Abstract
Background
The frequency of transfer of genes encoding resistance to antimicrobial agents was determined by conjugation in ESBL-producing and/or fluoroquinolone or aminoglycoside resistant Enterobacteriaceae clinical isolates at a tertiary care center in Lebanon. In addition, the role of tra genes encoding transferases in mediating conjugation was assessed.
Methods
Conjugation experiments were done on 53 ESBL-producing and/or fluoroquinolone resistant E. coli and K. pneumoniae and ESBL-producing S. sonnei isolates. Antimicrobial susceptibility testing on parent and transconjugant isolates, and PCR amplifications on plasmid extracts of the resistance-encoding genes: blaCTX-M-15 with the ISEcp1 insertion sequence, the aac(6')-lb-cr and qnrS genes, as well as tra encoding transferases genes were done. Random amplified polymorphic DNA (RAPD) analysis was performed to demonstrate whether conjugative isolates are clonal and whether they are linked epidemiologically to a particular source.
Results
Antimicrobial susceptibility testing on transconjugants revealed that 26 out of 53 (49%) ESBL-producing Enterobacteriaceae were able to transfer antimicrobial resistance to the recipients. Transfer of high-level resistance to the transconjugants encoded by the blaCTX-M-15 gene downstream the ISEcp1 insertion sequence against 3rd generation cephalosporins, and of low-level resistance against ciprofloxacin, and variable levels of resistance against aminoglycosides encoded by aac(6')-lb-cr gene, were observed in transconjugants. tra encoding transferase genes were detected exclusively in conjugative isolates.
Conclusion
In conclusion, the frequency of transfer of antimicrobial resistance in non clonal Enterobacteriaceae at the tertiary care center by conjugation was 49%. Conjugation occurred in isolates expressing the tra encoding transferase genes. Multiple conjugative strains harboring the plasmid encoded antimicrobial resistant genes were circulating in the medical center. Molecular epidemiology analysis showed that conjugative isolates are neither clonal nor linked to a particular site and transfer of antimicrobial resistance is by horizontal transfer of plasmids.
Background
Plasmid-encoded Extended-spectrum β-lactamases (ESBL) are increasingly spreading among Enterobacteriaceae clinical isolates throughout the world due mostly to their presence on highly conjugative plasmid. Surveys that were done in Canada, Greece, United Kingdom and Italy showed an association between the CTX-M type ESBL and resistance to other antimicrobial agents [1]. This was explained by a number of findings showing that blaCTX-M genes are commonly found on large plasmids that often carry other genes conferring resistance to other antimicrobial agents including aminoglycosides, fluoroquinolones, chloramphenicols, tetracyclins and others (particularly, blaOXA-1, blaTEM-1, tetA, aac(6')-lb-cr) [2,3]. CTX-M-15, one of the CTX-M enzymes which has an increased catalytic activity against ceftazidime[3], is now found worldwide especially in E. coli isolates from France, Canada and the United Kingdom [3-6].
pC15-1a, one of the plasmids responsible for the spread of the blaCTX-M-15 gene, was found associated with the Canadian outbreak, and its complete sequence was reported by Boyd et.al [4]. This plasmid was also found to harbor in addition to the blaCTX-M-15 gene, the aminoglycoside modifying enzyme encoding gene, aac(6')-lb-cr, which confers resistance to two unrelated antimicrobial agent classes, the aminoglycosides and the fluoroquinolones, by acetylating these drugs. The mechanism of resistance to fluoroquinolones encoded by aac(6')-lb-cr gene is different from that induced by other fluoroquinolones resistance encoding genes such as qnr genes [7]. In addition the role of plasmid mediated tra genes encoding transferase proteins was assessed.
