Abstract
Hemizygous deletions of the chromosome 22q11.2 region result in the 22q11.2 deletion syndrome also referred to as DiGeorge, Velocardiofacial or Shprintzen syndromes. The phenotype is variable but commonly includes conotruncal cardiac defects, palatal abnormalities, learning and behavioral problems, immune deficiency, and facial anomalies. Four distinct highly homologous blocks of low copy number repeat sequences (LCRs) flank the deletion region. Mispairing of LCRs during meiosis with unequal meiotic exchange is assumed to cause the recurrent and consistent deletions. The proximal LCR is reportedly located at 22q11.2 from 17.037 to 17.083 Mb while the distal LCR is located from 19.835 to 19.880 Mb. Although the chromosome breakpoints are thought to localize to the LCRs, the positions of the breakpoints have been investigated in only a few individuals. Therefore, we used high resolution oligonucleotide-based 244K microarray comparative genomic hybridization (aCGH) to resolve the breakpoints in a cohort of 20 subjects with known 22q11.2 deletions. We also investigated copy number variation (CNV) in the rest of the genome. The 22q11.2 breaks occurred on either side of the LCR in our subjects, although more commonly on the distal side of the reported proximal LCR. The proximal breakpoints in our subjects spanned the region from 17.036 to 17.398 Mb. This region includes the genes DGCR6 (DiGeorge syndrome critical region protein 6) and PRODH (proline dehydrogenase 1), along with three open reading frames that may encode proteins of unknown function. The distal breakpoints spanned the region from 19.788 to 20.122 Mb. This region includes the genes GGT2 (gamma-glutamyltransferase-like protein 2), HIC2 (hypermethylated in cancer 2), and multiple transcripts of unknown function. The genes in these two breakpoint regions are variably hemizygous depending on the location of the breakpoints. Our 20 subjects had 254 CNVs throughout the genome, 94 duplications and 160 deletions, ranging in size from 1 kb to 2.4 Mb. The presence or absence of genes at the breakpoints depending on the size of the deletion plus variation in the rest of the genome due to CNVs likely contribute to the variable phenotype associated with the 22q11.2 deletion or DiGeorge syndrome.
Introduction
Hemizygous deletions of the chromosome 22q11.2 region typically result in conotruncal cardiac anomalies, moderate immune deficiency, developmental delay and behavioral problems, facial dysmorphia, palatal dysfunction and feeding difficulties (Kobrynski and Sullivan, 2007; Ben-Shachar et al., 2008; Shprintzen, 2008). The clinical phenotype is variable and severity of symptoms range from mild, near-normal to life threatening. Stochastic, environmental and genetic modifiers likely account for the wide variation seen in 22q11.2 deletion syndrome (also known as DiGeorge, Velocardiofacial or Shprintzen syndrome), but no mechanistic explanation currently exists.
The 22q11.2 deletion syndrome results most commonly from a 3-Mb deletion of the chromosome 22q11.2 region (about 90% of cases); less common is a 1.5-Mb deletion (about 7%) (Carlson et al., 1997; Kates et al., 2005). Four distinct highly homologous blocks of low copy number repeats (LCRs) flank the deletion region (Shaikh et al., 2000) which can lead to mispairing of the LCRs during meiosis resulting in unequal meiotic exchange and hypothesized to cause the deletion (Shaikh et al., 2000; Baumer et al., 2004). The proximal LCR located in the 22q11.2 region resides between 17.037 and 17.083 Mb (based on the human genome release version, hg17) and the distal LCR is located between 19.835 and 19.880 Mb. Each LCR is ∼40 kb in length. Although the chromosome breakpoints associated with the 22q11.2 deletion are reported to localize to the LCRs, actual breakpoints have been investigated in only a few individuals (Mantripragada et al., 2004; Urban et al., 2006).
Resolution of genetic techniques to identify alterations range from 500 kb to 1 Mb in size using microsatellites (Carlson et al., 1997); approximately 25 kb or less for high resolution CGH arrays (Mantripragada et al., 2004); several hundred nucleotides for tiling CGH arrays (Urban et al., 2006) and single nucleotide changes accomplished by sequencing breakpoint junctions (Shaikh et al., 2000). Whole genome microarrays are now available commercially for high resolution analysis of copy number variation (CNV).
