Abstract
An immunofluorescence approach was used to directly examine meiotic recombination events in 483 pachytene spermatocytes from 11 male rhesus monkeys. Specifically, we examined the nuclear localization patterns of the DNA mismatch repair protein MLH1, known from analyses of other mammalian species to be a useful marker of meiotic cross-overs. Our results indicated that rhesus pachytene spermatocytes contain approximately 40 cross-overs per cell, corresponding to about one cross-over per chromosome. The chromosomal distribution of these exchanges was consistent with data from human and mouse males but, surprisingly, the overall number of foci was lower, and the number of ‘exchangeless’ bivalents higher, than reported for either humans or mice.
Introduction
Homologous recombination plays a crucial role in the first meiotic division, tethering homologous chromosomes and thereby facilitating their segregation at anaphase I. An important byproduct of this process is the generation of recombinant chromosomes, yielding genetic variation and from a practical standpoint providing the basis for genetic linkage analysis. Genetic maps are now available for a variety of mammalian species, including humans (Matise et al., 2003, 2007; Kong et al., 2004), but relatively little linkage information is available for non-human primates (Cox et al., 2006; Rogers et al., 2006; Jasinska et al., 2007).
Recently, an alternative to genetic linkage analysis has become available that makes it possible to directly study the recombination process as it occurs. This approach relies on immunofluorescence methodology to analyze localization patterns of recombination machinery proteins at different sub-stages of meiotic prophase. For example, antibodies against strand invasion proteins (e.g., DMC1, RAD51) can be used to monitor events occurring immediately after formation of meiotic double strand breaks (DSBs), antibodies against the mismatch repair proteins MSH4 and MSH5 to assess formation of double Holliday junctions and, most importantly in the context of cross-overs, antibodies against MLH1 and MLH3 to estimate the number and distribution of recombination events (Cohen et al., 2006). Originally utilized to analyze recombination in mice (Anderson et al., 1999), MLH1 and/or MLH3 analyses have also now been used to construct whole-genome- and chromosome-specific recombination maps in human males and females (Lynn et al., 2002; Tease et al., 2002; Lenzi et al., 2005; Sun et al., 2006).
We recently initiated studies to apply this methodology to the rhesus monkey (Macaca mulatta), a non-human primate that has been an important biomedical model for humans. In this report, we summarize analyses of meiotic recombination in nearly 500 spermatocytes from 11 rhesus males. Not unexpectedly, we find that the basic ‘rules’ of meiosis apply to rhesus males as they do to other organisms that have been appropriately studied. For example, in most cells we observed at least one MLH1 focus (exchange) per chromosome; the number of MLH1 foci per chromosome was constrained, with smaller chromosomes typically having 1–2 foci and larger chromosomes 2–3 foci; and consistent with interference, the foci were non-randomly spaced along the length of the chromosomes. Further, similar to previous results from a series of human males (Hassold et al., 2004) and different strains of male mice (Koehler et al., 2002), we identified significant variation in the number of MLH1 foci per cell both within and among different rhesus males. However, there were also some surprising differences between rhesus males and other mammalian species for which MLH1 data have been reported. Most importantly, we observed a high proportion of ‘exchangeless’ chromosomes (i.e., chromosomes lacking MLH1 foci), and overall, the number of MLH1 foci per genome was substantially lower than anticipated from previous studies of human and mouse males. Thus, it seems clear that, among mammalian males, the number and distribution of cross-over events is subject to species-specific regulation.
Materials and methods
Sample population and sample processing
Animals were maintained at the California National Primate Research Center (CNPRC) at the University of California, Davis either in individual indoor cages (VandeVoort et al., 1992) or in outdoor group field cages (Capitanio et al., 2006). The CNPRC is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Testes from sexually mature rhesus macaques (Macaca mulatta) were obtained from animals assigned to surgery or necropsy for other purposes. Animals were anesthetized with 10 mg/kg Ketamine prior to delivery to surgery or necropsy. For surgical procedures, anesthesia with inhaled Isofluorane gas was initiated prior to testis removal. For necropsy, animals were anesthetized with pentobarbital prior to testes removal. Testes were immediately placed in normal saline and delivered to the laboratory. An approximate 1 cm3 piece of tissue was dissected from the center of the testis for isolation of tubules.
