Abstract
Objective
Mucin genes MUC2, MUC5AC and MUC5B have been identified as major gel-forming mucins in the middle ear (ME). This study compared polymorphisms in MUC2, MUC5AC and MUC5B genes in otitis media (OM) patients and controls.
Design
Cross-sectional case-control study. Patients age 6 months to 14 years undergoing routine tympanostomy tube (TT) insertion for recurrent otitis media (RecOM) or chronic otitis media with effusion (OME) were compared to control patients with no history of OM undergoing an unrelated procedure.
Methods
Genomic DNA extracted from peripheral blood samples was analyzed by Southern blots using gene-specific probes to determine sizes of MUC2 and MUC5AC genes. Polymerase chain reaction (PCR) was used to determine the size of MUC5B genes.
Results
Twenty RecOM patients, twenty OME patients, and forty control patients were analyzed. There were no statistically significant differences in polymorphism expression identified between groups for MUC2 and MUC5B genes. OME patients were more likely than Controls or RecOM patients to carry the longer MUC5AC-b allele.
Conclusion
Differences in mucin polymorphisms have been demonstrated in other diseases. In otitis media, OME patients are more likely than Controls or RecOM to carry the longer MUC5AC-b allele. MUC5AC is a primary innate defense mechanism for ME epithelium and alterations in protein structure have the potential to affect that defense and predispose patients to disease. This study demonstrates that the properties of post-transcriptionally modified MUC5AC deserve further study and specific pathogen-host interactions studies should explore the impact of this polymorphism.
Keywords: mucin gene, otitis media, polymorphism
INTRODUCTION
Otitis media (OM) is the most common diagnosis in pediatric patients who visit physicians for illness in the United States.1 Despite the prevalence of otitis media, its potential for morbidity, the enormous health care expenditures resulting from its treatment, the frequent need for surgical intervention and the increasing therapeutic challenges imposed by antimicrobial resistance, much is still unknown about molecular processes that are important in OM pathogenesis. Particularly, though it has been well-documented that genetic variations among individuals have a significant impact on OM susceptibility2,3, there has been very little investigation into specific genetic traits that might be important in this disease process.
Mucins are a family of large glycoproteins and 19 unique human mucin genes have been described. In the middle ear, mucins provide important protective functions for the underlying epithelium including mechanical protection and pathogen eradication through clearance mechanisms and interaction with the host immune system. However, variation in the quantity and character of middle ear secretions, and specifically mucin secretion, are known to be important in the pathophysiologic mechanisms of otitis media as well.4 Mucins are the only component of middle ear effusions responsible for its viscosity and over-production of highly viscous mucins contribute to abnormal mucociliary clearance in the middle ear (ME), which, in turn, causes pathology such as chronic otitis media and hearing loss.5 Investigations designed to increase our knowledge of middle ear mucin regulation may develop meaningful new intervention strategies for otitis media by incorporating a concept of limiting or modulating mucin production by middle ear epithelium. Despite this understanding of the importance of mucin in otitis media, very little has been done to investigate specific cellular, molecular or genetic mechanisms that may be related to differences in ME mucin production. This study, utilizing a patient cohort that is part of a broader investigation of OM pathogenesis, assessed mucin gene polymorphisms in patients with a diagnosis of OM and compared this data to a control population to investigate the impact of mucin gene polymorphisms in OM.
MATERIALS AND METHODS
Subject Recruitment and Enrollment
IRB approval was obtained from Children’s Hospital of Wisconsin prior to initiating this investigation and informed consent was obtained for all patients participating in this investigation.
Consecutive children in the senior author’s (JEK) pediatric otolaryngology practice who were scheduled to undergo tympanostomy tube (TT) placement for otitis media after clinical evaluation and were assessed by a clinical research nurse were eligible for entry into this study as the Study Group. At total of 40 patients were enrolled in the Study Group, 20 patients in the Study Group had a diagnosis of OME and 20 patients had a diagnosis of RecOM as defined below. Children having other surgical procedures that required the starting of an IV but without a significant history of otitis media were eligible for entry into this study as the Control Group. There were 40 patients in the Control Group. Children in the Control Group had one or fewer episodes of OM per year of life per parent history. The Study Group patients were enrolled first. Upon completion of this portion of the proposal, the Control Group patients were enrolled with an attempt to match the Study Group patients with respect to gender. Ethnicity and age were used as secondary considerations for matching.
Definitions
OME: Any patient with persistent middle ear effusion for greater than three month duration.
RecOM: Any patient with 3 or more episodes of acute OM over a 6 month period with resolution between episodes and middle ear fluid that does not persist greater than 3 months duration.
Absence of history of otitis media (Control Group): One or fewer episodes of otitis media requiring a visit to a health professional per 12 month period of life.
Inclusion criteria
Study Group: Patients age 6 months to 14 years scheduled for TT placement with a diagnosis of RecOM or OME.
Control Group: Patients age 6 months to 14 years scheduled for outpatient surgery (e.g. hernia repair) without a history of RecOM or OME.