Array comparative genomic hybridization (aCGH) using high resolution oligonucleotide-based microarrays allow breakpoint resolution of approximately 10 kb and in some regions of the genome, higher resolution depending on local probe density. In addition, recent studies of the human genome have found a greater than expected level of copy number variability which may contribute to genetic diversity and/or human disease (Barrett et al., 2004; Conrad et al., 2006; Fiegler et al., 2006; Khaja et al., 2006; Komura et al., 2006; Perry et al., 2006; Redon et al., 2006; Sebat et al., 2007; Stranger et al., 2007). We evaluated a cohort of 20 subjects with known 22q11.2 deletions using high resolution aCGH. Our goal was to more precisely localize the proximal and distal 22q11.2 deletion breakpoints in our cohort of subjects using this technology and assess the location, size and number of CNVs in this classical cytogenetic disorder.
Materials and methods
Subjects
Our study group consisted of 20 anonymous subjects (sex was recorded to facilitate aCGH) with known 22q11.2 deletions who had been referred for cytogenetic diagnosis. IRB approval was obtained for this retrospective study. Diagnosis was determined by GTG-band conventional cytogenetic analysis and/or fluorescence in situ hybridization (FISH).
Methods
DNA was isolated from peripheral blood using a Puregene DNA isolation kit (Qiagen Inc., Valencia, CA) following the manufacturer's protocol. The Agilent 244K human whole genome microarray was used with an average distance of 6.4 kb between probes (Agilent Technologies, Santa Clara, CA). Comparison genomic DNA was obtained commercially (Promega, Madison, WI) and matched for sex. Detailed methods for genomic DNA preparation, labeling, hybridization and scanning can be found at website, http://www.home.agilent.com.
Copy number variants were identified with CGH Analytics software using the Aberration Detection Method 2 (ADM-2) analysis algorithm (Agilent Technologies, Santa Clara, CA) as previously described (Butler et al., 2008). The ADM-2 quality-weighted interval score algorithm identifies aberrant intervals (e.g., CNVs) in a sample that has consistently high or low log ratios based on their statistical score. ADM-2 uses an iterative procedure to find all genomic intervals with a score above a user-specified statistical threshold value (e.g., threshold set to a minimum of 6 with the minimum number of probes required in a region of 3 and the minimum absolute average log ratio of 0.25). The score represents the deviation of the weighted average of the normalized log ratios from its expected value of zero and incorporates quality information about each probe measurement.
All 22q11.2 breakpoints were validated internally and externally by quantitative microsphere suspension hybridization (QMH) assay (Newkirk et al., 2006) and/or quantitative RT-PCR using 60-mer probes from the proximal breakpoint region at 16,966,717, 16,966,867, 16,926,643, and 16,927,361 and the distal breakpoint region at 20,251,402. The version of aCGH analytics used for this analysis was based on information from the human genome release hg17. There was an addition of approximately 6 kb in nucleotide sequence in the 22q11.21 region as the sequence was refined for human genome release hg18. However, this additional sequence did not impact on our conclusions.
We also examined CNVs outside the 22q11.2 region. In our study, CNVs were included only if they met the statistical parameters described above. Copy number variants outside the 22q11.2 region were compared to CNVs reported in the human genome (see http://projects.tcag.ca/variation/).
