Abstract
A cell-free expression system using an Escherichia coli extract was adapted for large-scale expression and purification of mammalian membrane proteins. The system was tested with a set of human membrane proteins of different sizes, numbers of transmembrane domains, oligomeric arrangements, and native membrane locations. Tens of milligrams of protein were readily expressed and purified from an overnight cell-free reaction. Both reaction ‘mode A’ (proteins were expressed as precipitant) and ‘mode B’ (proteins were expressed in the presence of mild detergents to keep them soluble) were investigated. The combination of ‘mode B’ and the right detergents, used in the subsequent extraction and purification steps, is critical for obtaining properly folded proteins (CX32 and VDAC1) that can be crystallized and diffracted (VDAC1). The E. coli cell-free system is capable of efficient expression of many mammalian membrane proteins. However, fine-tuning of the system, especially to facilitate proper protein folding, will be required for each specific target.
Key Words: Cell-free expression, Escherichia coli, Membrane protein, X-ray structural analysis
Introduction
Cell-free protein synthesis was first used to understand the process of protein translation; especially to elucidate the nucleotide makeup of codons [Nirenberg and Matthaei, 1961]. Since then, it has become an important tool for studying protein expression and folding. In the post-genomic era, cell-free protein synthesis has found many applications in the area of genomics, proteomics, and synthetic biology. Recent developments of the cell-free platform have allowed for increased productivity and various post-translational modifications. The ease of manipulating the reaction components (energy source, labeled amino acids, lipids, detergents, etc.) and conditions (temperature, ionic strength, and redox environment, etc.) makes cell-free protein synthesis particularly amenable to automation and miniaturization, enabling application to the fields of protein arrays, in vitro evolution and multiplexed real-time labeling, among others [He, 2008; Katzen et al., 2005].
One of the important applications of cell-free protein synthesis is in the area of membrane protein production. About 30% of the genes in a genome are predicted to code for membrane proteins. However, to date, relatively few (∼200) unique membrane protein structures have been solved by X-ray crystallography. This discrepancy is largely due to the fact that large-scale production of membrane proteins in living cells is a challenging task. Expression of these membrane proteins frequently results in cell toxicity and/or disruption of the host cell membrane. In principle, cell-free expression can potentially overcome these obstacles. Several membrane proteins have been shown to be efficiently expressed and correctly folded using a cell-free platform. These include the bacterial transmembrane multidrug transporter EmrE [Chen et al., 2007; Elbaz et al., 2004], the bacterial mechanosensitive channel MscL [Berrier et al., 2004], several mammalian G-protein-coupled receptors [Klammt et al., 2007], and the plant solute transporter AtPPT1 [Nozawa et al., 2007].
Theoretically, extracts from any cell type can be used in a cell-free platform. The most common cell-free protein synthesis systems are derived from cell extracts of Escherichia coli, wheat germ, rabbit reticulocytes, and insect cells. Alternatively, a system derived from complete in vitroreconstitution of known elements, in replacement of cell extracts, has also been developed [Shimizu et al., 2001]. Among these, the E. coli cell-free system is the most simple, robust, highly productive, and cost-effective platform. The system has been used successfully to solve a multidrug transporter by X-ray crystallography [Chen et al., 2007]. It would be interesting to see if the platform can be used for the large-scale expression and subsequent structure determination of mammalian membrane proteins, whose structures are few and far between, despite their vast importance in drug discovery.
Results
Mammalian Membrane Proteins Are Efficiently Expressed in the E. coli Cell-Free System
To validate the usefulness of the E. coli expression system for the production of mammalian membrane proteins, we selected several human membrane proteins as model systems that differ in size, number of transmembrane domain, oligomeric arrangement, and native membrane location. These include the lysosome-located LAPTM4A, the hexameric connexin CX32, the large 12-TM glucose transporter GLUT4, and the mitochondria-located, voltage-dependent anion channel VDAC1 (table 1). Except for VDAC1, which is a beta-barrel protein, all these proteins are alpha helical.
Table 1.
