Abstract
In recently generated B6.56R anti-DNA autoantibody transgenic mice, it was noted that a substantial faction of the B-cells that had “avoided” DNA-reactivity had done so through the rearrangement and usage of the endogenous, non-targeted HC allele. This suggested that rearrangement at the second HC locus might be an important mechanism through which self-reactive B-cells might successfully revise their initial antigen specificity. To test the importance of this mechanism in B-cell tolerance, we generated B6.56R/56R mice that possessed the “56R” anti-DNA heavy chain transgene inserted into both HC loci. These transgenic homozygotes developed higher titers of anti-DNA antibodies, with an expanded population of B220low, MHC-Class IIlo B-cells, enriched for CD21low, CD23low pre-plasmablasts. The analysis of hybridomas from these mice revealed that the only avenue by which these B-cells could avoid DNA-reactivity was through the use of the editor light chains, Vk20 or Vk21. Hence, in addition to LC editing, rearrangement and usage of the second HC locus/allele constitutes an important “safety valve” for B-cells whose primary BCR confers DNA-reactivity. In contrast to these tolerance mechanisms, editing the first rearranged HC locus (through HC replacement) and somatic mutations appear to be less frequently employed to edit/revise self-reactive B cells.
Keywords: Lupus, Animal models, Anti-DNA, Immunoglobulin, Receptor-editing
INTRODUCTION
Newly generated B-lymphocytes are known to undergo a meticulous censoring program through which self-reactive B-cells are removed from the immune system. This is an important ritual since the persistence of potentially self-reactive B-cells and their subsequent activation can lead to autoimmunity. The mechanisms through which self-reactive B-cells are censored have been extensively studied, particularly through the use of BCR transgenic (Tg) models. It is now apparent that several checkpoints exist to maintain B-cell tolerance, as reviewed elsewhere (1).
Studies in BCR transgenic models have demonstrated that B-cells with strong self-reactivity are censored in the bone marrow largely through receptor–editing and deletion (1–5). Elegant work by several investigators has shown that secondary rearrangement at the light chain (LC) locus is a particularly effective mechanism that B-cells commonly employ to veto self-reactivity (6–9). Examples of receptor revision at the heavy chain (HC) locus have also been documented (10–12). B-cells that have weaker degrees of self-reactivity have been shown to be rendered “anergic” or functionally incapacitated (1, 13, 14). Additional tolerance events in the germinal center have also been uncovered (15–17). Hence, a rich panoply of central and peripheral mechanisms exists to censor self-reactive B-cells from the re-circulating immune system.
It has also become apparent that the genetic background can substantially impact how effectively anti-self B-cells are censored. Of relevance to this report, studies using the “3H9/56R” anti-DNA immunoglobulin (Ig) Tg model have shed light on how different genetic backgrounds might impact B-cell tolerance to DNA. Thus, whereas anti-DNA B-cells are effectively tolerized in the normal BALB/c background, tolerance is breached when the same Tg is bred onto the MRL/lpr background (18, 19). Interestingly, on the C57BL/6 (B6) background, an intermediate degree of breach in tolerance is noted, as reported elsewhere (20–22).
Thus, when the “56R” anti-DNA Ig Tg (with an arginine residue at position 56 in the CDR2 region of the “3H9” anti-DNA Ig HC) is bred onto the B6 background, these mice develop modest titers of autoantibodies to DNA as early as 4–6 mo of age. Potentially, B-cells from these anti-DNA HC-only Tg mice could potentially preclude DNA-reactivity in a couple of different ways – rearrangement and usage of the endogenous HC allele, pairing of the Tg HC with a LC partner that vetoes DNA-binding, mutation of the Tg HC so that it loses affinity for DNA, and replacement of the targeted HC by an endogenous VH gene. When hybridomas were previously derived from B6.56R mice to determine the molecular makeup of anti-DNA and non-anti-DNA Abs, it was intriguing to note that a substantial faction of the B-cells that had “avoided” DNA-reactivity had done so through the rearrangement and usage of endogenous (i.e., non-targeted) HC locus (21). Most of the remaining B-cells were apparently not DNA-reactive because they had paired the Tg 56R HC with a non-DNA binding LC partner, a mechanism that has been well documented in this Tg model (6, 20).
The above findings suggested that rearrangement at the second (non-targeted) HC locus might be an important mechanism through which self-reactive B-cells might successfully revise their initial antigen specificity. However, these observations did not indicate if this particular molecular mechanism was absolutely essential for thwarting autoimmunity. Conversely, when B-cells did not have the option of rearranging/using a second HC locus, it was not apparent whether LC editing alone was sufficient for “revising” all self-reactivity. To investigate this, we generated a mouse strain, referred to as B6.56R/56R that possessed the “56R” anti-DNA HC Tg inserted into both HC loci. If these B6.56R/56R mice were to exhibit a greater degree of autoreactivity compared to Tg-hemizygous B6.56R/+ mice, this would suggest that rearrangement at the alternate HC locus and usage of the second HC allele plays an additional role in maintaining self-tolerance. On the other hand, if both the hemizygous and homozygous Tg mice exhibited similar degrees of autoreactivity, this would indicate that other mechanisms (e.g., LC editing, mutation of Tg HC, etc) may be sufficient to thwart autoimmunity, without the need for any additional contribution from rearrangement at the alternate HC locus.
Materials and Methods
Mice
“56R” anti-DNA HC-only site-directed Tg mice were a kind gift from Dr. Martin Weigert (University of Chicago) (6). These mice were backcrossed onto the C57BL/6 (B6) background, as recently described (20, 21). “B6.56R/+” mice are hemizygous for the 56R anti-DNA Ig HC Tg, whereas “B6.56R/56R” mice bear the Tg knocked into both the HC loci. Double Tg mice were identified by PCR analysis using JH1/JH2 intron specific PCR primers that will amplify DNA from the non-targeted locus, but not the Tg, and confirmed by flow cytometry (based on the absence on any staining for surface IgMb). B6.56R/56R mice were bred to B6.56R/+ mice to generate both the hemizygous and Tg homozygous mice. All mice used for this study were bred and housed in a specific pathogen free colony, at UT Southwestern Medical Center Department of Animal Resources, Dallas, TX. Comparable numbers of male and female mice were pooled for all experiments, as no significant sex differences were noted in the phenotypes studied.
