Abstract
DNA transposition has contributed significantly to evolution of eukaryotes and prokaryotes. Insertion sequences (IS) are the simplest prokaryotic transposons and are divided into families based on their organization and transposition mechanism. Here, we describe a link between transposition of IS608 and ISDra2, both members of the IS200/IS605 family which uses obligatory single-stranded (ss) DNA intermediates, and the host replication fork. Replication direction through the IS plays a crucial role in excision: activity is maximal when the “top” IS strand is located on the lagging-strand template. Excision is stimulated upon transient inactivation of replicative helicase function or inhibition of Okazaki fragment synthesis. IS608 insertions also exhibit an orientation preference for the lagging-strand template and insertion can be specifically directed to stalled replication forks. An in silico genomic approach provides evidence that dissemination of other IS200/IS605 family members is also linked to host replication.
INTRODUCTION
DNA transposition involves movement of discrete DNA segments (transposons) from one genomic location to another. It occurs in all kingdoms of life and has contributed significantly to evolution of eukaryotes and prokaryotes. Transposable elements can represent a significant proportion of their host genomes (Biemont and Vieira, 2006). They have been particularly well-studied in bacteria where they are major motors of broad genome remodelling, play an important role in horizontal gene transfer and can sequester and transmit a variety of genes involved in accessory cell functions such as resistance to antimicrobial agents, catabolism of unusual compounds, and pathogenicity, virulence or symbiosis. They are also important as genetic tools in identifying specific gene regulatory regions by insertion and are being developed as delivery systems for gene therapy applications.
A variety of structurally and mechanistically distinct enzymes (transposases) have evolved to carry out transposition by several different pathways (Turlan and Chandler, 2000; (Curcio and Derbyshire, 2003). They all possess an endonuclease activity allowing them to cleave, excise and insert transposon DNA into a new location. Depending on the system (Curcio and Derbyshire, 2003), different types of nucleophile can be used by transposases to attack a phosphorus atom of a backbone phosphodiester bond and cleave DNA. These include: water (generally activated by enzyme-bound metal ions); a hydroxyl group at the 5' or 3' end of a DNA strand; or a hydroxyl-group of an amino acid of the transposase itself, such as serine or tyrosine.
Many mobile DNA elements move using a "cut-and-paste" mechanism by excision of a double-stranded copy from one genomic location and insertion at another. Recently, a family of bacterial insertion sequences (IS), the IS200/IS605 family, has been found which uses a completely different pathway and an unusual transposase with a catalytic tyrosine (a Y1 transposase).
Studies of one member, IS608 (Fig. 1A), provided a detailed picture of their transposition (Ton-Hoang et al., 2005; Ronning et al., 2005; Guynet et al., 2008; Barabas et al., 2008). In vitro, this requires single strand (ss) DNA substrates and is strand-specific: only the “top” strand is recognised by the element-encoded transposase, TnpA, and is cleaved and transferred while the “bottom” strand does not transpose. Excision of the top strand as a transposon circle with joined left and right ends is accompanied by rejoining of the DNA flanks. The circle junction then undergoes TnpA-catalysed integration into an ssDNA target in a sequence-specific reaction. Insertion involves transfer of both the 5’ and 3’ ends of the single strand circle junction into the ssDNA target. The left (5’) IS608 end always inserts specifically just 3’ of the tetranucleotide, 5’-TTAC-3’ (Kersulyte et al., 2002) which is also essential for subsequent transposition (Ton-Hoang et al., 2005).
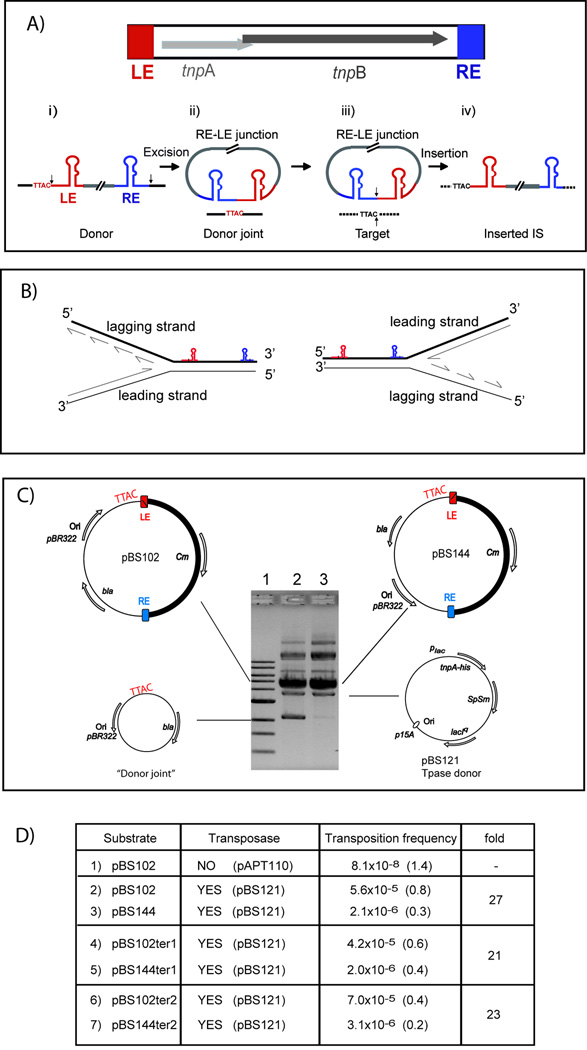
Figure 1. IS608 excision depends on the direction of replication.
(A) IS608 organisation and a simplified transposition model: grey arrows: tnpA and tnpB orfs; red and blue boxes, left (LE) and right (RE) ends (colour code retained throughout). i) Schematised single-stranded IS608 showing secondary structures of LE and RE, the flanking TTAC and cleavage positions at the ends (vertical black arrows). ii) Excision and ssDNA circle formation with an RE-LE junction and a sealed donor joint (black line) retaining TTAC. iii) TnpA brings together the transposon junction with a new TTAC-carrying target (dotted black line). Vertical black arrows: points of cleavage and strand transfer. iv) IS608 insertion into the target.
(B) Orientation of the IS608 derivative with respect to replication direction: The disposition of the IS608 active (top) strand with respect to replication direction is shown when the fork approaches from one direction (left) when it is part of the lagging strand template or the other (right) when it is part of the leading strand. This is described in more detail in Supplementary Material: Fig. S1 and legend.
(C) Excision measured directly in vivo by the appearance of “donor joint” plasmids, deleted for the IS608 derivative: Left: pBS102 with the active IS608 strand as part of the lagging strand template. Right: pBS144 with the active IS608 strand as part of the leading strand template. Ori, pBR322 origin of replication; Cm, bla, SpSm, chloramphenicol, β-lactamase and, streptomycin/spectinomycin resistance genes; Plac, lac promoter; tnpA-his, C-terminal his6-tagged tnpA gene. Directions of DNA replication and transcription are indicated. 0.8 % agarose gel showing separation of plasmid DNA from overnight cultures of strains carrying pBS102 + pBS21 (lane 2) and pBS144 + pBS121 (lane 3). Lane 1, 1 kb standard.
(D) Mating-out assays: Left hand column, transposon donor plasmid; middle, presence or absence of TnpA (relevant plasmids in parentheses); right, measured transposition frequencies (standard error in parentheses; n >3). Plasmids pBS102ter1 and pBS144ter1 carry a single set of origin proximal terminators; pBS102ter2 and pBS144ter2 carry two sets of terminators flanking both transposon ends (Supplementary Experimental Procedures).
The obligatorily single-stranded nature of IS200/IS605 transposition in vitro raises the possibility that it is limited in vivo by the availability of its ssDNA substrates. A number of cellular processes generate or occur using ssDNA including: DNA repair, natural transformation, conjugative plasmid transfer, single-stranded phage infection, and replication (where the DNA serving as the template for Okazaki fragment synthesis on the lagging strand of the replication fork is single stranded).
Here we investigate the link between IS608 transposition and the availability of ssDNA during replication in vivo. Our results demonstrate that transposition of the IS200/IS605 family is closely integrated into the host cell cycle and takes advantage of the presence of ssDNA on the lagging strand template at the replication fork for dissemination. We also show that IS608 transposition is affected by perturbing the fork: transitory inactivation of crucial replication proteins increased excision from the lagging strand template, and stalling of the fork resulted in insertions directed to the lagging strand of the blocked fork.