Human membrane proteins used in this study: they differ in size, numbers of transmembrane domains, oligomeric arrangements, and native membrane locations
| Protein | MW (kDa) | Number of TM | Sub cellular location |
|---|---|---|---|
| LAPTM4A | 28 | 4 | lysosomal membrane |
| CX32 | 32 | 4 | plasma membrane |
| GLUT4 | 55 | 12 | plasma membrane |
| VDAC1 | 36 | − | outer mitochondrial membrane |
The expression system was adapted from the E. coli Expressway™ Milligram Cell-Free Expression System (Invitrogen, Carlsbad, Calif., USA). Several modifications were made to accommodate the specific characteristics of membrane proteins, and to support amino acid labeling techniques for X-ray crystallography (see Materials and Methods).
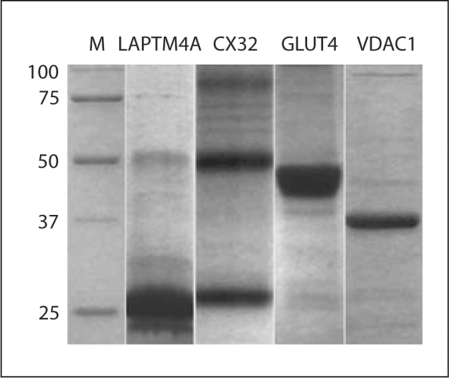
All mammalian membrane proteins included in this study were expressed at relatively high levels (fig. 1). The protein yields, when extracted with fos-choline 14 detergent, were about 0.5–1.0 mg/ml of cell-free reactions. The yields were comparable to those reported previously [Kigawa et al., 1999; Morita et al., 2003]. Our typical large-scale reaction mixture is about 100 ml. Therefore, we could readily obtain over 50 mg of highly purified proteins per overnight reaction. Because all the cell-free components (buffers, extracts, and DNA) can be prepared in large batches, the cell-free system can provide large quantity of protein in a very short period of time, facilitating fast and extensive screening of conditions for 3D crystal growth.
Fig. 1.
Expression and purification of human membrane proteins using the E. coli cell-free expression system. After the cell-free reaction, proteins were extracted with Fos-choline 14 detergent, and purified using a Ni-NTA column. Note the multimeric forms (1, 2 and 3× of MW) of CX32.
The Two Modes of Cell-Free Reactions Affect Yield and Quality of Membrane Proteins
In a typical cell-free reaction, membrane proteins are expressed as precipitates, which will then be solubilized and extracted, with appropriate detergents, after the reaction is finished. Recently, it has been shown that a mild detergent can be added to the reaction to keep the protein in solution during the reaction. The two schemes are referred to as ‘mode A’ and ‘mode B’, respectively [Klammt et al., 2006].
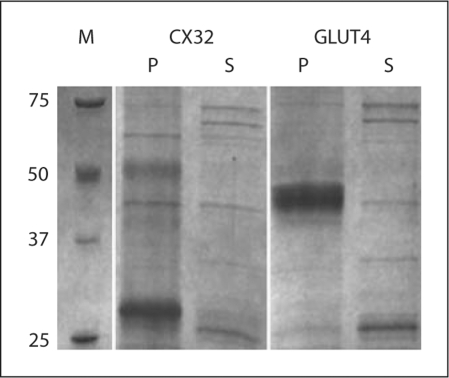
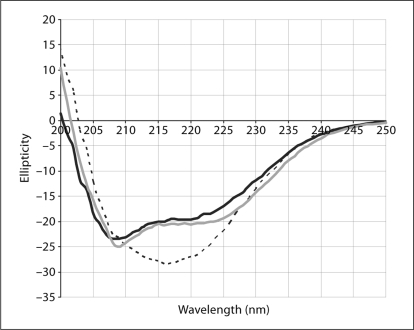
In ‘mode A’, all of the membrane protein targets were, as predicted, expressed as insoluble precipitates (fig. 2). The lipid-like fos-choline detergents (phospholipid analogs) are very efficient for the extraction/solubilization of precipitated membrane proteins. Therefore, they can be used as controls to accurately estimate the total expression level. Unfortunately, these detergents cause denaturation or misfolding of the purified proteins. We have extensively screened about 50 different commercially available detergents, but found no detergent, other than fos-cholines, capable of extracting our mammalian membrane protein targets expressed in ‘mode A’. Although the fos-choline-extracted membrane proteins can be easily purified to high degrees and form monodisperse species of the right apparent molecular weight, the quality of the protein is not suitable for protein crystallization. Far-UV circular dichroism (CD) spectra indicated that fos-choline irreversibly denatures VDAC1 (fig. 3). We postulate that it also denatures all other membrane protein targets. Attempts to exchange fos-cholines with other milder detergents, such as β-NG (n-nonyl-β-D-glucoside), β-DM (n-decyl-β-D-maltoside), β-DDM (n-dodecyl-β-D-maltoside), Cymal 6 (6-cyclohexyl-1-hexyl-β-D-maltoside), and LDAO (n-dodecyl-n,n-dimethylamine-n-oxide), for example, to refold the protein correctly, were not successful.
