Abstract
Background
MCF2L2, ADIPOQ and SOX2 genes are located in chromosome 3q26-27, which is linked to diabetic nephropathy (DN). ADIPOQ and SOX2 genetic polymorphisms are found to be associated with DN. In the present study, we first investigated the association between MCF2L2 and DN, and then evaluated effects of these three genes on the development of DN.
Methods
A total of 1177 type 1 diabetes patients with and without DN from the GoKinD study were genotyped with TaqMan allelic discrimination. All subjects were of European descent.
Results
Leu359Ile T/G variant in the MCF2L2 gene was found to be associated with DN in female subjects (P = 0.017, OR = 0.701, 95%CI 0.524-0.938) but not in males. The GG genotype carriers among female patients with DN had tendency decreased creatinine and cystatin levels compared to the carriers with either TT or TG genotypes. This polymorphism MCF2L2-rs7639705 together with SNPs of ADIPOQ-rs266729 and SOX2-rs11915160 had combined effects on decreased risk of DN in females (P = 0.001).
Conclusion
The present study provides evidence that MCF2L2, ADIPOQ and SOX2 genetic polymorphisms have effects on the resistance of DN in female T1D patients, and suggests that the linkage with DN in chromosome 3q may be explained by the cumulated genetic effects.
Background
The global increase in diabetes prevalence has been accompanied by a rise in a number of patients with diabetic nephropathy (DN). This complication affects 30-40% of the patients with type 1 (T1DM) and type 2 diabetes (T2DM), and has become the most common single cause of end-stage renal disease (ESRD) [1,2]. Evidence has demonstrated that DN has genetic components [3,4]. Therefore, identification of susceptibility or resistance genes in DN will provide better knowledge of pathomechanism and future therapies in this disease [5,6].
With the approaches of genome wide scan and linkage analyses, researchers have searched for the linkage to DN along chromosomes. The linkage to DN in chromosome 3q has been evidenced repeatedly in many ethnic groups, and indicated that there reside the susceptibility and/or resistant genes for DN in this chromosomal arm [7-13]. The adiponectin (ADIPOQ) gene is located in chromosome 3q27. Vionnet et al. have analyzed 14 genes including the ADIPOQ gene selected from chromosome 3q and found that a promoter polymorphism rs17300539 (-11391A/G) in the ADIPOQ gene is associated with DN in Danish T1DM patients [14]. We have searched for the susceptibility and/or resistance genes for DN in chromosome 3q with a positional candidate SNP genotyping approach. Our data indicate that the ADIPOQ promoter polymorphism rs266729 (-11377C/G) is associated with DN in female T1DM patients [15]. Another gene encoded by sex-determining region Y-box 2 (SOX2) is located in chromosome 3q26.3. There is unique tag SNP rs11915160 in UTR-3' in this intron-less gene. We have recently found that this polymorphism is associated with DN in female subjects with T1DM [16].
MCF.2 cell line derived transforming sequence-like 2 (MCF2L2) is a guanine-nucleotide exchange factor (GEF) from Rho family. Evidence has demonstrated that GEFs from Rho family are signaling molecules responsible for Rho protein activity. Over-activation of Rho protein is a common component involved in the pathogenesis of diabetes and several cardiovascular disorders, including hypertension, coronary and cerebral vasospasm and atherosclerosis [17,18]. Takeuchi et al. have demonstrated that MCF2L2 genetic polymorphisms confer the risk susceptibility to T2DM [19]. The MCF2L2 gene is located in chromosome 3q27, and resides between locations of the ADIPOQ and SOX2 genes. In the present study, we first genotyped genetic polymorphisms of the MCF2L2 gene and then performed multiplex gene association analysis for MCF2L2, ADIPOQ and SOX2 to evaluate their genetic effects on the development of DN. The data may provide useful information to better understand the linkage in chromosome 3q with DN.
