the DNA damage response proteins BRCA1 and RAD51 in S phase foci that assemble in the absence of exogenous DNA damaging agents [4]. Thus, FANCD2 could have a function in the repair of DNA detected by the advancing replication fork. Further, the assembly of FANCD2 foci in response to ICLs is associated with passage through S phase [150]. While FANCI also assembles into nuclear foci and can form a complex with FANCD2 [9,10], little is known about the specific localization of FANCI or its behavior during the cell cycle. Thus, the localization of FANCD2 remains the best characterized, and may therefore be the most informative, of the FA proteins.
Since FANCD2 spontaneously assembles into S phase foci in cultured cells without exposure to DNA damaging agents, an obvious question is whether FANCD2 is present at the advancing replication fork under these conditions. As measured by colocalization with PCNA, which is a marker for the replication fork, FANCD2 foci are not present at the replication fork during an unperturbed S phase [12,42]. Thus, FANCD2 is not a constitutive component of the replication fork. Further, measurements of the progression of transformed FA-D2 cells, or their counterparts corrected by exogenous expression of FANCD2, through S phase following double thymidine synchronization suggests that FANCD2 is not essential for DNA replication [151].
5.2. ATR regulates FANCD2 monoubiquitination and the cellular response to replication stress
Two related checkpoint kinases, ATM (ataxia-telangiectasia mutated) and ATR (ATM and RAD3-related) coordinate the cellular response to DNA damage (reviewed in [152,153]). While ATM is primarily involved in the response to DNA double-strand breaks (DSBs), ATR is central to the response to replication stress or DNA damage that impedes DNA replication.
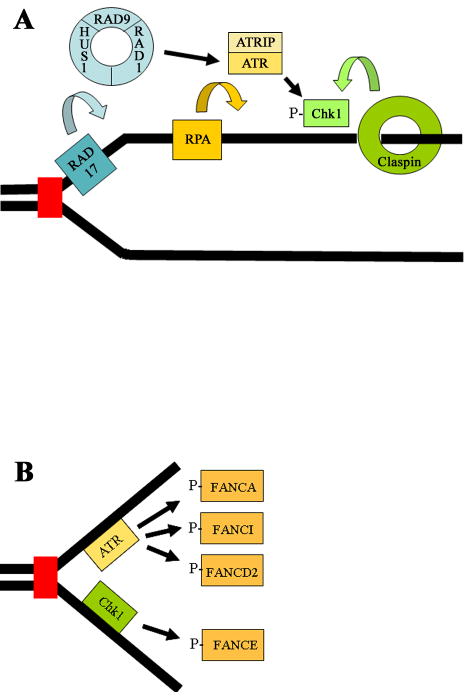
ATR is activated by a network of proteins, summarized in Fig. 4A, that recognizes replication stress or abnormal DNA structures present at the replication fork ([152]). Many of these proteins are associated with chromatin. RPA binds to single-strand DNA present at the blocked replication fork and ATR is recruited by the binding of its partner, ATRIP, to RPA [154]. The constitutive chromatin-associated protein RAD17 is required for ATR activation and acts as a clamp loader for the RAD9-RAD1-HUS1 complex [155,156]. The RAD9-RAD1-HUS1 complex forms a ring that can slide along DNA to act as a damage sensor required for the activation of ATR. Activated ATR phosphorylates a number of DNA damage response proteins, including BLM, BRCA1, and RAD17 [155,157,158]. ATR also phosphorylates, and activates, the Chk1 protein kinase. Chk1, like ATR, is critical for the cellular response to replication stress [153,159]. ATR-dependent phosphorylation of Chk1 requires claspin [152,160]. Similar to the RAD9-RAD1-HUS1 complex, claspin also forms a ring-like structure and may respond to changes in DNA structure [161].
Figure 4. Schematic of the ATR-Chk1 pathway and possible targets for phosphorylation in the FA pathway.
RAD17 functions as a clamp loader for the RAD9-RAD1-HUS1 trimer. This trimer acts as a ring that can slide on DNA to act as a sensor of DNA damage, and activates ATR in response to replication stress or blockage of replication forks by DNA damage. RPA is a single-strand DNA binding protein that independently recruits the ATRIP-ATR complex through direct binding of RPA to ATRIP. ATR then activates the downstream Chk1 kinase by phosphorylating it. Claspin also forms a ring-like structure that can slide on DNA and which is required for phosphorylation of Chk1 by ATR. ATR also phosphorylates a large number of other DNA damage response proteins, including RAD17 and RAD9. (B) ATR or Chk1-dependent phosphorylation of multiple FA proteins, including FANCA, FANCE, FANCDD2, and FANCDI, may regulate their recruitment to sites of replication stress. Phosphorylation of FANCA, FANCI, and FANCD2 by ATR ultimately upregulates monoubiquitination of FANCD2 and FANCI in response to replication stress or DNA damage. The stability of FANCE, a member of the FA nuclear core complex, is regulated by Chk1-mediated phosphorylation.
ATR is required for FANCD2 monoubiquitination in response to DNA damage or inhibition of DNA replication [11,162]. This observation further supports the possibility that FA proteins may have a role at the replication fork (for further discussion see “The Genetic and Molecular Basis of Fanconi Anemia” de Winter and Joenje, this issue). Additionally, ATR colocalizes with FANCD2 following exposure of cells to ICLs [11,163], and ATR is required for the assembly of FANCD2 foci in response to replication stress or DNA damage but not for the baseline assembly of S phase foci [11]. These results indicate that S phase and DNA damage/replication stress-induced FANCD2 foci may have distinct functions. While ATR can directly phosphorylate FANCD2 [164], the mechanism by which ATR regulates FANCD2 monoubiquitination is still not well understood. A recent study suggests that ATR-dependent phosphorylation of FANCI, and not FANCD2 itself, is critical for monoubiquitination of both proteins [165] and thus their recruitment to chromatin. ATR-dependent phosphorylation of FANCA at S1449 also appears to have an important role in regulating FANCD2 monoubiquitination [166]. FANCA that has been phosphorylated at S1449 is associated with chromatin. A summary of FA proteins that are phosphorylated by ATR or Chk1, presumably at sites of replication stress, is shown in Fig. 4B.
