Abstract
The phagocyte NADPH oxidase, belonging to the NADPH oxidase family (Nox), is dedicated to the production of bactericidal reactive oxygen species. The enzyme catalytic center is the cytochrome b558, formed by 2 subunits, Nox2 (gp91-phox) and p22-phox. Cytochrome b558 activation results from a conformational change induced by cytosolic regulatory proteins (p67-phox, p47-phox, p40-phox and Rac). The catalytic subunit is Nox2, while p22-phox is essential for both Nox2 maturation and the membrane anchorage of regulatory proteins. Moreover, it has been shown to be necessary for novel Nox activity. In order to characterize both p22-phox topology and cytochrome b558 conformational change, 6 monoclonal antibodies were produced against purified cytochrome b558. Phage display epitope mapping combined with a truncation analysis of recombinant p22-phox allowed the identification of epitope regions. Some of these antibodies almost completely inhibited in vitro reconstituted NADPH oxidase activity. Data analysis identified antibodies that recognized epitopes involved in either Nox2 maturation or Nox2 activation. Moreover, flow cytometry analysis and confocal microscopy performed on stimulated neutrophils showed that the monoclonal antibody 12E6 bound preferentially active cytochrome b558. These monoclonal antibodies provided novel and unique probes to investigate maturation, activation and activity, not only of Nox2 but also of novel Nox.
Key Words: Cytochrome b558, p22-phox conformation, Monoclonal antibody, Phagocyte NADPH oxidase, Nox
Introduction
Neutrophil cytochrome b558 is a membrane heterodimer composed of Nox2 (gp91-phox) and p22-phox. This is the catalytic center of the phagocyte NADPH oxidase complex responsible for the production of reactive oxygen species in response to inflammatory stimuli [1,2]. The NADPH oxidase is a multisubunit enzyme complex which is inactive and compartmentalized between cytosol and membrane in resting neutrophils. In stimulated cells, cytosolic regulatory proteins (p47-phox, p67-phox, p40-phox and the monomeric G protein Rac) translocate to the membrane cytochrome b558, inducing a cytochrome b558 conformational change and electron transfer from the NADPH to the molecular oxygen. Resulting superoxide anion radicals are precursors for various toxic oxygen metabolites implicated in the defense against pathogens [1,2,3,4,5,6]. A defect in NADPH oxidase activity causes chronic granulomatous disease (CGD), a rare genetic disorder characterized by severe and recurrent infections due to the inability of phagocytes to contribute to the killing of microorganisms [7,8].
The structure of cytochrome b558 has not yet been solved. Computational analysis of primary sequence of Nox2 and p22-phox suggests that both subunits contain transmembrane domains (6 and 2–4, respectively). The cytochrome b558 catalytic subunit is Nox2. It binds NADPH and flavin adenine dinucleotide in the last 280 residues of its C-terminal tail and possesses 4 histidine residues that coordinate 2 hemes involved in the last electron transfer step leading to the production of superoxide anions [9]. Even though no catalytic activity has been described for p22-phox, it is an essential component of cytochrome b558, as its absence in myeloid cells leads to the degradation of Nox2. It has been previously shown that the incorporation of hemes was required for both the p22-phox/Nox2 dimer formation in the endoplasmic reticulum and the maturation of cytochrome b558[10]. The p22-phox subunit has 2 major functions. It binds and stabilizes Nox proteins, and serves as a membrane anchor for cytosolic regulatory factors. It was recently shown that p22-phox also associates with Nox1, Nox3 and Nox4 [11,12], suggesting a central role of p22-phox in the cellular production of reactive oxygen species.
At the moment, the topology and functional domains within p22-phox are not clearly characterized. P22-phox was proposed to contain 2–4 membrane-spanning segments according to the primary sequence analysis by computational methods [13]. However, experimental evidence obtained with both specific antibodies directed against the cytosolic parts of p22-phox and cytochrome b558 proteolytic digestion [13] suggest (1) the presence of 2 transmembrane domains and (2) the cytosolic localization of the N- and C-terminal regions of the protein. The C-terminal tail of p22-phox contains a proline-rich region involved in interactions with SH3 domains of p47-phox and also p67-phox [14,15]. Phage display epitope mapping of specific monoclonal antibodies (mAbs) has identified 2 tertiary structures within the cytosolic parts of p22-phox. The first is composed of residues 29TAGRF33 and 183PQVNPI188 corresponding to the mAb 44.1 epitope, and the second is composed of 51LLEYPRG57 and 77VKLFGP82 corresponding to the mAb NS5 [13].