Our results also suggest that insertion and excision of the related, ISDra2, also depends on the lagging strand template in its host, the radiation-resistant bacterium Deinococcus radiodurans, and that this dependency can be abolished with irradiation. We have extended our analysis to a number of related IS200/IS605 elements in a variety of sequenced bacterial genomes. The results of this in silico analysis are also consistent with a strong bias of insertion into the lagging strand template in these organisms.
Together, the results demonstrate the importance of the lagging strand template for IS608 and ISDra2 activity and suggest that all IS200/IS605 family members have evolved a mode of transposition that exploits ssDNA at the replication fork.
RESULTS
IS608 excision depends on the direction of replication
To investigate whether replication direction affects IS608 transposition, we used a plasmid assay in E. coli to monitor the excision step of transposition (Ton-Hoang et al., 2005). In this assay, the IS-carrying plasmids included an IS608 derivative in which the tnpA and tnpB genes (Fig. 1A) was replaced by a chloramphenicol resistance (CmR) cassette (Fig. 1C). In one case, the active (top) IS608 strand was located in the lagging strand template (pBS102), and in the second, the replication origin was inverted (Fig. 1B and C), placing the transposionally active top strand on the leading strand template (pBS144). A second compatible plasmid supplied TnpA in trans under control of plac (Fig. 1C). After overnight growth, IS excision was monitored by detecting reclosed donor backbone molecules from which the IS had been deleted.
As shown in Figure 1C, when the IS608 active strand was located on the lagging strand template, the donor backbone species could be clearly identified along with the parental plasmid and the plasmid used to supply transposase (lane 2). However, when the replication origin was inverted, and the active IS strand was located on the leading strand, formation of the excised donor backbone species was only barely detectable (lane 3).
This effect of replication direction on IS608 transposition was also observed in mating out assays (Galas and Chandler, 1982) that measure overall transposition frequency. Transposition was monitored by following movement of IS608 from a non-mobilizable donor plasmid into a conjugative plasmid. When the IS608 active strand was on the lagging strand template (Fig. 1D, line 2), the transposition frequency was 5.6 × 10−5 but when it was on the leading strand template, the frequency dropped 27-fold to 2.1 × 10−6 (line 3). To ensure that this was not due to possible changes in transcription resulting from the inversion introduced during cloning to switch the orientation of the replication origin (where bla was inverted together with ori), transcriptional terminators (Simons et al., 1987) were inserted on either one (lines 4–5) or both (lines 6–7) sides of the IS608 derivatives to insulate them from impinging transcription. In these cases, the observed effect of replication direction on transposition frequency remained unchanged.
Effect of mutant primase and replicative helicase on IS excision
Since IS608 excision is sensitive to replication direction, we asked whether it was affected by perturbation of the replication fork. We used two temperature sensitive replication mutants, dnaG308ts, encoding a mutant DNA primase, DnaG (Weschler and Gross, 1971) and dnaB8ts, encoding a mutant of the essential replication fork DNA helicase, DnaB (Carl, 1970). Replication in these mutants occurs at 30°C but is interrupted after a shift to the restrictive temperature of 42°C. Inhibition of either DnaG or DnaB activity is expected to increase the amount of ssDNA at the fork, principally on the lagging strand template (Louarn, 1974; Fouser and Bird, 1983; Belle et al., 2007; and see Discussion).
To test the effect of inhibiting DnaG and DnaB activities, we used a genetic screen in which a β-lactamase gene is interrupted by an IS608 derivative (pAM1, Fig. S2; Materials and Methods). TnpA-catalysed precise excision (using TnpA provided in trans) results in reconstitution of the β-lactamase gene (Fig. 2A) and the appearance of ampicillin resistant (ApR) colonies.
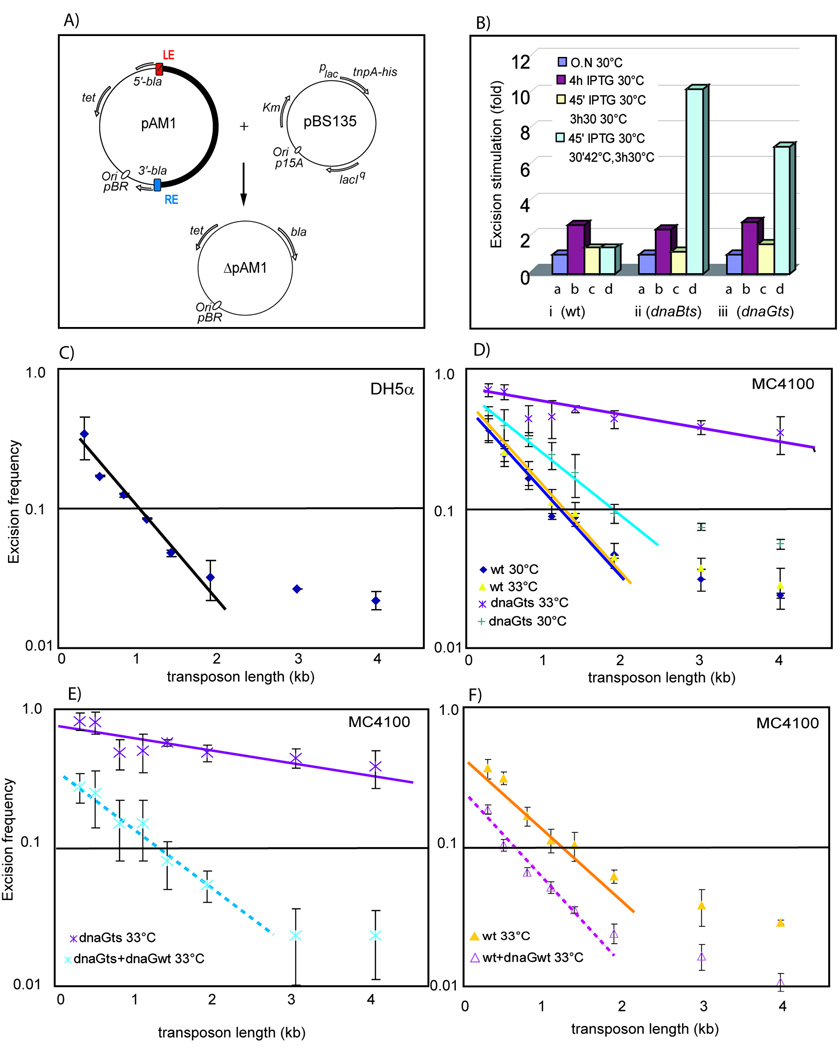
Figure 2. Excision frequency: effects of dnaBts and dnaGts mutants and IS Length.
A) Schematic of the excision assay: The relevant features of the generic IS-carrying donor plasmid, pAM1, and the product following IS excision, ΔpAM1, are shown together with the plasmid used to supply transosase, pBS135. These are described in detail in Supplementary Material Fig. S1 and legend.
(B) Transitory inactivation of DnaB or DnaG. Stimulation factors for excision from wildtype, dnaBts and dnaGts strains: i, ii and iii respectively. Values are normalised to those of an overnight culture without IPTG at 30°C for each strain (0.8×10−2, 0.6×10−2 and 2.2×10−2 corresponding to columns a); overnight cultures diluted at 30°C with IPTG for 4 h (columns b); TnpA induction at 30°C (45 min) and incubation at 30°C (3 h 30 min) without IPTG (columns c); TnpA induction at 30°C (45 min) followed by a shift to 42°C without IPTG (30 min) and then at 30°C (3 h) (columns d).
(C) Effect of IS length on excision frequency. IS608 derivatives are 0.3; 0.5; 0.8; 1.1; 1.4; 1.9; 3 and 4 kb long. The curve shows results, expressed as ApRTpRKmR/ TcRKmR using DH5a at 37°C. Error bars are sd of >4 independent experiments.
(D) Effect of dnaG on IS length-dependant excision. MC4100, 30°C (dark blue) and 33°C (yellow); MC4100dnaGts, 30°C (light blue) and MC4100dnaGts+dnaGwt, 33°C (purple).
(E) Effect of DnaGwt overexpression on excision frequency in dnaGts strains. MC4100dnaGts, 33°C (purple) and MC4100dnaGts+dnaGwt, 33°C (light blue).
(F) Effect of DnaGwt overexpression on excision frequency in wildtype strains. MC4100, 33°C (yellow) and MC4100+dnaGwt, 33°C (purple).