Methods
Subjects
The subjects in Genetics of Kidney Diseases in Diabetes (GoKinD) study were collected by the Juvenile Diabetes Research Foundation in collaboration with the Joslin Diabetes Centre, George Washington University, and the United States Centres for Diabetes Control and Prevention [20]. The GoKinD collection consists of both singletons and trios, however, only singletons were included in the present study. In this GoKinD cohort, ~8% of total subjects were Americans of Black, Asian, Hispanic or Indian descents. In order to avoid the error caused by the subjects from different races, this small proportion of subjects were excluded from analyses. Subsequently, a total of 599 (female 357/male 242) T1DM patients without DN and 578 (265/313) T1DM patients with DN were included, and all subjects were of European descent. Presence of DN was defined either by persistent proteinuria (urinary albumin excretion rate >300 mg/24 h) in two out of three consecutive measurements (at least one month apart), or ESRD (not due to condition other than diabetes). Absence of DN was considered as persistent normal albuminuria (< 30 mg/24 h) despite duration of T1DM for at least 15 years. Clinical parameters of all subjects were collected by the GoKinD study [20] and are represented in Table 1. Sample collection of GoKinD study was approved by the local ethics committees. Data and material transfer agreement was completed prior to the study.
Table 1.
Clinical material
| T1DM without DN | T1DM with DN | |||||
|---|---|---|---|---|---|---|
| All | Female | Male | All | Female | Male | |
| N | 599 | 357 | 242 | 578 | 265 | 313 |
| Age (years) | 40 ± 8 | 40 ± 9 | 40 ± 8 | 44 ± 6 | 44 ± 7 | 45 ± 6 |
| Duration of diabetes (years) | 26 ± 8 | 26 ± 8 | 26 ± 8 | 32 ± 8 | 32 ± 8 | 32 ± 8 |
| HbA1c (%) | 7.5 ± 1.1 | 7.5 ± 1.1 | 7.4 ± 1.1 | 7.4 ± 1.9 | 7.4 ± 2.1 | 7.4 ± 1.7 |
| BMI (kg/m2) | 26.0 ± 4.4 | 25.7 ± 4.7 | 26.6 ± 3.7 | 25.7 ± 5.3 | 25.2 ± 5.7 | 26.2 ± 4.8 |
| Creatinine (mg/dL) | 0.9 ± 0.2 | 0.8 ± 0.1 | 1.0 ± 0.1 | 2.1 ± 1.9 | 1.9 ± 1.6 | 2.3 ± 2.0 |
| Cystatin (mg/L) | 0.8 ± 0.1 | 0.8 ± 0.1 | 0.8 ± 0.1 | 2.2 ± 1.7 | 2.2 ± 1.6 | 2.4 ± 1.7 |
| Cholesterol (mg/dL) | 185.0 ± 31.3 | 188.7 ± 30.9 | 179.7 ± 31.2 | 186.3 ± 45.9 | 190.3 ± 47.3 | 183.1 ± 44.5 |
| HDL (mg/dL) | 59.1 ± 15.9 | 64.8 ± 15.5 | 51.0 ± 12.4 | 53.6 ± 17.3 | 59.3 ± 18.2 | 48.9 ± 15.0 |
| Systolic BP (mm Hg) | 118 ± 12 | 116 ± 12 | 122 ± 12 | 131 ± 19 | 130 ± 20 | 133 ± 18 |
| Diastolic BP (mm Hg) | 71 ± 8 | 70 ± 7 | 74 ± 8 | 74 ± 11 | 72 ± 11 | 75 ± 11 |
All data are means ± SD. T1DM = Type 1 diabetic patients, DN = diabetic nephropathy.
Marker selection and genotyping
Examined SNPs in the MCF2L2 gene were selected based upon the information from International HapMap Project, dbSNP databases http://www.ncbi.nlm.nih.gov/SNP/. Haploview (ver4.1) was employed to visualize linkage disequilibrium (LD) and haplotype block structures (r2 > 0.8, LOD threshold 3.0). Three SNPs i.e. rs684846, rs35069869 and rs35368790 in the MCF2L2 gene are reported to be associated with T2DM in a Japanese population [19]. Latest information from dbSNP database indicates that SNP rs684846 resides at 3'-flanking sequence but not within the MCF2L2 gene region. Therefore, this polymorphism was excluded from our study.
Rs7639705 is a non-synonymous SNP and causes amino acid change from Leu to Ile at the position 359 of MCF2L2 cDNA sequence. This polymorphism is a tag marker for LD block 8 (8 kb) in the population of European Caucasians (CEU, Haploview ver4.1). In the Japanese study, however, SNP rs7639705 was not included. In the present study, we validated this polymorphism with 32 subjects (64 chromosomes) randomly selected from the GoKinD study cohort, and then included it for genotyping analysis.