In the present study, we report the production and characterization of 6 novel mAbs (12E6, 17A2, 16G7, 16G6, 13D4 and 13C4) obtained after injection of a mixture of cytochrome b558 purified from resting and PMA-stimulated neutrophils. These novel mAbs were epitope mapped by phage display analysis and were shown to recognize various regions in the C-terminal domain of p22-phox. These mAbs displayed different and specific properties with regard to their ability to detect native and/or denatured p22-phox and their capacity to inhibit the production of superoxide in vitro. Moreover, mAb 12E6 bound a conformational epitope localized in the polyproline region of p22-phox. Interestingly, it better recognized cytochrome b558 in its active form, suggesting that the target epitope was unmasked during activation. Information resulting from a computational analysis of phage display, results by the program FINDMAP and a visual comparison of phage display sequences and the gp91phox sequence suggest a potential complex epitope for mAb 12E6 shared between p22-phox and Nox2. These mAbs provide novel and unique probes for investigation of the maturation, activation and activity, not only of cytochrome b558 but also of novel Nox.
Materials and Methods
Materials
Chemical reagents used in this study were obtained from the following sources: enhanced chemiluminescence (ECL) Western blotting detection reagents (Amersham Pharmacia Biotech, Uppsala, Sweden); PMA, arachidonic acid (Sigma Chemicals, St. Louis, Mo., USA); n-octyl glucoside (Roche Diagnostics, Meylan, France); Alexa Fluor® 488 F(ab′)2 fragment of goat anti-mouse IgG (H+L) (Molecular Probes Europe BV, Leiden, The Netherlands); Goat F(ab′)2 fragment anti-mouse IgG (H+L)-phycoerythrin (Immunotech-Beckman Coulter, Marseille, France); horseradish peroxidase-conjugated rabbit anti-mouse IgG (γ-specific) (Open Biosystems, Huntsville, Ala., USA); pCDNA3.1-V5/HisB (Invitrogen, Cergy Pontoise, France).
Purification of Cytochrome b558 from Resting or Stimulated Neutrophils
Cytochrome b558 was purified from the plasma membranes of 1010 resting or PMA-stimulated neutrophils after 3 chromatographic steps (ion exchange, heparin-agarose and Sephacryl S300) as previously described [16]. The purified cytochrome b558 solubilized in 0.1% (w/v) Triton X-100 buffer was quantified by the reduced minus oxidized difference spectra using an absorption coefficient of 106 mM−1 cm−1 for the Soret band at 426 nm [16].
Production of mAbs to Cytochrome b558
Mice immunization and mAb production were performed by the ‘BET analyses’ company (Marseille, France) and Biotem company (Le Rivier d'Apprieu, France), respectively. Briefly, Balb/c mice were immunized by 3 intraperitoneal injections (interval of 14 days) of a mixture of 15 μg cytochrome b558 purified from resting neutrophils (306 μl) and 15 μg cytochrome b558 purified from PMA-stimulated neutrophils (158 μl) diluted with 1 volume of Freund's complete adjuvant (464 μl). Antigens were solubilized in 0.1% (w/v) Triton X-100 [17]. Resulting mAbs were purified from ascitic fluid after isotype identification (Dr. J. C. Renversez, DBI, CHU Grenoble, France). The isotype for 16G7, 12E6 and 13C4 was IgG1, the isotype for 17A2 was IgG2a, the isotype for 16G6 was IgG2b and the isotype for 13D4 was IgM.
Purification of Immunoglobulins from Ascitic Fluids
Ascitic fluids were centrifuged at 10,000 g for 10 min at 4°C. IgG were purified from the supernatant of centrifugation on protein A Sepharose. Depending on the Ig isotype, protein A Sepharose was equilibrated with either 50 mM sodium borate pH 8.9 containing 3 M NaCl for IgG1 purification or 20 mM Na2HPO4 pH 7 for IgG2 purification. After extensive washes with the equilibration buffer, IgG were eluted with 0.1 M glycine pH 3 [17]. Purified Ig were dialyzed against PBS and stored at −20°C. IgG purification was controlled by SDS-PAGE. IgM were purified by gel filtration on Sephacryl S-300 [18]. Purified antibodies (protein 1 mg/ml) were dialyzed against PBS and stored at −20°C.