Other components of the ATR pathway also play a role in regulating FANCD2 monoubiquitination, further linking the control of FANCD2 behavior to the machinery that senses and responds to replication stress. For example, RAD17 and RAD9 are required for FANCD2 monoubiquitination in response to DNA damage or replication stress [167]. RPA is also required for FANCD2 monoubiquitination and assembly into nuclear foci in response to DNA damage [11]. Additionally, HCLK2 has been identified as an ATR-interacting protein that is required for DNA damage-induced FANCD2 monoubiquitination [168]. Interestingly, it has recently been reported that FANCM, and FAAP24, interact with HCLK2 independently of the FA core complex and are required for activation of ATR-mediated signaling [169]. Thus, FANCM and FAAP24 may have a role in coordinating damage recognition with FANCD2 monoubiquitination. Another mechanism by which HCLK2 could affect FANCD2 monoubiquitination is through a role in stabilizing the Chk1 protein kinase against proteasomal-dependent degradation [168]. Two different reports have suggested that Chk1 may regulate DNA damage-induced FANCD2 monoubiquitination and assembly into nuclear foci [167,170]. These effects may be related to destabilization of FANCE following its phosphorylation by Chk1 [170]. It has also been reported that claspin is required for FANCD2 monoubiquitination [167].
A cell-free system based upon extracts from Xenopus eggs can be used to analyze the cell cycle-dependent regulation of DNA damage responses [171]. Experiments which utilize this system have further suggested that FANCD2 is regulated by the ATR pathway and potentially implicate FANCD2 in replication stress responses [172]. The recruitment of FANCD2 to chromatin is dependent upon initiation of DNA replication, and requires ATR and ATRIP [172].
5.3. Potential functions of FA proteins at the replication fork
The recruitment of FANCD2 to blocked replication forks [12,42] through ATR-dependent regulation of FANCD2 monoubiquitination [11] strongly suggests a potential role for monoubiquitinated FANCD2 at the replication fork. Defects in dealing with replication stress could potentially underlie the pathogenesis of FA. Possible functions include roles in restarting stalled replication forks, preventing collapse of blocked replication forks, and S phase checkpoint regulation. Potential functions for FANCD2 at the replication fork are shown schematically in Fig. 5.
Figure 5. Schematic diagram of possible roles of FANCD2 at replication forks.
FANCD2, and perhaps other FA proteins, could have a role in restarting stalled replication forks, as symbolized here by the re-engagement of DNA polymerases at the replication fork. FA proteins could also have a role in preventing the collapse of stalled replication forks, as symbolized here, by stabilizing the association of DNA polymerases with the replication fork. FA proteins may have roles in the intra-S phase checkpoint, which primarily blocks new initiation of replication following DNA damage. Finally, FANCD2 and other FA proteins may simply be recruited to repair DNA damage encountered by the replication fork.
HR is required to restart stalled replication forks (reviewed in [173,174]). FA proteins have been implicated in HR [84,86,89], consistent with a possible role for FA proteins in restarting stalled replication forks. It should be noted, however, that while FANCD1/BRCA2 has an integral role in DNA double-strand break-initiated (DSB) HR, components of the FA pathway have a more modest role in this process [86,175]. Since DSBs form at stalled replication forks [173,176], the modest role of FA pathway proteins in DSB-initiated HR might suggest that the FA pathway and monoubiquitinated FANCD2 do not have an essential role in restarting stalled replication forks. In support of this possibility, cells that lack FANCD2 and their genetically corrected counterparts show the same level of bulk DNA synthesis following release from replication arrest in early S phase [4]. Further, measurements of nucleotide incorporation using Xenopus cell-free extracts similarly suggest that neither the FA nuclear core complex nor FANCD2 is required to restart stalled replication forks [177].
Another possible function of the FA-BRCA pathway at the replication fork could be in preventing the collapse of stalled forks. Experiments with Xenopus egg extracts, which were described above, support a possible function for monoubiquitinated FANCD2 in preventing the collapse of stalled replication forks. Immunodepletion of FANCD2 or FANCA from the extracts results in the accumulation of DSBs in chromatin, interpreted as an indicator of collapsed replication forks, during unperturbed replicative DNA synthesis [172]. Also, depletion of FANCL or FANCD2 from Xenopus egg extracts decreases the efficiency of DNA replication following treatment with camptothecin or MMC [177]. It is unclear though whether the FA pathway is required to repair the resulting DNA damage or to restart collapsed replication forks.
Work with cells derived from FA patients suggests that chromatin structures that are difficult to replicate may be another potential source of endogenous replication stress to which the FA pathway responds. Deficiency for the FA nuclear core complex, or for FANCD2, results in increased chromosome breakage at common fragile sites in the presence of low doses of the replication inhibitor, APH [12]. It is believed that such fragile sites are structurally distinct regions of the chromatin where replication forks are particularly vulnerable to stalling or collapse [12]. G quadruplexes are four-stranded DNA structures that form from guanine-rich sequences, and are associated with genetic instability. FANCJ unwinds such structures in vitro and may thereby act to prevent the collapse of replication forks [178]. It has also been reported that FANCD1/BRCA2 has a role in stabilizing stalled replication forks and preventing their collapse [43].
Certain FA proteins also have a role in S phase checkpoints. Defects in the intra-S phase checkpoint are frequently measured as radioresistant DNA synthesis following the exposure of cells to IR [179]. There is a staged order to which origins of replication are fired. The major component of decreased DNA synthesis following treatment with IR is inhibition of the initiation of DNA replication at origins that would normally be fired next (reviewed in [174,180]). Inhibition of elongation of origins that have already been fired also contributes to the intra-S phase checkpoint [179,181].