As shown in Figure 2Bi, ii and iii for the wildtype, dnaB and dnaG mutants respectively, after overnight growth at 30°C (a permissive temperature) without TnpA induction, the frequency of ApR colonies was low for wildtype and mutant hosts (columns a). Induction of TnpA expression resulted in a nearly 3-fold increase in excision with little difference between wildtype, dnaBts and dnaGts strains (columns b). However, when the growth protocol was modified to include a 30 min temperature shift to 42°C (and further incubation of 3 hrs at 30°C to allow replication to recover), excision increased about ten- and seven-fold in the dnaB and dnaG mutants respectively compared to the wildtype (columns d). Omitting the 42°C step resulted in indistinguishable basal levels for wildtype and mutant hosts (columns c).
Thus, inactivation of DnaB helicase function or inhibition of initiation of Okazaki fragment synthesis with a dnaGts mutation stimulated the excision step of transposition, consistent with the notion that ssDNA at the replication fork is a substrate for IS excision.
Effect of transposon size on excision
If excision occurs at the replication fork and requires ssDNA, it seemed possible that IS length might influence excision frequency since the probability that both IS ends are within the single-stranded region of the lagging strand template should decrease with increasing IS length. We therefore examined the effect of IS length in the excision assay using a set of IS608 derivatives with varying spacing between the LE and RE and found that excision decreased strongly as a function of increasing IS length and that these frequencies were strongly modified in a strain carrying a dnaGts mutation.
Increasing the IS length from 0.3 to 2 kb, resulted in a 7–10 fold decrease in excision frequency with a log-linear relationship, consistent with the notion that excision occurs more efficiently when both ends are located within the short single lagging strand region of about 1.5–2 kb upstream of the first complete Okazaki fragment at the replication fork (see Johnson and O'Donnell, 2005 for review). As the IS length was further increased, excision decreased only slightly, at least up to a transposon length of 4 kb (Fig. 2C) with a possible inflection between 1.5 – 2 kb.
The length of ssDNA on the lagging strand template depends on the initiation frequency of Okazaki fragment synthesis in turn determined by DnaG. Progressive inactivation of DnaG activity by growth of the dnaGts mutant at increasing but sublethal temperatures should reduce this frequency and increase the mean length of ssDNA at the replication fork upstream of the first complete Okazaki fragment. We therefore analysed excision of the IS608-derived transposons in wildtype and dnaGts mutants at different temperatures (Fig. 2D). While profiles were indistinguishable for the wildtype strain at 30°C and 33°C, the dnaGts mutant exhibited a generally higher excision frequency at 30°C and showed a lower length dependent slope revealing that the replication fork is affected by the mutation even at the normal permissive temperature. However, an inflection still appeared to occur. At 33°C, excision increased significantly, particularly for the longer transposons.
To further examine this, the wildtype dnaG allele was cloned downstream of the tnpAIS608 gene in the TnpA-providing plasmid so that both were under control of the same promoter (Materials and Methods). When this plasmid, pBS179, was introduced into the dnaGts strain, it clearly suppressed the dnaGts defect at 33°C (Fig 2E). Moreover, when introduced into the wildtype dnaG strain, the excision frequencies were even further reduced and the length dependant slope was increased (Fig. 2F).
IS608 insertion into the E. coli chromosome
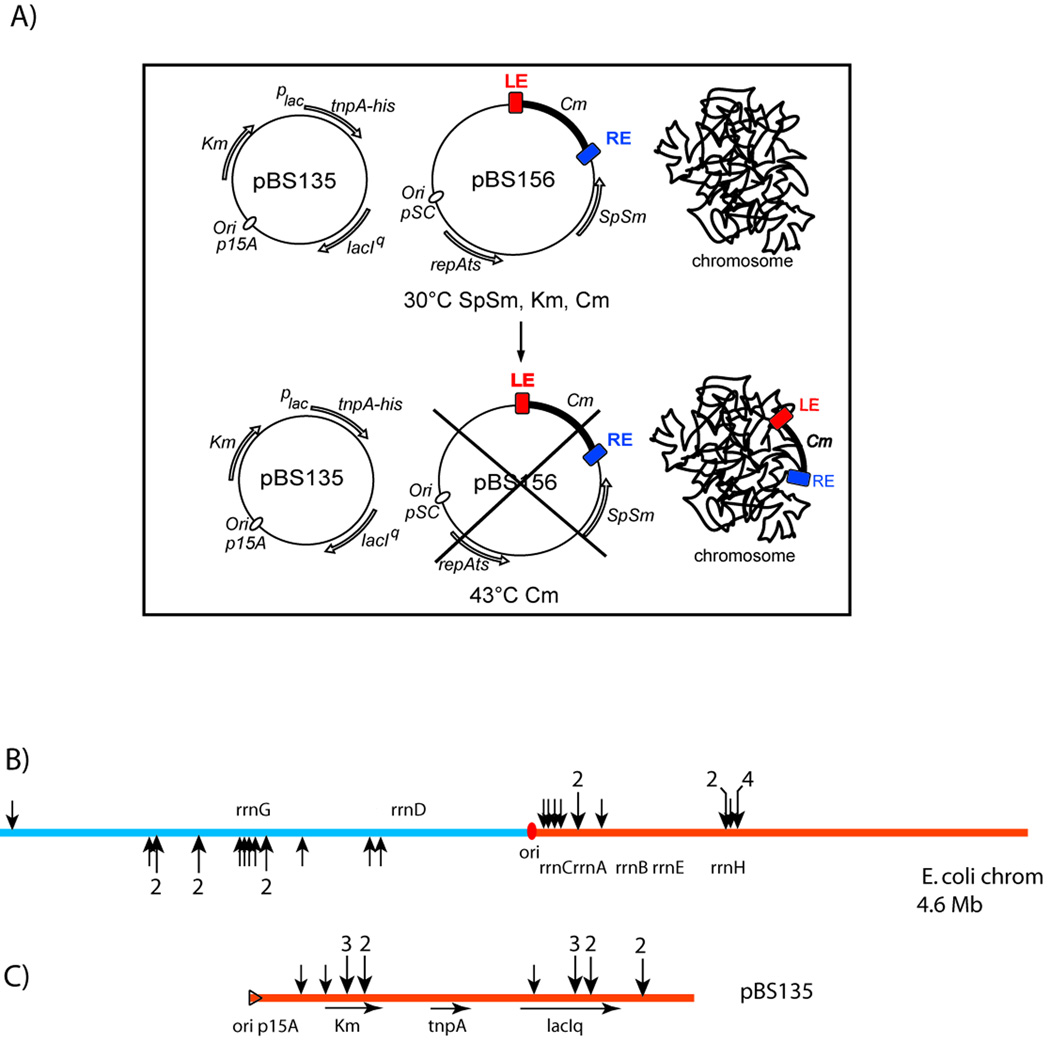
The circular E. coli chromosome replicates bidirectionally from the replication origin, oriC. If IS608 insertions target the lagging strand template, they should occur in one orientation on one side of ori, and in the opposite orientation on the other. To test this, we isolated IS608 insertions in the E. coli chromosome using a temperature sensitive plasmid as the IS donor, and supplying TnpA in trans (Fig 3A). Insertions were localised by an arbitrary PCR procedure (Materials and Methods) followed by DNA sequencing. We observed a dramatic skew in strand specificity of IS608 insertion relative to the origin of replication, oriC.
Figure 3. Insertions into the E. coli chromosome and into a resident plasmid.
A) Experimental system: The temperature sensitive transposon donor plasmid, pBS156 (middle) and the plasmid used for supplying TnpA, pBS135 (left), are shown together with a schematic of the host chromosome (right). Growth at the non-permissive temperature results in loss of pBS156 and IS insertion into the chromosome or pBS135. This is described in detail in Supplementary Material Fig. S1 and legend.
(B) Chromosomal insertions. The chromosome is shown linearised at its terminus. Red spot, origin of replication, ori; rrn, ribosomal RNA genes; black arrows, positions of insertions (Table S1). Multiple insertions are indicated by a number above the larger arrows. Replication occurs in both directions into the left and right replicores blue and orange respectively: the lagging strand template of the left (bottom) and right (top) replicores. Fisher Exact Test gives a p-value of 7.122e−6(C) Plasmid insertions. Replication of the p15A-based plasmid occurs from left to right. Orange triangle, origin of replication; KmRtnpA and lacIQ, kanamycin resistance, transposase and lac repressor genes respectively; arrows indicate their direction of transcription. Vertical arrowheads show the position of the insertions obtained from the same experiments as in (A) above.
Insertions either in the left or right replicores were in the orientation expected for transposition into the lagging strand template (Fig. 3B). It is interesting to note that while insertions were distributed around the chromosome, many appeared in the vicinity of the highly transcribed rRNA genes (Table S1).