Exclusion criteria
Immunologic abnormality, intrinsic or pharmacologic
Anatomic or physiologic defect of the ear
Syndrome associated with OM (e.g. Down syndrome, cleft palate)
History of sensorineural hearing loss
Chronic mastoiditis, cholesteatoma or other complications associated with OM except for conductive hearing loss
Tympanic membrane perforation
Major medical conditions or undergoing treatment for chronic illness
Patients were enrolled and matched for gender. Fisher Exact Test and Chi-square testing was used to assess differences between Study and Control Groups. Secondary analysis was performed matching patients for ethnicity.
Sample collection
After induction of general anesthesia a 4-cc sample of venous blood was obtained from the arm of all patients enrolled in both the Study and Control Groups. The blood sample was kept on ice until transportation to the laboratory for DNA isolation, but less than one hour transpired between obtaining the samples and transport to the laboratory.
Isolation of genomic DNA (Blood samples)
Genomic DNA from anticoagulated peripheral blood was then extracted with the Puregene Isolation Kit (Gentra Systems, Minneapolis, MN). The DNA was resuspended in Puregene hydration solution at a concentration of 100μg/ml and stored at −20°C until analyzed for mucin genetic polymorphisms. Each blood sample generated genomic DNA with 2 alleles for mucin gene analysis.
DNA Sequencing
SMUC41 and Jer47 fragments obtained from PCR reaction were purified with QIAEX II gel extraction kit (Qiagen, Valencia, CA) and sequenced at the Protein and Nucleic Acid (PNA) Core Facility at the Medical College of Wisconsin.
Southern Blot/PCR studies to assess mucin gene polymorphism
A total of 5 to 7μg of DNA was utilized from each sample and treated with HinfI or PvuII in a final volume of 25μl, as recommended by the manufacturer (GIBCO-BRL, Life Technologies Ltd., Paisley, UK). The HinfI fragments were separated by agarose electrophoresis (0.8% agarose in 0.89M Tris, 0.1M borate, 0.002M ethylenediaminetetraacetic acid [EDTA] buffer, pH 8.3 [TBE]) on 20 x 25cm gels with 30 samples per gel (Horizon 20:25 apparatus; GIBCO-BRL) for 24 h at 2 V/cm. The PvuII fragments were separated using 0.5% agarose, 1x TBE, and run at 2V/cm for 24 h, followed by a complete change of the tank buffer and continued electrophoresis at 1.2 V/cm for a further 19 h. Three kinds of markers were applied to each gel: Raoul markers (Appligene, Durham, UK), 1-kb ladder (GIBCO-BRL), and lamda HindIII (GIBCO-BRL), and were detected with ethidium bromide. The Raoul markers were also visible after transfer because they are revealed with the MUC probes. Three DNA samples with alleles of known size were also used as further size standards, with two of these samples being mixed and loaded in the same well.
After electrophoresis, the markers were visualized by post-staining with ethidium bromide. After measuring the migration of the marker brands, the gels were depurinated (0.25M HCI for 30 min), denatured (1.5M NaCl, 0.5M NaOH for 30 min), neutralized (0.5M Tris-HCI, 0.001M EDTA, pH 7.2, for 30 min), and the digested DNA was then transferred onto Hybond N+ membranes (Amersham Pharmacia Biotech, Buckinghamshire, UK) by capillary blotting overnight or vacuum blotting (Vacugene XL; Amersham Pharmacia Biotech) for 2h, all as recommended by the manufacturers. The DNA was then fixed onto the filters by baking at 80°C for at least 2h.
Probing and washing of the filters was conducted by standard procedures. The MUC genes were detected using tandem repeat cDNA probes: SMUC41 for MUC2 6, JER47 for MUC5AC.7 A total of 25ng was labeled by random primed labeling using the Multiprime DNA labeling kit (Amersham Pharmacia Biotech). Filters were prehybridized in 200ml of 6x saline sodium citrate (SSC) (20xSSC: 3M NaCl, 0.3M trisodium citrate), 5x Denhardts (100xDenhardts: 2% w/v Ficoll, 2% w/v polyvinylpyrrolidone, 2% w/v bovine serum albumin [BSA]), 0.5% w/v sodium dodecyl sulfate (SDS) in a shaking water bath at 65°C for approximately 4 hours, and 500μg of sonicated Herring sperm DNA and labeled probe boiled together for 5 min will be added to the prehybridization solution for overnight hybridization. The filters were washed down in several changes of SSC, with a final stringent wash of 0.1x SSC, 0.1% SDS at 65°C for 10 min. Autoradiography was carried out with Fuji HRE-30 Medical X-ray film and Fuji screens (FG8) for 1 to 7d at −70°C. The HinfI filters were sequentially probed for MUC2 and MUC5AC.