ATM coordinates the intra-S phase checkpoint in response to IR by phosphorylating a large number of substrates, such as Chk2, BRCA1, and NBS1 [174,180]. FANCD2, FANCD1/BRCA2, and FANCN/PALB2 are FA proteins that are required for this intra-S phase checkpoint [151,182-184]. ATM-dependent phosphorylation of FANCD2 at S222, T691 and S717 are all involved in the intra-S phase checkpoint following treatment with IR [151,164]. ATR coordinates intra-S phase checkpoints in response to other types of DNA damage [174]. It has been reported that ATR, FANCD2, FANCC, and FANCG are required for an intra-S phase checkpoint that responds to ICLs [163].
Importantly, it is not clear that defective S phase checkpoints are central to the FA phenotype. For example, not all FA proteins are required for S phase checkpoints. It has been reported that FANCC is not required for the intra-S phase checkpoint in cells exposed to IR [151]. Even more confounding, while FANCD2 is required for S phase checkpoints in human cells, it is not required for the S phase checkpoint response of mouse embryonic fibroblasts to IR or ICLs [185]. One possible explanation for these discrepancies is that FA proteins could potentially participate in the intra-S phase checkpoint by assembling into a complex of DNA repair proteins that impedes elongation rather than initiation of replication. The extent of this impediment might vary depending on the type of DNA damage, the timepoint, and the particular FA protein that is deficient. Also, it should be noted that a defective IR-induced intra-S phase checkpoint in FA cells is not strictly linked to sensitivity to IR [183].
It has been suggested that the FA pathway may coordinate various steps involved in the repair of ICLs, including NER, TLS, and HR [186]. An involvement of different FA proteins at specific steps in the repair of ICLs might explain the need for the large number of FA proteins. In support of this possibility, it has recently been reported that the FA nuclear core complex, but not FANCD2, may be associated with TLS [88]. In contrast, FANCD1/BRCA2 and FANCN/PALB2, but not other FA proteins, appear to have an integral role in DSB-initiated HR [89,184]. Thus, FA proteins could potentially play a role in the repair of lesions detected by the advancing replication fork, including endogenous DNA damage, rather than being required for processes related to DNA replication itself. In this case, ATR-dependent regulation of FANCD2 monoubiquitination in response to DNA damage and the colocalization of FANCD2 with blocked replication forks may simply reflect a mechanism of recruitment.
6. Conclusion
Here we have discussed four potential sources of endogenous DNA damage, or endogenous stresses, to which Fanconi anemia (FA) proteins may respond. The underlying hypothesis is that deficiency for any of the thirteen known FA genes may result in the clinical phenotypes associated with this disease due to a defective response to one, or several, of these stresses. There is abundant evidence that each of these endogenous stresses or DNA damage may be relevant to the pathophysiology of FA. It is our opinion that a defective response to replication stress is most central to FA, since such a deficiency might sensitize the cell to the other stresses discussed, such as ICLs resulting from lipid peroxidation or oxidation of bases in DNA. As discussed, it appears that a defect in telomere shortening does not initiate FA. In the future, determining the specific function of FA proteins at the replication fork will be critical for understanding the pathophysiology of FA.
Acknowledgments
We are grateful to Dr. Wei Du for assistance with the preparation of figures. This work was supported in part by NIH R01 grants HL076712 (Q.P.) and HL085587 (P.R.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 2.Jacquemont C, Taniguchi T. The Fanconi anemia pathway and ubiquitin. BMC Biochem. 2007;8 1:S10. doi: 10.1186/1471-2091-8-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi T, D'Andrea AD. The Fanconi anemia protein, FANCE, promotes the nuclear accumulation of FANCC. Blood. 2002;100:2457–2462. doi: 10.1182/blood-2002-03-0860. [DOI] [PubMed] [Google Scholar]
- 6.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, Hoatlin ME, Joenje H, Wang W. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 7.Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, Ling C, Rooimans MA, Bier P, Hoatlin M, Pals G, de Winter JP, Wang W, Joenje H. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- 8.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, Huang TT. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 11.Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 13.Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, Belanger C, Bell R, Berry S, Bogden R, Chen Q, Davis T, Dumont M, Frye C, Hattier T, Jammulapati S, Janecki T, Jiang P, Kehrer R, Leblanc JF, Mitchell JT, McArthur-Morrison J, Nguyen K, Peng Y, Samson C, Schroeder M, Snyder SC, Steele L, Stringfellow M, Stroup C, Swedlund B, Swense J, Teng D, Thomas A, Tran T, Tranchant M, Weaver-Feldhaus J, Wong AK, Shizuya H, Eyfjord JE, Cannon-Albright L, Tranchant M, Labrie F, Skolnick MH, Weber B, Kamb A, Goldgar DE. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 14.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erkko H, Xia B, Nikkila J, Schleutker J, Syrjakoski K, Mannermaa A, Kallioniemi A, Pylkas K, Karppinen SM, Rapakko K, Miron A, Sheng Q, Li G, Mattila H, Bell DW, Haber DA, Grip M, Reiman M, Jukkola-Vuorinen A, Mustonen A, Kere J, Aaltonen LA, Kosma VM, Kataja V, Soini Y, Drapkin RI, Livingston DM, Winqvist R. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 16.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 17.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D'Andrea AD. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 18.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, Wurm M, Batish SD, Lach FP, Yetgin S, Neitzel H, Ariffin H, Tischkowitz M, Mathew CG, Auerbach AD, Rahman N. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 19.Levitus M, Rooimans MA, Steltenpool J, Cool NF, Oostra AB, Mathew CG, Hoatlin ME, Waisfisz Q, Arwert F, de Winter JP, Joenje H. Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood. 2004;103:2498–2503. doi: 10.1182/blood-2003-08-2915. [DOI] [PubMed] [Google Scholar]