In Lebanon, 96% of ESBL-producing E.coli and K.pneumoniae clinical isolates from various sources harbored blaCTX-M-15 and a number of these isolates carried also aac(6')-lb-cr gene [8]. The encoding genes of the two enzymes were found on pC15-1a, and other related plasmids. The blaCTX-M-15 gene located on these plasmids was found downstream of the ISEcp1 insertion sequence implicated in its expression [9]. A recent study[10] done on Shigella sonnei isolates revealed that 4 ESBL-producers had a plasmid harboring blaCTX-M-15 gene with the ISEcp1 but had no resistance to floroquinolones.
Based on the results of these studies and others, we aimed at determining the frequency of conjugation in 53 ESBL-producing and/or fluoroquinolone resistant E.coli and K.pneumoniae and 4 ESBL-producing S.sonnei isolates from a major tertiary care center, and confirming transfer of resistance to recipients by antimicrobial susceptibility testing and PCR amplification of the encoding resistance genes, ISEcp1-blaCTX-M-15, aac(6')-lb-cr and qnrS. In addition the role of plasmid encoded tra genes encoding transferase proteins in the transfer of antimicrobial resistance was investigated.
Methods
Bacterial strains
Fifty-three clinical isolates of ESBL producing E. coli (n = 25), K. pneumoniae (n = 24) and S. sonnei (n = 4) were collected at the Medical center and identified to the species level using the API 20E (Biomerieux, Marcy L'Etoile, France). These isolates were used as donor or parent strains in conjugation experiments.
Conjugation experiments
Parental strains and J53 E.coli were conjugated in Luria Bertani broth (Becton, Dickinson and company-BBL®) using 1:10 ratio. The transconjugants were selected using Mueller-Hinton II (Becton, Dickinson and company-BBL®) agar plates containing 100 mg/l sodium azide plus 2 mg/l cefotaxime[8].
Antimicrobial susceptibility testing
MICs of the antibiotics ceftazidime, cefpodoxime, cefotaxime, ciprofloxacin, gentamicin and kanamicin, for the parental strains and the transconjugants were determined using the E-test strips (AB Biodisk, Solna, Sweden). ESBLs detection was done by the combination disk method according to the CLSI guidelines [5], using disks containing ceftriaxone or ceftazidime with clavulanic acid (30/10 μg) and disks containing ceftazidime (30 μg) or ceftriaxone (30 μg) alone. ESBLs production was confirmed by a zone of inhibition for the combination disk that is at least 5 mm larger than that of the cephalosporin alone.
Plasmid extraction
Qiagen plasmid mini kit (GmbH, Hilden, Germany) was used to extract plasmids from the parental strains and their transconjugants according to manufacturers' specifications.
Polymerase Chain Reaction (PCR)
Primers were selected to amplify the aac(6')-lb-cr [11] and qnrS [12] genes, and the sequence containing the ISEcp1 insertion sequence and part of the blaCTX-M-15 gene (CTX-MA3 (5'-ACY TTA CTG GTR CTG CAC AT-3'), and the forward primer of the mobilizing insertion sequence ISEcp1U1 (5'-AAA AAT GAT TGA AAG GTG GT-3) as described previously [13] as well as the whole bla CTX-M-15 gene using the following primers: CTX-M-15-F (5'-GGTTAAAAAATCACTGCGTC-3') and CTX-M-15-R (5'-TTACAAACCGTCGGTGACGA-3'). The bla CTX-M-15 gene primers aligned with positions 3-876 of accession number FJ815288. Primers of all genes were used for PCR amplification on the extracted plasmids from the parents and the transconjugants. In addition primers amplifying the tra genes encoding the transferase proteins TraM, TraY, TraI to OriT, [14,15] and TraD [16] was performed on conjugative and non-conjugative isolates. PCR products were run on 1.5% agarose gel, stained with ethidium visualized under U.V. light and photographed using the dig-it-doc it software and Olympus digital camera. Sequence analysis was performed on all amplicons in a previous study [8].
Random amplified polymorphic DNA (RAPD)
RAPD analysis [8] was performed using the Ready-To-Go kit (GE healthcare, Buckinghamshire, UK ) according to manufacturer's specifications. Gels were analyzed by the BIONUMERICS (Applied Maths, Sint-Martens-Latem, Belgium).