Isolation of Human Neutrophils from Fresh Blood
Human neutrophils were isolated from citrated venous blood of healthy volunteers after informed consent, diluted twice in PBS (137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4 pH 7.3) containing 1% (w/v) tri-sodium citrate, using a 33% (v/v) Hypaque-Ficoll gradient. After 20 min centrifugation at 800 g at 20°C, the pellet was submitted to a hypotonic lysis for 5–15 min on ice. After 5 min centrifugation at 350 g at 4°C, the neutrophil pellet was collected and washed once in PBS [17].
Triton X-100 Soluble Extract from Control and X910CGD Neutrophils
Neutrophils from healthy donors and X910CGD patients (107 cells) were suspended in 100 μl of Triton X-100 lysis buffer (3.5 mM MgCl2, 1% (w/v) Triton X-100, 10 mM Hepes pH 7.4) containing a mixture of protease inhibitors (1.8 μM leupeptin, 1.5 μM pepstatin) and incubated for 20 min on ice. Then the mixture was centrifuged at 12,000 g for 30 min at 4°C. The supernatant corresponding to the Triton X-100 soluble extract was collected and stored at −80°C until further use [17].
Neutrophil Stimulation by PMA
Purified neutrophils (5 × 106 cells/ml) were stimulated by PMA [130 nM in 0.05% (v/v) DMSO] for 10 min at 37°C. A control experiment was performed by incubating cells with 0.05% (v/v) DMSO alone for 10 min at 37°C. The reaction was stopped by adding ice-cold PBS. Cells were centrifuged at 350 g for 8 min at 4°C and resuspended in PBS for flow cytometry analysis or confocal microscopy.
Flow Cytometry
Purified neutrophils resuspended in PBS (107 cells/ml) were fixed with 1% (w/v) paraformaldehyde for 15 min on ice. Cells were washed once with PBS and then resuspended at the concentration of 107 cells/ml in PBS/BSA/CaCl2 [PBS containing 0.2% (w/v) BSA and 0.5 mM CaCl2] containing 0.01% (w/v) saponin for the solubilization. Incubation was performed for 10 min on ice. Cells (5 × 105) were then incubated on ice for 30 min with 5 μg of mouse monoclonal Ig (irrelevant IgG or specific IgG) diluted in 100 μl of PBS/BSA/CaCl2 buffer containing 0.01% (w/v) saponin [19]. Cells were washed twice in 500 μl of PBS/BSA/CaCl2 buffer containing 0.01% (w/v) saponin and resuspended in 150 μl of the phycoerythrin-conjugated goat anti-mouse antibody diluted 1: 200 in PBS/BSA/CaCl2 buffer containing 0.01% (w/v) saponin. After 30 min incubation on ice, cells were washed twice with 500 μl PBS/BSA/CaCl2 buffer containing 0.01% (w/v) saponin before being resuspended in 500 μl of PBS/BSA/CaCl2. Fluorescence intensity (FL2) of the phycoerythrin-labeled polymorphonuclear neutrophils was measured on a FACScalibur (Becton Dickinson) cytometer [19].
Immunoprecipitation Experiments
Neutrophil membranes were prepared as described previously [20]. Crude membrane proteins were adjusted at a concentration between 2 and 3 mg protein/ml in 100 mM Hepes pH 7.2, containing 100 mM KCl, 10 mM NaCl, 1 mM EDTA, 20% (v/v) glycerol and the mixture of protease inhibitors (1.8 μM leupeptin, 1.5 μM pepstatin, 10 μM TLCK). Membrane proteins were then solubilized in presence of 68 mM n-octyl glucoside for 20 min on ice followed by centrifugation at 200,000 g for 1 h at 4°C (Rotor 50 Ti, Beckman L-60 ultracentrifuge) [20]. The resulting supernatant corresponded to the membrane soluble extract. Soluble extract (200 μg) was diluted twice in PBS and incubated with protein G agarose (40 μl) for 1 h at 4°C to remove nonspecific binding. The flow through was then incubated with 2 μg specific mAb (12E6, 16G7 or 16G6) or irrelevant mAb for 4 h at 4°C. Then protein G agarose was added to each sample for a further 1-hour incubation at 4°C. After 3 washes with first PBS containing 0.5 M NaCl, then PBS with 0.1% Triton X-100 and finally PBS, the immunoprecipitates were solubilized in Laemmli buffer containing 5% (v/v) mercaptoethanol [21] and analyzed by Western blot by using a secondary antibody directed against the heavy chain of mAb and coupled to peroxidase.