The position of the well was marked onto the autoradiographs by using luminescent marks (Globug X-ray marking solution; Radleys, Saffron Walden, UK) to line up the filter. The autoradiographs were recorded using a UVP image analysis system (Cambridge, UK) and the alleles were sized in comparison with markers using the Gelworks Image Analysis software package (UVP). The standard curve was plotted using the MUC alleles of known size as well as the Raoul markers, the other markers being used as an extra check for gel quality. MUC2 size was analyzed in relationship to “bins” of 0.5kb. MUC5AC, which shows simpler allelic variation, was analyzed as two size class alleles as described previously8 that were named MUC5AC*a and MUC5AC*b (6.6 and 7.4kb, respectively).
The intronic MUC5B polymorphism was examined by polymerase chain reaction (PCR) using primers that amplify across the entire VNTR region. Each PCR reaction for MUC5B contained 2μl 10x PCR buffer (500mM KC1, 100mM Tris-HC1 pH 9.0, 15mM MgC12, 0.1% Triton X-100), 2μl 2mM deoxynucleotide triphosphates, 1μM sense and antisense primers, 0.5μl (0.25μg) DNA, 0.1μl (0.5U) Taq polymerase (Promega, Madison, WI), and 3μl 50% (v/v) glycerol, with added water to a final volume of 20μl. After a 3-min denaturation at 94°C, 30 cycles were performed consisting of a 30-s annealing at 64°C, 2-min extension at 70°C, and a final extension of 6 min at 72°C (sense primer, 5’-AGTGTGCAGTGACTGGCGAG-3’ nt 3967–3986 in Genbank Y09788; antisense primer, 5’-CTAGAGT-TGCAGGTGGCAGG-3’ nt 4655–4674 in Genbank Y09788). The PCR products were electrophoresed on 2% agarose gels and visualized with ethidium bromide staining under ultraviolet light. The number of tandem repeats was determined utilizing the overall size in base pairs identified on the PCR gel. Mean allele length of MUC2 was assessed by 95% confidence interval.
RESULTS
A total of 80 patients were enrolled in this study. Forty Study patients were enrolled with twenty patients having a diagnosis of RecOM and twenty with a diagnosis of OME. Forty Control patients without a significant history of OM were also enrolled. Two patients in the RecOM group were excluded due to experimental error in DNA extraction from their peripheral blood samples. The patients were similar, but not identical with respect to demographic characteristics (Table I). The proportion of male and female patients in the otitis media patients compared with the Controls was similar (p=0.677). There were differences in the median age in months (ANOVA Rank Sum, p < 0.001). There did exist discrepancies in the overall ethnic representations of the groups.