We also obtained insertions into the TnpA donor plasmid in the same experiment. In contrast to the chromosome, this plasmid replicates unidirectionally and the IS608 insertion pattern was completely (Fig. 3C) different. All occurred into only one strand, the lagging strand template, a result which has strong statistical support (Fig. 3 legend).
In all cases both plasmid and chromosome insertions occurred 3' to a TTAC target, as expected. As these are distributed equally on both strands of the chromosome and the TnpA donor plasmid (data not shown), the observed strand biases for insertion cannot be explained by a bias in target sequence distribution.
Targeting stalled plasmid replication forks: Tus-Ter system
Since our accumulating data suggested that IS608 targets ssDNA at the replication fork, we asked whether insertion could also be observed into forks stalled at a pre-defined location. For this, we used the E. coli Tus/Ter system in which a Ter site, when bound by the protein Tus, strongly reduces replication fork progression through Ter in the non-permissive orientation (Ternp) (see Bierne et al., 1994) but not in the opposite, permissive, orientation (Terp) (Neylon et al., 2005).
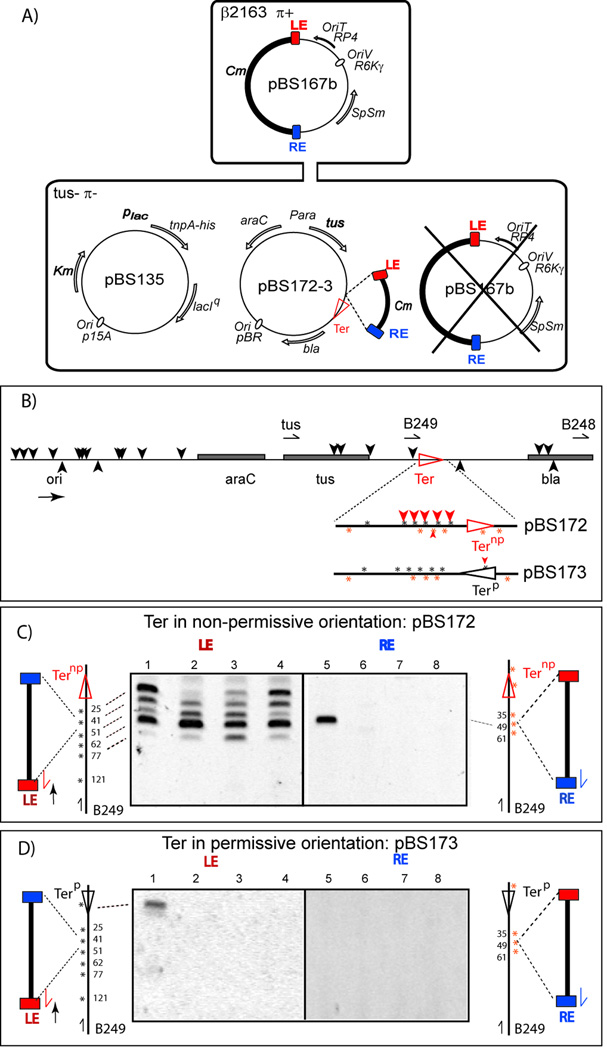
The assay (Fig. 4A and Fig. S1) used a suicide conjugation system (Demarre et al., 2005) in which the unidirectionally replicating target plasmid was carried by a recipient strain with an inactivated chromosomal tus gene. The target plasmid carried a functional tus gene and a Ter site in either the permissive or non-permissive orientation (Figs. 4B and Supplementary Experimental Procedures). The plasmid-based tus gene was transitorily induced only for the duration of the experiment to avoid plasmid loss. We also inserted a stretch of DNA with several spaced copies of the TTAC target tetranucleotide on both strands upstream of the Ter sites.
Figure 4. IS608 insertions into Tus/Ter stalled replication forks.
(A) Experimental design: The suicide conjugative IS-carrying donor plasmid, pBS167b, is shown (top). It cannot propagate in the recipient and IS insertions were monitored into the Ter-carrying target plasmid, pBS172-3 as described in detail in Supplementary Material (Fig. S1 and legend) and plasmids in Supplementary Experimental Procedures.
(B) Insertions with Ter in permissive and non-permissive orientations. Replication from ori is from left to right; araC repressor, tus and bla genes, grey boxes; *, Ter proximal TTAC sequences; horizontal red arrowhead, Ternp (non-permissive, pBS172; horizontal black arrowhead Terp (permissive, pBS173); black vertical arrowheads, IS608 insertions; red vertical arrowheads, multiple insertions upstream of Ternp or an insertion inside of Terp. The position of the oligonucleotides used to localise insertions are also shown.
(C) Detailed analysis of insertions in theTernp region. PCR amplification products were separated on a 5% acrylamide gel. The distribution and orientation of insertions close to Ter were determined in 4 independent experiments (LE: lanes 1–4; RE: lanes 5–8). Distribution of TTAC on the lagging strand and leading strand template are shown in the cartoon (left and right respectively).
(D) Detailed analysis of insertions in theTerp region. Identical experiment as shown in (C) using Terp.
Mapping the IS insertion sites revealed that all occurred at TTAC sequences and were distributed over the entire length of both target plasmids, largely in an orientation expected for insertion into the lagging strand template (Fig. 4B black arrowheads). The overall distribution was the same regardless of Ter site orientation. As for the other target plasmids used here, we confirmed that TTAC sequences were distributed roughly equally on both DNA strands (data not shown).
An additional set of insertions was observed in the target plasmid carrying Ternp when compared to that with Terp (Fig. 4B and C). To precisely map these we used PCR analysis with primers complementary to a sequence upstream of the Ter sites and either LE (for lagging strand insertions) or RE (for leading strand insertions). When the target plasmid carried Ternp, amplification products from four independent experiments (Fig. 4C, lanes 1–4) revealed insertions adjacent to the Ter site on the lagging strand template (Fig. 4B, filled red arrowheads; Fig 4C) although occasionally an insertion was observed on the opposite strand (lane 5). Although there was a degree of variability in the insertion distribution between experiments (compare lanes 1–4), a consistently strong signal was obtained from the TTAC site located at 63 bp from Ternp, suggesting that this is the preferred insertion site. However, insertions were also observed at the closest site, located only 26 bp from Ter (Fig. 4C, lanes 1, 3 and 4).
With Terp, no strong Tus-dependent insertions were observed (Fig. 4D). In four independent experiments we only once isolated an insertion in this region (in the plasmid population from experiment 1, lane 1) and this had occurred into a TTAC site within the Ter sequence itself rather than immediately upstream.
Thus the lagging strand of replication forks blocked by Tus is a preferential target for IS608 insertions and these can occur in close proximity to the Tus binding site, Ter.
Another IS200/IS605 family member: ISDra2
ISDra2 is an IS200/IS605 family member from the highly radiation resistant D. radiodurans. It has a similar organisation to IS608 and inserts specifically 3' to a TTGAT pentanucleotide (Islam et al., 2003). Like IS608, it transposes using ssDNA intermediates (Pasternak et al., 2010). We had observed specific stimulation of ISDra2 transposition upon γ- or UV-irradiation (Mennecier et al., 2006; Pasternak et al., 2010) related to the availability of ssDNA generated during radiation-induced fragmentation and reassembly of the host genome, called extended synthesis dependent strand annealing (ESDSA; Zahradka et al., 2006).