Results
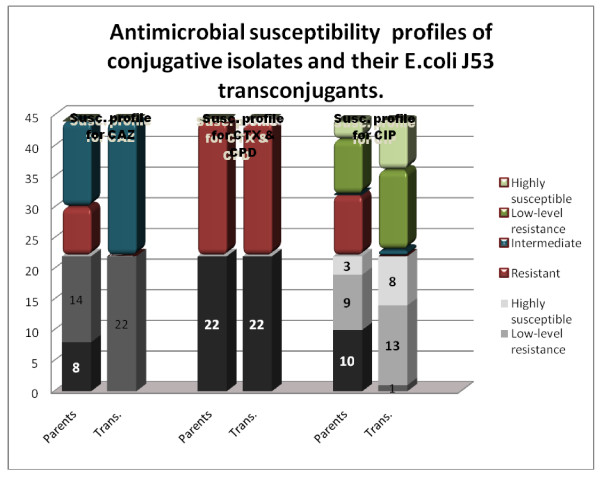
Our data showed that 26 of 53 (49%) E. coli and K. pneumoniae isolates were able to transfer resistance to recipients (Table 1). E-test done on 22 of the conjugative isolates showed that all of them expressed high-level resistance (with MIC of more than 256 μg/ml) to cefotaxime and cefpodoxime and were able to transfer this high-level resistance to the recipient strain. Eight and 14 of 22 revealed high-level resistance and intermediate resistance (MIC values between 16 and 32 μg/ml) to ceftazidime respectively; twenty one of the transconjugants expressed intermediate resistance and 1 revealed low-level resistance to ceftazidime (MIC equals to 8 μg/ml). Concerning resistance profile to ciprofloxacin, 10 and 9 out of the 22 isolates revealed high-level resistance (MIC of more than 32 μg/ml) and low-level resistance (MIC values ranging between 0.064 and 0.25 μg/ml, slightly higher than the MIC for the J53 E.coli before conjugation which is 0.047 μg/ml) respectively. Eight of the 10 resistant and 5 out of the 9 isolates with low-level resistance transferred low-level resistance against ciprofloxacin to the recipient strain. Only one transconjugant revealed intermediate resistance to ciprofloxacin with an MIC of 3 μg/ml (Figure 1, Table 1, 2).
Table 1.
MIC (μg/ml) of the antimicrobial agents, ceftazidime (CAZ), cefotaxime (CTX), cefpodoxime (CPD), ciprofloxacin (CIP), gentamicin (GEN) and kanamicin (KAN), and PCR amplification results of the resistance-encoding genes on selected conjugative isolates and their corresponding E. coli J53 (sodium azide-R) transconjugants, in comparison to their RAPD profiles.