Results
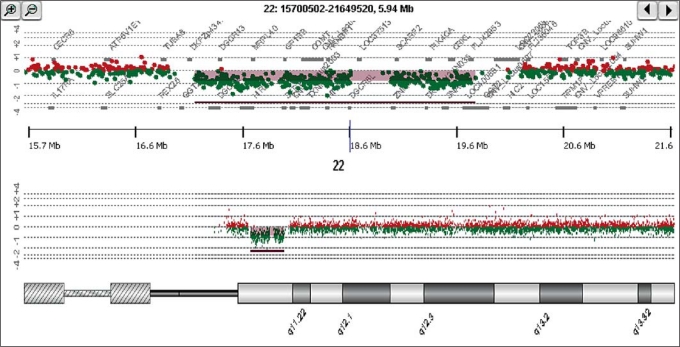
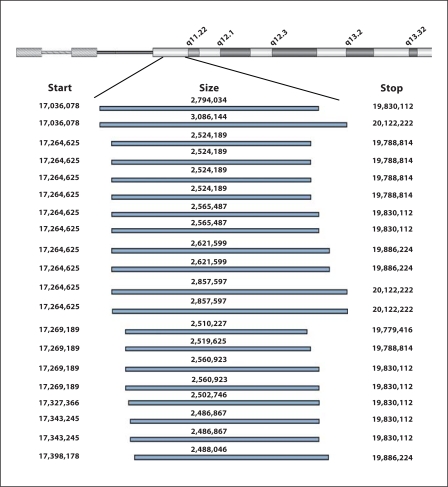
We analyzed 20 subjects with known deletions of 22q11.2 using the Agilent 244K whole genome microarray (see Fig. 1 for a representative example). All microarrays used for these analyses met the excellent or good criteria as determined by the Feature Extraction software v 9.5.1 (Agilent Inc.). The spatial resolution of the probes in the 22q11.2 region averages 8 kb; however, the probes are spaced more closely in many areas allowing for more accurate identification of breakpoints than in previous analyses of typical deletions of 22q11.2. Deletions in these 20 subjects ranged from 2.49 to 3.09 Mb and involved the LCRs located at 17.037 and 19.835 Mb. The proximal 22q11.2 LCR location was 17,037,083 to 17,083,237 (hg17) (http://genome.ucsc.edu). The proximal 22q11.2 breakpoint was distal to the proximal LCR in 18 subjects (Fig. 2) with ten breaks occurring at 17,264,625, four at 17,269,189, two at 17,343,245 and one each at 17,327,366 and 17,398,178. Two of our subjects had breakpoints proximal to the proximal LCR at 17,036,078. The distance between the proximal breakpoints from 17,036,078 to 17,398,178 is 362,199 bp. The distal 22q11.2 LCR location was 19,835,382 to 19,880,148 (hg17). The distal 22q11.2 breakpoint was proximal to the distal LCR in 14 subjects (Fig. 2) with eight breaks occurring at 19,830,112, five at 19,788,814, and one at 19,779,416. Six subjects had breakpoints distal to the distal LCR; three at 19,886,224 and three at 20,122,222. The distance between the distal breakpoints from 19,779,416 to 20,122,222 was 342,806 bp.
Fig. 1.
Representative example of array comparative genomic hybridization (aCGH) using the Agilent 244K array showing a chromosome 22q11.2 deletion in one of our subjects.
Fig. 2.
Results of analysis of subjects with 22q11.2 deletion syndrome using the Agilent 244K array. Breakpoint positions (start and stop) are shown on the ends and total deletion size is shown in the middle of bar.
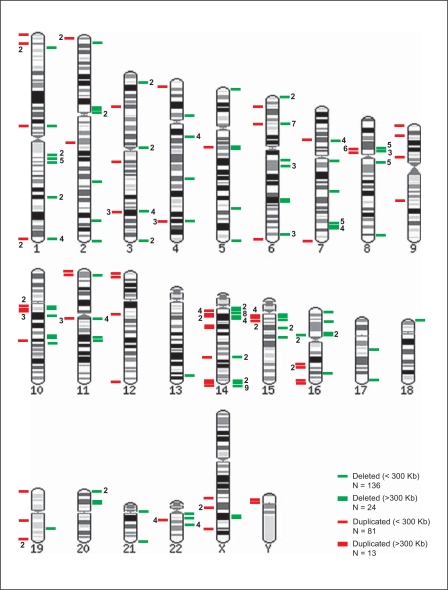
We examined the complete genomes of our subjects to identify CNVs in chromosomes other than the typical chromosome 22q deletion seen in our DiGeorge syndrome subjects and compared the CNVs to those reported in the human genome literature (Barrett et al., 2004; Sebat et al., 2004; Conrad et al., 2006; Khaja et al., 2006; Komura et al., 2006; Redon et al., 2006; Scherer et al., 2007). We observed 254 CNVs, 94 duplications and 160 deletions in our subjects (Fig. 3), ranging in size from 400 bp to 1.5 Mb. The smallest detectable duplication was 400 bp and the largest was 1.36 Mb. The smallest detectable deletion was 400 bp and the largest was 2.4 Mb not including the 22q11.2 deletions. Most of the regions containing CNVs in our subjects have been reported previously to contain copy number variation in apparently healthy individuals (Perry et al., 2006; Redon et al., 2006) (for summary, see http://projects.tcag.ca/variation/).
Fig. 3.