Genotyping experiments were performed with TaqMan allelic discrimination technique. SNP genotyping assays that incorporate minor groove binding (MGB) probes were purchased from Applied Biosystems (ABI 7300, Foster City, USA). The assay ID numbers and PCR protocol are available either on request or from ABI database. For quality control, the subjects (cases and controls) were distributed randomly on each PCR plate. The negative (Universal-mixture blanks) controls were included on each plate. Genotyping experiments in 20-25% of samples were performed in duplicate, and the duplication accuracy was calculated to be 99%.
Gene-gene interaction analyses
Generalized multifactor dimensionality reduction (GMDR) is a nonparametric and genetic model-free alternative to linear or logistic regression program for detection of gene-gene or gene-environmental interactions [21,22]. We employed this program to investigate gene-gene interactions concerning MCF2L2-rs7639705, AdipoQ-rs266729 and Sox2-rs11915160. This program provides a number of parameters including cross-validation consistency, testing balanced accuracy and empirical P-values to assess each selected interaction. The GMDR analyses were performed separately in female and male subjects. Logistic regression models were used for confirmation of the data from GMDR analyses.
Statistical analyses
Hardy-Weinberg equilibrium (HWE) for individual loci was assessed using the Pearson Chi-square (χ2) statistic. A 2 × 2 contingency table was used for test of the differences of allele frequencies between cases and controls. The test with additive model and Cochran-Armitage χ2 trend test for the combined equal additive effects were used to identify differences in genotype distribution between cases and controls. Odds ratios (OR) and 95% confidence intervals (CI) were calculated to test the relative risk for association. Statistical powers were calculated using software of PowerSampleSize (PS version 2.1.31). The sample sizes of cases and controls was sufficient to detect association with 80% power (5% level) if allele frequencies 0.15 and 0.20. Tests for association between genotypes and quantitative traits were performed by using ANOVA for normally distributed traits, or alternatively Kruskal-Wallis analysis of ranks for traits with non-normal distributions. Normal probability plots were created and parameter distributions were transformed to natural logarithm when needed to improve the skew-ness and for obtaining a normal distribution before performing statistical analysis of the relevant phenotypes. Analyses were performed using BioMed Data Program (BMDP ver1.12, Cork, Ireland) and Statistica (ver7.1, StatSoft, Tulsa, OK, USA).
Results
Single gene analyses
We began with analysis of SNPs rs35069869, rs35368790 and rs7639705 in the MCF2L2 gene. Independent segregation of alleles for these studied SNPs were tested in the GoKinD population and they were kept in HWE (P > 0.05). Genetic association analyses with Armitage's trend test, additive and dominant models were performed and data are summarized in Table 2. In SNP rs7639705 (T1165G, Leu359Ile), the G allele frequency in the patients with DN compared to the patients without DN was significantly lower in females (16.5% vs. 22.0%, P = 0.017, OR = 0.701) but not in males (19.7% vs. 22.4%, P = 0.275). Comparison analysis of genotype distribution indicated a significant association between this polymorphism and DN among females (P = 0.018, OR = 0.757). The same analyses in SNPs rs35069869 and rs35368790 between cases (T1DM with DN) and controls (T1DM without DN) were done and no statistically significant association was found.
Table 2.
Association between the MCF2L2 genetic polymorphisms and diabetic nephropathy
| SNP ID Hit Orientation |
Group | Genotype Frequency | Allele Frequency | Association Significance P-value/Odds ratio (95% CI) |
|||
|---|---|---|---|---|---|---|---|
| rs35069869 | Women | AA | AG | GG | A | G | |
| A/G | T1D without DN | 174 (0.489) | 153 (0.430) | 29 (0.081) | 0.704 | 0.211 | |
| T1D with DN | 135 (0.509) | 109 (0.411) | 21 (0.079) | 0.715 | 0.285 | 0.661 a | |
| Men | |||||||
| T1D without DN | 125 (0.519) | 101 (0.419) | 15 (0.062) | 0.728 | 0.272 | ||
| T1D with DN | 150 (0.479) | 137 (0.438) | 26 (0.083) | 0.698 | 0.302 | 0.273 a | |
| rs35368790 | Women | GG | GC | CC | G | C | |
| G/C | T1D without DN | 168 (0.471) | 151 (0.423) | 38 (0.106) | 0.682 | 0.318 | |
| T1D with DN | 134 (0.511) | 107 (0.408) | 33 (0.105) | 0.716 | 0.284 | 0.204 a | |
| Men | |||||||
| T1D without DN | 107 (0.442) | 110 (0.455) | 25 (0.103) | 0.669 | 0.331 | ||
| T1D with DN | 146 (0.466) | 134 (0.428) | 33 (0.105) | 0.681 | 0.319 | 0.696 a | |
| rs7639705 | Women | TT | TG | GG | T | G | |
| T1165G | T1D without DN | 213 (0.603) | 125 (0.354) | 15 (0.043) | 0.780 | 0.220 | |
| Leu359Ile | T1D with DN | 188 (0.712) | 65 (0.246) | 11 (0.042) | 0.835 | 0.165 | 0.017/0.701 (0.524-0.938) a 0.018/0.757 b |
| Men | |||||||
| T1D without DN | 157 (0.651) | 73 (0.303) | 11 (0.046) | 0.803 | 0.197 | ||
| T1D with DN | 183 (0.590) | 115 (0.371) | 12 (0.039) | 0.776 | 0.224 | 0.275 a | |
T1D = type 1 diabetes, DN = diabetic nephropathy. Association significances = a. test for allele frequency difference (additive model), and b. for Armitage's trend.