Table I.
Numbers and demographics of patients enrolled.
| Patient Group (n) | Gender | Median Age (months) | Ethnicity (n) |
|---|---|---|---|
| Controls (40) | M = 27 F = 13 |
67.91 | W = 21 AA = 12 H = 4 O = 3 |
| RecOM (18) | M = 10 F = 8 |
24.70 | W = 15 AA = 1 H = 1 O = 1 |
| OME (20) | M = 13 F = 7 |
46.62 | W = 17 AA = 0 H = 1 O = 2 |
(W: white, AA: African american, H: hispanic, O: other, M: Male, F: Female). There existed no differences between groups with respect to gender (p = 0.677). There were differences in the median age in months (ANOVA Rank Sum, p < 0.001). There did exist discrepancies in the overall ethnic representations of the groups.
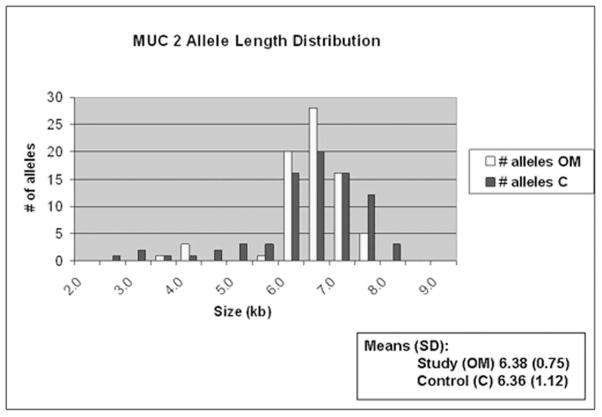
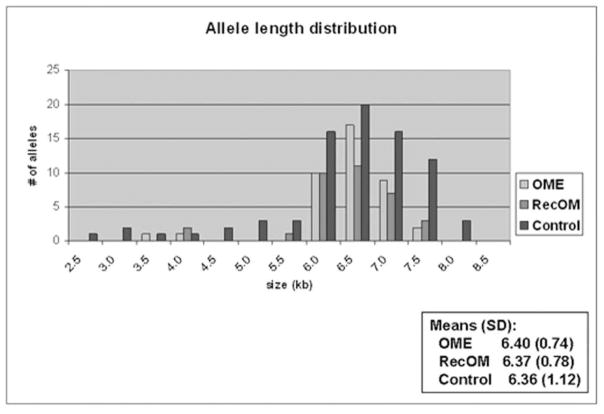
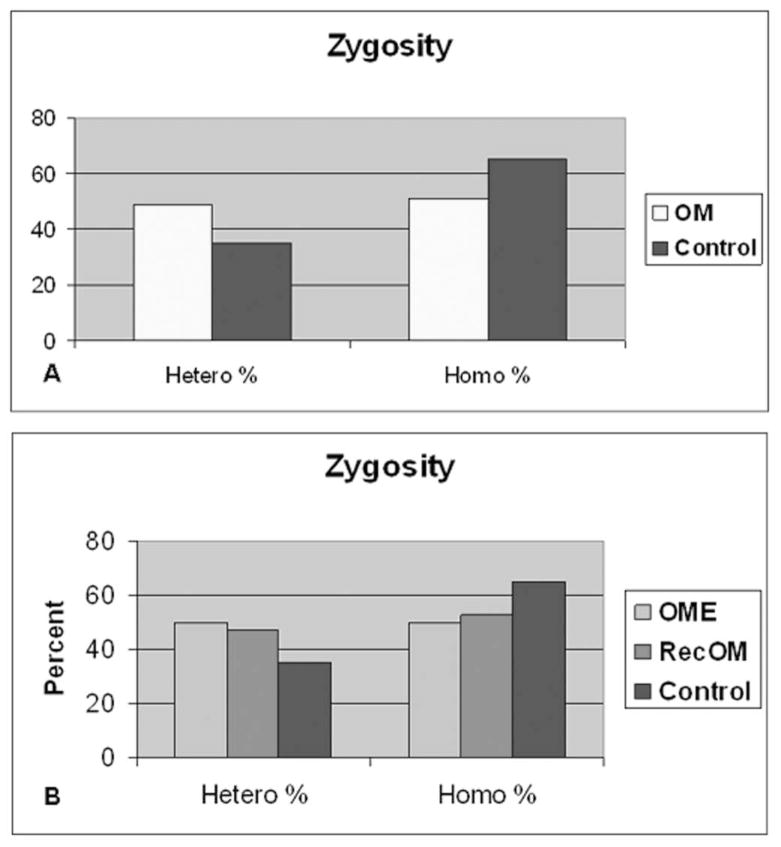
Figure 1 demonstrates a representative autoradiograph utilizing Southern blot techniques to assess MUC2 gene length in patients. Analysis of genomic DNA provides two alleles for assessment in each patient. Patients demonstrated both homozygous (single band) and heterozygous (double band) patterns of allele length. MUC2 polymorphisms are possible within a broad distribution of lengths depending upon the number of tandem repeats and varied from approximately 2.5kb to 8.0kb (Figure 2) in this series of patients. There was a greater overall spread in allele length in the control patients. The most commonly represented allele length was between 6.5 and 7.0kb in both Study patients and Control patients. The mean allele length for Study patients was 6.39kb and 6.36 for Control patients. Breakdown of the Study patients into RecOM and OME groups (Figure 3) did not reveal any significant differences between the subgroups in the Study population or between either of the subgroups and the Control population. There was a greater percentage of patients in the Control population that exhibited homozygous mucin gene length compared with the Study population (65% compared with 51%), however, this was not statistically significant (chi-square p=0.33) (Figure 4A). Similar rates of homozygous patients were found in both the RecOM and OME patients (53% and 50% respectively).
Fig. 1.
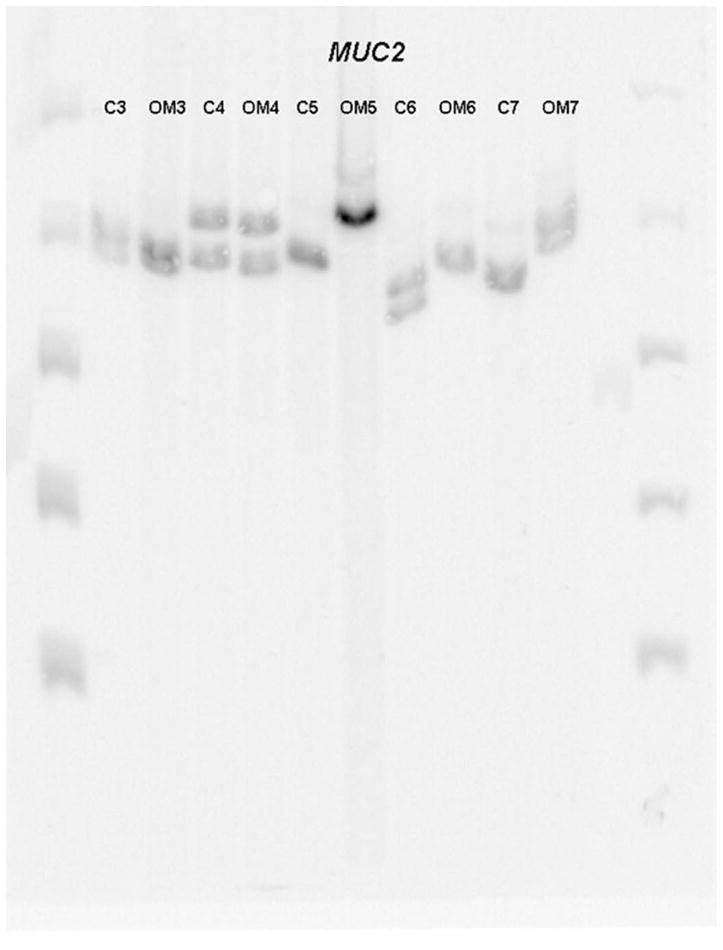
Autoradiograph demonstrating MUC2 allele length.