- 20.Carreau M, Alon N, Bosnoyan-Collins L, Joenje H, Buchwald M. Drug sensitivity spectra in Fanconi anemia lymphoblastoid cell lines of defined complementation groups. Mutat Res. 1999;435:103–109. doi: 10.1016/s0921-8777(99)00041-5. [DOI] [PubMed] [Google Scholar]
- 21.Kupfer GM, D'Andrea AD. The effect of the Fanconi anemia polypeptide, FAC, upon p53 induction and G2 checkpoint regulation. Blood. 1996;88:1019–1025. [PubMed] [Google Scholar]
- 22.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 23.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Povirk LF, Shuker DE. DNA damage and mutagenesis induced by nitrogen mustards. Mutat Res. 1994;318:205–226. doi: 10.1016/0165-1110(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 25.Palom Y, Kumar G Suresh, Tang LQ, Paz MM, Musser SM, Rockwell S, Tomasz M. Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbamoyl mitomycin C and mitomycin C. Chem Res Toxicol. 2002;15:1398–1406. doi: 10.1021/tx020044g. [DOI] [PubMed] [Google Scholar]
- 26.Grompe M, D'Andrea A. Fanconi anemia and DNA repair. Hum Mol Genet. 2001;10:2253–2259. doi: 10.1093/hmg/10.20.2253. [DOI] [PubMed] [Google Scholar]
- 27.Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol. 1993;21:731–733. [PubMed] [Google Scholar]
- 28.Heinrich MC, Hoatlin ME, Zigler AJ, Silvey KV, Bakke AC, Keeble WW, Zhi Y, Reifsteck CA, Grompe M, Brown MG, Magenis RE, Olson SB, Bagby GC. DNA cross-linker-induced G2/M arrest in group C Fanconi anemia lymphoblasts reflects normal checkpoint function. Blood. 1998;91:275–287. [PubMed] [Google Scholar]
- 29.Chandra S, Levran O, Jurickova I, Maas C, Kapur R, Schindler D, Henry R, Milton K, Batish SD, Cancelas JA, Hanenberg H, Auerbach AD, Williams DA. A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Mol Ther. 2005;12:976–984. doi: 10.1016/j.ymthe.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Gallmeier E, Calhoun ES, Rago C, Brody JR, Cunningham SC, Hucl T, Gorospe M, Kohli M, Lengauer C, Kern SE. Targeted disruption of FANCC and FANCG in human cancer provides a preclinical model for specific therapeutic options. Gastroenterology. 2006;130:2145–2154. doi: 10.1053/j.gastro.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Saadatzadeh MR, Bijangi-Vishehsaraei K, Hong P, Bergmann H, Haneline LS. Oxidant hypersensitivity of Fanconi anemia type C-deficient cells is dependent on a redox-regulated apoptotic pathway. J Biol Chem. 2004;279:16805–16812. doi: 10.1074/jbc.M313721200. [DOI] [PubMed] [Google Scholar]
- 32.Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DA, Sale JE, Yamazoe M, Bishop DK, Takata M, Takeda S, Watanabe M, Swenberg JA, Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 33.Berneburg M, Lehmann AR. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv Genet. 2001;43:71–102. doi: 10.1016/s0065-2660(01)43004-5. [DOI] [PubMed] [Google Scholar]
- 34.Daya-Grosjean L, Sarasin A. The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skin tumors. Mutat Res. 2005;571:43–56. doi: 10.1016/j.mrfmmm.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Tischkowitz MD, Hodgson SV. Fanconi anaemia. J Med Genet. 2003;40:1–10. doi: 10.1136/jmg.40.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alter BP. Cancer in Fanconi anemia, 1927-2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 37.Grillari J, Katinger H, Voglauer R. Contributions of DNA interstrand cross-links to aging of cells and organisms. Nucleic Acids Res. 2007;35:7566–7576. doi: 10.1093/nar/gkm1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scharer OD. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 39.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 40.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 41.Bakkenist CJ, Drissi R, Wu J, Kastan MB, Dome JS. Disappearance of the telomere dysfunction-induced stress response in fully senescent cells. Cancer Res. 2004;64:3748–3752. doi: 10.1158/0008-5472.CAN-04-0453. [DOI] [PubMed] [Google Scholar]
- 42.Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, Ananth S, Davies A, Masson JY, Moses R, West SC, de Winter JP, Ashworth A, Jones NJ, Mathew CG. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Genet. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 43.Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fridovich I. Oxygen toxicity: a radical explanation. J Exp Biol. 1998;201:1203–1209. doi: 10.1242/jeb.201.8.1203. [DOI] [PubMed] [Google Scholar]
- 45.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuzin B, Roberts I, Peunova N, Enikolopov G. Nitric oxide regulates cell proliferation during Drosophila development. Cell. 1996;87:639–649. doi: 10.1016/s0092-8674(00)81384-7. [DOI] [PubMed] [Google Scholar]
- 47.Regulski M, Stasiv Y, Tully T, Enikolopov G. Essential function of nitric oxide synthase in Drosophila. Curr Biol. 2004;14:R881–882. doi: 10.1016/j.cub.2004.09.068. [DOI] [PubMed] [Google Scholar]
- 48.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 49.Poli G, Leonarduzzi G, Biasi F, Chiarpotto E. Oxidative stress and cell signalling. Curr Med Chem. 2004;11:1163–1182. doi: 10.2174/0929867043365323. [DOI] [PubMed] [Google Scholar]