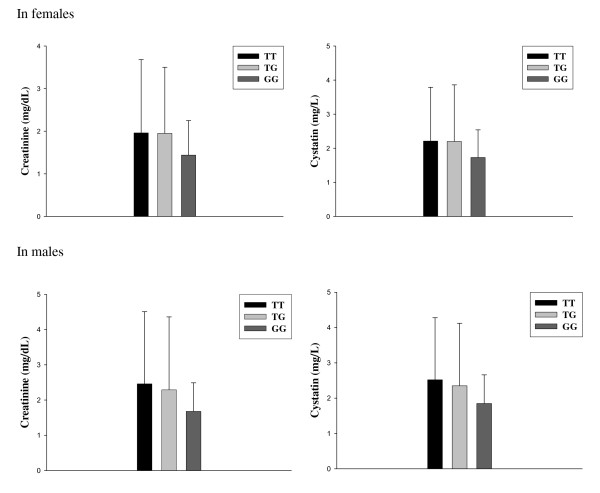
We further detected the association between phenotypes and genotypes of SNP rs7639705 (T1165G, Leu359Ile) in female T1DM patients with or without DN. Figure 1 represents variations of the creatinine and cystatin levels in female and male T1DM patients with DN according to the genotypes of SNP rs7639705. In females, the DN patients carrying TT and TG genotypes had similar levels of creatinine (1.96 ± 1.55 and 1.95 ± 1.72 mg/mL) and cystatin (2.20 ± 1.66 and 2.21 ± 1.58 mg/L) in mean values, while the carriers with GG genotype had tendency decreased mean values of creatinine and cystatin levels (1.44 ± 0.81 mg/mL; 1.73 ± 1.09 mg/L) compared to the carriers with TT and TG genotypes. But, the differences were not statistically significant in both creatinine and cystatin levels (P = 0.074 and 0.058) mainly due to the higher standard deviations. Among male T1DM patients with DN, the GG genotype carriers also showed tendency decreased but not significant mean values of creatinine (1.68 ± 0.57 mg/mL vs. 2.29 ± 2.07 and 2.46 ± 2.05 mg/mL) and cystatin (1.85 ± 0.87 mg/L vs. 2.35 ± 1.77 and 2.52 ± 1.76 mg/L) levels compared to the TT and TG genotype carriers. There was no difference in creatinine and cystatin levels among T1DM patients without DN in both females and males who carry three different genotypes (data not shown).
Figure 1.
Variation of creatinine and cystatin levels among female and male type 1 diabetes patients with diabetic nephropathy according to the genotypes of Leu359Ile variant in the MCF2L2 gene. Untransformed mean values (± SD) for creatinine and cystatin levels in female and male type 1 diabetes patients with DN are shown. The patients with DN carrying GG genotype had tendency decreased levels of creatinine and cystatin compared the carriers with TT and TG genotypes. But, the differences were not statistically significant mainly due to the higher standard deviations.