Immunophenotyping of mice
Splenocytes were depleted of red blood cells using tris ammonium chloride, and single-cell suspensions were prepared for culture or flow cytometric analysis, as described previously (21, 23). Sera and monoclonal antibody (mAb) culture supernatants were screened for antibody-reactivity to ssDNA, dsDNA, and histone/DNA complexes by ELISA, as described elsewhere (21, 23). All serum samples were diluted with serum dilution buffer (2% BSA, 3mM EDTA, 0.05% Tween20, 0.1% gelatin). A positive control serum sample derived from a B6.Sle1z.Sle3z mouse (23; which bears the IgMb/IgG2ab allotype), or a serum sample derived from a seropositive B6.Sle1z.56R mouse (which bears the IgMa/IgG2aa allotype) was included as a test standard. Sera with reactivities stronger than the test standard were diluted further and re-assayed one more time. For the allotype-specific ELISA, the above assays were repeated with one modification: instead of using anti-IgM or anti-IgG second-step antibodies, biotin-coupled anti-IgMa, anti-IgMb, anti-IgG2aa, or anti-IgG2ab antibodies were used, followed by avidin-coupled alkaline phosphatase.
B-cell sorting and culture
Spleens were removed from anesthetized mice. Splenocytes were depleted of red blood cells using ACK lysis buffer (Biosource, Rockville, MD) and single-cell suspensions were prepared as for Flow cytometry. Cells were stained with anti-B220, and sorted based on the level of B220 expression, using BD FACSAria™ (BD Bioscience). B220high and B220intermediate cells were collected and cultured at 1 × 106 cells/ml for 48 hours in DMEM medium supplemented with 10% fetal bovine serum (FBS), 25 mM Hepes, 2 mM L-glutamine, antibiotics, 50 µM 2-mercaptoethanol, with or without LPS (20 ug/ml). Culture supernatants were analyzed for antibody levels by ELISA.
Assessment of nephritis
Mice were monitored at 6 mo and 9–12 mo of age for evidence of clinical nephritis. Urine was collected over a 24-h period using metabolic cages, and the total amount of urinary protein was assayed using a Comassie-based kit (Pierce, Rockford, IL). The level of blood urea nitrogen (BUN) was measured using a commercially available assay (Sigma Chemicals, St. Louis, MO). For the analysis of kidney sections, at least 100 glomeruli were examined per section by light microscopy, for evidence of hypertrophy, proliferative changes, crescent formation, hyaline deposits, fibrosis/sclerosis, and basement membrane thickening, and graded semi-quantitatively, as detailed previously (23).
Hybridomas
Spleens were removed aseptically from 9–12 mo old seropositive Tg mice, and the splenocytes were fused to the SP2/0 fusion partner, as described before (21, 24). All wells with single colonies were tested for Ab production 7–10 days post-fusion. Cells from positive wells were subcloned once more. Ab concentration was determined using a Coomassie PLUS protein assay kit (Pierce, Rockford, IL), and an isotype specific ELISA, as described above.
Sequence analysis and statistics
The HC and LC sequences were amplified and sequenced from the cDNA of the hybridomas. The primers and conditions for HC and LC RT-PCR were determined as described previously (21, 24). Specifically, the HC was amplified using a 5' primer that hybridizes to all VH genes (5'-AGGT(G/C)(A/C)A(A/G)CTGCAG(G/C)AGTC(A/T)GG-3') and 3’ primers to Cµ (5'-CAGGGGGCTCTCGCAGGAGACGAGG-3') and Cγ (5'-GGACAGGGATCCAGAGTTCC-3'), as described previously (24). Hence, this should allow the amplification of Tg as well as endogenous sequences, including those with non-productive rearrangements and edited sequences with VH replacements or D-invasions. The Ig-κ LC primers used have the potential to amplify most Ig-κ gene families including Vκ1, Vκ2, Vκ4, Vκ9, Vκ19, Vκ20, Vκ21, Vκ23, and Vκ38c, based on our previous work (21, 24 and data not shown). All antibody sequences were “blasted” against the public “IgBlast” database of mouse Ig sequences at NIH/NCBI (http://www.ncbi.nlm.nih.gov/igblast/), to determine the closest germline gene of origin. Comparing these sequences to the deposited repertoire of germline genes allowed us to identify residues that were potentially somatically mutated. The CDR position and numbering scheme adopted matched that used by the NCBI IgBlast database. The usage frequencies of individual genes were compared using Chi square tests. Where applicable, the Fisher’s exact test was applied. Inter-group comparisons of phenotypes between the two Tg strains were carried out using a one-tailed Student’s t-test (SigmaStat; Jandel Scientific). Complete sequence information is available from the authors.