Fig. 2.
Membrane proteins are expressed as insoluble precipitate in ‘mode A’. They are found predominantly in the pelleted fraction of the cell-free reaction. M = Marker; P = pellet; S = supernatant.
Fig. 3.
Far-UV CD spectra of the VDAC1 protein in the ‘mode A’ reaction where VDAC1 was expressed as a precipitate and subsequently solubilized with Fos-choline detergents. The spectrum of VDAC1 ‘mode A’ (black solid line) is similar to that of SDS/denatured spec previously reported [Shao et al., 1996], and is similar to those of a high alpha-helical content protein such as LAPTM4A (control, grey solid line). In contrast, a CD spectrum of VDAC1 in ‘mode B’ (black dotted line) shows high beta-sheet content, indicating proper folding.
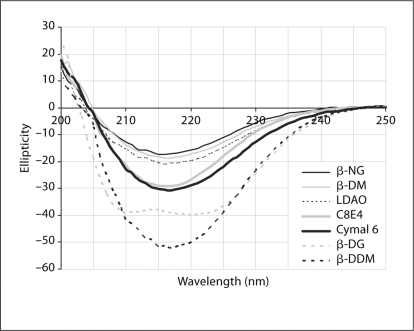
In ‘mode B’, a mild detergent is added to the cell-free reaction to help keep the membrane proteins in soluble forms. We have found that several polyoxyethylene detergents can be used for this purpose: Brij-35 (polyoxyethyleneglycol dodecyl ether, C12E23), Brij-58 (polyoxyethylene glycol hexadecyl ether, C16E20), and other Brij analogs with shorter tails such as polyoxyethylene(8)dodecyl ether (C12E8) and polyoxyethylene(9)dodecyl ether (C12E9). Up to 1% of these detergents can be added to the cell-free reaction without negative effects on the overall yield. After ‘mode B’ reaction, the proteins can be extracted and purified with the same detergent used in the reaction. Unlike in ‘mode A’, the proteins can also be extracted and purified with quite a few other detergents. In figure 3, we examine the circular dichroism spectra of VDAC1 expressed in ‘mode B’ in the present of 1% Brij-35, and subsequently extracted with a variety of secondary detergents. Because VDAC1 is a beta barrel protein, a CD spectrum of high beta-sheet content indicates proper folding, while a spectrum of high helix content indicates misfolding. In this experiment, certain detergents such as β-NG, β-DM, β-DDM, Cymal 6, LDAO, and C8E4 (tetraethylene glycol monooctyl ether), facilitate correct folding while other detergents such as β-DG (n-dodecyl-β-D-glucoside), β-OG (n-octyl-β-D-glucoside), Fos-cholines, CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate), and Triton X-100 (α-[4-(1,1,3,3-tetramethylbutyl)phenyl]-ω-hydroxy-poly(oxy-1,2-ethanediyl)) result in misfolded structures (fig. 4). Although Brij-35 is a detergent with a very low CMC (potentially causing incomplete exchange), we have determined with thin-layer chromatography (TLC) that the exchange reactions from Brij-35 to the secondary detergents were complete (data not shown).