Testicular tissue was processed as outlined in Peters et al. (1997), but with a few modifications. In short, the testes were removed from the animal, and rinsed briefly in 1× PBS. The tunica was removed from each retrieved testis and the seminiferous tubules were placed in a hypotonic extraction buffer, teased apart with forceps, and incubated at 35°C for about 45 min. Small sections of tubule were excised and macerated in 25 μl of a sucrose solution (100 mM). 10 μl portions of this slurry were spread onto 1% PFA-coated slides and dried overnight at room temperature in a humid chamber. The following day, the slides were washed for 2 min in 0.04% Photoflo-200 (Kodak) and air dried. The slides were stored at −20°C and shipped on dry ice from CNPRC to Washington State University.
Immunostaining
The slides were immunostained using the protocol of Tease and Hulten (2004), with minor modifications. Briefly, the slides were washed in 1× ADB for 20 min. CREST anti-human antiserum (Fisher Scientific) and mouse anti-human MLH1 antibody (BD Pharmingen), were diluted 1:1000 and 1:75, respectively, in 1× ADB. 60 μl of this primary mix was spread across the slides, which were then sealed with a glass coverslip, secured with rubber cement and incubated overnight at 37°C in a humid chamber. The rubber cement was carefully removed and the slides were soaked in 1× ADB to allow the coverslips to float off. Rabbit polyclonal anti-human SYCP3 antibody (Novus Biologicals) was diluted 1:150 in 1× ADB. 60 μl of this primary antibody mix was added to the slides. The slides were covered in parafilm and incubated for 2 h at 37°C in a humid chamber. Upon removal, the parafilm was slowly peeled off and the slides were washed twice in 1× ADB for 20 min on a shaker. AMCA-labeled donkey anti-human CREST (Jackson ImmunoResearch) and fluorescein-labeled donkey anti-mouse MLH1 (Jackson ImmunoResearch) were diluted 1:100 and 1:75, respectively, in 1× ADB, and 60 μl of the mix was placed on the slides. A glass coverslip secured with rubber cement was applied to each slide and the slides were incubated overnight at 37°C in a humid chamber. The rubber cement was carefully removed and the slides were soaked in 1× ADB to allow the coverslips to float off. Rhodamine-labeled donkey anti-rabbit SYCP3 (Jackson ImmunoResearch) was diluted 1:100 in 1× ADB and 60 μl of the solution was deposited on the slides. Parafilm was placed over the slides and the slides were incubated for 45 min at 37°C in a humid chamber. The parafilm was slowly peeled off and the slides were washed twice in 1× PBS for 15 min. FluoroGuard Antifade Reagent (BioRad Laboratories) was dropped onto the slides and the slides were coverslipped for subsequent microscopic analysis.
Fluorescence in situ hybridization (FISH)
Chromosome-specific FISH was performed on pachytene cells that had been analyzed for MLH1. Whole chromosome paint probes specific to HSA1, HSA6, HSA9, and HSA16 (MP Biomedicals) were used to identify homologous rhesus chromosomes 1, 4, 15 and 20, respectively. Briefly, the slides were washed in a 3:1 methanol:acetic acid solution at room temperature for 5 min, washed in a 2× SSC, 0.1% NP-40 solution at 37°C for 5 min, and dehydrated in an ethanol series (75, 90, 100%) at room temperature for 2 min at each concentration. A mix containing 10 μl each of two different probes was added to the slides and a glass coverslip applied. The slides were denatured at 75°C for 5 min and then incubated at 37°C overnight in a humid chamber. The rubber cement was carefully removed and the slides were soaked in the following wash solution to allow the coverslips to float off. The slides were washed in a 0.5× SSC, 0.1% SDS solution at 45°C for 5 min. The slides were drained of excess liquid, stained with Vectashield™ (Vector Laboratories), and coverslipped for subsequent microscopic analysis. Typically, slides were first probed with paints specific to HSA6 and HSA9 and then probed a second time with paints specific to HSA1 and HSA16.
Analysis
A Zeiss Axiophot epifluoresence microscope connected to a CCD camera and computer with Applied Spectral Imaging Quips Path Vysion Smart-Capture VP 1.4 Software (Digital Scientific Ltd.) was employed to collect data. Images of fully synapsed pachytene stage cells with obvious MLH1 signals were captured and the cells examined by two independent observers. Because the sex chromosomes synapse and recombine asynchronously from the autosomes, MLH1 foci were scored for autosomal bivalents, but not for the XY pair. To make measurements of the length of the synaptonemal complexes, we used MicroMeasure software (www.biology.colostate.edu/MicroMeasure); as for the scoring of MLH1 foci, we only analyzed autosomal synaptonemal complexes.