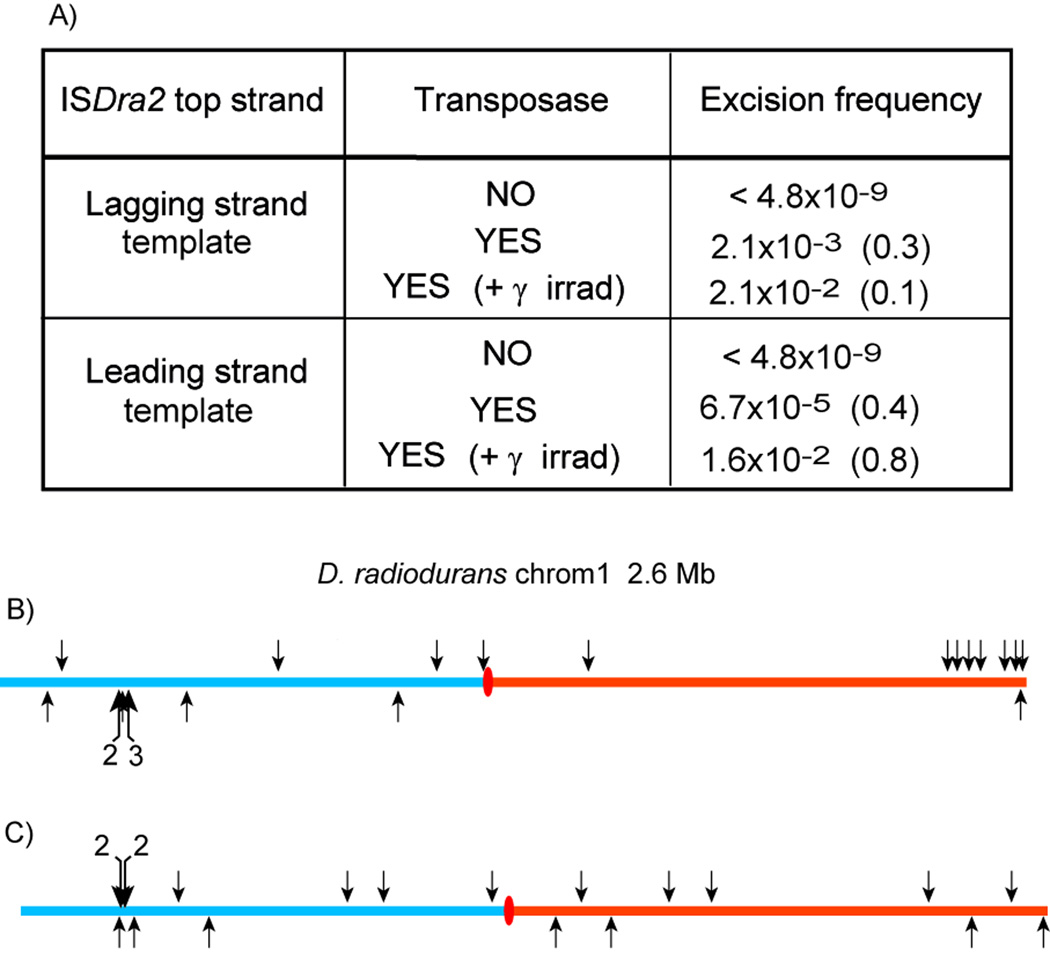
To determine if spontaneous ISDra2 transposition is influenced by host chromosome replication, we measured ISDra2 excision in D. radiodurans as a function of replication direction through the IS. D. radiodurans replication is thought to be bidirectional (Hendrickson and Lawrence, 2006). We constructed D. radiodurans strains with an ISDra2 derivative, ISDra2-113 (Pasternak et al., 2010), placed in one or the other orientation at the same chromosomal location (Supplementary Experimental Procedures). Excision of these ISs reconstitutes a functional TcR gene. Consistent with our results with IS608, no excision (< 4.8 × 10−9; Fig. 5A) was detected without transposase whereas in the presence of TnpAISDra2, relatively high excision levels (2.1 × 10−3) were observed when the IS derivative was in one orientation but was reduced about 30-fold to 6.7 × 10−5 in the other (Fig. 5A).
Figure 5. Excision and Insertion of ISDra2-113 in Deinococcus radiodurans.
(A) ISDra2-113 excision frequencies depend whether the active (top) ISDra2-113 strand is located on the lagging or leading strand template. The middle column indicates if TnpA was provided and whether the cells were subjected to γ radiation. Strain constructions are presented in Supplementary Experimental Procedures.
(B) Spontaneous ISDra2-113 insertions into the D. radiodurans chromosome I. The origin (ori, red ellipse) and direction of replication of the D. radiodurans chromosome was assumed to be that proposed by (Hendrickson and Lawrence, 2006). All insertions occurred 3’ to TTGAT pentanucleotide sequences. Detailed mapping is presented in Table S2. Fisher Exact Test gave a p-value of 0.01108. Multiple insertions are indicated by a number above the larger arrows.
(C) ISDra2113 insertions into the D. radiodurans chromosome I following γ-irradiation. Detailed mapping is presented in Table S3. Fisher Exact Test gave a p-value of 0.1344 suggesting that the pattern is random.
However, the orientation bias of excision was lost in γ-irradiated D. radiodurans strains: after γ-irradiation, excision of ISDra2-113 reached the same and maximal level of about 2 × 10−2 for both orientations relative to the direction of replication (Fig. 5A). Since there should be no strand bias in generating ssDNA following irradiation during ESDSA, the lack of excision and insertion bias in irradiated D. radiodurans suggests that insertion targets are no longer limited to those rendered accessible during the replication.
Finally, we isolated 23 independent spontaneous insertions of ISDra2-113 in D. radiodurans chromosome I by scoring for TcR CmR colonies. All occurred into a TTGAT target and exhibited an orientation bias (18/23) on the lagging strand template (Fig. 5B and Table S2) without irradiation. In contrast, 21 insertions isolated from γ-irradiated D. radiodurans occurred essentially equally on both strands (Fig. 5C and Table S3).
Genomic analysis reveals orientation bias in other IS200/IS605 family members
In view of the strong orientation bias observed for IS608 and ISDra2 insertions, we wondered if similar patterns might be found in other bacterial genomes as vestiges of members of the IS200/IS605 family occur widely (ISfinder: www-is.biotoul.fr). We identified several genomes with a significant number of copies of various members of this family and determined the orientation of insertion relative to the origin of replication (as defined by GC skew). The genomic ISs exhibited a strong orientation bias supporting our notion that the lagging strand template provides the most attractive ssDNA for IS targeting.
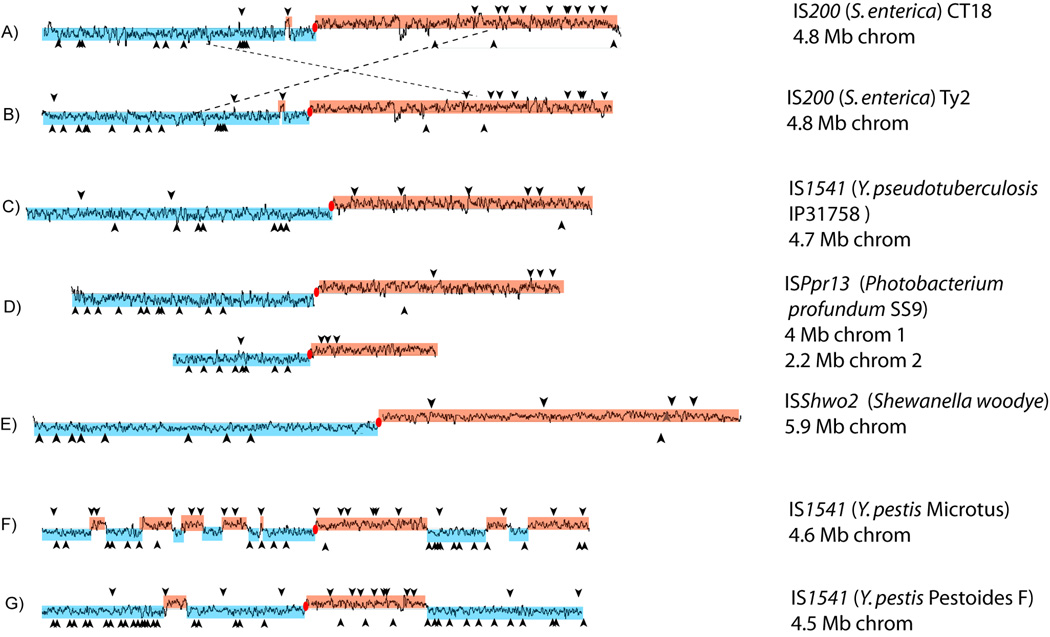
The results for representative bacterial genomes are shown in Fig. 6. For example, S. enterica CT18 carries 27 full copies of IS200 (Deng et al., 2003; Fig. 6A). Thirteen of these are on one replicore and 14 on the other while all but five occur in the orientation expected for insertion into the lagging strand template. Another example is Yersinia pseudotuberculosis (IP31758) (Chain et al., 2004; Fig 6C) with 16 full IS1541 copies (9 in the left and 7 in the right replicore), 13 of these are in an orientation consistent with insertion into the lagging strand template. Photobacterium profundum SS9 (Vezzi et al., 2005; Fig 6D) carries 28 ISPpr13 copies distributed between its two chromosomes with 26 in the expected orientation; and Shewanella woodyi with 13 copies of ISShwo2 and all except one are in the expected orientation (Karpinets et al., 2009; Fig. 6E). The Fisher Exact Test provided strong statistical support (Fig. 6 legend).