Fig. 4.
Far-UV CD spectra of the VDAC1 protein obtained by the ‘mode B’ reaction where VDAC1 was expressed in the presence of Brij 35 and subsequently exchanged to a secondary detergent. Several detergents can be used while maintaining the high beta-sheet content, such as β-NG, β-DM, LDAO, C8E4, Cymal 6, and β-DDM. In contrast, some detergents such as β-DG can denature the protein after the exchange step. CD spectra with β-OG, Foscholines, CHAPS and Triton X-100 are similar to that of β-DG and are omitted in the graph for clarity.
Mammalian Membrane Proteins Expressed in E. coli Cell-Free Extract Can Be Properly Folded and Crystallized
We further investigated CX32 and VDAC1 to find out whether the mammalian membrane protein targets were properly folded and/or functional. The gap junction protein connexin is a hexamer in its native state [Maeda et al., 2009]. In figure 1, it is clear that some oligomeric forms of CX32 are readily detectable on protein gels. CX32 hemichannels of doughnut shape with dark-stained centers are readily seen under electron microscope, indicating that the cell-free expressed CX32 monomers are folded properly and form correct higher-order structures (data not shown).
As shown in a previous section, the CD spectra of VDAC1 indicated that it was properly folded when expressed in ‘mode B’ (fig. 4). Protein crystals were readily obtained for VDAC1 in various crystallographic trials (fig. 5). Currently, we are testing several small amphiphile detergents to be used as the secondary detergent in mode B scheme. These detergents have been shown to facilitate stabilization and improved X-ray diffraction resolution of several membrane proteins [Zhang et al., 2007, 2008]. The structures of VDAC1 have recently been determined by NMR and X-ray crystallography [Bayrhuber et al., 2008; Hiller et al., 2008; Ujwal et al., 2008]. In all of these structures, the protein was solubilized from inclusion body in an in vivo expression system. It will be very interesting to compare the structure obtained from the cell-free system against those from the in vivo expression mentioned above.
Fig. 5.

VDAC1 crystals. The human VDAC1 was expressed and purified in ‘mode B’ in the presence of Brij-35 and exchanged to NG on Ni-NTA resin. The protein was further purified with ultracentrifugation and gel-filtration. Purified protein was mono-dispersed with high beta-sheet content. Crystals were obtained under PEG 550 MME, 100 m M calcium chloride, 100 m M bicine buffer pH 8.5, 0.3% β-NG.
Discussion
The known structures of membrane protein are few and far between despite their abundance and their important roles in many cellular processes as well as their immense potentials in drug discovery. The major reason for this paucity is the difficult task of producing relatively large quantities of properly folded, biologically active membrane proteins, via cell-based expression systems. Cell-free protein synthesis has significant potential in this area. The cell-free system using E. coli extract provides a robust and relatively cheap platform that is amenable to scaling up, to produce the large quantities of proteins often required for structural determination.
We have shown that all of the mammalian membrane protein targets examined in this paper are efficiently produced with high yield. Our large-scale cell-free reactions routinely produce more than 50 mg of purified protein in a 100-ml overnight reaction. In our experience, the scale, speed and consistency of the cell-free platform is far superior to conventional in vivo expression.
‘Mode A’ can be used successfully for small-membrane proteins like the small bacterial drug transporter, EmrE, which was fully functional and yielded high-quality crystals [Chen et al., 2007]. Most membrane proteins having a molecular mass >30 kDa are usually expressed as insoluble precipitates using ‘mode A’, and cannot be extracted with, or exchanged to, a proper detergent that allows the protein to be properly refolded. Here, fos-choline detergents can efficiently solubilize and extract the membrane proteins, but they are usually denatured or misfolded in an irreversible manner. Generally, this material is not suitable for biophysical studies, and extensive screening for alternative detergents that can be used for solubilization and extraction has yielded few positive results. Screening of detergents that can exchange with fos-cholines to facilitate refolding are generally also unsuccessful. In our view, ‘mode A’ expression can be used successfully for small membrane proteins possessing a relatively simple fold, but not for larger and more complicated targets.