Results
We analyzed the number and location of MLH1 foci on each of the 20 autosomal chromosomes in pachytene stage spermatocytes. In total, we analyzed 483 cells from 11 rhesus males (see Fig. 1 for representative images); the results from individual animals are summarized in Table 1. The overall mean number of MLH1 foci per cell was 39.0 ± 3.0. However, there was significant variation among individual males (F = 11.2; P < 0.0001), with mean values ranging from a low of 36.8 ± 2.0 (MMU 34596) to a high of 41.6 ± 2.6 (MMU 28317). The variation was independent of the age of the animal, as we saw no obvious relationship between recombination and age (Table 1).
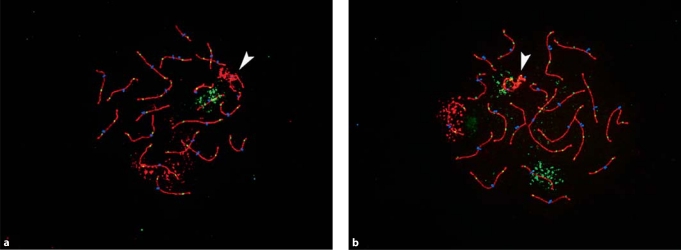
Fig. 1.
Representative images of rhesus male pachytene preparations. Antibodies detect the synaptonemal complex protein SYCP3 (in red) and the DNA mismatch repair protein MLH1 (in green) and CREST antiserum-positive signals (in blue) detect centromeric regions. (a) Rhesus chromosomes 1 (in red; homologous to HSA1) and 20 (in green; homologous to HSA16) were identified using FISH paint probes. (b) Rhesus chromosomes 4 (in red; homologous to HSA6) and 15 (homologous to HSA9) were similarly identified. In each image, an arrowhead denotes the XY bivalent.
Table 1.
Summary of MLH1 analyses
| ID | Age (year.month) | Number of cells | Mean MLH1 foci ± SD | Range |
|---|---|---|---|---|
| MMU 34966 | 4.5 | 89 | 38.4 ± 2.5 | 32–44 |
| MMU 34814 | 4.7 | 51 | 38.1 ± 2.4 | 32–44 |
| MMU 34200 | 5.3 | 29 | 40.9 ± 2.2 | 37–45 |
| MMU 33659 | 5.6 | 47 | 40.1 ± 3.7 | 29–46 |
| MMU 28317 | 13.3 | 38 | 41.6 ± 2.6 | 35–46 |
| MMU 27432 | 14.7 | 45 | 37.2 ± 3.1 | 31–43 |
| MMU 26823 | 15.3 | 7 | 38.1 ± 2.2 | 35–41 |
| MMU 26806 | 15.8 | 29 | 39.0 ± 3.2 | 30–48 |
| MMU 26072 | 16.7 | 45 | 39.3 ± 2.8 | 35–46 |
| MMU 34596 | 17.6 | 39 | 36.8 ± 2.0 | 32–41 |
| MMU 24197 | 19.7 | 64 | 39.6 ± 2.8 | 32–45 |
Figure 2 shows the pooled data from all 483 cells. Each of the 20 rhesus autosomes is either metacentric or sub-metacentric; thus, assuming that each chromosome arm contains at least one exchange (MLH1 focus), we would anticipate a minimum of 40 MLH1 foci in pachytene cells. From Fig. 2 it is clear that this was not the case. Of the 483 cells analyzed, the majority (275/483 = 56.9%) contained 39 or fewer MLH1 foci and, in over 10% of cases (57/483 = 11.8%), no more than 35 foci were identified. In most of the cells with 35 or fewer foci, one arm was ‘exchangeless’, while the other arm contained at least one MLH1 focus. However, there were also a surprisingly high number of cells in which we were unable to identify an MLH1 focus on either arm. Specifically, 66 cells had one bivalent that lacked an MLH1 focus, 13 had two such bivalents and two cells had three such bivalents. Altogether over 15% of cells (81/483 = 16.8%) contained one or more exchangeless chromosome pairs.
Fig. 2.
Distribution of the number of MLH1 foci/cell for 483 pachytene spermatocytes from 11 rhesus males.