RESULTS
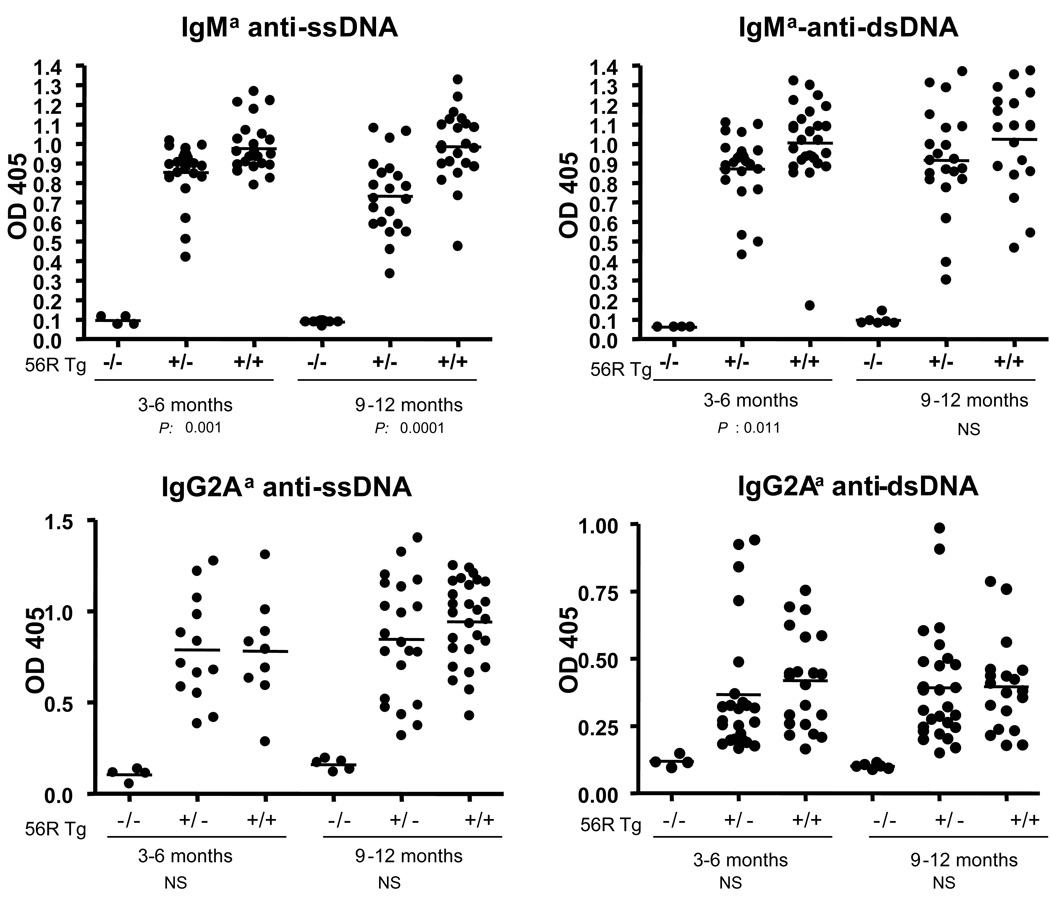
Interestingly, B6.56R/56R homozygous mice exhibited elevated levels of IgMa anti-ssDNA and anti-dsDNA Abs, relative to B6.56R/+ hemizygous controls, as displayed in Fig. 1. This serological difference was apparent as early as 3–6 mo of age, and became more pronounced with age. However, there were no significant differences in the titers of IgG (IgG2Aa) Tg-encoded anti-DNA Abs between the hemizygous and homozygous Tg mice (Fig. 1). There were also no gender differences in these phenotypes (data not shown). Finally, there was no evidence of renal disease in these strains, reproducing earlier results in B6.56R/+ mice (21; data not shown).
Figure 1. Serum levels of IgM and IgG Abs to nuclear antigens in B6.56R/+ and B6.56R/56R Tg mice.
Allotype-specific IgM and IgG Abs to ssDNA, dsDNA were measured by ELISA. Both 56R homozygous and hemizygous Tg male and female mice (pooled) were analyzed at the ages of 3–6 mo and 9–12 mo. Each dot represents a single mouse. The horizontal lines represent the mean levels of serum autoantibodies in each group of mice. The indicated P values represent the result of a Student’s t-test comparison of B.56R/56R versus B6.56R/+ mice. OD405 readings for the positive control sera ranged from 1.2–1.5. NS = not significant
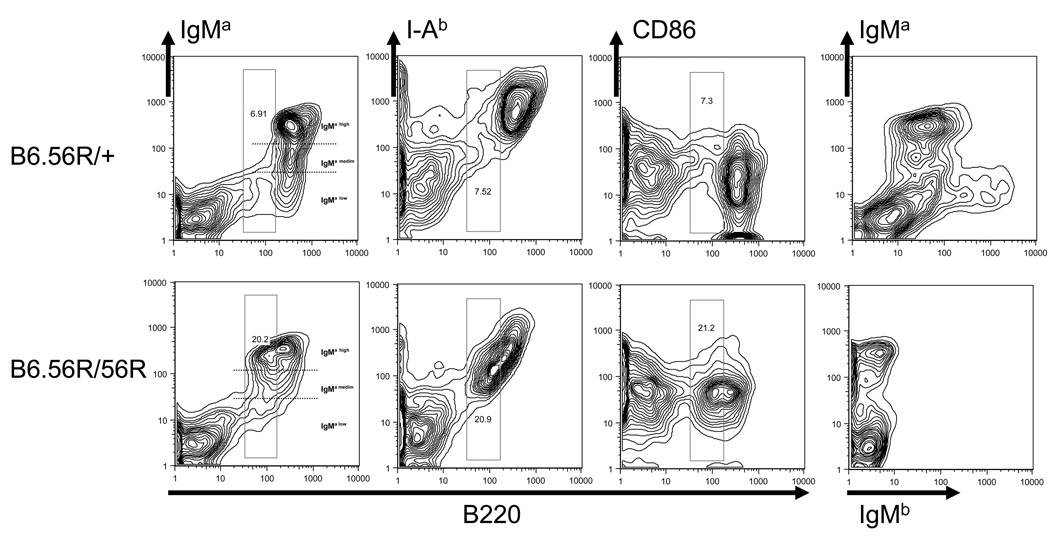
Next, we examined the cellular makeup of the lymphoid organs in B6.56R/56R and B6.56R/+ mice. Both strains exhibited similar splenic weights and cell numbers (Table 1). Likewise, both strains exhibited similar numbers of CD4 and CD8 T-cells, as well as B-cells (Table 1). Most of the B-cells in B6.56R/+ spleens exhibited high surface levels of IgMa (i.e., with high mean fluorescent intensity), and also exhibited low grade staining for the endogenous HC, IgMb (Fig. 2, Table 1). In contrast, almost all the B-cells in B6.56R/56R Tg mice expressed higher staining intensity for IgMa with the absence of staining for endogenous IgMb, as one might expect (Fig. 2). If the Tg B-cells were segregate based on their surface levels of IgMa into IgMa-high, IgMa-medium and IgMa-low B-cells, as demonstrated in Fig. 2, B6.56R/56R mice possessed significantly higher percentages of IgMa-high B-cells but lower percentages of IgMa-low B-cells, compared to B6.56R/+ mice (Table 1, Fig. 2).
Table 1.