Multi-gene analyses
We have recently genotyped SNPs rs266729 and rs11915160, respectively, in the ADIPOQ and SOX2 genes in the same cohort of GoKinD study. Data indicate that these two polymorphisms have an interactive association effect with DN in female T1DM patients [15,16]. In the present study, we employed the GMDR analyses to assess the impact of combinations with the model of MCF2L2 (rs7639705)-SOX2 (rs11915160)-ADIPOQ (rs266729) in 622 female subjects. Table 3 presents the gene-gene interaction and impact of MCF2L2, SOX2 and ADIPOQ genetic polymorphisms in DN among female T1DM patients from GoKinD population. Data were obtained from GMDR analyses without covariate adjustment. There was a significant three-gene model (empirical P-value = 0.001). The model had the cross-validation consistency of 10/10 and testing accuracy of 56.2%. The significance was confirmed by a logistic regression model (P < 0.001). The analyses with two-gene model i.e. MCF2L2 together with either ADIPOQ or SOX2 genetic polymorphisms were also performed, and no significant interaction of MCF2L2-ADIPOQ or MCF2L2-SOX2 (P = 0.377, 0.623). The GMDR analysis with the same models studied in females was also performed in male subjects and no significant data were observed.
Table 3.
MCF2L2, Sox2 and AdipoQ gene-gene interaction and impact on diabetic nephropathy in female type 1 diabetic patients
| Best combination | Cross-validation consistency | Testing accuracy (%) | P-value |
|---|---|---|---|
| MCF2L2 (rs7639705) - Sox2 (rs11915160) - AdipoQ (rs266729) | 10/10 | 0.562 | 0.001 |
| MCF2L2 (rs7639705) - Sox2 (rs11915160) | 10/10 | 0.513 | 0.623 |
| MCF2L2 (rs7639705) - AdipoQ (rs266729) | 10/10 | 0.545 | 0.377 |
P-values were based upon 1000 permutations.
Discussion
We have used a positional candidate gene research approach to search for the susceptibility or resistance genes in DN. Three candidates including MCF2L2, ADIPOQ and SOX2 in chromosome 3q26-27 are selected for the genotyping analyses. The MCF2L2 gene has the distances of 1,463 kb and 3,415 kb, respectively, to the location of SOX2 and ADIPOQ. In the present study, we found that SNP rs7639705 (T1165G Leu359Ile) in the MCF2L2 gene is significantly associated with decreased risk of DN in female T1DM patients in the GoKinD population. The patients carrying GG genotype have tendency decreased creatinine and cystatin levels compared to the TT and TG carriers. Serum creatinine and cystatin levels are useful for prediction of the impact on renal function measurement and prognostic stages of DN. Therefore, the data suggest that MCF2L2 may have resistant genetic effects on DN.
Takeuchi et al. have reported that two intronic polymorphisms i.e. rs35069869 and rs35368790 in the MCF2L2 gene are associated with T2DM in a Japanese population [19], but the Leu359Ile polymorphism is not included in their study. Interestingly, FARP2 encoded for FERM, pleckstrin domain protein 2 is another Rho GEF in the same family. Savage et al. have demonstrated that rs757978 in the FARP2 gene is associated with ESRD in T1DM, and the G allele frequency of this polymorphism in T1DM patients with ESRD was lower than the control subjects (0.090 vs 0.118, P = 0.008) [23]. Rs757978 is a non-synonymous polymorphism causing amino acid change from Thr to Ile at the code 260 of FARP2 cDNA. This is similar to what we have found in the MCF2L2 gene. Furthermore, the recent biological studies have indicated that GEFs of Rho family may regulate GTPase Cdc42 activity and subsequently contribute to the processes of macro-and/or micro-vascular diseases [24,25]. Therefore, we have a hypothesis that GEFs of Rho family may play an important role in diabetic micro-vascular complications. At the present stage, information of MCF2L2 and FARP2 in patho-physiology is very limited. Further investigation is necessary to investigate the direct effect of the Rho family GEFs, particularly MCF2L2 and FARP2, on the development of DN or indirect influence via variation of GTPase activity.