Figure 6. Orientation of IS200/IS605 family members in bacterial genomes.
Overall GC skew (G-C/G+C) is indicated in blue and orange. Black line: calculated GC skew using Artemis (http://www.sanger.ac.uk/Software/Artemis/) with a 10 kb window. Origin of replication, red ellipse. Individual ISs, vertical arrow heads: above or below the genome depending on their orientation. A)S. enterica (typhi) CT18 (NC_003198); IS200, 709 bp (Fisher’s Exact Test p-value, 0.002054) B)S. enterica (typhi) Ty2 (NC_004631); IS200, 709 bp (p-value, 0.004799). C)Y. pseudotuberculosis IP31758 (NC_009708); IS1541, 708–709 bp (p-value, 0.02144). D)P. profundum SS9 (NC_006371 and NC_006370); ISPr13, 596 bp (chromosome 1: p-value, 0.0005139; chromosome 2: p-value, 0.002806). E)S. woodyi (NC_010506); ISShwo2, 613 bp. (p-value, 0.02297) F)Y. pestis Microtus 91001 (NC_005810); IS1541, 711 bp. (p-value, 1.139e-06) G)Y. pestis Pestoides F (NC_009381); IS1541D, 711 bp (p-value, 2.479e-06). In F) and G) p-values were calculated using the potential number of target sequences taking into account the regions of GC skew inversion.
Interestingly, the distribution of the 42 IS1541 copies in Y. pestis (Microtus) (Fig 6F) and the 50 copies of it in Y. pestis (Pestoides F) (Fig 6G) are clearly different and show complex pattern with no apparent relationship to the replication direction. However, these Y. pestis strains revealed aberrant GC skews resulting from a series of inversions and rearrangements throughout the chromosome (Song et al., 2004; Garcia et al., 2007). When these are taken into account, an astoundingly close correlation emerges between GC skew and IS1541 orientation.
Similar reasoning may explain those insertions that initially appear in contradiction with a lagging strand template targeting preference. For example, of the five IS200 insertions in S. enterica strain CT18 oriented in the opposite direction (Figure 6A), one is located in a short chromosome region with an inverted GC-skew compared to the neighbouring sequences. This implies that this region has undergone inversion and that the associated IS insertion likely had occurred prior to this event. The insertion pattern observed in S. enterica (Ty2) (Deng et al., 2003; Fig. 6B) is quite similar to that of S. enterica CT18; both carry an identical number of IS200 copies. The differences in IS200 distribution between these two strains can be entirely explained by a large inter-replicore inversion between S. enterica (Ty2) and S. enterica CT18.
These genomic results strongly support the idea that insertion into the lagging strand template of the chromosomes of their host is a general characteristic of IS200/IS608 family members.
DISCUSSION
The transposition pathway for members of the IS200/IS605 IS family occurs using ssDNA substrates and intermediates. In vitro IS excision requires both transposon ends to be single stranded, and insertion of the excised single-stranded circular transposon intermediate requires access to a ssDNA target (Guynet et al., 2009). We now demonstrate that excision and insertion of two family members, IS608 and ISDra2, occur preferentially in the lagging strand DNA template in vivo.
Excision is dependent on replication direction through the IS: it is high when the active IS strand is on the lagging strand template but substantially lower when part of the leading strand template. We also examined insertions of a plasmid-localized IS608 into normally replicating E. coli chromosomes and spontaneous insertions of ISDra2 into D. radiodurans chromosome I. They were largely directed to the lagging strand template resulting in a skew of strand-specific insertion on either side of the replication origin in these bidirectionally replicating chromosomes. For a unidirectionally replicating E. coli plasmid, insertions occurred into only one strand. Importantly, the orientation effect for ISDra2 insertion was abolished when transposition was triggered by γ-irradiation, accompanied by an increase in transposition frequency, consistent with the observation that γ-irradiation induces a repair pathway resulting in massive amounts of ssDNA with no obvious strand bias.
IS608 insertions into the E. coli chromosome were fairly evenly distributed but a significant number occurred in the highly transcribed rrn genes (which are oriented in the sense of replication) suggesting that high transcription levels might also provide accessible ssDNA for IS608 insertion, e.g. by generating R-loops or by affecting replication fork passage. Replication in E.coli has been estimated to be approximately 20-fold faster than is transcription (800 nt/s versus 20 to 50 nt/s; Kornberg and Baker, 1992). Replication forks may stall at tRNA and other highly expressed genes, possibly because transcription complexes collide “head-on” with the forks. Replication forks also stall upon co-directional encounters with RNA polymerase (Elias-Arnanz and Salas, 1997) (Mirkin et al., 2006) possibly due to a trapped RNA polymerase not readily displaced from DNA by fork progression.
That ssDNA at the replication fork facilitates IS608 transposition is reinforced by studies which perturb the fork using temperature sensitive DnaG (primase) or DnaB (helicase) mutants. DnaG inactivation prevents initiation of Okazaki fragment synthesis, increasing the average length of ssDNA upstream of the first complete Okazaki fragment on the lagging strand template. (Louarn, 1974; Fouser and Bird, 1983). DnaB inactivation results in accumulation of large amounts of ssDNA mostly likely arising from degradation of both the nascent DNA and leading strand template or from uncoupled leading-strand synthesis (Belle et al., 2007). We observed a significant stimulation of IS608 excision after transitory inactivation of both dnaGts and dnaBts mutants.
If excision occurs at the replication fork and requires ssDNA, the probability that both ends are within the single stranded region of the lagging strand template should decrease with increasing IS length and thus the size of the IS should influence excision frequency exactly as we observe. Moreover, the efficiency of excision was generally higher in the dnaGts mutant even at the permissive growth temperature of 30°C and the slope of the curve was less steep. At the sublethal temperature of 33°C, excision was even higher and the length dependence even less marked. This is consistent with an increase in ssDNA length of on the lagging strand template resulting from a lower Okazaki fragment initiation frequency in the dnaGts mutant even at temperatures permissive for growth and suggests that the slope is a function of the ssDNA length available on the lagging strand template.
We also investigated the effect of DnaG overproduction. Expression of a cloned wildtype dnaG gene not only suppressed the dnaGts phenotype but resulted in an even more pronounced length dependence in the wildtype background suggesting that, as observed in vitro (Zechner et al., 1992; Sanders et al., 2010), DnaG concentration controls the frequency of initiation and Okazaki fragment size. More importantly, this result would suggest that DnaG is not saturating at the normal replication fork in vivo.
While the slopes of the curves presumably reflect the length of ssDNA on the lagging strand template, the explanation for the apparent inflection of the curves for the longer transposons is less clear. It is possible, for example, that we are observing effects of two phenomena: an initial probability that both ends are in a single-stranded form and also the probability of both ends finding each other. Further analysis is required to determine the exact reason behind this behaviour.
Although, for simplicity, we describe the lagging strand template as single stranded at the fork, it is important to note that it is not naked but protected by proteins such as single strand binding protein (Ssb; Shereda et al., 2008). This implies that the transposition machinery can access the DNA through the protecting protein and raises the question of whether TnpA can recognise Ssb or other components of the replisome. Experiments to investigate this are in progress.
We also used the natural Tus/Ter replication fork termination system (Neylon et al., 2005; Kaplan and Bastia, 2009) to examine whether blocked forks might also attract IS608 insertions. The Tus–Ter complex forms a transient barrier to the replicative helicase, DnaB, when a fork arrives in the non-permissive direction (Neylon et al., 2005). When provided with appropriate target tetranucleotides, Tus-dependent IS608 insertions readily occurred close (26–77 nt) to the Ter site on the lagging strand template, consistent with nucleotide resolution mapping of the terminated nascent DNA in vitro and in vivo showing that the final lagging-strand primer sites are arrested 50–70 nucleotides upstream of Ter (Hill and Marians, 1990; Mohanty et al., 1998).
While our data are consistent with the idea that stalled forks favor IS608 insertion, we do not yet know whether the ssDNA substrates are directly available at blocked forks or are generated during their processing, e.g. during replication restart or repair of ds breaks caused by replication arrest (Michel et al., 1997; Bierne and Michel, 1994).
To determine whether other IS200/IS605 family members might locate suitable ssDNA substrates for transposition, we annotated several complete bacterial genomes for ISs and identified several that harbour multiple copies of these family members. In the majority, these ISs showed a similar orientation bias to IS608 and ISDra2 relative to replication direction. Thus, targeting the ssDNA available on the lagging strand template appears to be a general theme among IS200/IS605 family members.