In ‘mode B’, the synthesized proteins are kept in a soluble form, using a mild detergent such as Brij-35 which does not inhibit the cell-free reaction. The Brij-35 detergent can be readily exchanged with several other detergents commonly used for membrane protein crystallization trials, such as β-OG, β-NG, β-DM, β-DDM, Cymal 6, LDAO, and C8E4. Using the beta-barrel protein, VDAC1, as an example, we have demonstrated that E. coli cell-free expression is capable of producing properly folded mammalian membrane protein targets. VDAC1 protein crystals were readily obtained using cell-free-produced materials. Being an open system, one can introduce small amphiphile detergents [Zhang et al., 2007] during protein translation in vitro, for example, to improve the stability and possibly the resolution of subsequent crystals.
The merits of cell-free protein synthesis for the production of membrane proteins have been well recognized. Cell-free systems adapted for this task will be continuously improved, to obtain large quantities of properly folded and biologically active proteins suitable for structural determination. It is highly unlikely that a one-size-fits-all cell-free system can be used for many membrane protein targets. However, the open nature of cell-free protein synthesis will enable rapid validation of novel modifications tailored to specific targets of interest.
Materials and Methods
Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, Mo., USA) unless otherwise stated. Detergents were purchased from Anatrace (Maumee, Ohio, USA). Full-length cDNA clones of membrane protein genes were obtained from the Mammalian Gene Collection (OpenBioSystems, Huntsville, Ala., USA) and cloned into the pIVEX-2.3d vector (Invitrogen, Carlsbad, Calif., USA). Several modular modifications were constructed from the basic pIVEX-2.3d, to accommodate various affinity tags and protease cleavage sites, at either N- or C-terminus, to facilitate different protein purification schemes. Linear DNA fragments could also be used in the cell-free protein expression reactions and were obtained by PCR amplification using the same templates and included the T7 promoter, ribosome binding site, His-tag at either the N- or C-terminus, optional protease cleavage sites, and ORF and T7 terminator sequences. Circular DNA fragments were preferred because they could readily be propagated and maintained. To support large-scale cell-free reactions, milligram quantities of DNA were isolated with fermentor-grown E. coli and with either a GenElute HP Plasmid Mega-prep Kit (Sigma-Aldrich, St. Louis, Mo., USA) or the QIAfilter Plasmid Giga Kit (Qiagen, Valencia, Calif., USA) using the instructions provided by the manufacturers.
Production of Cell-Free Lysate
E. coli BL21 cells were transformed with pAR1219 (a pBR322-based vector that expresses T7 RNA polymerase under control of the IPTG-inducible lac UV5 promoter) and grown in a 100-liter fermentor with appropriate antibiotics. Induction with 1 mM IPTG was done at OD 0.7–1.0, and harvested at OD 3.5–4.0. For a good lysate, it is important that the cells maintain a fast doubling time of about 30 min. Cells were harvested via centrifugation (8,000 g) and resuspended in 5 liters of cold S30 buffer (10 mM Tris-acetate, pH 8.0, 14 mM magnesium acetate, 60 mM potassium acetate, 1 mM DTT). The concentrated cells were then recentrifuged (8,000 g) and the pellet kept at −80°C.
To make cell-free extracts, 200 ml of S30 buffer was added to 200 g of frozen cells and stirred at 4°C until the cells were completely resuspended. DTT was added to 1 mM final concentration. The suspended cells were then passed though a M-110L microfluidizer processor (Microfluidics, Newton, Mass., USA). Broken cells were centrifuged at 30,000 g for 40 min. The supernatant (cell-free lysate) was collected and snap frozen with liquid nitrogen and kept at −80°C. Each batch of lysate was tested for efficiency by expressing green fluorescent protein and other known membrane proteins, like EmrE, in control reactions.