Several genetic linkage analyses have predicted that a chromosomal region on 3q is linked to diabetes and DN [7-13]. Therefore, researchers have searched for the susceptibility and/or resistance genes of DN in the region of this chromosomal arm. He et al. have recently genotyped 3072 tag SNPs) spanning a 28 Mb region around chromosome 3q22 and found that rs1866813 is associated with DN. However, this is an intergenic polymorphism and no gene conferring the susceptibility risk to DN has been identified [26]. Vionnet et al. have selected and studied 14 candidate genes including AGTR1, PTX3, IL12A, SLC2A2, TNFSF10, ECE2, THPO, EHHADH, HRG, KNG1, AdipoQ, SST, PPP1R2 and APOD from chromosome 3q24-29 and found that the AdipoQ promoter polymorphism rs17300539 is associated with DN in T1DM patients in Danish, but not in French and Finnish populations [14]. We have studied the candidate genes including TRPC1 (3q22-24), MME (3q25), SOX2 (3q26.3), MCF2L2 (3q27) and ADIPOQ (3q27). We have found that the AdipoQ promoter polymorphism-11377C/G is associated with DN in female subjects from the GoKinD population [15]. This polymorphism alters the sequence in one of transcription factor SP1 binding sites and causes reduction of ADIPOQ promoter activity [27,28]. Moreover, the investigators in the field of stem cell research have amply demonstrated that four transcription factors, including Oct3/4, Sox2, Klf4 and c-Myc, play an important role in induction of pluripotent stem cells from somatic cells ([29-32]. We have recently performed a genetic study of the Sox2gene in the same GoKinD cohort as studied in the MCF2L2 and AdipoQ genes, and found that Sox2 genetic polymorphism is associated with DN in female T1DM subjects [16]. The MCF2L2 gene has the distances of 146 kb and 341 kb, respectively, to the location of Sox2 and AdipoQ. In order to evaluate whether MCF2L2 has gene-gene interaction with ADIPOQ and Sox2 in term of association with DN, we have performed gene-gene interaction analysis. The results suggest that MCF2L2 has combined genetic effects with both ADIPOQ and Sox2 on DN among female subjects but not with either ADIPOQ or Sox2 alone. The linkage of chromosome 3q to DN is most likely determined by the multiplex loci, including these three genes.
Evidence has indicated that gender is an important risk factor for the development and progression of DN. This disease appears less frequently in females than in males with a ratio of approximately 1:1.3 [33]. Previously, Pettersson-Fernholm et al. have demonstrated that the AT2 genetic polymorphisms confer risk susceptibility to the development of DN in male but not female T1DM subjects of Finnish population [34]. In the present study, we have demonstrated that MCF2L2, AdipoQ and Sox2 genetic polymorphisms have protective effects on DN in female T1DM patients. The genetic association of MCF2L2, AdipoQ and Sox2 genetic polymorphisms with DN in male T1DM patients is not observed. The pattern of tendency decreased creatinine and cystatin levels in DN patients carrying GG genotype compared to the TT and TG carriers is seen in both females and males. Therefore, the genotypic and phenotypic differences in gender should be taken into our consideration in genetic and functional analyses of DN.
Conclusions
The present study provides the first evidence that the MCF2L2 Leu359Ile polymorphism is associated with decreased risk of DN in female T1DM patients. MCF2L2 together with ADIPOQ and Sox2 genetic polymorphisms have combined genetic effects on DN in females, which reveals that the linkage of chromosome 3q to DN may be explained by the accumulated effects of multi-genes.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DZ performed genotyping experiments and data analysis. SE, KB and HFG conceived of the study and also evaluated the data. HFG participated in experiments and data analysis. All authors prepared and approved the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Dongying Zhang, Email: dongying.zhang@ki.se.
Suad Efendic, Email: suad.efendic@ki.se.
Kerstin Brismar, Email: kerstin.brismar@ki.se.
Harvest F Gu, Email: harvest.gu@ki.se.
Acknowledgements
The authors wish to thank all subjects included in GoKinD study for their participation. The GoKinD sample collection was supported by the Juvenile Diabetes Research Foundation in collaboration with the Joslin Diabetes Center and George Washington University, and by the United States Centers for Diabetes Control and Prevention. This work was supported by Family Erling-Persson Foundation, Loo and Hans Osterman Foundation and Stig och Gunborg Westmans Foundation.
References
- Hasslacher C, Ritz E, Wahl P, Michael C. Similar risks of nephropathy in patients with type I or type II diabetes mellitus. Nephrol Dial Transplant. 1989;4:859–63. doi: 10.1093/ndt/4.10.859. [DOI] [PubMed] [Google Scholar]
- U.S. Renal Data System, USRDS 2007 Annual Data Report. Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD. 2007.