- 50.Massey V, Strickland S, Mayhew SG, Howell LG, Engel PC, Matthews RG, Schuman M, Sullivan PA. The production of superoxide anion radicals in the reaction of reduced flavins and flavoproteins with molecular oxygen. Biochem Biophys Res Commun. 1969;36:891–897. doi: 10.1016/0006-291x(69)90287-3. [DOI] [PubMed] [Google Scholar]
- 51.Loschen G, Azzi A, Richter C, Flohe L. Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett. 1974;42:68–72. doi: 10.1016/0014-5793(74)80281-4. [DOI] [PubMed] [Google Scholar]
- 52.Forman HJ, Kennedy JA. Role of superoxide radical in mitochondrial dehydrogenase reactions. Biochem Biophys Res Commun. 1974;60:1044–1050. doi: 10.1016/0006-291x(74)90418-5. [DOI] [PubMed] [Google Scholar]
- 53.McCord JM, Fridovich I. The utility of superoxide dismutase in studying free radical reactions. II. The mechanism of the mediation of cytochrome c reduction by a variety of electron carriers. J Biol Chem. 1970;245:1374–1377. [PubMed] [Google Scholar]
- 54.Li H, Poulos TL. Structure-function studies on nitric oxide synthases. J Inorg Biochem. 2005;99:293–305. doi: 10.1016/j.jinorgbio.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 55.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 56.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 57.Pfeiffer P, Goedecke W, Obe G. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis. 2000;15:289–302. doi: 10.1093/mutage/15.4.289. [DOI] [PubMed] [Google Scholar]
- 58.Shukla A, Gulumian M, Hei TK, Kamp D, Rahman Q, Mossman BT. Multiple roles of oxidants in the pathogenesis of asbestos-induced diseases. Free Radic Biol Med. 2003;34:1117–1129. doi: 10.1016/s0891-5849(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 59.Shi H, Hudson LG, Ding W, Wang S, Cooper KL, Liu S, Chen Y, Shi X, Liu KJ. Arsenite causes DNA damage in keratinocytes via generation of hydroxyl radicals. Chem Res Toxicol. 2004;17:871–878. doi: 10.1021/tx049939e. [DOI] [PubMed] [Google Scholar]
- 60.Maier CM, Chan PH. Role of superoxide dismutases in oxidative damage and neurodegenerative disorders. Neuroscientist. 2002;8:323–334. doi: 10.1177/107385840200800408. [DOI] [PubMed] [Google Scholar]
- 61.Kirkman HN, Gaetani GF. Mammalian catalase: a venerable enzyme with new mysteries. Trends Biochem Sci. 2007;32:44–50. doi: 10.1016/j.tibs.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 62.Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 63.Nakamura H. Thioredoxin and its related molecules: update 2005. Antioxid Redox Signal. 2005;7:823–828. doi: 10.1089/ars.2005.7.823. [DOI] [PubMed] [Google Scholar]
- 64.Rikans LE, Hornbrook KR. Lipid peroxidation, antioxidant protection and aging. Biochim Biophys Acta. 1997;1362:116–127. doi: 10.1016/s0925-4439(97)00067-7. [DOI] [PubMed] [Google Scholar]
- 65.Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A. Protein carbonylation in human diseases. Trends Mol Med. 2003;9:169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 66.Krishnamurthy N, Haraguchi K, Greenberg MM, David SS. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 2008;47:1043–1050. doi: 10.1021/bi701919u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niedernhofer LJ, Daniels JS, Rouzer CA, Greene RE, Marnett LJ. Malondialdehyde, a product of lipid peroxidation, is mutagenic in human cells. J Biol Chem. 2003;278:31426–31433. doi: 10.1074/jbc.M212549200. [DOI] [PubMed] [Google Scholar]
- 68.Kadlubar FF, Anderson KE, Haussermann S, Lang NP, Barone GW, Thompson PA, MacLeod SL, Chou MW, Mikhailova M, Plastaras J, Marnett LJ, Nair J, Velic I, Bartsch H. Comparison of DNA adduct levels associated with oxidative stress in human pancreas. Mutat Res. 1998;405:125–133. doi: 10.1016/s0027-5107(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 69.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA cross-links induced by alpha,beta-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caulfield JL, Wishnok JS, Tannenbaum SR. Nitric oxide-induced interstrand cross-links in DNA. Chem Res Toxicol. 2003;16:571–574. doi: 10.1021/tx020117w. [DOI] [PubMed] [Google Scholar]
- 71.Joenje H, Arwert F, Eriksson AW, de Koning H, Oostra AB. Oxygen-dependence of chromosomal aberrations in Fanconi's anaemia. Nature. 1981;290:142–143. doi: 10.1038/290142a0. [DOI] [PubMed] [Google Scholar]
- 72.Schindler D, Hoehn H. Fanconi anemia mutation causes cellular susceptibility to ambient oxygen. Am J Hum Genet. 1988;43:429–435. [PMC free article] [PubMed] [Google Scholar]
- 73.Cohen-Haguenauer O, Peault B, Bauche C, Daniel MT, Casal I, Levy V, Dausset J, Boiron M, Auclair C, Gluckman E, Marty M. In vivo repopulation ability of genetically corrected bone marrow cells from Fanconi anemia patients. Proc Natl Acad Sci U S A. 2006;103:2340–2345. doi: 10.1073/pnas.0510613103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pagano G, Degan P, d'Ischia M, Kelly FJ, Nobili B, Pallardo FV, Youssoufian H, Zatterale A. Oxidative stress as a multiple effector in Fanconi anaemia clinical phenotype. Eur J Haematol. 2005;75:93–100. doi: 10.1111/j.1600-0609.2005.00507.x. [DOI] [PubMed] [Google Scholar]
- 75.Futaki M, Igarashi T, Watanabe S, Kajigaya S, Tatsuguchi A, Wang J, Liu JM. The FANCG Fanconi anemia protein interacts with CYP2E1: possible role in protection against oxidative DNA damage. Carcinogenesis. 2002;23:67–72. doi: 10.1093/carcin/23.1.67. [DOI] [PubMed] [Google Scholar]
- 76.Kruyt FA, Hoshino T, Liu JM, Joseph P, Jaiswal AK, Youssoufian H. Abnormal microsomal detoxification implicated in Fanconi anemia group C by interaction of the FAC protein with NADPH cytochrome P450 reductase. Blood. 1998;92:3050–3056. [PubMed] [Google Scholar]