Lymphocyte composition of spleen and peritonal cavity1
| B6.56R/+ (N=11) | B6.56R/56R (N=13) | P value | |
|---|---|---|---|
| Spleen | |||
| Weight (mg) | 109 ± 15.9 | 104.5 ± 27.6 | NS |
| Total cell numbers (×106) | 68 ± 20.9 | 66.3 ± 14.5 | NS |
| Total CD4+ (×106) | 18.7 ± 2.4 | 20.9 ± 4.5 | NS |
| % of all splenocytes | 12.7 ± 0.6 | 13.6 ± 0.9 | NS |
| Total CD8+ (×106) | 18.4 ± 1.9 | 14.9 ± 3.9 | 0.017 |
| % of all splenocytes | 12.5 ± 0.6 | 10.1 ± 0.8 | 0.015 |
| CD4:CD8 ratio | 1.0 | 1.4 | NS |
| CD4:%CD69+ | 17.2 ± 3.4 | 17.5 ± 7.3 | NS |
| CD8:%CD69+ | 18.4 ± 1.9 | 14.9 ± 3.9 | 0.003 |
| Total B cells (×106) | 41.9 ± 0.5 | 44.6 ± 1.9 | NS |
| % of all splenocytes | 28.5 ± 3.1 | 29.4 ± 2 | NS |
| IgMa (mfi) | 218.4 ± 23.9 | 215 ± 23.7 | NS |
| %IgMa+ | 69.1 ± 6.2 | 95.4 ± 1.7 | 0.0001 |
| %IgMa high | 48.7 ± 4.6 | 56.6 ± 4.9 | 0.0009 |
| %IgMa medium | 21.6 ± 2.7 | 26.9 ± 2.8 | 0.0003 |
| %IgMa low | 19.7 ± 2.9 | 11.9 ± 2.4 | 0.0001 |
| %IgMb+ | 14.2 ± 2.7 | 0.8 ± 0.7 | 0.0001 |
| % B1a cells | 3.9 ± 1.9 | 4.7 ± 2.2 | NS |
| Among IgMa cells | |||
| % follicular B-cells | 42.5 ± 4 | 39.8 ± 1.5 | NS |
| % MZ B-cells | 44.3 ± 4.2 | 48.2 ± 1.6 | NS |
| % CD21− CD23− B-cells | 17 ± 4 | 10.8 ± 1.3 | NS |
| % Igλ + | 8.2 ± 1.2 | 10.4 ± 1.3 | NS |
| I-Ab (mfi) | 803 ± 100.6 | 687.9 ± 60.7 | NS |
| CD86 (mfi) | 46.3 ± 2.7 | 143.8 ± 12.9 | 0.0001 |
| Peritoneal cavity | |||
| Among B220+ cells | |||
| % B1a | 31.9 ± 11.8 | 8.9 ± 6.4 | 0.0001 |
| % B1b | 37.1 ± 8.2 | 44.41 ± 17.7 | NS |
| % B2 | 31 ± 7.4 | 46.8 ± 19.8 | 0.023 |
Mice of each strain were examined at 9–12-mo age. Total cell numbers and percentage of splenocyte subsets shown are mean ± SEM.
The P-values shown pertain to one-tailed t-test comparisons of the 2 strains. NS means "not significant". mfi = geometric mean fluorescence intensity.
Figure 2. Splenic B-cell phenotypes in B6.56R/+ and B6.56R/56R Tg mice.
Splenocytes from 9–12 mo old B6.56R/+ (TOP) and B6.56R/56R mice (BOTTOM) were analyzed by flow cytometry for the expression of B220 vs IgMa, IgMb, CD86 and I-Ab, respectively. IgMa B-cells were further divided arbitrarily into three groups as demarcated by the dotted lines: IgMa-high B-cells, IgMa-medium B-cells and IgMa-low B-cells; the frequencies of B-cells with different surface IgMa levels are summarized in Table 1. The boxes drawn indicate the expanded population of B-cells in B6.56R/56R Tg mice that were B220int, I-Abint, CD86hi, relative to the B6.56R/+ controls. The profiles shown are representative of data obtained from 8 mice per stain.
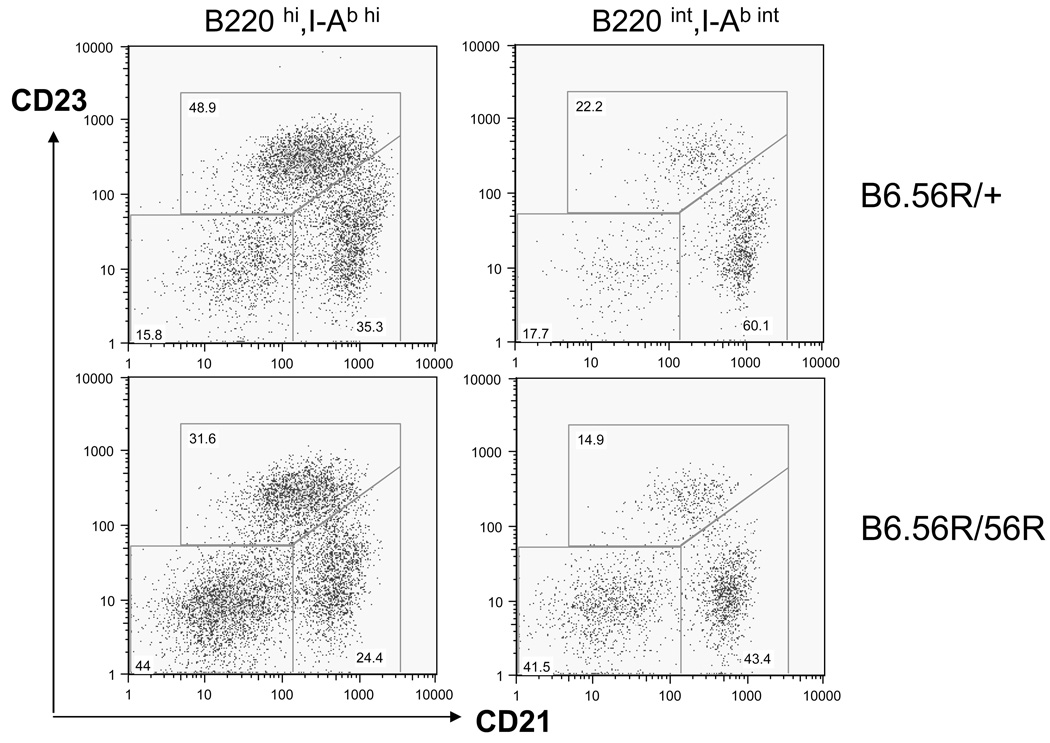