| Isolate number (parents and transconjugants) | MIC of antimicrobial agents | Resistance-encoding genes | RAPD profile | |||||||
| CAZ | CPD | CTX | CIP | GEN | KAN | aac (6')lb-cr | ISEcp1-blaCTX-M-15 | qnrS. | ||
| E. coli J53 | 0.19 | 0.064 | 0.75 | 0.047 | 0.5 | 1.5 | - | - | - | |
| E. coli 1 | >256 | >256 | >256 | >32 | 96 | 12 | + | + | + | E-d |
| E. coli 1 T | 32 | >256 | >256 | 3 | 16 | 6 | + | + | + | |
| E. coli 3 | 16 | >256 | >256 | 0.125 | 32 | 12 | + | + | - | E-j |
| E. coli 3 T | 16 | >256 | >256 | 0.125 | 16 | 8 | + | + | - | |
| E. coli 6 | 24 | >256 | >256 | >32 | 1.5 | 16 | + | + | - | E-l |
| E. coli 6 T | 16 | >256 | >256 | 0.125 | 0.75 | 6 | + | + | - | |
| E. coli 8 | 16 | >256 | >256 | >32 | 128 | 32 | + | + | - | E-r |
| E. coli 8 T | 16 | >256 | >256 | 0.094 | 12 | 8 | + | + | - | |
| E. coli 11 | 24 | >256 | >256 | >32 | >256 | 64 | + | + | - | E-l |
| E. coli 11 T | 16 | >256 | >256 | 0.125 | 16 | 16 | + | + | - | |
| E. coli 12 | 24 | >256 | >256 | >32 | 64 | 24 | + | + | - | E-l |
| E. coli 12 T | 12 | >256 | >256 | 0.125 | 16 | 4 | + | + | - | |
| E. coli 14 | 16 | >256 | >256 | 0.094 | 16 | 3 | + | + | - | E-b |
| E. coli 14 T | 8 | >256 | >256 | 0.047 | 1.5 | 6 | + | + | - | |
| E. coli 15 | 24 | >256 | >256 | >32 | 1.5 | 24 | + | + | - | E-q |
| E. coli 15 T | 24 | >256 | >256 | 0.125 | 1.5 | 4 | + | + | - | |
| K. pneumoniae 1 | >256 | >256 | >256 | >32 | 128 | 48 | + | + | - | K-e |
| K. pneumoniae 1 T | 32 | >256 | >256 | 0.125 | 16 | 8 | + | + | - | |
| K. pneumoniae 2 | 32 | >256 | >256 | 0.25 | >256 | 32 | + | + | - | K-o |
| K. pneumoniae 2 T | 16 | >256 | >256 | 0.125 | 12 | 8 | + | + | - | |
| K. pneumoniae 13 | 96 | >256 | >256 | 0.5 | 1.5 | 4 | + | + | - | K-g |
| K. pneumoniae 13 T | 24 | >256 | >256 | 0.047 | 0.5 | 4 | + | + | - | |
| S. sonnei 1 | 12 | >256 | >256 | 0.047 | 2 | 8 | - | + | - | S-a |
| S. sonnei 1 T | 12 | >256 | >256 | 0.047 | 0.5 | 1.5 | - | + | - | |
| S. sonnei 3 | 12 | >256 | >256 | 0.047 | 2 | 8 | - | + | - | S-a |
| S. sonnei 3 T | 12 | >256 | >256 | 0.047 | 0.5 | 1.5 | - | + | - | |
| S. sonnei 4 | 12 | >256 | >256 | 0.047 | 2 | 8 | - | + | - | S-a |
| S. sonnei 4 T | 12 | >256 | 96 | 0.047 | 0.5 | 1.5 | - | + | - | |
Figure 1.
Antimicrobial susceptibility profiles done on the 22 conjugative isolates (parents) and their transconjugants for the antimicrobial agents, ceftazidime (CAZ), cefotaxime (CTX), cefpodoxime (CPD) and ciprofloxacin (CIP).
Table 2.
RAPD profiles of non-conjugative isolates.
| Isolate number | RAPD profile |
| E. coli 2 | E-e |
| E. coli 4 | E-r |
| E. coli 5 | E-f |
| E. coli 7 | E-r |
| E. coli 9 | E-s |
| E. coli 10 | E-k |
| E. coli 13 | E-q |
| E. coli 16 | E-l |
| E. coli 17 | E-k |
| E. coli 18 | E-m |
| E. coli 19 | E-c |
| E. coli 23 | E-j |
| E. coli 21 | E-d |
| E. coli 22 | E-c |
| E. coli 23 | E-j |
| K. pneumoniae 3 | K-o |
| K. pneumoniae 4 | K-m |
| K. pneumoniae 7 | K-g |
| K. pneumoniae 8 | K-g |
| K. pneumoniae 12 | K-n |
| K. pneumoniae 14 | K-g |
| K. pneumoniae 16 | K-g |
| K. pneumoniae 21 | K-n |
| K. pneumoniae 22 | K-h |
| K. pneumoniae 24 | K-i |
PCR detection confirmed the transfer of the blaCTX-M-15 gene together with the ISEcp1 insertion sequence and the aac(6')-lb-cr gene from the parental strains to their recipients. Only one isolate harbored the qnrS gene and was able to transfer it together with the aac(6')-lb-cr gene to its recipient that expressed intermediate resistance to ciprofloxacin (Table 1). PCR amplification of the tra genes was positive exclusively in the conjugative isolates.