Fig. 2.
MUC2 allele length distribution comparing Study patients to Controls.
Fig. 3.
MUC2 allele length distribution comparing with breakdown of OME and RecOM patients.
Fig. 4.
MUC 2 zygosity of allele length. 4A represents all Study (OM) patients compared with Controls (p=0.326) and 4B represents Study patients broken down into OME and RecOM. OME patients demonstrated no difference compared with Controls (p= 0.471).
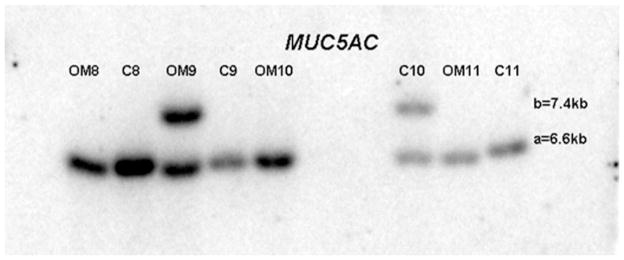
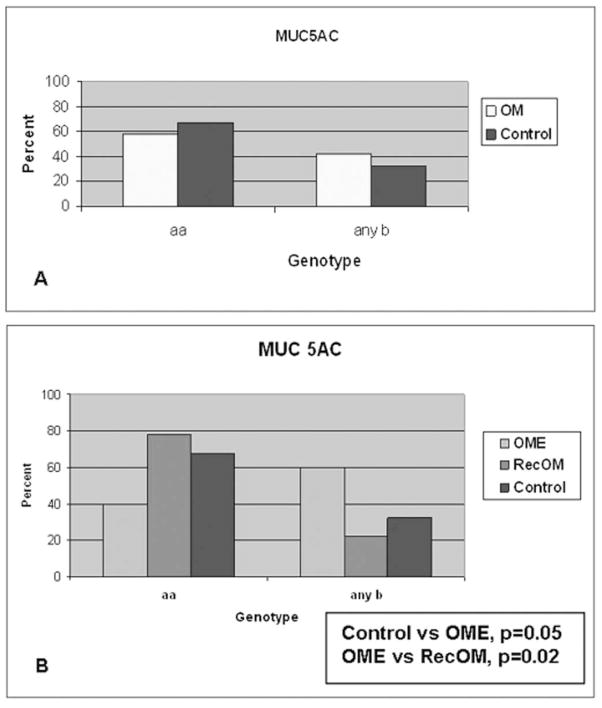
Figure 5 demonstrates a representative autoradiograph utilizing Southern blot techniques to assess MUC5AC gene length in patients. The MUC5AC gene exhibits only two polymorphisms associated with variable numbers of tandem repeats at 6.6kb (designated “a”) and 7.4kb (designated “b”). Patients demonstrated both homozygous (single band) and heterozygous (double band) patterns of allele length (Figure 5). Only three combinations are therefore possible for these patients: homozygous aa, homozygous bb or heterozygous ab. Figure 6 demonstrates that a higher percentage of patients contained an allele (either ab or bb) in the Study (42%) population compared with Controls (33%). This higher percentage in the Study population was completely dependent upon the findings that the longer b allele length was significantly more prevalent in the OME population (60%) compared with theRecOM (22%, p = 0.025). There was no significant difference between the Controls (33%) and RecOM population (22%, p = 0.54). There was difference between Controls and OME populations approached significance (p =0.055). In performing a post-enrollment analysis of the data matching Study and Control populations with respect to race; the prevalence of the b allele in the OME population persisted compared with RecOM population (p = 0.025).
Fig. 5.
Autoradiograph demonstrating MUC5AC allele length.
Fig. 6.
MUC 5AC zygosity of allele length. 6A represents all Study (OM) patients compared with Controls with no differences. 6B represents Study patients broken down into OME and RecOM with significant differences between OME patients and RecOM and Controls.