When activation markers were examined, it was interesting that B6.56R/56R B-cells expressed significantly higher levels of CD86 (B7-2) as illustrated in Fig. 2, and quantitated in Table 1. Interestingly, 2 clusters of B-cells were discerned based on their surface levels of B220 and I-Ab – B220int, I-Ab-int B-cells (bearing intermediate levels of both surface molecules) and B220hi, I-Ab-hi B-cells, with the former being boxed in Fig. 2. Interestingly, B220int, I-Ab-int B-cells were significantly expanded in B6.56R/56R mice (Table 2). When these 2 populations of B-cells were further sub-gated, it was apparent that the expanded population of B220int I-Ab-int B-cells in B6.56R/56R spleens were skewed towards being CD21−CD23− in surface phenotype, compared to B220hi I-Ab-hi B-cells from the same spleens (Table 2, Fig. 3). The possibility that the expanded population of B-cells in the Tg homozygous mice might include pre-plasmablasts is supported by their increased production of Abs in culture (Fig. 4). When all B-cells were pooled, the B6.56R/56R and B6.56R/+ spleens did not differ significantly in their overall fractions of B-cells that were follicular, marginal-zone, or B1a in phenotype (Table 1). When peritoneal B-cells were examined, almost all B6.56R/56R B-cells lacked CD5; in contrast, a third of the peritoneal B-cells in B6.56R/+ mice were CD5-positive, i.e., B1a in phenotype (Table 1), suggesting that endogenous IgMb expression was required for selection into the B1a pool, as noted in our earlier work (21).
Table 2.
Comparsion of B220hi and B220int B cells1
| B220 hi | B220 int | P value | |
|---|---|---|---|
| B6.56R/56R (n=12) | |||
| % of all B-cells | 57 ± 4.3 | 42.9 ± 4.32 | |
| mean size | 72.5 ± 1.0 | 64.5 ± 1.3 | 0.0001 |
| % FO | 39.0 ± 1.2 | 21 ± 2.2 | 0.0001 |
| % MZ | 56.2 ± 1.3 | 36.7 ± 3.3 | 0.0001 |
| % CD21−,CD23− | 4.1± 0.7 | 41.4 ± 4.7 | 0.0001 |
| I-Ab (mfi) | 883.1 ± 48.9 | 405.5 ± 39.6 | 0.0001 |
| CD86 (mfi) | 133.9 ± 12 | 122.3 ± 9.9 | NS |
| B6.56R/+ (n=11) | |||
| % of all B-cells | 65.2 ± 3.7 | 34 ± 3.82 | |
| mean size | 71.7 ± 6.6 | 71.4 ± 0.4 | NS |
| % FO | 39.8 ± 2.2 | 21.7 ± 2.8 | 0.0001 |
| % MZ | 46.9 ± 2.0 | 37 ± 2.0 | 0.0012 |
| % CD21−,CD23− | 11.8 ± 0.9 | 38.5 ± 3.8 | 0.0001 |
| I-Ab (mfi) | 1050 ± 78.4 | 407.4 ± 39.6 | 0.0001 |
| CD86 (mfi) | 24.9 ± 4.9 | 37.5 ± 8 | NS |
Splenic cells from both strains were assayed by flow cytometry and B-cells were distinguished based on surface B220 levels. Tabulated are the surface phenotype and subset distribution data pertaining to these 2 B-cell types. P values pertain to comparisons of these 2 B-cell types using the Student’s t-test. NS means not significant.
The percentage of B-cells that typed as B220int I-Ab-int were significantly different in the 2 strains (P < 0.01).
Figure 3. Surface phenotypes of B220int I-Ab-int and B220hi I-Ab-hi B-cells from B6.56R/56R and control spleens.
Splenic B-cells from 56R Tg mice were first gated based on surface levels of B220 and I-Ab, as indicated in Figure 2, and then analyzed from their surface CD21/CD23 profiles. Shown plots are representative of data obtained from 11–12 mice each, as detailed in Table 3.
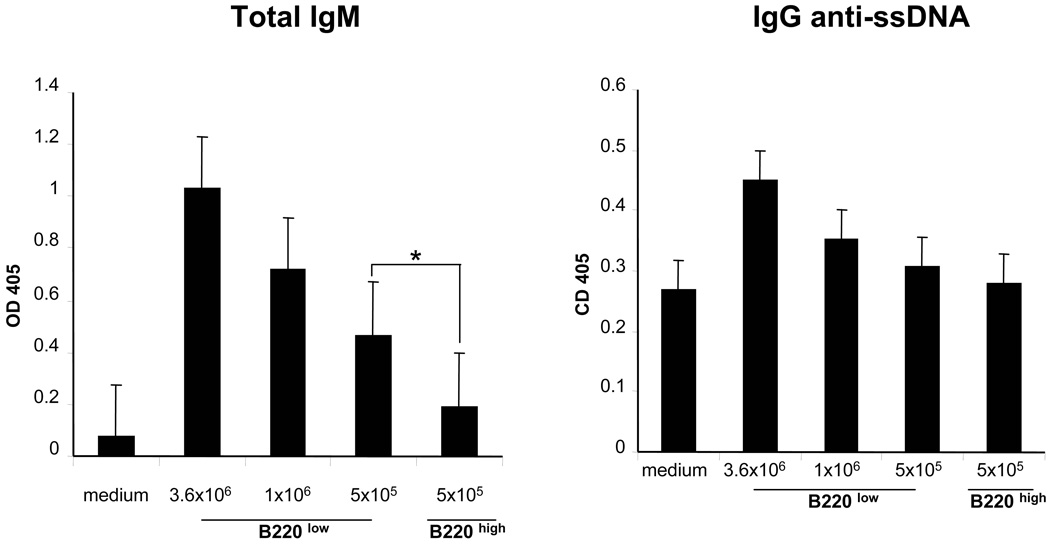
Figure 4. In vitro antibody production by B220int and B220hi B-cells from B6.56R/56R spleens.