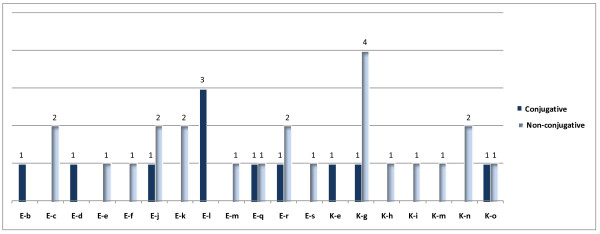
RAPD analysis (Table 1, Figures 2A,B, 3) showed that E. coli and K. pneumoniae conjugative isolates are not clonal. Multiple strains are common to both conjugative and non-conjugative isolates. S. sonnei isolates are clonal. Epidemiological data in conjunction with molecular analysis confirmed that transfer of resistance among conjugative strains is merely by horizontal transfer of antimicrobial resistance plasmid encoded determinants and that strains were linked to multiple sources.
Figure 2.
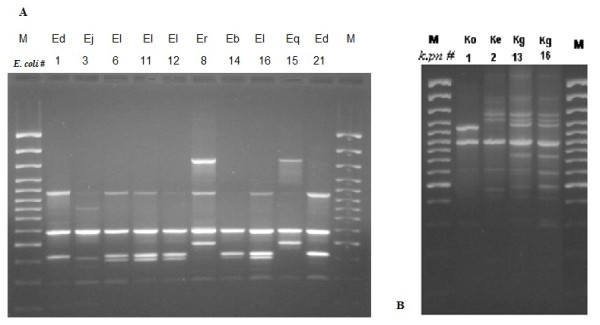
A Representative RAPD gel on E. coli isolates (RAPD types Ed, Ej, El Er Eb El Eq, Ed with corresponding numbers in Table 1&2). M: 100 bp ladder. B Representative RAPD gel on K. pneumoniae isolates (RAPD types Ko, Ke, Kg, with corresponding numbers in Tables 1&2)
Figure 3.
Histogram showing the frequency of conjugative and non-conjugative isolates corresponding to each RAPD profile.
Discussion
In this study, we determined the frequency and mode of transfer of resistance to antimicrobial agents by conjugation, among a number of multi-drug resistant Enterobacteriaceae isolates. These are all ESBL producing and/or fluoroquinolone and aminoglycoside resistant. Most of these isolates harbor the pC15-1a plasmid, that carries the blaCTX-M-15 gene in addition to the aminoglycoside modifying enzyme aac(6')lb-cr gene which also confers some resistance to the fluoroquinolone ciprofloxacin as demonstrated in a previous study [8]. The MDR region is [8] indeed harbored on the pC151a or the pCTX15 plasmids since first, the plasmids characterization done in the previous study showed that the hpa1 digestion profile was related to the pC151a or the pCTX15 plasmids and second the PCR based detection of the bla CTX-M-15 and the aac6'lb-cr genes on transferable plasmids in transconjugants were on pC15-1a or the pCTX-15 plasmids.
Molecular epidemiology analysis confirmed that conjugative isolates are not clonal and transfer of antimicrobial resistance was acquired through horizontal transfer of plasmids. Besides conjugative strains were linked epidemiologically to multiple sources. No single conjugative strain harboring the plasmid encoded antimicrobial resistant genes was circulating in the medical center.