Replication direction and therefore identification of the lagging strand is generally implied from GC skew, the preference for G over C on the leading strand thought to be the result of differential repair (Lobry, 1996; Grigoriev, 1998). More strictly speaking, we observed that IS orientation was correlated with the GC skew of the region into which they were inserted rather than with replication direction per se suggesting that the insertions predated the genome rearrangements whose scars are revealed by changes in GC skew. This has two important implications: either that transposition is infrequent or that transposition events become genetically fixed in the population.
IS200/IS605 family members are not alone in showing asymmetric strand preference in insertion. Other transposable elements such as Tn7 and IS903 also appear to do so (Peters and Craig, 2001) (Hu and Derbyshire, 1998). Tn7 is targeted to replication forks during conjugative plasmid transfer and inserts in a specific orientation. The transposon-encoded TnsE protein is instrumental in targeting the transpososome to the junction between single and double stranded DNA by interaction with the β-clamp component of the replication apparatus (Parks et al., 2009; Chandler, 2009). Insertion of Tn7 into a replication fork likely occurs within the dsDNA covered by an Okazaki fragment. IS903 insertion bias in the conjugative F plasmid might also reflect targeting to the conjugative replication fork. It is worthwhile noting that IS10 and IS50 have also exploited host replication: both are activated by transient formation of hemimethylated DNA following fork passage (Roberts et al., 1985; Yin et al., 1988; Dodson and Berg, 1989).
However, there are major mechanistic differences between Tn7, IS903 and members of the IS200/IS605 family, suggesting that different pathways are at play. Perhaps most importantly, Tn7 transposes using a dsDNA intermediate in contrast to the ssDNA species used by IS608 and ISDra2. For Tn7 and many other transposons with dsDNA intermediates, strand transfer into a dsDNA target occurs using the 3’OH groups on each complementary strand at each end. Insertion of the first strand into a single strand target would leave a fatal break. Thus, whereas an overarching theme in DNA transposition may be the exploitation of replication forks, different elements do so in different ways.
The unique excision and insertion properties of the IS200/IS605 family may make them useful tools for probing ssDNA structures in vivo. The relationship between excision and IS length might be used to determine the effect of various factors on the state of the replication fork in vivo. For example, treatments leading to reduced fork velocity, Okazaki fragment synthesis, uncoupling of lagging from leading strand synthesis or simply forks blocked by different factors could all be probed using an excision assay as an experimental readout. We are aware that the topology of the replication fork of small, multicopy plasmids may differ in some respects from that of the chromosome, and this system may also be used to explore these possible differences. We are at present testing these possibilities experimentally.
MATERIALS AND METHODS
Bacterial strains and media
Bacterial strains are listed in Supplementary Experimental Procedures. E. coli cultures were grown in Luria broth supplemented where necessary with: chloramphenicol (Cm, 10 or 30 µg/ml); kanamycin (Km, 20 µg/ml); ampicillin (Ap, 100 µg/ml); gentamycin (Gm, 5µg/ml); spectinomycin (Sp 30 µg/ml) and streptomycin, (Sm, 20 µg/ml); tetracycline (Tc, 15 µg/ml) or 2,6-Diaminoheplanediole acid (DAP, 0.006%). D. radiodurans media, and growth conditions were as described (Pasternak et al., 2010; Bonacossa de Almeida et al., 2002)
Plasmids
Details of plasmid constructions are available upon request; schematics of plasmids used are shown in Supplementary Experimental Procedures. The dnaG-carrying plasmid pBS179 was constructed by inserting a DNA fragment carrying the wildtype dnaG allele with its natural ribosome binding site isolated by PCR using flanking oligonucleotides, dnaGN and dnaGC into the SphI site of transposase supplying plasmid downstream of the tnpA gene placing both genes under control of plac. The clone was verified by sequencing and complements the dnaGts allele.
Excision assay with pAM1 derivatives
Effect of dnaG308 and dnaB8 on excision
E. coli MG4100 wildtype, dnaG308 and dnaB8 mutant strains were grown overnight at 30°C in LB+KmTc, the permissive temperature, cultures were diluted at 30°C, and grown with 0.5 mM IPTG for 4 h or 45 min. IPTG was removed by centrifugation and cells were resuspended in pre-warmed medium at 42°C for 30 min and incubated at 30°C for 3 h. As a control, the 42°C step was omitted. Cells were harvested and dilutions plated on LA+KmTc and LA+KmTcAp.
Effect of transposon length
E. coli DH5α harbouring pBS135 and pAM1-derivative plasmids was grown overnight in Luria Broth supplemented with Tc and Km at 37°C. Cultures were diluted in fresh medium at 37°C with TcKm and 0.5 mM IPTG to induce TnpA expression. Cells were harvested after 5 h and dilutions plated on LA+KmTc and LA+KmTcAp.
Effect of transposon length in the dnaG308 mutant strain
overnight cultures of MC4100 and MC4100 dnaGts were grown at 30°C and diluted in fresh medium at 30°C or 33°C containing TcKm and 0.5 mM IPTG. Cells were harvested after 5 h and dilutions plated on LA+KmTc and LA+KmTcAp at 30°C.
Tus/Ter experiments
Overnight donor and recipient strains were grown in LB+DAP+SpSmCm and LB+GmKmAp respectively. 0.5% Glucose was added to fully repress the para used to drive Tus expression. Cultures were diluted in fresh LB medium without antibiotics (with DAP for donor cells). At OD600 of 0.5, donor cells were incubated without agitation, the recipient was diluted to an OD600 of 0.15 and TnpA was induced with 0.5 mM IPTG. After 60 min Tus was induced for 30 min by addition of 0.08% arabinose. Donor and recipient strains were mixed, incubated for 2 h and plated on Cm-containing LA plates supplemented with 0.5% Glucose. Bulk plasmid DNA was isolated and the distribution of insertions in the population was analysed by PCR using 6 sets of primers: B248, B249, tus as forward primers and LE, RE as reverse primers (Supplemental Experimental Procedures) to determine the insertion orientation. PCR amplification used Phusion DNA Polymerase (Finnzyme) in GC buffer as follows: 30 s 98°C, 35 cycles of 10 s 98°C, 30 s 64°C, ∼1 min (72°C). 50 ng of bulk plasmid DNA were used for each reaction.
Sequencing IS608 insertions in the E.coli chromosome
IS608 insertions into the E.coli chromosome and coresident plasmid pBS135 were sequenced by arbitrary PCR as described (Guynet et al., 2009).
Measurement of in vivo spontaneous, and γ-induced excision frequencies of ISDra2-113
Excision frequencies were determined using individual CmR TcS colonies purified from strains GY14302 or GY14303 (Supplemental Experimental Procedures) grown with or without TnpA expression in trans as described (Pasternak et al., 2010). The mean and standard deviations were calculated from six independent experiments.
ISDra2 insertion sites were mapped by arbitrary-primed (AP)-PCR using DyNazyme EXT DNA polymerase (Finnzymes) as described (Supplementary Experimental Procedures).
Statistical Analysis of Insertion orientation
This was performed using the Fisher exact test with the “R” statistical package (http://www.r-project.org/) taking into account the number of potential target sites on the leading and lagging strands. For the Yersinia species the number of potential target sites and of insertion sites was calculated taking into account GC skew inversion.