- Freedman BI, Tuttle AB, Spray BJ. Familial predisposition to nephropathy in African-Americans with non-insulin-dependent diabetes mellitus. Am J Kidney Dis. 1995;25:710–3. doi: 10.1016/0272-6386(95)90546-4. [DOI] [PubMed] [Google Scholar]
- Osterholm AM, He B, Pitkaniemi J, Albinsson L, Berg T, Sarti C, Tuomilehto J, Tryggvason K. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int. 2007;71:140–5. doi: 10.1038/sj.ki.5001933. [DOI] [PubMed] [Google Scholar]
- Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic factors in diabetic nephropathy. Clin J Am Soc Nephrol. 2007;2:1306–16. doi: 10.2215/CJN.02560607. [DOI] [PubMed] [Google Scholar]
- Yamagishi S, Fukami K, Ueda S, Okuda S. Molecular mechanisms of diabetic nephropathy and its therapeutic intervention. Curr Drug Targets. 2007;8:952–9. doi: 10.2174/138945007781386884. [DOI] [PubMed] [Google Scholar]
- Moczulski DK, Rogus JJ, Antonellis A, Warram JH, Krolewski AS. Major susceptibility locus for nephropathy in type 1 diabetes on chromosome 3q: results of novel discordant sib-pair analysis. Diabetes. 1998;47:1164–9. doi: 10.2337/diabetes.47.7.1164. [DOI] [PubMed] [Google Scholar]
- Bowden DW, Colicigno CJ, Langefeld CD, Sale MM, Williams A, Anderson PJ, Rich SS, Freedman BI. A genome scan for diabetic nephropathy in African Americans. Kidney Int. 2004;66:1517–26. doi: 10.1111/j.1523-1755.2004.00915.x. [DOI] [PubMed] [Google Scholar]
- Chistiakov DA, Savost'anov KV, Shestakova MV, Chugunova LA, Samkhalova MSh, Dedov II, Nosikov VV. Confirmation of a susceptibility locus for diabetic nephropathy on chromosome 3q23-q24 by association study in Russian type 1 diabetic patients. Diabetes Res Clin Pract. 2004;66:79–86. doi: 10.1016/j.diabres.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Freedman BI, Bowden DW, Sale MM, Langefeld CD, Rich SS. Genetic susceptibility contributes to renal and cardiovascular complications of type 2 diabetes mellitus. Hypertension. 2006;48:8–13. doi: 10.1161/01.HYP.0000227047.26988.3e. [DOI] [PubMed] [Google Scholar]
- Osterholm AM, He B, Pitkaniemi J, Albinsson L, Berg T, Sarti C, Tuomilehto J, Tryggvason K. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int. 2007;71:140–5. doi: 10.1038/sj.ki.5001933. [DOI] [PubMed] [Google Scholar]
- McKnight AJ, Maxwell AP, Sawcer S, Compston A, Setakis E, Patterson CC, Brady HR, Savage DA. A genome-wide DIABETIC NEPHROPATHYA microsatellite association screen to identify chromosomal regions harboring candidate genes in diabetic nephropathy. J Am Soc Nephrol. 2006;17:831–6. doi: 10.1681/ASN.2005050493. [DOI] [PubMed] [Google Scholar]
- Rogus JJ, Poznik GD, Pezzolesi MG, Smiles AM, Dunn J, Walker W, Wanic K, Moczulski D, Canani L, Araki S, Makita Y, Warram JH, Krolewski AS. High-density single nucleotide polymorphism genome-wide linkage scan for susceptibility genes for diabetic nephropathy in type 1 diabetes: discordant sibpair approach. Diabetes. 2008;57(9):2519–26. doi: 10.2337/db07-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vionnet N, Tregouët D, Kazeem G, Gut I, Groop PH, Tarnow L, Parving HH, Hadjadj S, Forsblom C, Farrall M, Gauguier D, Cox R, Matsuda F, Heath S, Thévard A, Rousseau R, Cambien F, Marre M, Lathrop M. Analysis of 14 candidate genes for diabetic nephropathy on chromosome 3q in European populations: strongest evidence for association with a variant in the promoter region of the adiponectin gene. Diabetes. 2006;55(11):3166–74. doi: 10.2337/db06-0271. [DOI] [PubMed] [Google Scholar]