Conjugation experiments in this study have shown that about 50 percent of the Enterobacteriaceae isolates were able to transfer resistance to cefotaxime, which is a significantly high prevalence rate of transfer of resistance among ESBL producing multi-drug resistant isolates. As demonstrated, conjugative isolates harbored the plasmid encoded tra genes encoding the transferase proteins TraM responsible for mating aggregation, TraY directing the nicking enzyme TraI to OriT, [14] TraI inducing nicking and unwinding [15] and TraD involved in pumping the DNA into the recipient cells [16] and hence were able to transfer resistance.
All the isolates were resistant to the 3rd generation cephalosporins, and conjugative ones were able to transfer resistance against these antimicrobial agents to the recipients. Most of the isolates were able to transfer high-level resistance against cefotaxime and cefepodoxime to the transconjugants which revealed MIC equal or above 256. It was found however, that the MIC of ceftazidime for most of the donor isolates was less than that usually reported for CTX-M-15 producing isolates (between 128 and 256 μg/ml) (1). CTX-M-15 differs from its relative CTX-M-3 by a single amino-acid substitution, which increases its activity against ceftazidime[17]. Most of the bla¬CTX-M-15 genes encountered in ESBL- producing isolates are located on plasmids downstream an ISEcp1 insertion sequence which harbors the -10 and -35 promoter sequences (TTGACA and TAAACT respectively) essential for the high expression of the gene and hence, the increased activity against ceftazidime and other third generation cephalosporins. These were detected in a previous study [8]. However, CTX-M-15-producing strains with low activity against ceftazidime were previously reported in many countries including UK and the United Arab Emirates [5,8]. A study that was done by Wooford et. al on the UK isolates, showed that there is a number of CTX-M-15-producing E. coli for which ceftazidime had MIC values equal or less than 32 μg/ml, which is lower than the usually reported for the CTX-M-15 producing Enterobacteriaceae (between 128 and 256 μg/ml) [18]. Genotypic characterization by Woodford et al.[5] demonstrated that the blaCTX-M-15 gene was located on a plasmid downstream the ISEcp1; however, an IS26 insertion element was present within the terminal inverted repeat of ISEcp1, separating blaCTX-M-15 from its usual promoter located in the ISEcp1. This genotypic characteristic was thought to be responsible for the lower blaCTX-M-15 expression, and hence, the decreased ceftazidime MIC for the UK and the UAE strains. We suspect that the same genotypic feature is also responsible for the level of resistance against ceftazidime that is lower than expected in our CTX-M-15 producing strains.
All the transconjugants on the other hand, had intermediate resistance to ceftazidime. The seven isolates that were resistant to ceftazidime with MIC above 96 were unable to transfer this resistance to the recipients, for which the MIC of ceftazidime did not cross 32 μg/ml. This could have resulted from the presence of chromosomally-mediated β-lactamase genes in the parental strains that could not be transferred to the recipients.
With respect to the transfer of resistance to ciprofloxacin, it was noticed that all except one of the transconjugants revealed low-level resistance, resulting from the transfer of aac(6')-lb-cr. This gene has an aminoglycoside-modifying activity, and crossed the antimicrobial agents' class boundaries by its activity also against the fluoroquinolones. Although the degree of resistance conferred is small, this gene was shown to act additively with other genes that confer resistance against fluoroquinolones, like the plasmid-mediated qnr genes, or chromosomal mutations. Despite the relatively low activity of the aac(6')-lb-cr gene product against fluoroquinolones, it could play an important role in facilitating the selection of strains with mutations in the chromosomal gyrA, gyrB, and parC genes, among a bacterial population that is exposed to be fluoroquinolone resistant[19].