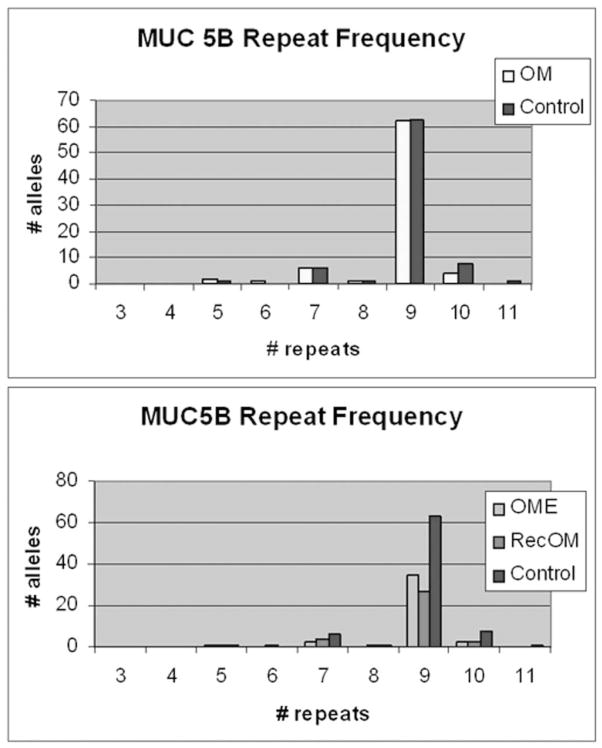
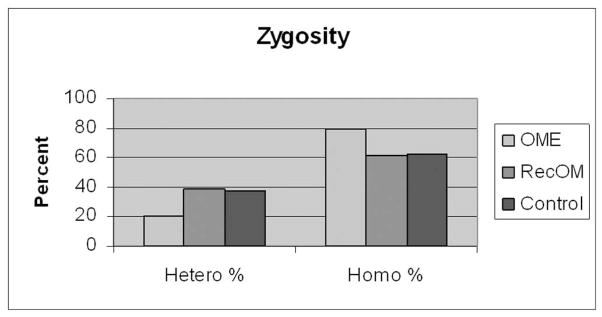
Figure 7 demonstrates a representative PCR gel assessing MUC5B allele length. The number of tandem repeats is directly correlated to the base pair (bp) length on PCR. Figure 8 demonstrates the distribution of number of repeats in the VNTP region for MUC5B and there were no significant differences between Control or Study patients with or without breaking out the OME and RecOM patients. Figure 9 assessed the zygosity of the groups and did demonstrate a higher rate of homozygous allele length in the OME (80%) patients compared with the RecOM patients (61%) and Control patients (63%). This was not statistically significant, however (p=0.339 by Chi-square).
Fig. 7.
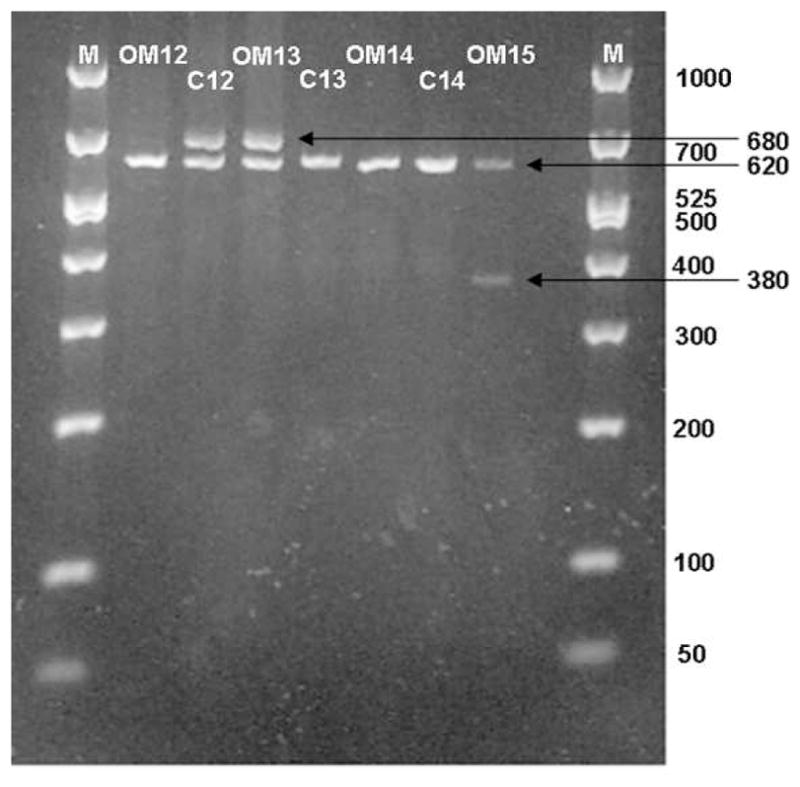
PCR gel demonstrating MUC5B allele length in control (C) and study (OM) patients.
Fig. 8.
MUC5B number of tandem repeats comparing Study patients to Controls.
Fig. 9.
MUC 5B zygosity of allele length. OME patients demonstrated larger percentage of homozygous alleles compared with RecOM and Control patients.
DISCUSSION
The response to infection and inflammation in any individual is controlled by a variety of factors. An individual’s genotype must be considered as one important factor and genetic polymorphism among a group of individuals may markedly influence disease incidence and severity. Recent investigations have determined that the genes MUC1, MUC2, MUC4 and MUC5AC and MUC5B appear to all play some role in chronic otitis media with effusion (OME).5 The gel-forming mucins, MUC2, MUC5AC, and MUC5B are of particular interest in OM given their ability to affect ME fluid viscosity. This ME fluid viscosity, in turn, modulates mucociliary clearance and ultimately impacts the formation of persistent ME effusion, OME, hearing loss and the need for surgical intervention. Given this, these particular mucin genes were the focus of this investigation.