Cell-Free Protein Synthesis
Membrane proteins were expressed using a lysate based originally on the E. coli Expressway™ Milligram Cell-Free Expression system (Invitrogen, Carlsbad, Calif., USA). Modifications include: (1) the addition of 1–2% Brij-35 to solubilize the membrane proteins during the cell-free reaction, (2) the addition of any amino acids for labeling, like seleno-methionine at 2 mM, and (3) in some cases the direct incorporation of lipid or bicelles. Brij-58 and other brij analogs can also be used. The final concentration of circular or linear DNA template in the cell-free reaction was 5–10 μg/ml. Our cell-free reaction is composed of two components: the reaction buffer and lysate. The lysate, which includes the T7 polymerase, comprises 40% of the volume. The reaction buffer consists of 230 mM potassium glutamate, 58 mM Hepes-KOH, pH 7.5, 2% PEG 8,000, 80 mM ammonium acetate, 13.6 mM magnesium acetate, 1.2 mM ATP, 0.88 mM UTP, 0.88 mM CTP, 0.88 mM GTP, 0.65 mM cyclic adenosine monophosphate (cAMP), 1.7 mM dithiothreitol (DTT), 0.034 mg/ml folinic acid, 30 mM 3-phosphoglycerate, 2 mM of all the amino acids except for cysteine and methionine, which have a concentration of 1.5 mM, 0.17 mg/ml tRNA, 2 mM sodium oxalate, and 3.33 units of RNaseOUT™ ribonuclease inhibitor (Invitrogen, Carlsbad, Calif., USA). After a 2-hour incubation, the reaction is supplemented with additional components from the feed buffer, which consists of 58 mM Hepes-KOH, pH 8.0, 2 mM calcium chloride, 14 mM magnesium acetate, 230 mM potassium glutamate, 0.35 mM cAMP, 0.3 mM NAD, 1.7 mM DTT, 0.034 mg/ml folinic acid, 2 mM sodium oxalate, 30 mM glucose-6-phosphate, and the same concentration of amino acids as in the reaction buffer.
For screening, typical reaction sizes are approximately 1–2 ml (2–4 ml after feeding). For larger scale, the reactions were conducted in a volume of 45–90 ml (90–180 ml after feeding) using tissue culture flasks (Nalgene, Rochester, N.Y., USA). Reactions were incubated at 37°C with moderate shaking (80–100 rpm) overnight.
Protein Extraction and Purification
With a large-scale cell-free reaction of 180 ml, buffer A (20 mM Tris HCl, pH 8.0, 20 mM NaCl) was added to the overnight reaction to make up a final volume of 250 ml. In some cases, a second detergent can be introduced to enhance the extraction yield of protein from the cell-free reaction. Although this process is protein specific, the addition of a secondary detergent can increase the yield of soluble membrane protein very significantly. The reaction was centrifuged at 38,000 g for 20 min, and the supernatant was used for further purification.
Purification of his-tagged proteins was accomplished using Ni-NTA resin by either gravity flow or FPLC. The crude lysate was washed with several volumes of buffer A (20 mM Tris HCl, pH 8.0, 20 mM NaCl), followed by a high-salt wash (20 mM Tris HCl, pH 8.0, 200 mM NaCl) and 30 mM imidazole wash (20 mM Tris 8.0, 20 mM NaCl, 30 mM imidazole), before elution with 300 mM imidazole (20 mM Tris 8.0, 20 mM NaCl, 300 mM imidazole). Detergents at proper concentrations were present in all purification steps. Purified proteins were desalted with a desalting column and concentrated with amicon ultrafilters (Millipore, Billerica, Mass., USA) to an appropriate concentration (10–30 mg/ml) for further biophysical analysis. In some cases, the protein can be ultra-centrifuged with an Optima TLX ultracentrifuge (Beckman Coulter, Fullerton, Calif., USA) at 370,000 g. The ultracentrifugation step greatly reduces any aggregates and improves homogeneity. The N-terminal his-tag leader sequence was removed by overnight digestion with thrombin, factor Xa, or enterokinase depending on the template construct used. The final purified protein can be used for crystallization trials or biophysical analysis. For many of our samples, protein identity can be verified to within 1% of the expected molecular weight by MALDI-TOF mass spectrometry, or with N-terminal sequencing.