- 77.Park SJ, Ciccone SL, Beck BD, Hwang B, Freie B, Clapp DW, Lee SH. Oxidative stress/damage induces multimerization and interaction of Fanconi anemia proteins. J Biol Chem. 2004;279:30053–30059. doi: 10.1074/jbc.M403527200. [DOI] [PubMed] [Google Scholar]
- 78.Hadjur S, Ung K, Wadsworth L, Dimmick J, Rajcan-Separovic E, Scott RW, Buchwald M, Jirik FR. Defective hematopoiesis and hepatic steatosis in mice with combined deficiencies of the genes encoding Fancc and Cu/Zn superoxide dismutase. Blood. 2001;98:1003–1011. doi: 10.1182/blood.v98.4.1003. [DOI] [PubMed] [Google Scholar]
- 79.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 80.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 81.Cumming RC, Lightfoot J, Beard K, Youssoufian H, O'Brien PJ, Buchwald M. Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat Med. 2001;7:814–820. doi: 10.1038/89937. [DOI] [PubMed] [Google Scholar]
- 82.Mukhopadhyay SS, Leung KS, Hicks MJ, Hastings PJ, Youssoufian H, Plon SE. Defective mitochondrial peroxiredoxin-3 results in sensitivity to oxidative stress in Fanconi anemia. J Cell Biol. 2006;175:225–235. doi: 10.1083/jcb.200607061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 84.Hirano S, Yamamoto K, Ishiai M, Yamazoe M, Seki M, Matsushita N, Ohzeki M, Yamashita YM, Arakawa H, Buerstedde JM, Enomoto T, Takeda S, Thompson LH, Takata M. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. Embo J. 2005;24:418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 86.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peng M, Litman R, Xie J, Sharma S, Brosh RM, Jr, Cantor SB. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. Embo J. 2007;26:3238–3249. doi: 10.1038/sj.emboj.7601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mirchandani KD, McCaffrey RM, D'Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7:902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 90.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 91.Vaisman A, Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. Embo J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 93.Mazurek A, Berardini M, Fishel R. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J Biol Chem. 2002;277:8260–8266. doi: 10.1074/jbc.M111269200. [DOI] [PubMed] [Google Scholar]
- 94.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 95.Bagby GC., Jr Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 96.Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, Hanenberg H, Auerbach AD. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 97.Lensch MW, Rathbun RK, Olson SB, Jones GR, Bagby GC., Jr Selective pressure as an essential force in molecular evolution of myeloid leukemic clones: a view from the window of Fanconi anemia. Leukemia. 1999;13:1784–1789. doi: 10.1038/sj.leu.2401586. [DOI] [PubMed] [Google Scholar]
- 98.Cumming RC, Liu JM, Youssoufian H, Buchwald M. Suppression of apoptosis in hematopoietic factor-dependent progenitor cell lines by expression of the FAC gene. Blood. 1996;88:4558–4567. [PubMed] [Google Scholar]
- 99.Rani R, Li J, Pang Q. Differential p53 engagement in response to oxidative and oncogenic stresses in Fanconi anemia mice. Cancer Res. 2008;68:9693–9702. doi: 10.1158/0008-5472.CAN-08-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liebetrau W, Budde A, Savoia A, Grummt F, Hoehn H. p53 activates Fanconi anemia group C gene expression. Hum Mol Genet. 1997;6:277–283. doi: 10.1093/hmg/6.2.277. [DOI] [PubMed] [Google Scholar]
- 101.Liebetrau W, Buhner M, Hoehn H. Prototype sequence clues within the Fanconi anaemia group C gene. J Med Genet. 1995;32:669–670. doi: 10.1136/jmg.32.8.669-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liebetrau W, Runger TM, Mehling BE, Poot M, Hoehn H. Mutagenic activity of ambient oxygen and mitomycin C in Fanconi's anaemia cells. Mutagenesis. 1997;12:69–77. doi: 10.1093/mutage/12.2.69. [DOI] [PubMed] [Google Scholar]
- 103.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 104.Freie B, Li X, Ciccone SL, Nawa K, Cooper S, Vogelweid C, Schantz L, Haneline LS, Orazi A, Broxmeyer HE, Lee SH, Clapp DW. Fanconi anemia type C and p53 cooperate in apoptosis and tumorigenesis. Blood. 2003;102:4146–4152. doi: 10.1182/blood-2003-03-0971. [DOI] [PubMed] [Google Scholar]
- 105.Houghtaling S, Granville L, Akkari Y, Torimaru Y, Olson S, Finegold M, Grompe M. Heterozygosity for p53 (Trp53+/-) accelerates epithelial tumor formation in fanconi anemia complementation group D2 (Fancd2) knockout mice. Cancer Res. 2005;65:85–91. [PubMed] [Google Scholar]
- 106.Liu TX, Howlett NG, Deng M, Langenau DM, Hsu K, Rhodes J, Kanki JP, D'Andrea AD, Look AT. Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell. 2003;5:903–914. doi: 10.1016/s1534-5807(03)00339-3. [DOI] [PubMed] [Google Scholar]
- 107.Klebanoff SJ. S.J. Phagocytic cells: products of oxygen metabolism. In: Gallin JI, Goldstein IM, Synderman R, editors. Inflammation: basic principles and clinical correlates. Raven; New York: 1988. pp. 391–444. [Google Scholar]
- 108.Adams DO, Hamilton TA. Macrophages as destructive cells in host defense. In: Gallin JI, Goldstein IM, Synderman R, editors. Inflammation: basic principles and clinical correlates. Raven; New York: 1992. pp. 637–662. [Google Scholar]
- 109.Shacter E, Beecham EJ, Covey JM, Kohn KW, Potter M. Activated neutrophils induce prolonged DNA damage in neighboring cells. Carcinogenesis. 1988;9:2297–2304. doi: 10.1093/carcin/9.12.2297. [DOI] [PubMed] [Google Scholar]
- 110.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 111.Tak PP, Zvaifler NJ, Green DR, Firestein GS. Rheumatoid arthritis and p53: how oxidative stress might alter the course of inflammatory diseases. Immunol Today. 2000;21:78–82. doi: 10.1016/s0167-5699(99)01552-2. [DOI] [PubMed] [Google Scholar]