Human chromosome ideograms showing regions of copy number variation (deletion and/or duplication) observed in high resolution (244K) whole genome hybridization (aCGH) analysis of 20 subjects with 22q11.2 deletion syndrome excluding the 22q11.2 deletion seen in this cytogenetic syndrome. Deletions are shown in green on the right side of each chromosome ideogram and duplications in red on the left. Numbers indicate the frequency of copy number variants (CNVs) observed in the subjects studied.
Discussion
The breakpoints which bookend the 22q11.2 deletion leading to the 22q11.2 deletion syndrome (DiGeorge, Velocardiofacial or Shprintzen syndromes) approximate the LCRs more closely than the breakpoints of the 15q11–q13 deletion that results in Prader-Willi syndrome (PWS) or Angelman syndrome (Butler et al., 2008). The proximal deletion breakpoints in chromosome 15 in PWS spanned 1.5 Mb (for typical Type I deletion) to 0.5 Mb (for typical Type II deletion) (Butler et al., 2008) while in our subjects the proximal and distal breakpoints on 22q11 spanned approximately 350 kb each. The breaks in our subjects occurred on either side of the LCR, though it occurred more commonly on the distal side of the proximal LCR and on the proximal side of the distal LCR. This suggests that the forces which produce a break after pairing of the LCRs are applied on both sides of the paired LCR, but perhaps with greater intensity internal to the proximal LCRs, resulting in loss of material between the LCRs but leaving intact adjacent sequences. The process which produces recombination errors is not clearly delineated but the 22q11 region has recently received intense scrutiny producing clear evidence of the involvement of nonhomologous recombination involving the LCRs (Shaikh et al., 2000, 2007).
In our 20 subjects, the minimal common deleted region was 2,381,238 bp based on the location of the most distal point for the proximal deletion breakpoint and the most proximal point for the distal breakpoint. Genes and transcripts within the proximal and distal variable regions are listed within the distal variable region in Table 1. Genes within the proximal variable breakpoint region from 17,036,078 to 17,398,178 include DGCR6 (DiGeorge syndrome critical region protein 6) and PRODH (proline dehydrogenase 1) along with three transcripts with unknown function. Genes within the distal variable breakpoint region from 19,779,416 to 20,122,222 include GGT2 (Gamma-glutamyltransferase-like protein 2), HIC2 (hypermethylated in cancer 2), and multiple transcripts of unknown function. The genes in these two regions are variably hemizygous depending on the individual subject's breakpoints. DGCR6 is expressed in all adult and fetal tissues thus far examined (Edelmann et al., 2001) but no function has been determined. The PRODH gene, deleted in 16 of our 20 subjects studied, may be important in the 30% of 22q deletion patients with schizophrenia (Murphy et al., 1999; Kates et al., 2005). Missense mutations within PRODH are associated with schizophrenia and schizoaffective disorder (Paterlini et al., 2005; Prasad et al., 2008; Willis et al., 2008). GGT is an enzyme which plays a role in the metabolism of glutathione and facilitates amino-acid transport. Patients with cholestasis usually have increased serum GGT concentrations (OMIM: http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim). HIC2 is a homologue of HIC1 which is a candidate tumor suppressor gene. Investigation of the genes in these variably deleted regions is essential to the discovery of contributing factors in the inconsistent phenotype of the 22q11.2 deletion syndrome.
Table 1.