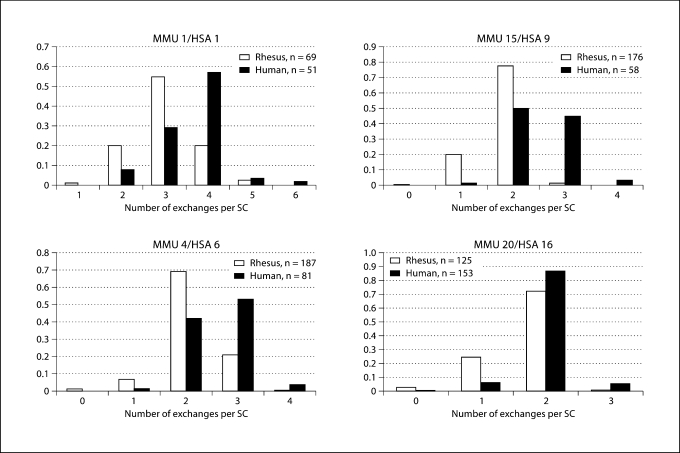
In a second set of studies, we analyzed the number and location of MLH1 foci on individual chromosomes. We chose different-sized rhesus chromosomes known to be homologous to individual human chromosomes. Specifically, we analyzed two ‘large’ rhesus chromosomes (1 and 4, homologous to human chromosomes 1 and 6, respectively), one ‘medium-sized’ chromosome (15, homologous to human chromosome 9) and one ‘small’ chromosome (20, homologous to human chromosome 16). We compared the results of the analysis of rhesus cells with equivalent data from our ongoing studies of MLH1 localization in human male pachytene preparations (Hassold et al., 2004). For each of the four comparisons (Fig. 3), the mean number of MLH1 foci was significantly lower for the rhesus chromosome than for its human counterpart (for MMU1 vs HSA1, t = 9.3; P < 0.0001; for MMU4 vs HSA6, t = 11.9, P < 0.0001; for MMU15 vs HSA9, t = 16.4, P < 0001; for MMU20 vs HSA16, t = 9.4, P < 0.001). Further, the reductions appeared to be of relatively similar magnitude for each of the four chromosomes; i.e, for MMU1 a 17% reduction (from 3.6 exchanges for HSA1 to 3.0 exchanges), for MMU4 a 19% reduction (from 2.6 for HSA6 to 2.1), for MMU15 a 28% reduction (from 2.5 for HSA to 1.8) and for MMU20 a 15% reduction (from 2.0 for HSA16 to 1.7).
Fig. 3.
Comparison of the number of MLH1 foci/bivalent for four rhesus chromosomes and their homologous human counterparts.
In subsequent studies, we examined the relationship between synaptonemal complex length and the number of MLH1 foci in individual spermatocytes. Previous studies in other mammals (e.g., mouse males and females, human males; see Lynn et al., 2002) indicate that the SC ‘measures’ genetic length, since there appears to be a simple linear relationship between the total length of the SCs and the number of MLH1 foci in a cell. As shown in Fig. 4, this relationship appears to hold for rhesus males as well. In analyses of 12 representative cells from three different males, we observed a linear relationship between the number of MLH1 foci/cell and the combined length of the 20 autosomal SCs (r2 = 0.32; P = 0.05).
Fig. 4.
Correlation of MLH1 counts per cell and the total length of the autosomal SCs in 12 cells from three different males; cells were selected to provide a wide range of MLH1 values.
Discussion
The purpose of the present study was two-fold: first, to estimate the genome-wide level of meiotic recombination in rhesus males and to compare our estimate with the limited data from genetic linkage analysis and second, to assess the number and location of exchanges (MLH1 foci) on rhesus chromosomes, and to ask whether their distribution conforms to properties identified in previous cytological studies of mammalian males.
In our analyses of 483 cells from 11 rhesus males, we observed an overall mean number of MLH1 foci per cell of 39.0. Assuming that 1 MLH1 focus = one cross-over = 50 cM, this implies a genome-wide genetic length of approximately 1950 cM. This estimate is nearly identical to the 2048 cM value recently reported in a first generation genetic linkage study of rhesus animals (Rogers et al., 2006), but there are at least two caveats to this comparison. First, the linkage data of Rogers and colleagues (2006) almost certainly underestimates the true level of recombination in rhesus, since distal chromosome regions were under-represented in the available marker set; indeed, the authors suggested that the map was likely truncated by at least 500 cM. Second, the purpose of the Rogers et al. study (2006) was to generate a sex-averaged map; i.e., one based on analyses of both male and female meioses. In most mammalian species, there are significant sex-specific differences in recombination, with higher levels occurring in females; e.g., in humans the female:male ratio is approximately 1.6:1 (Matise et al., 2007). Assuming that rhesus follows this pattern, the 2048 cM value provided by the linkage analysis is, in essence, based on two separate sets of data, one with a lower (male) and one with a higher (female) level of recombination. Thus, one aspect of the linkage analysis likely underestimated genetic length and the other likely inflated it, making it difficult to compare our data with the linkage map. Clearly, in future linkage analyses it will be useful to separately analyze male and female meiotic events; similarly, analyses of the number and distribution of MLH1 foci in rhesus oocytes will be instructive and we are presently undertaking such studies.