Splenic B-cells were sorted based on surface B220 levels from B6.56R/56R mice and cultured at indicated cell numbers (per well) with LPS (20 ug/ml) for 48 h, after which culture supernatants were assayed for antibody levels by ELISA. Each bar represents the mean ± SEM of three experiments.
Shown in Table 3 are the HC and LC gene compositions noted in B6.56R/56R B-cells, juxtaposed to previously reported data from B6.56R/+ control (21). As is evident from Table 3, DNA-binding B-cells from both the B6.56R/+ and B6.56R/56R mice utilized the Tg HC, predominantly or exclusively, paired with a spectrum of LC partners, notably Vk20, Vk21 and Vk38c. When non-ANA B-cells were next examined (Table 3, bottom), interesting molecular differences were noted. More than 80% of the non-DNA binding B-cells from B6.56R/56R spleens possessed BCRs composed of the 56R HC Tg paired with either Vk20 or Vk21, both of which have previously been shown to be effective at vetoing DNA-binding (6). In contrast, about 70% of the non-ANA binding B-cells from B6.56R/+ spleens apparently “avoided” DNA-binding through the use of endogenous HC genes (21, Table 3). Hence, in the absence of an alternative (non targeted) HC locus, B-cells expressing a potentially self-reactive BCR HC appear to “avoid” self-reactivity exclusively through the use of DNA-vetoing LC partners. Very few of these B-cells showed evidence of mutations in the 56R Tg or evidence of HC replacement or D-invasion into the Tg (Table 3).
Table 3.
Heavy chain and light chain uasge analysis in hybridomas1
| B6.56R/+ | B6.56R/56R | P value (Chi-sqr) | ||
|---|---|---|---|---|
| ANA | Total cells (N) | 62 | 69 | |
| Heavy Chain amplified | ||||
| Endogenous HC (%) | 29 | 0 | 0.001 | |
| 56R Tg HC (%) | 71 | 100 | 0.001 | |
| % of 56R HC Tg with mutations | 2 | 5 | ns | |
| Tg HC invasion (N) | 2 | 1 | ns | |
| IgG in isotype (%) | 4 | 13 | 0.001 | |
| LC usage among B-cells with Tg HC | ||||
| %Vk4 | 5 | 5 | ns | |
| %Vk9 | 21 | 3 | 0.002 | |
| %Vk19 | 20 | 5 | 0.004 | |
| %Vk20 | 6 | 36 | 0.001 | |
| %Vk21 | 0 | 18 | 0.003 | |
| %Vk38c | 28 | 20 | ns | |
| %Other Vk | 20 | 13 | ns | |
| Jκ Usage among B-cells with Tg HC | ||||
| %Jk1,Jk2 | 40 | 28 | ns | |
| %Jk4,Jk5 | 60 | 72 | ns | |
| non-ANA | Total cells (N) | 27 | 21 | |
| Heavy Chain amplified | ||||
| Endogenous HC (%) | 70 | 0 | 0.001 | |
| 56R Tg HC (%) | 30 | 100 | 0.001 | |
| % of 56R HC Tg with mutations | 4 | 0 | ns | |
| Tg HC invasion (N) | 1 | 0 | ns | |
| IgG in isotype (%) | 11 | 5 | ns | |
| LC usage among B-cells with Tg HC | ||||
| %Vk9 | 19 | 6 | ns | |
| %Vk20 | 52 | 31 | ns | |
| %Vk21 | 0 | 56 | 0.001 | |
| %Vk38c | 6 | 0 | ns | |
| %Other Vk | 23 | 7 | ns | |
| Jκ Usage among B-cells with Tg HC | ||||
| %Jk1,Jk2 | 25 | 63 | 0.009 | |
| %Jk4,Jk5 | 75 | 37 | 0.009 |
Whereas the fusion data for the B6.56R/56R spleen was generated in this study, the data shown for the B6.56R/+ mouse has been recently reported elsewhere (21). "ns" = not significant
It was somewhat perplexing to note that whereas some B-cells expressing Vk20 paired with the Tg HC were DNA-reactive, others were not (Table 3). Interestingly, when the bt20 Vk20 germline gene was recombined to Jk4 it was associated with DNA-reactivity (when paired with the 56R HC); when the same Vk germline gene was recombined to Jk2, it was apparently not associated with DNA-reactivity (Fisher’s exact test P < 0.002). However, this difference alone is unlikely to account for the differential DNA reactivity, based on previous reports (20, 21). Likewise, it was intriguing that whereas some of the B-cells bearing the 56R HC Tg paired with the Vκ21 LC gene did not bind DNA, others clearly did (Table 3). When we compared these 2 specificities of B-cells, there were no significant differences in Jκ or Vκ germline gene usage between the DNA-binding and non-DNA binding B-cells. The specificity differences were not due to any somatic mutations since all Vκ21-expressing clones exhibited no mutations in the Tg HC or the LC partners (data not shown). Also, we could not amplify any Ig lambda from these B-cells. Currently, we do not know if these Vκ21-expressing B-cells differ in the expression of any additional LC partners that our PCR primers might have failed to amplify.
DISCUSSION
Developing pre-B-cells first re-arrange their HC locus during early B-cell development. In the event that the first VH gene randomly selected for rearrangement confers self-reactivity when paired with a LC partner, several molecular events occurring at different stages of B-cell development have the potential to revise the molecular makeup of the initial BCR so that it is no longer self-reactive. These non-mutually exclusive options include:
rearrangement at the alternate HC locus and usage of the alternate HC allele
pairing of the HC with an appropriate LC partner (either generated through primary recombination or through LC editing), so that DNA-reactivity is precluded, as discussed previously (6, 19)
editing the first HC allele so that it is no longer self-reactive (e.g., through VH replacement), as reported by Weigert and colleagues (10), and
mutation of the HC or the paired LC gene so that DNA-reactivity is abrogated.