Polymorphisms in the genes that regulate mucin production have been well described.7–11 The clinical importance of these polymorphisms in mucin genes is less well understood. Mucin genes have a common feature of having large tandem repeat regions that can make up to 50% of the polypeptide. Much of the polymorphism in human mucin genes is found in the DNA sequence responsible for the tandem repeat regions leading to differing numbers of these tandem repeats in the polypeptide.7,8 This is commonly referred to as variable numbers of tandem repeats (VNTR). Concentrating on these regions, it has been demonstrated that VNTR polymorphism of the MUC1 mucin gene impacts an individual’s susceptibility to H. pylori gastritis, and longer MUC2 mucin genes protect atopic individuals from developing asthma.8,9 The possibility of mucin gene VNTR polymorphism contributing to the severity or incidence of otitis media has not been investigated and this study investigated these potential differences.
MUC2 and MUC5AC mucin gene polymorphisms occur in an exon corresponding to a highly glycosylated portion of the protein. As such, these polymorphisms have the potential for clinically significant implications resulting from a different length glycoprotein. Polymorphisms may lead to different glycoprotein folding properties or variable levels of glycosylation. Clinically, these changes may alter mucin function by altering middle ear fluid viscosity, mucociliary flow, bacterial or viral adherence, and bacterial- or viral-host interactions. While MUC5B polymorphism occurs in an intron, it could still affect protein folding, as well as binding of transcription factors and ultimately expression of the gene.
The findings from this investigation are particularly interesting in that the longer MUC5AC allele was found in significantly more patients with OME compared with Control patients. There was also a marked difference between OME and RecOM patients with the RecOM patients more closely resembling those of the Control population. MUC5AC is one of the primary mucins secreted from goblet cells and goblet cell hypertrophy is characteristic of chronic disease in respiratory epithelium including that of patients with OME. As such, MUC5AC has been implicated as the major mucin involved in the pathogenesis of chronic OME. MUC5AC has also been shown to be regulated by inflammatory cytokines know to be important in the pathogenesis of OM.12,13
Stimulation of goblet cells to produce MUC5AC significantly increases the viscosity of fluid in respiratory epithelium. The finding that patients with OME are more likely to demonstrate a longer length MUC5AC mucin may indicate that during periods of inflammation and goblet cell stimulation these patients are predisposed to produce mucin of even higher viscosity, leading to a higher incidence of abnormal mucociliary clearance, middle ear fluid stasis and ultimately chronic disease. The implications of these findings are significant in that they would indicate that, if these findings can be replicated in a broader fashion, that there exists a genetic marker which could be utilized to predict which patients are more likely to develop chronic OME when exposed to OM pathogens or inflammation within the ear.
It should also be mentioned that there did exist differences in zygosity for MUC2 and MUC5B genes. OME patients were more likely to be heterozygous for MUC2 gene length and homozygous for MUC5B length compared with Controls. The differences did not reach statistical significance, but this is likely related to relatively small patient numbers. Although there is not an obvious biochemical benefit in being homozygous or heterozygous for mucin gene polymorphism expression there are certainly many examples in which zygosity in gene expression is linked to disease. In the case of mucin expression and OM this may certainly be the case and warrants further study.
There are several other limitations with this study which warrant mentioning. First, the subjects enrolled, both in the Study and Control were not prospectively followed with respect to diagnosis of OM and follow-up of OM. Although patient histories of OM were corroborated with primary care physician records, future studies should focus on prospectively monitoring and controlling for the treatment and diagnosis of OM in study patients utilizing the 2004 Practice Guidelines published for the diagnosis and treatment of patients with OM.14,15 This would likely decrease the possibility of overlap between the Study and Control populations. However, despite the current study design, the finding of differences between OME patients and RecOM and Control patients in MUC5AC likely increases the chance that these differences would be corroborated in a study more tightly controlled on diagnosis. Larger studies should also be considered for future investigations to reassess some of the trends identified in this study to ensure that the study’s power was not the primary determining factor in preventing a statistical difference from being demonstrated. A larger study would also allow for more detailed analysis of many of the other variables known to impact OM incidence including daycare attendance, socioeconomic status and family history as well as allow for design to ensure that patient ethnicity are controlled during design. Given the nature of the current work as a preliminary investigation, the study was not powered to adequately assess each of these factors. However, the current work will provide guidance for future mucin gene – otitis media association studies which are currently being designed.
CONCLUSION
There exist differences in mucin gene polymorphisms between patients with OME and patients without a history of OM. Patients with OME were significantly more likely to have a longer allele length for the MUC5AC mucin gene. MUC5AC is a primary innate defense mechanism for ME epithelium and alterations in protein structure have the potential to affect that defense and predispose patients to disease. This study demonstrates that the properties of post-transcriptionally modified MUC5AC deserve further study and specific pathogen-host interactions studies should explore the impact of this polymorphism. The availability of high-throughput genotyping and the development of large collaborative clinical networks creating well-characterized patient populations with DNA repositories should facilitate identification of additional genotypes that predispose children to this very common disease. These strategies will undoubtedly impact the evaluation and treatment of OM in the years to come.