Gel Filtration
Purified protein (1–2 ml) was loaded on a gel filtration column (highload 16/60 with superdex 200 resin, GE Healthcare, Waukesha, Wisc., USA) and filtrated with buffer A in the present of the appropriate detergent. A mixture of gel filtration standards (Biorad Laboratories, Hercules, Calif., USA) were used to estimate the apparent molecular weights. Protein peaks were collected and analyzed on SDS-PAGE.
Crystallography
Purified proteins of concentration about 20 mg/ml were set up on crystal trays to screen for crystallization conditions using a combination of precipitants, including polyethylene glycol of various lengths, different salts, and different buffers. VDAC1 protein crystals appear at the following conditions: PEG 550 MME, 100 mM CaCl2, and 100 mM bicine pH 8.2–9.2.
Acknowledgements
This work was supported by a grant from the NIH Roadmap (GM073197) and the Skaggs Chemical Biology Foundation. We thank Dr. Yen-Ju Chen for his support and valuable discussion.
References
- Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrier C, Park KH, Abes S, Bibonne A, Betton JM, Ghazi A. Cell-free synthesis of a functional ion channel in the absence of a membrane and in the presence of detergent. Biochemistry. 2004;43:12585–12591. doi: 10.1021/bi049049y. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Pornillos O, Lieu S, Ma C, Chen AP, Chang G. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci USA. 2007;104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz Y, Steiner-Mordoch S, Danieli T, Schuldiner S. In vitro synthesis of fully functional EmrE, a multidrug transporter, and study of its oligomeric state. Proc Natl Acad Sci USA. 2004;101:1519–1524. doi: 10.1073/pnas.0306533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M. Cell-free protein synthesis: applications in proteomics and biotechnology. N Biotechnol. 2008;25:126–132. doi: 10.1016/j.nbt.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F, Chang G, Kudlicki W. The past, present and future of cell-free protein synthesis. Trends Biotechnol. 2005;23:150–156. doi: 10.1016/j.tibtech.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T, Yokoyama S. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999;442:15–19. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- Klammt C, Schwarz D, Eifler N, Engel A, Piehler J, Haase W, Hahn S, Dotsch V, Bernhard F. Cell-free production of G protein-coupled receptors for functional and structural studies. J Struct Biol. 2007;158:482–493. doi: 10.1016/j.jsb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Klammt C, Schwarz D, Lohr F, Schneider B, Dotsch V, Bernhard F. Cell-free expression as an emerging technique for the large scale production of integral membrane protein. FEBS J. 2006;273:4141–4153. doi: 10.1111/j.1742-4658.2006.05432.x. [DOI] [PubMed] [Google Scholar]
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- Morita EH, Sawasaki T, Tanaka R, Endo Y, Kohno T. A wheat germ cell-free system is a novel way to screen protein folding and function. Protein Sci. 2003;12:1216–1221. doi: 10.1110/ps.0241203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MW, Matthaei JH. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci USA. 1961;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa A, Nanamiya H, Miyata T, Linka N, Endo Y, Weber AP, Tozawa Y. A cell-free translation and proteoliposome reconstitution system for functional analysis of plant solute transporters. Plant Cell Physiol. 2007;48:1815–1820. doi: 10.1093/pcp/pcm150. [DOI] [PubMed] [Google Scholar]
- Shao L, Kinnally KW, Mannella CA. Circular dichroism studies of the mitochondrial channel, VDAC, from Neurospora crassa. Biophys J. 1996;71:778–786. doi: 10.1016/S0006-3495(96)79277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping P, Abramson J. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Horst R, Geralt M, Ma X, Hong WX, Finn MG, Stevens RC, Wuthrich K. Microscale NMR screening of new detergents for membrane protein structural biology. J Am Chem Soc. 2008;130:7357–7363. doi: 10.1021/ja077863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew Chem Int Ed Engl. 2007;46:7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]