When we recently derived B-cell hybridomas from B6 and B6.Sle2z congenics bearing the 56R anti-DNA Tg, the first 2 of the above mechanisms were evidently more commonly employed, compared to the latter options (21). Surprisingly, in that study, the vast majority of the B6.56R-derived non-DNA binding B-cells had apparently “avoided” DNA-reactivity through the rearrangement of the non-targeted HC locus (i.e., by adopting the first mechanism listed above). Though it was clear that this mechanism was commonly employed to maintain B-cell tolerance (at least on the B6 background), it was not apparent if this mechanism was absolutely essential. This study, designed to directly test this, illustrates that B-cells that are engineered to lack a non-rearranged alternate HC locus are more likely to remain autoreactive, presumably because the only available mechanism through which they can revise their initial BCR is through LC editing.
Previous studies using Tg-hemizygous 3H9/56R/76R mice (with progressively increasing avidity of the Tg for DNA) have shown that (a) increasing the B-cell avidity for DNA has a profound influence on the degree and nature of B-cell editing, and (b) the availability of a non-targeted HC locus is not sufficient to prevent autoimmunity in there mice (25, 26). Although the latter mechanism of tolerance by itself may not be sufficient at re-establishing tolerance, it appears that it may contribute incrementally in this direction, based on findings from the present work. When operative, the non-DNA binding HC substitute appears to originate predominantly from de novo rearrangement of the non-targeted locus, rather than through VH replacement at the targeted HC locus (21), and this difference may relate to the relative efficiencies of these two processes.
The phenotype of the B-cells observed in the Tg-homozygous mice points to a couple of interesting observations. First, the co-presence of the pre-arranged HC Tg at both loci did not alter total B-cell numbers, consistent with previous reports in HC Tg-homozygous mice (27). Also, the total IgM levels (i.e., IgMa + IgMb) on B-cells did not differ between the Tg-hemizygous and Tg-homozygous mice (Fig. 2, Table1), again in-keeping with previous reports in the literature (27). Presently, we do not know if the increased numbers of IgMa-hi B-cells in Tg-homozygous mice might arise in part from expression of HC from both alleles, but this appears to be unlikely based on earlier reports in other Ab-Tg-homozygous mice (27).
Though Tg-hemizygous and Tg-homozygous mice did not differ significantly in B-cell numbers or surface levels of total IgM, it was interesting that the Tg-homozygous mice exhibited an expanded population of B220int I-Ab-int B-cells. Since these B-cells were skewed toward being CD21−ve CD23−ve (Table 2), they may comprise of memory B-cells and/or pre-plasmablasts. Expansions of similar populations of B-cells have been noted in other autoimmune mouse models (28, 29). Consistent with this, sorted B220int I-Ab-int B-cells from Tg-homozygous mice tended to elaborate more ANAs in culture (Fig 4). Based on these findings, we envisage the following model. Precluding B-cells from rearranging and using the alternate HC allele may augment the pool of autoreactive B-cells, as observed in the Tg-homozygous mice. Continuous stimulation of these B-cells by self DNA might skew these cells towards plasmacytoid and memory B-cell differentiation, a scenario which is consistent with the B-cell phenotypes and elevated autoantibody levels seen in these mice.
Based on these observations and previous reports in the literature, we propose that newly developing B-cells (in a non-Tg, physiological setting) may employ 2 main mechanisms to revise their BCR if the initial HC/LC combination turned out to be self-reactive - rearrangement at the second HC locus or LC editing. In a context where anti-self B-cells have the luxury of rearranging a second HC locus, as well as editing the LC partner, as in the case in B6.56R/+ mice, the former mechanism is used by 70% of the B-cells that might have potentially been DNA-reactive (21, Table 3). Precluding the former tolerance mechanism from operating (as engineered in the B6.56R/56R double knock-in mice) may amplify the degree of autoimmunity that ensues, as demonstrated in the present study. In these mice, editing at the LC locus using strong DNA-vetoing LC such as Vk20 and Vk21 appear to be the only mechanism available for precluding DNA-reactivity, a finding that resonates well with previous reports in Tg-hemizygous mice by Weigert and colleagues (6). Hence, in addition to LC editing, rearrangement and usage of the second HC locus constitutes an important “safety valve” for B-cells whose primary BCR confers DNA-reactivity. In contrast to these mechanisms, editing the first rearranged HC locus (through HC replacement) and somatic mutation do not appear to be commonly employed for editing/revising self-reactive B cells.