Acknowledgments
This work was supported by the National Institutes of Health [grant number NIDCD DC007903 to JEK]; and the Department of Otolaryngology and Communication Sciences, Medical College of Wisconsin.
Footnotes
Portions of this manuscript were presented at the 9th International Symposium on Recent Advances in Otitis Media in St. Pete, FL on June 5, 2007.
None of the authors of this manuscript have any financial or non-financial competing interests to disclose.
References
- 1.Rosenfeld RM, Casselbrant ML, Hannley MT. Implications of the AHRQ evidence report on acute otitis media. Otolaryngol Head Neck Surg. 2001;125:439–448. doi: 10.1067/mhn.2001.119326. [DOI] [PubMed] [Google Scholar]
- 2.Casselbrant ML, Mandel EM. Genetic susceptibility to otitis media. Curr Opin Allergy Clin Immunol. 2005;5(1):1–4. doi: 10.1097/00130832-200502000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Segade F, Daly KA, Allred D, et al. Association of the FBXO11 gene with chronic otitis media with effusion and recurrent otitis media: the Minnesota COME/ROM Family Study. Arch Otolaryngol Head Neck Surg. 2006;132(7):729–733. doi: 10.1001/archotol.132.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ubell ML, Kerschner JE, Wackym PA, Burrows A. MUC2 expression in human middle ear epithelium of otitis media patients. Arch Otolaryngol Head Neck Surg. 2008;134(1):39–44. doi: 10.1001/archoto.2007.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerschner JE. Mucin gene expression in human middle ear epithelium. American Laryngological, Rhinological and Otological Society Thesis Laryngoscope. 2007;117(9):1666–1676. doi: 10.1097/MLG.0b013e31806db531. [DOI] [PubMed] [Google Scholar]
- 6.Gum JR, Byrd JC, Hicks JW, Toribara NW, Lamport DT, Kim YS. Molecular cloning of human intestinal mucin cDNAs. J Biol Chem. 1989;264:6480–6487. [PubMed] [Google Scholar]
- 7.Guyonnet Duperat V, Audie JP, Debailleul V, et al. Characterization of the human mucin gene MUC5AC: a consensus cysteine-rich domain for 11p15 mucin genes? Biochem J. 1995;305:211–219. doi: 10.1042/bj3050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinall LE, Fowler JC, Jones AL, et al. Polymorphism in human mucin genes in chest disease: possible significance of MUC2. Am J Respir Cell Mol Biol. 2000;23:678–686. doi: 10.1165/ajrcmb.23.5.4176. [DOI] [PubMed] [Google Scholar]
- 9.Vinall LE, King M, Novelli M, et al. Altered expression and allelic association of the hypervariable membrane mucin MUC1 in Heliobacter pylori gastritis. Gastroenterology. 2002;123:41–49. doi: 10.1053/gast.2002.34157. [DOI] [PubMed] [Google Scholar]
- 10.Vinall LE, Hill AS, Pigny P, et al. Variable number tandem repeat polymorphism of the mucin genes located in the complex 11p15.5. Hum Genet. 1998;102:357–366. doi: 10.1007/s004390050705. [DOI] [PubMed] [Google Scholar]
- 11.Imbert Y, Foulks GN, Brennan MD, et al. MUC1 and estrogen receptor alpha gene polymorphisms in dry eye patients. Exp Eye Res. 2008 Jun 20; doi: 10.1016/j.exer.2008.05.019. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Smirnova MG, Birchall JP, Pearson JP. In vitro study of IL-8 and goblet cells: possible role of IL-8 in the aetiology of otitis media with effusion. Acta Oto-Laryngologica. 2002;122(2):146–152. doi: 10.1080/00016480252814144. [DOI] [PubMed] [Google Scholar]
- 13.Kerschner JE, Meyer TK, Burrows A. Chinchilla middle ear epithelial mucin gene expression in response to inflammatory cytokines. Arch Otolaryngol Head Neck Surg. 2004;130:1163–1167. doi: 10.1001/archotol.130.10.1163. [DOI] [PubMed] [Google Scholar]
- 14.American Academy of Pediatrics Subcommittee on Management of Acute Otitis Media. Diagnosis and management of acute otitis media. Pediatrics. 2004;113(5):1451–65. doi: 10.1542/peds.113.5.1451. [DOI] [PubMed] [Google Scholar]
- 15.Rosenfeld RM, Culpepper L, Doyle KJ, et al. American Academy of Pediatrics Subcommittee on Otitis Media with Effusion. American Academy of Family Physicians. American Academy of Otolaryngology Head and Neck Surgery. Clinical practice guideline: Otitis media with effusion. Otolaryngol Head Neck Surg. 2004;130(5 Suppl):S95–118. doi: 10.1016/j.otohns.2004.02.002. [DOI] [PubMed] [Google Scholar]