Supplementary Material
Acknowledgements
We would like to thank members of the “Mobile Genetic Elements” group, A. Bailone, P. Polard and D. Lane for discussions, G. Coste and B. Marty for expert technical assistance, L. Lavatine for guiding us through the mysteries of statistics, A. Varani for identifying a representative oligonucleotide sequence used in the PCR mapping for the D. radiodurans genome and the Institut Curie for the use of the 137Cs irradiation system. This work was supported by: intramural funding from the CNRS (France); in its later stages by ANR grant Mobigen (MC and SS); European contract LSHM-CT-2005-019023 (MC); and the CEA and EDF (France) (SS). At the NIH, this work was supported by the Intramural Program of the NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barabas O, Ronning DR, Guynet C, Hickman AB, Ton-Hoang B, Chandler M, Dyda F. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell. 2008;132:208–220. doi: 10.1016/j.cell.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle JJ, Casey A, Courcelle CT, Courcelle J. Inactivation of the DnaB Helicase Leads to the Collapse and Degradation of the Replication Fork: a Comparison to UV-Induced Arrest 10.1128/JB.00408-07. J Bacteriol. 2007;189:5452–5462. doi: 10.1128/JB.00408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemont C, Vieira C. Genetics: Junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- Bierne H, Ehrlich SD, Michel B. Flanking sequences affect replication arrest at the E. coli terminator TerB in vivo. J Bacteriol. 1994;176:4165–4167. doi: 10.1128/jb.176.13.4165-4167.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Michel B. When replication forks stop. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- Bonacossa de Almeida C, Coste G, Sommer S, Bailone A. Quantification of RecA protein in D. radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol Genet Genomics. 2002;268:28–41. doi: 10.1007/s00438-002-0718-x. [DOI] [PubMed] [Google Scholar]
- Carl PL. E. coli mutants with temperature-sensitive synthesis of DNA. Mol Gen Genet. 1970;109:107–122. doi: 10.1007/BF00269647. [DOI] [PubMed] [Google Scholar]
- Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, et al. Insights into the evolution of Y. pestis through whole-genome comparison with Y. pseudotuberculosis. Proc Natl Acad Sci U S A. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. Clamping down on transposon targeting. Cell. 2009;138:621–623. doi: 10.1016/j.cell.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM. The outs and ins of transposition: from Mu to Kangaroo. Nat Rev Mol Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, Mazel D. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate E. coli host strains. Res Microbiol. 2005;156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Deng W, Liou SR, Plunkett G, 3rd, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. Comparative genomics of S. enterica serovar Typhi strains Ty2 and CT18. J Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson KW, Berg DE. Factors affecting transposition activity of IS50 and Tn5 ends. Gene. 1989;76:207–213. doi: 10.1016/0378-1119(89)90161-3. [DOI] [PubMed] [Google Scholar]
- Elias-Arnanz M, Salas M. Bacteriophage phi29 DNA replication arrest caused by codirectional collisions with the transcription machinery. Embo J. 1997;16:5775–5783. doi: 10.1093/emboj/16.18.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouser L, Bird RE. Accumulation of ColE1 early replicative intermediates catalyzed by extracts of E. coli dnaG mutant strains. J Bacteriol. 1983;154:1174–1183. doi: 10.1128/jb.154.3.1174-1183.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas DJ, Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J Mol Biol. 1982;154:245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- Garcia E, Worsham P, Bearden S, Malfatti S, Lang D, Larimer F, Lindler L, Chain P. Pestoides F, an atypical Yersinia pestis strain from the former Soviet Union. Adv Exp Med Biol. 2007;603:17–22. doi: 10.1007/978-0-387-72124-8_2. [DOI] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guynet C, Achard A, Hoang BT, Barabas O, Hickman AB, Dyda F, Chandler M. Resetting the site: redirecting integration of an insertion sequence in a predictable way. Mol Cell. 2009;34:612–619. doi: 10.1016/j.molcel.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guynet C, Hickman AB, Barabas O, Dyda F, Chandler M, Ton-Hoang B. In vitro reconstitution of a single-stranded transposition mechanism of IS608. Mol Cell. 2008;29:302–312. doi: 10.1016/j.molcel.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Hendrickson H, Lawrence JG. Selection for chromosome architecture in bacteria. J Mol Evol. 2006;62:615–629. doi: 10.1007/s00239-005-0192-2. [DOI] [PubMed] [Google Scholar]
- Hill TM, Marians KJ. E. coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci U S A. 1990;87:2481–2485. doi: 10.1073/pnas.87.7.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WY, Derbyshire KM. Target choice and orientation preference of the insertion sequence IS903. J Bacteriol. 1998;180:3039–3048. doi: 10.1128/jb.180.12.3039-3048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam SM, Hua Y, Ohba H, Satoh K, Kikuchi M, Yanagisawa T, Narumi I. Characterization and distribution of IS8301 in the radioresistant bacterium D. radiodurans. Genes Genet Syst. 2003;78:319–327. doi: 10.1266/ggs.78.319. [DOI] [PubMed] [Google Scholar]
- Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Kaplan DL, Bastia D. Mechanisms of polar arrest of a replication fork. Mol Microbiol. 2009;72:279–285. doi: 10.1111/j.1365-2958.2009.06656.x. [DOI] [PubMed] [Google Scholar]
- Karpinets TV, Obraztsova AY, Wang Y, Schmoyer DD, Kora GH, Park BH, Serres MH, Romine MF, Land ML, Kothe TB, et al. Conserved synteny at the protein family level reveals genes underlying Shewanella species' cold tolerance and predicts their novel phenotypes. Funct Integr Genomics. 2009 doi: 10.1007/s10142-009-0142-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Velapatino B, Dailide G, Mukhopadhyay AK, Ito Y, Cahuayme L, Parkinson AJ, Gilman RH, Berg DE. Transposable element ISHp608 of H. pylori: nonrandom geographic distribution, functional organization, and insertion specificity. JBacteriol. 2002;184:992–1002. doi: 10.1128/jb.184.4.992-1002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA Replication. New York: W.H.Freeman; 1992. [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. MolBiolEvol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Louarn JM. Size distribution and molecular polarity of nascent DNA in a temperature-sensitive dna G mutant of E. coli. Mol Gen Genet. 1974;133:193–200. doi: 10.1007/BF00267668. [DOI] [PubMed] [Google Scholar]
- Mennecier S, Servant P, Coste G, Bailone A, Sommer S. Mutagenesis via IS transposition in D. radiodurans. Mol Microbiol. 2006;59:317–325. doi: 10.1111/j.1365-2958.2005.04936.x. [DOI] [PubMed] [Google Scholar]
- Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. Embo J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Castro Roa D, Nudler E, Mirkin SM. Transcription regulatory elements are punctuation marks for DNA replication. Proc Natl Acad Sci U S A. 2006;103:7276–7281. doi: 10.1073/pnas.0601127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Sahoo T, Bastia D. Mechanistic studies on the impact of transcription on sequence-specific termination of DNA replication and vice versa. J Biol Chem. 1998;273:3051–3059. doi: 10.1074/jbc.273.5.3051. [DOI] [PubMed] [Google Scholar]
- Neylon C, Kralicek AV, Hill TM, Dixon NE. Replication termination in E. coli: structure and antihelicase activity of the Tus-Ter complex. Microbiol Mol Biol Rev. 2005;69:501–526. doi: 10.1128/MMBR.69.3.501-526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AR, Li Z, Shi Q, Owens RM, Jin MM, Peters JE. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell. 2009;138:685–695. doi: 10.1016/j.cell.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak C, Ton-Hoang B, Coste G, Bailone A, Chandler M, Sommer S. Irradiation-induced D. radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genetics. 2010;6:e1000799. doi: 10.1371/journal.pgen.1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes Dev. 2001;15:737–747. doi: 10.1101/gad.870201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Hoopes BC, McClure WR, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol Cell. 2005;20:143–154. doi: 10.1016/j.molcel.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Song Y, Tong Z, Wang J, Wang L, Guo Z, Han Y, Zhang J, Pei D, Zhou D, Qin H, et al. Complete genome sequence of Y. pestis strain 91001, an isolate avirulent to humans. DNA Res. 2004;11:179–197. doi: 10.1093/dnares/11.3.179. [DOI] [PubMed] [Google Scholar]
- Ton-Hoang B, Guynet C, Ronning DR, Cointin-Marty B, Dyda F, Chandler M. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. Embo J. 2005;24:3325–3338. doi: 10.1038/sj.emboj.7600787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlan C, Chandler M. Playing second fiddle: second-strand processing and liberation of transposable elements from donor DNA. Trends Microbiol. 2000;8:268–274. doi: 10.1016/s0966-842x(00)01757-1. [DOI] [PubMed] [Google Scholar]
- Vezzi A, Campanaro S, D'Angelo M, Simonato F, Vitulo N, Lauro FM, Cestaro A, Malacrida G, Simionati B, Cannata N, et al. Life at depth: P. profundum genome sequence and expression analysis. Science. 2005;307:1459–1461. doi: 10.1126/science.1103341. [DOI] [PubMed] [Google Scholar]
- Weschler J, Gross J. E. coli mutants temperature sensitive for DNA synthesis. Molec gen Genetics. 1971;113:273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- Yin JC, Krebs MP, Reznikoff WS. Effect of dam methylation on Tn5 transposition. J MolBiol. 1988;199:35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]
- Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M. Reassembly of shattered chromosomes in D. radiodurans. Nature. 2006;443:569–573. doi: 10.1038/nature05160. [DOI] [PubMed] [Google Scholar]
- Zechner EL, Wu CA, Marians KJ. Coordinated leading- and lagging-strand synthesis at the E. coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J Biol Chem. 1992;267:4045–4053. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.