- 112.Umeda T, Hino O. Molecular aspects of human hepatocarcinogenesis mediated by inflammation: from hypercarcinogenic state to normo- or hypocarcinogenic state. Oncology. 2002;62 1:38–42. doi: 10.1159/000048274. [DOI] [PubMed] [Google Scholar]
- 113.Suematsu N, Tsutsui H, Wen J, Kang D, Ikeuchi M, Ide T, Hayashidani S, Shiomi T, Kubota T, Hamasaki N, Takeshita A. Oxidative stress mediates tumor necrosis factor-alpha-induced mitochondrial DNA damage and dysfunction in cardiac myocytes. Circulation. 2003;107:1418–1423. doi: 10.1161/01.cir.0000055318.09997.1f. [DOI] [PubMed] [Google Scholar]
- 114.Maini RN, Taylor PC. Anti-cytokine therapy for rheumatoid arthritis. Annu Rev Med. 2000;51:207–229. doi: 10.1146/annurev.med.51.1.207. [DOI] [PubMed] [Google Scholar]
- 115.Young NS. Hematopoietic cell destruction by immune mechanisms in acquired aplastic anemia. Semin Hematol. 2000;37:3–14. doi: 10.1016/s0037-1963(00)90026-x. [DOI] [PubMed] [Google Scholar]
- 116.Varfolomeev EE, Ashkenazi A. Tumor necrosis factor: an apoptosis JuNKie? Cell. 2004;116:491–497. doi: 10.1016/s0092-8674(04)00166-7. [DOI] [PubMed] [Google Scholar]
- 117.Ventura JJ, Cogswell P, Flavell RA, Baldwin AS, Jr, Davis RJ. JNK potentiates TNF-stimulated necrosis by increasing the production of cytotoxic reactive oxygen species. Genes Dev. 2004;18:2905–2915. doi: 10.1101/gad.1223004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rosselli F, Sanceau J, Gluckman E, Wietzerbin J, Moustacchi E. Abnormal lymphokine production: a novel feature of the genetic disease Fanconi anemia. II. In vitro and in vivo spontaneous overproduction of tumor necrosis factor alpha. Blood. 1994;83:1216–1225. [PubMed] [Google Scholar]
- 119.Rosselli F, Sanceau J, Wietzerbin J, Moustacchi E. Abnormal lymphokine production: a novel feature of the genetic disease Fanconi anemia. I. Involvement of interleukin-6. Hum Genet. 1992;89:42–48. doi: 10.1007/BF00207040. [DOI] [PubMed] [Google Scholar]
- 120.Schultz JC, Shahidi NT. Tumor necrosis factor-alpha overproduction in Fanconi's anemia. Am J Hematol. 1993;42:196–201. doi: 10.1002/ajh.2830420211. [DOI] [PubMed] [Google Scholar]
- 121.Dufour C, Corcione A, Svahn J, Haupt R, Poggi V, Beka'ssy AN, Scime R, Pistorio A, Pistoia V. TNF-alpha and IFN-gamma are overexpressed in the bone marrow of Fanconi anemia patients and TNF-alpha suppresses erythropoiesis in vitro. Blood. 2003;102:2053–2059. doi: 10.1182/blood-2003-01-0114. [DOI] [PubMed] [Google Scholar]
- 122.Haneline LS, Li X, Ciccone SL, Hong P, Yang Y, Broxmeyer HE, Lee SH, Orazi A, Srour EF, Clapp DW. Retroviral-mediated expression of recombinant Fancc enhances the repopulating ability of Fancc-/- hematopoietic stem cells and decreases the risk of clonal evolution. Blood. 2003;101:1299–1307. doi: 10.1182/blood-2002-08-2404. [DOI] [PubMed] [Google Scholar]
- 123.Li J, Sejas DP, Zhang X, Qiu Y, Nattamai KJ, Rani R, Rathbun KR, Geiger H, Williams DA, Bagby GC, Pang Q. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117:3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sejas DP, Rani R, Qiu Y, Zhang X, Fagerlie SR, Nakano H, Williams DA, Pang Q. Inflammatory reactive oxygen species-mediated hemopoietic suppression in Fancc-deficient mice. J Immunol. 2007;178:5277–5287. doi: 10.4049/jimmunol.178.8.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang X, Sejas DP, Qiu Y, Williams DA, Pang Q. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J Cell Sci. 2007;120:1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang QS, Eaton L, Snyder ER, Houghtaling S, Mitchell JB, Finegold M, Van Waes C, Grompe M. Tempol protects against oxidative damage and delays epithelial tumor onset in Fanconi anemia mice. Cancer Res. 2008;68:1601–1608. doi: 10.1158/0008-5472.CAN-07-5186. [DOI] [PubMed] [Google Scholar]
- 127.Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22:125–140. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 130.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 131.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 133.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 134.Grobelny JV, Godwin AK, Broccoli D. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J Cell Sci 113 Pt. 2000;24:4577–4585. doi: 10.1242/jcs.113.24.4577. [DOI] [PubMed] [Google Scholar]
- 135.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 136.Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 137.Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 138.Wu G, Jiang X, Lee WH, Chen PL. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63:2589–2595. [PubMed] [Google Scholar]
- 139.Nabetani A, Yokoyama O, Ishikawa F. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J Biol Chem. 2004;279:25849–25857. doi: 10.1074/jbc.M312652200. [DOI] [PubMed] [Google Scholar]
- 140.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 141.Varley H, Pickett HA, Foxon JL, Reddel RR, Royle NJ. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat Genet. 2002;30:301–305. doi: 10.1038/ng834. [DOI] [PubMed] [Google Scholar]
- 142.Ball SE, Gibson FM, Rizzo S, Tooze JA, Marsh JC, Gordon-Smith EC. Progressive telomere shortening in aplastic anemia. Blood. 1998;91:3582–3592. [PubMed] [Google Scholar]
- 143.Hanson H, Mathew CG, Docherty Z, Ogilvie C Mackie. Telomere shortening in Fanconi anaemia demonstrated by a direct FISH approach. Cytogenet Cell Genet. 2001;93:203–206. doi: 10.1159/000056985. [DOI] [PubMed] [Google Scholar]
- 144.Leteurtre F, Li X, Guardiola P, Le Roux G, Sergere JC, Richard P, Carosella ED, Gluckman E. Accelerated telomere shortening and telomerase activation in Fanconi's anaemia. Br J Haematol. 1999;105:883–893. doi: 10.1046/j.1365-2141.1999.01445.x. [DOI] [PubMed] [Google Scholar]