The qnr genes are other important plasmid-mediated genes that confer resistance to fluoroquinolones. qnrA is the first discovered and is found worldwide. qnrB and qnrS are recently emerging, and are still not as widespread [20]. Only one of our isolates, E. coli 1, was able to transfer intermediate-level resistance against ciprofloxacin to transconjugants, with an MIC in the transconjugant of 3 μg/ml; whereas in all the other transconjugants, ciprofloxacin had a maximum MIC value of 0.125 μg/ml. We suspected the presence of one of the plasmid-mediated qnr genes in E. coli 1 in addition to the aac(6')-lb-cr, and indeed, PCR amplification on the plasmid extract from this isolate confirmed the presence of both genes in the donor and the transconjugant. The higher MIC of ciprofloxacin in the E. coli 1 transconjugant compared to that of the transconjugants of the other isolates, leading to intermediate-level ciprofloxacin resistance is most probably due to the simultaneous transfer of both the qnrS and aac(6')-lb-cr genes to them, and hence, the coordinating action of 2 different fluoroquinolone-resistance mechanisms. It is known that the qnrS gene by itself does not confer intermediate-level resistance to ciprofloxacin [21,22].
In conclusion 49% of the E. coli and K. pneumoniae of non-clonal conjugative isolates in our medical center were able to transfer resistance to third generation cephalosporins in transconjugants as encoded by the blaCTX-M-15 gene along with its promoter in the ISEcp1 insertion element. In addition the isolates were also able to transfer a low-level resistance to ciprofloxacin and variable resistance to aminoglycosides, encoded by aac(6')-lb-cr gene. Conjugation occurred in isolates expressing the tra encoding transferase genes. Multiple conjugative strains harboring the plasmid encoded antimicrobial resistant genes were circulating in the medical center.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MH participated in designing the experiments, executing them, performing data analysis and writing the manuscript. MT K supervised conjugation and molecular experiments. JEC provided J53 E. coli and experimental procedures. GEF provided phenotypically identified bacterial isolates. GMM (senior author) designed experimental settings, finalized data analysis and the writing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Mohamad Harajly, Email: mh122@aub.edu.lb.
Marie-Therese Khairallah, Email: mk18@aub.edu.lb.
John E Corkill, Email: jcorkill@liv.edu.
George F Araj, Email: garaj@aub.edu.lb.
Ghassan M Matar, Email: gmatar@aub.edu.lb.
Acknowledgements
Thanks are due to the Faculty of Medicine Medical Practice Plan (MPP), American University of Beirut, for financial support.
References
- Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob.Agents Chemother. 2004;48(1):1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leflon-Guibout V, Jurand C, Bonacorsi S, Espinasse F, Guelfi MC, Duportail F, Heym B, Bingen E, Nicolas-Chanoine MH. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob.Agents Chemother. 2004;48(10):3736–3742. doi: 10.1128/AAC.48.10.3736-3742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch.Intern.Med. 2005;165(12):1430–1435. doi: 10.1001/archinte.165.12.1430. [DOI] [PubMed] [Google Scholar]
- Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. Complete Nucleotide Sequence of a 92-Kilobase Plasmid Harboring the CTX-M-15 Extended-Spectrum Beta-Lactamase Involved in an Outbreak in Long-Term-Care Facilities in Toronto, Canada. Antimicrob.Agents Chemother. 2004;48(10):3758–64. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. qnrB, another plasmid-mediated gene for quinolone resistance. 2006. pp. 1178–1182. [DOI] [PMC free article] [PubMed]
- Karisik E, Ellington MJ, Pike R, Warren RE, Livermore DM, Woodford N. Molecular characterization of plasmids encoding CTX-M-15 beta-lactamases from Escherichia coli strains in the United Kingdom. J.Antimicrob.Chemother. 2006;58(3):665–668. doi: 10.1093/jac/dkl309. [DOI] [PubMed] [Google Scholar]
- Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006;12(1):83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- Kanj SS, Corkill JE, Kanafani ZA, Araj GF, Hart CA, Jaafar R, Matar GM. Molecular characterisation of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. isolates at a tertiary-care centre in Lebanon. Clin.Microbiol.Infect. 2008;14(5):501–504. doi: 10.1111/j.1469-0691.2008.01964.x. [DOI] [PubMed] [Google Scholar]
- Poirel L, Decousser JW, Nordmann P. Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) beta-lactamase gene. Antimicrob.Agents Chemother. 2003;47(9):2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matar GM, Jaafar R, Sabra A, Hart CA, Corkill JE, Dbaibo GS, Araj GF. First detection and sequence analysis of the blaCTX-M-15 gene in Lebanese isolates of extended-spectrum-beta-lactamase-producing Shigella sonnei. Ann.Trop.Med.Parasitol. 2007;101(6):511–517. doi: 10.1179/136485907X193860. [DOI] [PubMed] [Google Scholar]
- Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6')-Ib-cr Encoding a Ciprofloxacin-Modifying Enzyme. Antimicrob.Agents Chemother. 2003;50(11):3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir V, Weill FX, Poirel L, Fabre L, Soussy CJ. Nordmann P. Prevalence of qnr genes in Salmonella in Francer. J. Antimicrob. Chemother. 2007;59:751–754. doi: 10.1093/jac/dkl547. [DOI] [PubMed] [Google Scholar]
- Saladin M, Bao Cao VT, Lambert T, Donay JL, Herrmann JL, Ould-Hocine Z, Verdet C, Delisle F, Philippon A, Arlet G. Diversity of CTX-M L-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 2002;209:161–168. doi: 10.1111/j.1574-6968.2002.tb11126.x. [DOI] [PubMed] [Google Scholar]
- Reygers U, Wessel R, Muller H, Hoffmann-Berling H. Endonuclease activity of Escherichia coli DNA helicase I directed against the transfer origin of the F factor. EMBO J. 1991;10(9):2689–2694. doi: 10.1002/j.1460-2075.1991.tb07812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler BA, Minkley EG Jr. Evidence that DNA helicase I and oriT site-specific nicking are both functions of the F Tral protein. J. Mol. Biol. 1998;204:205–209. doi: 10.1016/0022-2836(88)90609-2. [DOI] [PubMed] [Google Scholar]
- Panicker MM, Minkley EG Jr. DNA transfer occurs during a cell surface contact stage of F sex factor-mediated bacterial conjugation. J.Bacteriol. 1985;162(2):584–590. doi: 10.1128/jb.162.2.584-590.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Gniadkowski M, Nordmann P. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum-β-lactamase CTX-M-15 and of its structurally related β-lactamase CTX-M-3. J. Antimicrob. Chemother. 2002;50(A):1031–1034. doi: 10.1093/jac/dkf240. [DOI] [PubMed] [Google Scholar]
- Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, Johnson AP, Pike R, Warner M, Cheasty T, Pearson A, Harry S, Leach JB, Loughrey A, Lowes JA, Warren RE, Livermore DM. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J.Antimicrob.Chemother. 2004;54(4):735–743. doi: 10.1093/jac/dkh424. [DOI] [PubMed] [Google Scholar]
- Cattoir Vincent, Weill Francois-Xavier, Poirel Laurent, Fabre Lae¨titia, Soussy Claude-James, Patrice Nordmann. Prevalence of qnr genes in Salmonella in France. J.Antimicrob.Chemother. 2007;59:751–754. doi: 10.1093/jac/dkl547. [DOI] [PubMed] [Google Scholar]
- Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J.Antimicrob.Chemother. 2005;56(3):463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Friederichs S, de Jong A, Michael GB, Schwarz S. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J.Antimicrob.Chemother. 2006;58(1):18–22. doi: 10.1093/jac/dkl213. [DOI] [PubMed] [Google Scholar]
- Kassis-Chikhani N, Vimont S, Asselat K, Trivalle C, Minassian B, Sengelin C, Gautier V, Mathieu D, Dussaix E, Arlet G. CTX-M beta-lactamase-producing Escherichia coli in long-term care facilities, France. Emerg.Infect.Dis. 2004;10(9):1697–1698. doi: 10.3201/eid1009.030969. [DOI] [PMC free article] [PubMed] [Google Scholar]