Genes and transcripts within the proximal and distal variable 22q11.2 breakpoint regions
| Proximal gene/sequence | Name/function | Disease association | Ontologies |
|---|---|---|---|
| DGCR6 | DiGeorge critical region 6 | Lung and colon adenocarcinomas and mammary carcinomas, strongly induced in Burkitt's lymphoma |
|
| PRODH | Catalyzes conversion of proline to pyrroline-5-carboxylate | Mutations/deletions cause hyperprolinemia type I schizophrenia |
|
| AK129567 | Hypothetical LOC729444 | Unknown | |
| POM121L8p (BC090873) |
|
Expressed in testes | |
| Distal gene/sequence | Name/function | Disease association | Ontologies |
|---|---|---|---|
| POM121L8p (alias AL117485) | DKFZp434K191 | Unknown | Unknown |
| AIFM3 (alias FLJ30473) | AIFM3 (apoptosis inducing factor, mitochondrion associated, 3) | Unknown |
|
| FLJ42953 | Breakpoint cluster region pseudogene | Expressed in the subthalmic nucleus | GTPase activator activity, signal transduction – intracellular level |
| LOC376818 | Unknown | Unknown | |
| LOC284861 | Unknown | Unknown | |
| POM121L8p (BC090873) | Unknown | Unknown | |
| GGT2 | Gamma-glutamyltransferase 2 | Highly expressed in fetal and adult kidneys and liver | Initiates extracellular glutathione (gsh) breakdown; catalyzes the transfer of the glutamyl moiety of glutathione to amino acids and dipeptide acceptors (by similarity) |
| AK129567 | Hypothetical LOC729444 | Expressed in the testes and blood | |
| HIC2 | Hypermethylated in cancer 2 |
|
|
| RIMBP3 (BC035246) | RIMBP3 (RIMS binding protein 3) | Unknown | Involved in oxidoreductase activity and metabolic processes |
| RIMBP3 (AB051453) | RIMBP3 (RIMS binding protein 3); involved in oxidoreductase activity | Unknown | Involved in oxidoreductase activity and metabolic processes |
| BC03913 | Unknown | Unknown | |
| DQ571494 | Unknown | Unknown | |
| RIMBP3 (alias KIAA1666) | RIMBP3 (RIMS binding protein 3); involved in oxidoreductase activity | Unknown | Involved in oxidoreductase activity and metabolic processes |
Genes in the 22q11.2 common region have been extensively studied since the region was identified as the critical deletion which causes 22q deletion syndrome. However, there is still much to be learned about the molecular genetic function of sequences in this region. For example, recently, abnormal microRNA synthesis in mouse brain was linked to haploinsufficiency of DGCR8 (Stark et al., 2008). The authors presented evidence that the abnormal miRNA biogenesis contributed to behavioral and neuronal deficits associated with the microdeletion of the region homologous to human 22q11.2. The interaction of genetic, epigenetic and environmental factors which cause the widely variable phenotype associated with the deletion of 22q11.2 is still poorly understood.
The advent of new technology for analyzing copy number variation has led to the recent discovery of a surprising amount of deleted and duplicated genetic material in the human genome (Conrad et al., 2006; Khaja et al., 2006; Komura et al., 2006; Redon et al., 2006; Stranger et al., 2007). The estimated number of CNVs in a single genome ranges from a few to as many as 100, depending on the size of the CNV considered and the stringency of analysis. Interestingly, a recent report relates de novo CNVs to sporadic schizophrenia (Xu et al., 2008). Nevertheless, the phenotypic consequences of copy number variation are not yet clear. In our 20 subjects we observed 254 CNVs using fairly stringent selection criteria. The CNVs ranged in size from 400 bp to 2.4 Mb with 217 of them less than 300 kb and 37 greater than 300 kb. Most have been previously reported. Therefore, we observed an average of 12.7 CNVs per individual, 4.7 duplications and 8 deletions. Chromosome 14 had 37 CNVs, 20 of which occurred at 14q11.2 (Fig. 3). Chromosomes 1 and 8 had 22 and 21 CNVs, respectively (Fig. 3). These three chromosomes (i.e., 1, 8, and 14) had 80 CNVs or 31.5% of all CNVs. Chromosomes 13 and 18 had only a single CNV (deletion) each (Fig. 3). Several individuals had only four CNVs (in addition to the 22q11.2 deletion), but one individual had 25 CNVs detected using our selection criteria. We have been accumulating data on the range, size and distribution of CNVs in our pediatric population using the Agilent 244K platform (Yu et al., submitted). The numbers of CNVs seen in our cohort of subjects with 22q11.2 deletions, although variable, appear to be similar in number and distribution to the CNVs observed in our database containing more than 200 cases. Therefore, it does not appear that the size or frequency of CNVs in subjects with 22q11.2 deletions differs from the general pediatric population.
In summary, new high resolution techniques for examining the human genome are allowing unprecedented opportunities to define the chromosome breakpoints in individuals and to examine genotype-phenotype relationships. Several authors have recently noted the need for investigations aimed at determining the relationship between genotypic variation and the behavioral and physical features associated with the 22q deletion syndrome (Karayiorgou and Gogos, 2004; Ousley et al., 2007). The breakpoint variation of the 22q11.2 deletion between individuals in our study was not large, but the variable intervals contained known genes and as yet unstudied transcripts. Investigation of the genes located within these intervals and noting their presence or absence will be important in the search for genotype-phenotype correlations in future studies in this cytogenetic syndrome.