- 145.Callen E, Samper E, Ramirez MJ, Creus A, Marcos R, Ortega JJ, Olive T, Badell I, Blasco MA, Surralles J. Breaks at telomeres and TRF2-independent end fusions in Fanconi anemia. Hum Mol Genet. 2002;11:439–444. doi: 10.1093/hmg/11.4.439. [DOI] [PubMed] [Google Scholar]
- 146.Franco S, van de Vrugt HJ, Fernandez P, Aracil M, Arwert F, Blasco MA. Telomere dynamics in Fancg-deficient mouse and human cells. Blood. 2004;104:3927–3935. doi: 10.1182/blood-2003-10-3626. [DOI] [PubMed] [Google Scholar]
- 147.Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR. A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Spardy N, Duensing A, Hoskins EE, Wells SI, Duensing S. HPV-16 E7 reveals a link between DNA replication stress, fanconi anemia D2 protein, and alternative lengthening of telomere-associated promyelocytic leukemia bodies. Cancer Res. 2008;68:9954–9963. doi: 10.1158/0008-5472.CAN-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, Wang X, Houghtaling S, Grompe M, D'Andrea AD. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 150.Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 152.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 154.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 155.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen Y, Sanchez Y. Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair (Amst) 2004;3:1025–1032. doi: 10.1016/j.dnarep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 160.Lin SY, Li K, Stewart GS, Elledge SJ. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc Natl Acad Sci U S A. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sar F, Lindsey-Boltz LA, Subramanian D, Croteau DL, Hutsell SQ, Griffith JD, Sancar A. Human claspin is a ring-shaped DNA-binding protein with high affinity to branched DNA structures. J Biol Chem. 2004;279:39289–39295. doi: 10.1074/jbc.M405793200. [DOI] [PubMed] [Google Scholar]
- 162.Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA. Nbs1 is required for ATR-dependent phosphorylation events. Embo J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pichierri P, Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. Embo J. 2004;23:1178–1187. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ho GP, Margossian S, Taniguchi T, D'Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, Tashiro S, Elledge SJ, Takata M. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Collins NB, Wilson JB, Bush T, Thomashevski A, Roberts KJ, Jones NJ, Kupfer GM. ATR-dependent phosphorylation of FANCA on serine 1449 after DNA damage is important for FA pathway function. Blood. 2009;113:2181–2190. doi: 10.1182/blood-2008-05-154294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Guervilly JH, Mace-Aime G, Rosselli F. Loss of CHK1 function impedes DNA damage-induced FANCD2 monoubiquitination but normalizes the abnormal G2 arrest in Fanconi anemia. Hum Mol Genet. 2008;17:679–689. doi: 10.1093/hmg/ddm340. [DOI] [PubMed] [Google Scholar]
- 168.Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol. 2007;9:391–401. doi: 10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- 169.Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 170.Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D'Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27:3098–3108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Peng A, Lewellyn AL, Maller JL. DNA damage signaling in early Xenopus embryos. Cell Cycle. 2008;7:3–6. doi: 10.4161/cc.7.1.5157. [DOI] [PubMed] [Google Scholar]
- 172.Sobeck A, Stone S, Costanzo V, de Graaf B, Reuter T, de Winter J, Wallisch M, Akkari Y, Olson S, Wang W, Joenje H, Christian JL, Lupardus PJ, Cimprich KA, Gautier J, Hoatlin ME. Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol Cell Biol. 2006;26:425–437. doi: 10.1128/MCB.26.2.425-437.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 174.Andreassen PR, Ho GP, D'Andrea AD. DNA damage responses and their many interactions with the replication fork. Carcinogenesis. 2006;27:883–892. doi: 10.1093/carcin/bgi319. [DOI] [PubMed] [Google Scholar]
- 175.Ohashi A, Zdzienicka MZ, Chen J, Couch FJ. Fanconi anemia complementation group D2 (FANCD2) functions independently of BRCA2- and RAD51-associated homologous recombination in response to DNA damage. J Biol Chem. 2005;280:14877–14883. doi: 10.1074/jbc.M414669200. [DOI] [PubMed] [Google Scholar]
- 176.Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma -H2AX formation, and Mre11 relocalization. Proc Natl Acad Sci U S A. 2002;99:233–238. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang LC, Stone S, Hoatlin ME, Gautier J. Fanconi anemia proteins stabilize replication forks. DNA Repair (Amst) 2008;7:1973–1981. doi: 10.1016/j.dnarep.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc Natl Acad Sci U S A. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 181.Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang X, Andreassen PR, D'Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Godthelp BC, van Buul PP, Jaspers NG, Elghalbzouri-Maghrani E, van Duijn-Goedhart A, Arwert F, Joenje H, Zdzienicka MZ. Cellular characterization of cells from the Fanconi anemia complementation group, FA-D1/BRCA2. Mutat Res. 2006;601:191–201. doi: 10.1016/j.mrfmmm.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 184.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 185.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mirchandani KD, D'Andrea AD. The Fanconi anemia/BRCA pathway: a coordinator of cross-link repair. Exp Cell Res. 2006;312:2647–2653. doi: 10.1016/j.yexcr.2006.06.014. [DOI] [PubMed] [Google Scholar]