Nevertheless, if we assume that 1) when completed, the sex-averaged genetic linkage map of rhesus will, indeed, be approximately 500 cM longer than that reported by Rogers et al. (2006) and 2) the magnitude of the sex-specific difference in recombination in rhesus is similar to that identified in humans, then rhesus female- and male-specific maps of approximately 3100 and 1900 cM, respectively, seem reasonable. If so, the male map length would be virtually identical to our value of approximately 1900–2000 cM. This would recapitulate observations from humans in which MLH1-based assays and genetic linkage analyses are in good agreement in estimating male genome-wide genetic lengths of about 2500 cM.
However, if our MLH1 results accurately reflect genome-wide cross-over levels in rhesus males, an important question remains: given the similarity in the size of the genome between rhesus and humans (Gibbs et al., 2007), why is there so much less recombination in rhesus? The effect is presumably genome-wide as each of the four individual chromosomes we analyzed exhibited a similar 20–30% reduction in recombination in rhesus by comparison with human males (Fig. 3). Possibly there are more, or different, hot spots of recombination in the human genome or alternatively, there may be similar hot spots that are accessed differently between the two species. Clearly, in future cytological and linkage analyses of rhesus, it will be important to identify regions of the genome that are more or less likely to recombine, and to compare this information to data already available from humans.
In other analyses, we were interested in examining the number and location of MLH1 foci on individual rhesus chromosomes, and in asking whether they followed the basic ‘rules’ of meiosis. Many of the observations fit the expectations. For example, we found that in the vast majority of cells (402/483 = 83.2%) all autosomal synaptonemal complexes contained at least one MLH1 focus. Additionally, like previous studies of human males and mice (Lynn et al., 2002), the number of MLH1 foci per cell was correlated with the overall length of the SCs. Further, while we made no attempt to map the exact chromosomal locations of the MLH1 foci, their placement was consistent with two predictions. First, the majority of foci were distally located (Fig. 1); this is similar to genetic and cytological studies from both mice and humans, in which male, but not female exchanges are preferentially located in distal chromosome regions. Secondly, on SCs that contained two, three or more MLH1 signals, the foci were ‘spread out’ along the SC; this was most easily visualized on bivalents containing two foci, as the two foci were virtually always located on opposite arms (see Fig. 1). This spacing of MLH1 foci is consistent with positive interference, the highly evolutionarily conserved property of meiosis that promotes non-random positioning of homologous recombination events.
While our findings conformed to many a priori expectations, at least two observations were surprising. First, a majority of cells contained at least one bivalent in which one of the two arms was lacking an MLH1 focus. This is in sharp contrast to human and mouse males (Koehler et al., 2002; Lynn et al., 2002) in which chromosome arms lacking at least one MLH1 focus are a rarity. Even more surprising, however, was the high proportion of rhesus bivalents lacking a focus on either arm. Indeed, nearly 20% of all cells contained one or more such exchangeless chromosomes. In chiasmate species these exchangeless bivalents increase the risk of nondisjunction, since the homologs presumably travel independently to the metaphase plate and subsequently segregate randomly. Possibly, such cells are culled by checkpoint control mechanisms operating during pachytene and metaphase I in rhesus spermatogenesis. Alternately, similar to species such as Drosophila, an achiasmate segregation system (Hawley and Theurkauf, 1993) may operate in rhesus to ensure segregation of exchangeless bivalents.
However, if cells containing exchangeless chromosomes are not selectively eliminated and if there is no mechanism for segregating such chromosomes, our observations imply an extraordinarily high level – approaching 10% – of meiotic nondisjunction events in rhesus males. In future studies, it will be important to distinguish between these alternatives, an exercise that should be relatively straightforward. For example, we can compare the number of exchangeless chromosomes at two different timepoints during meiosis I – at pachytene, assaying the number of MLH1 foci (as described in this report) and at a later stage, diakinesis/metaphase I, examining the number of chiasmata. This will allow us to assess the efficiency of checkpoint control mechanisms at eliminating cells containing exchangeless chromosomes between pachytene and diakinesis/metaphase I. To monitor nondisjunction levels, we can take one of two approaches. First, we can examine chromosome preparations at meiosis II, allowing us to estimate the level of nondisjunctional errors occurring at meiosis I. Second, we can use chromosome-specific fluorescence in situ hybridization (FISH) to examine the overall level of meiotic nondisjunctional errors (meiosis I + meiosis II) for individual chromosomes (Hassold, 1998). Taken together, these analyses will allow us to determine whether nondisjunction levels are typical of other mammalian species (e.g., humans and mice, both with 1–2% levels of sperm disomy) or whether they do, indeed, approach the predicted 10% levels.