- Zhang D, Ma J, Brismar K, Efendic S, Gu HF. A single nucleotide polymorphism alters the sequence of SP1 binding site in the adiponectin promoter region and is associated with diabetic nephropathy among type 1 diabetic patients in the Genetics of Kidneys in Diabetes Study. J Diabetes Complications. 2009;23(4):265–72. doi: 10.1016/j.jdiacomp.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Gu HF, Alvarsson A, Efendic S, Brismar K. SOX2 Has Gender-Specific Genetic Effects on Diabetic Nephropathy in Samples From Patients With Type 1 Diabetes Mellitus in the GoKinD Study. Gender Medicine. 2009;6:555–564. doi: 10.1016/j.genm.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Loirand G, Scalbert E, Bril A, Pacaud P. Rho exchange factors in the cardiovascular system. Curr Opin Pharmacol. 2008;8:174–80. doi: 10.1016/j.coph.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Lezoualc'h F, Métrich M, Hmitou I, Duquesnes N, Morel E. Small GTP-binding proteins and their regulators in cardiac hypertrophy. J Mol Cell Cardiol. 2008;44:623–32. doi: 10.1016/j.yjmcc.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, Ochiai Y, Serizawa M, Yanai K, Kuzuya N, Kajio H, Honjo S, Takeda N, Kaburagi Y, Yasuda K, Shirasawa S, Sasazuki T, Kato N. Search for type 2 diabetes susceptibility genes on chromosomes 1q, 3q and 12q. J Hum Genet. 2008;53:314–24. doi: 10.1007/s10038-008-0254-6. [DOI] [PubMed] [Google Scholar]
- Mueller PW, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, Bucksa J, Gibson TB, Cordovado SK, Krolewski AS, Nierras CR, Warram JH. Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol. 2006;17:1782–90. doi: 10.1681/ASN.2005080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. Am J Hum Genet. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80(6):1125–37. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DA, Patterson CC, Deloukas P, Whittaker P, McKnight AJ, Morrison J, Boulton AJ, Demaine AG, Marshall SM, Millward BA, Thomas SM, Viberti GC, Walker JD, Sadlier D, Maxwell AP, Bain SC. Genetic association analyses of non-synonymous single nucleotide polymorphisms in diabetic nephropathy. Diabetologia. 2008;51(11):1998–2002. doi: 10.1007/s00125-008-1142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loirand G, Scalbert E, Bril A, Pacaud P. Rho exchange factors in the cardiovascular system. Curr Opin Pharmacol. 2008;8:174–80. doi: 10.1016/j.coph.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Lezoualc'h F, Métrich M, Hmitou I, Duquesnes N, Morel E. Small GTP-binding proteins and their regulators in cardiac hypertrophy. J Mol Cell Cardiol. 2008;44:623–32. doi: 10.1016/j.yjmcc.2008.01.011. [DOI] [PubMed] [Google Scholar]
- He B, Osterholm AM, Hoverfält A, Forsblom C, Hjörleifsdóttir EE, Nilsson AS, Parkkonen M, Pitkäniemi J, Hreidarsson A, Sarti C, McKnight AJ, Maxwell AP, Tuomilehto J, Groop PH, Tryggvason K. Association of genetic variants at 3q22 with nephropathy in patients with type 1 diabetes mellitus. Am J Hum Genet. 2009;84(1):5–13. doi: 10.1016/j.ajhg.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth N, langmann T, Scholmerich J, Schmitz G, Schaffler A. Identification of regulatory elements in the human adipose most abundant gene transcript-1 (apM-1) promoter: role of SP1/SP3 and TNF-alpha as regulatory pathways. Diabetologia. 2002;45:1425–33. doi: 10.1007/s00125-002-0895-5. [DOI] [PubMed] [Google Scholar]
- Bouatia-Naji N, Meyre D, Lobbens S, Seron K, Fumeron F, Balkau B, Heude B, Jouret B, Scherer PE, Dina C, Weill J, Froguel P. ACDC/adiponectin polymorphisms are associated with severe childhood and adult obesity. Diabetes. 2006;55:545–50. doi: 10.2337/diabetes.55.02.06.db05-0971. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–35. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–9. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney A, Tunbridge WM, Ireland JT, Watkins PJ. Mortality from diabetic nephropathy in the United Kingdom. Diabetologia. 1983;25:26–30. doi: 10.1007/BF00251892. [DOI] [PubMed] [Google Scholar]
- Pettersson-Fernholm K, Frojdo S, Fagerudd J, Tomas MC, Forsblom C, Wessman M, Groop PH. for the FinnDiane study group. The AT2 gene may have a gender-specific effect on kidney function and pulse pressure in type 1 diabetic patients. Kid Int. 2006;69:1880–4. doi: 10.1038/sj.ki.5000348. [DOI] [PubMed] [Google Scholar]