REFERENCES
- 1.Goodnow CC, Cyster JG, Hartley SB, Bell SE, Cooke MP, Healy JI, Akkaraju S, Rathmell JC, Pogue SL, Shokat KP. Self-tolerance checkpoints in B lymphocyte development. Adv. Immunol. 1995;59:279–368. doi: 10.1016/s0065-2776(08)60633-1. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Radic MZ, Erikson J, Camper SA, Litwin S, Hardy RR, Weigert M. Deletion and editing of B cells that express antibodies to DNA. J. Immunol. 1994;152:1970–1982. [PubMed] [Google Scholar]
- 3.Verkoczy LK, Martensson AS, Nemazee D. The scope of receptor editing and its association with autoimmunity. Curr. Opin. Immunol. 2004;16:808–814. doi: 10.1016/j.coi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Yarkoni Y, Fischel R, Kat I, Yachimovich CN, Eilat D. Peripheral B cell receptor editing may promote the production of high-affinity autoantibodies in CD22-deficient mice. Eur. J. Immunol. 2006;36(10):2755–2767. doi: 10.1002/eji.200636190. [DOI] [PubMed] [Google Scholar]
- 5.Yachimovich N, Mostoslavsky G, Yarkoni Y, Verbovetski I, Eilat D. The efficiency of B cell receptor (BCR) editing is dependent on BCR light chain rearrangement status. Eur. J. Immunol. 2002;32(4):1164–1174. doi: 10.1002/1521-4141(200204)32:4<1164::AID-IMMU1164>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Jiang Y, Prak EL, Radic M, Weigert M. Editors and editing of anti-DNA receptors. Immunity. 2001;15:947–957. doi: 10.1016/s1074-7613(01)00251-5. [DOI] [PubMed] [Google Scholar]
- 7.Casellas R, Shih TA, Kleinewietfeld M, Rakonjac J, Nemazee D, Rajewsky K, Nussenzweig MC. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- 8.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prak EL, Weigert M. Light Chain Replacement: A New Model for Antibody Gene Rearrangement. J. Exp. Med. 1995;182:541–548. doi: 10.1084/jem.182.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Nagy Z, Prak EL, Weigert M. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 1995;3:747–755. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 11.Sekiguchi DR, Eisenberg RA, Weigert M. Secondary heavy chain rearrangement: a mechanism for generating anti-double stranded DNA B-cells. J. Exp. Med. 2003;197:27–39. doi: 10.1084/jem.20020737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh K, Meffre E, Albesiano E, Farber A, Dines D, Stein P, Asnis SE, Furie RA, Jain RI, Chiorazzi N. Immunoglobulin heavy chain variable region gene replacement as a mechanism for receptor revision in rheumatoid arthritis synovial tissue B lymphocytes. J. Exp. Med. 2000;192:1151–1164. doi: 10.1084/jem.192.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrero M, Clarke SH. Low-affinity anti-Smith antigen B cells are regulated by anergy as opposed to developmental arrest or differentiation to B-1. J. Immunol. 2002;168(1):13–21. doi: 10.4049/jimmunol.168.1.13. [DOI] [PubMed] [Google Scholar]
- 14.Erikson J, Mandik L, Bui A, Eaton A, Noorchashm H, Nguyen KAT, Roark JH. Self-reactive B cells in nonautoimmune and autoimmune mice. Immunol. Res. 1998;17:49–61. doi: 10.1007/BF02786430. [DOI] [PubMed] [Google Scholar]
- 15.Paul E, Lutz J, Erikson J, Carroll MC. Germinal center checkpoints in B cell tolerance in 3H9 transgenic mice. Int. Immunol. 2004;16:377–384. doi: 10.1093/intimm/dxh035. [DOI] [PubMed] [Google Scholar]
- 16.Pulendran B, Kannourakis G, Nouri S, Smith KG, Nossal GJ. Soluble antigen can cause enhanced apoptosis of germinal-centre B cells. Nature. 1995;375(6529):331–334. doi: 10.1038/375331a0. [DOI] [PubMed] [Google Scholar]
- 17.Shokat KM, Goodnow CC. Antigen-induced B-cell death and elimination during germinal-centre immune responses. Nature. 1995;375(6529):334–338. doi: 10.1038/375334a0. [DOI] [PubMed] [Google Scholar]
- 18.Roark JH, Kuntz CL, Nguyen KA, Mandik L, Cattermole M, Erikson J. B cell selection and allelic exclusion of an anti-DNA Ig transgene in MRL-lpr/lpr mice. J. Immunol. 1995;154:4444–4455. [PubMed] [Google Scholar]
- 19.Brard F, Shannon M, Prak EL, Litwin S, Weigert M. Somatic mutation and light chain rearrangement generate autoimmunity in anti-single-stranded DNA transgenic MRL/lpr mice. J. Exp. Med. 1999;190:691–704. doi: 10.1084/jem.190.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekiguchi DR, Yunk L, Gary D, Charan D, Srivastava B, Allman D, Weigert MG, Luning Prak E. Development and selection of edited B cells in B6.56R mice. J. Immunol. 2006;176:6879–6887. doi: 10.4049/jimmunol.176.11.6879. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Li LN, Kirthi RK, Xie C, Lightfoot S, Zhou XJ, Kearney JF, Weigert MG, Mohan C. Lupus susceptibility genes may breach tolerance to DNA by impairing receptor editing of nuclear antigen reactive B-cells. J. Immunol. 2007;179:1340–1352. doi: 10.4049/jimmunol.179.2.1340. [DOI] [PubMed] [Google Scholar]
- 22.Fukuyama H, Nimmerjahn F, Ravetch JV. The inhibitory Fc gamma receptor modulates autoimmunity by limiting the accumulation of immunoglobulin G(+) anti-DNA plasma cells. Nat. Immunol. 2005;6:99–106. doi: 10.1038/ni1151. [DOI] [PubMed] [Google Scholar]
- 23.Mohan C, Morel L, Yang P, Watanabe H, Croker B, Gilkeson G, Wakeland EK. Genetic dissection of SLE pathogenesis: A recipe for nephrophilic autoantibodies. J. Clin. Invest. 1999;103:1685–1695. doi: 10.1172/JCI5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang ZY, Xie C, Chen C, Kreska D, Hsu K, Li LN, Zhou XJ, Mohan C. Pathogenic Profiles and Molecular Signatures of Antinuclear Autoantibodies Rescued from NZM2410 Lupus Mice. J. Eex. Med. 2004;199(3):381–398. doi: 10.1084/jem.20030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radic MZ, Mackle J, Mol C, Anderson WF, Weigert M. Residues that mediate DNA binding of autoimmune antibodies. J. Immunol. 1993;150:4966–4977. [PubMed] [Google Scholar]
- 26.Chen C, Li H, Tian Q, Beardall M, Xu Y, Casanova N, Weigert M. J Immunol. 2006;176:5183–5190. doi: 10.4049/jimmunol.176.9.5183. [DOI] [PubMed] [Google Scholar]
- 27.Sonoda E, Pewzner-Jung T, Schwers S, Taki S, Jung S, Eilat D, Rajewsky K. B cell development under the condition of allelic inclusion. Cell. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 28.Paul E, Nelde A, Verschoor A, Carroll MC. Follicular exclusion of autoreactive B cells requires FcRIIb. International Immunol. 2007 Feb 16;:1–9. doi: 10.1093/intimm/dxm002. 2007. [DOI] [PubMed] [Google Scholar]
- 29.Shi X, Xie C, Chang S, Zhou XJ, Tedder T, Mohan C. CD19 hyperexpression augments Sle1-induced humoral autoimmunity but not clinical nephritis. Arthritis & Rheum. 2007;56:3057–3069. doi: 10.1002/art.22825. [DOI] [PubMed] [Google Scholar]