Footnotes
This study was partially supported by a donation from the Fraternal Order of Eagles of the State of Kansas to D.C.B. and NICHD PO1HD 30329, NICHD RO1HD 41672 and the Hall Foundation of Kansas City to M.G.B.
References
- Barrett MT, Scheffer A, Ben-Dor A, Sampas N, Lipson D, et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc Natl Acad Sci USA. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer A, Riegel M, Schinzel A. Non-random asynchronous replication at 22q11.2 favours unequal meiotic crossovers leading to the human 22q11.2 deletion. J Med Genet. 2004;41:413–420. doi: 10.1136/jmg.2003.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, et al. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–221. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Fischer W, Kibiryeva N, Bittel DC. Array comparative genomic hybridization (aCGH) analysis in Prader-Willi syndrome. Am J Med Genet A. 2008;146A:854–860. doi: 10.1002/ajmg.a.32249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, et al. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet. 1997;61:620–629. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Stankiewicz P, Spiteri E, Pandita RK, Shaffer L, et al. Two functional copies of the DGCR6 gene are present on human chromosome 22q11 due to a duplication of an ancestral locus. Genome Res. 2001;11:208–217. doi: 10.1101/gr.143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H, Redon R, Andrews D, Scott C, Andrews R, et al. Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Gogos JA. The molecular genetics of the 22q11-associated schizophrenia. Brain Res Mol Brain Res. 2004;132:95–104. doi: 10.1016/j.molbrainres.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Kates WR, Antshel KM, Fremont W, Roizan N, Shprintzen R. Velo-Cardio-Facial syndrome. In: Butler MG, Meaney FJ, editors. Genetics of Developmental Disabilities. Boca Raton: Taylor and Francis group; 2005. pp. 383–418. [Google Scholar]
- Khaja R, Zhang J, MacDonald JR, He Y, Joseph-George AM, et al. Genome assembly comparison identifies structural variants in the human genome. Nat Genet. 2006;38:1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. Lancet. 2007;370:1443–1452. doi: 10.1016/S0140-6736(07)61601-8. [DOI] [PubMed] [Google Scholar]
- Komura D, Shen F, Ishikawa S, Fitch KR, Chen W, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantripragada KK, Tapia-Paez I, Blennow E, Nilsson P, Wedell A, Dumanski JP. DNA copy-number analysis of the 22q11 deletion-syndrome region using array-CGH with genomic and PCR-based targets. Int J Mol Med. 2004;13:273–279. [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:947–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- Newkirk HL, Rogan PK, Miralles M, Knoll JH. Determination of genomic copy number with quantitative microsphere hybridization. Hum Mutat. 2006;27:376–386. doi: 10.1002/humu.20312. [DOI] [PubMed] [Google Scholar]
- Ousley O, Rockers K, Dell ML, Coleman K, Cubells JF. A review of neurocognitive and behavioral profiles associated with 22q11 deletion syndrome: implications for clinical evaluation and treatment. Curr Psychiatry Rep. 2007;9:148–158. doi: 10.1007/s11920-007-0085-8. [DOI] [PubMed] [Google Scholar]
- Paterlini M, Zakharenko SS, Lai WS, Qin J, Zhang H, et al. Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia-related phenotypes in mice. Nat Neurosci. 2005;8:1586–1594. doi: 10.1038/nn1562. [DOI] [PubMed] [Google Scholar]
- Perry GH, Tchinda J, McGrath SD, Zhang J, Picker SR, et al. Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci USA. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad SE, Howley S, Murphy KC. Candidate genes and the behavioral phenotype in 22q11.2 deletion syndrome. Dev Disabil Res Rev. 2008;14:26–34. doi: 10.1002/ddrr.5. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DA, Eichler EE, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet Suppl. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, O'Connor RJ, Pierpont ME, McGrath J, Hacker AM, et al. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–491. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shprintzen RJ. Velo-cardio-facial syndrome: 30 years of study. Dev Disabil Res Rev. 2008;14:3–10. doi: 10.1002/ddrr.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban AE, Korbel JO, Selzer R, Richmond T, Hacker A, et al. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc Natl Acad Sci USA. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A, Bender HU, Steel G, Valle D. PRODH variants and risk for schizophrenia. Amino Acids. 2008;35:673–679. doi: 10.1007/s00726-008-0111-0. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]