In summary, our analyses provide evidence that, as in other mammalian species, immunofluorescence methodology can be used to examine recombination in the rhesus monkey. Our results indicate that rhesus pachytene spermatocytes contain approximately 40 cross-overs per cell, corresponding to a genome-wide genetic length of approximately 1900–2000 cM. The chromosomal distribution of these exchanges shares several features with other, previously analyzed mammalian species but, surprisingly, the overall number of foci is lower, and the number of exchangeless bivalents higher, than those reported for other mammalian males.
Footnotes
This work was supported by NIH grants HD042720 (to T.H.), ES013527 and HD037502 (to P.H.) and RR00169 (to C.V.).
References
- Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio JP, Kyes RC, Fairbanks LA. Considerations in the selection and conditioning of Old World monkeys for laboratory research: animals from domestic sources. ILAR J. 2006;47:294–306. doi: 10.1093/ilar.47.4.294. [DOI] [PubMed] [Google Scholar]
- Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr Rev. 2006;27:398–426. doi: 10.1210/er.2005-0017. [DOI] [PubMed] [Google Scholar]
- Cox LA, Mahaney MC, Vandeberg JL, Rogers J. A second-generation genetic linkage map of the baboon (Papio hamadryas) genome. Genomics. 2006;88:274–281. doi: 10.1016/j.ygeno.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Hassold TJ. Nondisjunction in the human male. Curr Top Dev Biol. 1998;37:383–406. doi: 10.1016/s0070-2153(08)60181-7. [DOI] [PubMed] [Google Scholar]
- Hassold T, Judis L, Chan ER, Schwartz S, Seftel A, Lynn A. Cytological studies of meiotic recombination in human males. Cytogenet Genome Res. 2004;107:249–255. doi: 10.1159/000080602. [DOI] [PubMed] [Google Scholar]
- Hawley RS, Theurkauf WE. Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet. 1993;9:310–317. doi: 10.1016/0168-9525(93)90249-h. [DOI] [PubMed] [Google Scholar]
- Jasinska AJ, Service S, Levinson M, Slaten E, Lee O, et al. A genetic linkage map of the vervet monkey (Chlorocebus aethiops sabaeus) Mamm Genome. 2007;18:347–360. doi: 10.1007/s00335-007-9026-4. [DOI] [PubMed] [Google Scholar]
- Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics. 2002;162:297–306. doi: 10.1093/genetics/162.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Barnard J, Gudbjartsson DF, Thorleifsson G, Jonsdottir G, et al. Recombination rate and reproductive success in humans. Nat Genet. 2004;36:1203–1206. doi: 10.1038/ng1445. [DOI] [PubMed] [Google Scholar]
- Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, et al. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am J Hum Genet. 2005;76:112–127. doi: 10.1086/427268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, et al. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 2002;296:2222–2225. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- Matise TC, Sachidanandam R, Clark AG, Kruglyak L, Wijsman E, et al. A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set. Am J Hum Genet. 2003;73:271–284. doi: 10.1086/377137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Rogers J, Garcia R, Shelledy W, Kaplan J, Arya A, et al. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics. 2006;87:30–38. doi: 10.1016/j.ygeno.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Sun F, Oliver-Bonet M, Liehr T, Starke H, Turek P, et al. Variation in MLH1 distribution in recombination maps for individual chromosomes from human males. Hum Mol Genet. 2006;15:2376–2391. doi: 10.1093/hmg/ddl162. [DOI] [PubMed] [Google Scholar]
- Tease C, Hulten MA. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet Genome Res. 2004;107:208–215. doi: 10.1159/000080599. [DOI] [PubMed] [Google Scholar]
- Tease C, Hartshorne GM, Hulten MA. Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet. 2002;70:1469–1479. doi: 10.1086/340734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeVoort CA, Tollner TL, Overstreet JW. Sperm-zona pellucida interaction in cynomolgus and rhesus macaques. J Androl. 1992;13:428–432. [PubMed] [Google Scholar]