Confocal Microscopy
Purified neutrophils (2 × 105 cells in 50 μl PBS) were incubated on 0.01% (w/v) poly-L-lysine-coated round glass cover slips for 15 min at 37°C. After 2 washes with PBS, cells were fixed with 4% (w/v) paraformaldehyde for 10 min at room temperature. Cover slips were rinsed twice with PBS and paraformaldehyde fluorescence was quenched by 50 mM NH4 Cl for 10 min at room temperature. After 2 washes with PBS, cells were permeabilized with 0.1% (w/v) Triton X-100 for 5 min at room temperature, followed by 2 washes with PBS. Then cells were incubated for 1 h at room temperature with 50 μl of mouse monoclonal Ig [p22-phox-specific Ig or irrelevant Ig; 5 μg in 50 μl PBS containing 1% (w/v) BSA]. The cells were washed 3 times for 10 min with PBS containing 1% (w/v) BSA prior to a 1-hour incubation with 100 μl of Alexa Fluor 488 donkey anti-mouse-Ig antibody [1: 500 in PBS containing 1% (w/v) BSA]. After 2 washes with PBS, cell nuclei were stained with 40 μl Hoechst 33258 (0.5 μg/ml). Samples were then mounted in 10 μl DABCO solution, sealed, and stored at 4°C in the dark. Fixed cells were imaged at room temperature using the inverted confocal and 2-photon laser-scanning microscope (LSM 510 NLO META; Carl Zeiss, Göttingen, Germany) equipped with a 40×/1.3 Plan-Neofluar oil immersion objective. The pinhole adjustment to 1 airy unit resulted in less than 0.7 μm optical slice at 488 nm excitation wavelength. The image plane was chosen to be near the equator of the cell body and nucleus. The Alexa Fluor 488 fluorescence was selected with NFT490 dichroic beamsplitter and LP505 long-pass filter. The Hoechst 33258 fluorescence was excited by 2-photon absorption of the 720-nm radiation of fs Ti: Sa laser (Tsunami; Spectra-Physics GmbH, Darmstadt, Germany) and selected with 390–465 nm emission band-pass filter. No significant photobleaching was induced during image acquisition in either detection channel.
Generation of a Recombinant Plasmid for the Expression of a p22-Phox-Truncated Protein Coupled to eGFP
CDNA3.1-V5/HisB was used to express p22 (1M-Q143)-GFP, a human p22-phox-truncated protein corresponding to amino acids 1M to 143Q fused with eGFP at the C-terminal part. Firstly, full-length eGFP was amplified with Pfu polymerase from the pTT3/eGFP plasmid with the following primers: 5′-GTTTCTCGAGATGATCGATGTGAGCAAGGGCGAGGAG-3′ and 5′-GTTTGGGCCCTCACTTGTACAGCTCGTCCATGCCG-3′. The nucleotides corresponding, respectively, to the restriction sites XhoI and ApaI are underlined. The purified PCR product was digested with XhoI and ApaI and ligated into linearized pCDNA3.1-V5/HisB vector to obtain the pCDNA/eGFP plasmid. Secondly, PCR fragments corresponding to the p22(1–143)-truncated form were obtained by using pEF/p22phox plasmid as matrix with the forward primer 5′-GTTTGGTACCGCGGCCGCATGGGGCAGATCGAGTG-3′ including a KpnI site (underlined) and the reverse primer 5′-CAAACTCGAGCTGCGGCCGCTCC-3′. The XhoI restriction site of the reverse primers is underlined. The purified PCR product was digested with KpnI and XhoI and ligated into linearized pCDNA/eGFP plasmid to obtain a plasmid encoding a p22-phox-truncated protein fused with eGFP. All inserts were verified by sequencing (Genome Express, Grenoble, France) on an Abi Prism automatic sequencer (Perkin Elmer, Courtaboeuf, France).
Stable Transfection of Mammalian Expression Plasmids
HEK293E cells were trypsinized and counted; 4 × 105 HEK293E cells were seeded in 6-well plates and allowed to grow 24 h to reach a 60% confluence in 2 ml of culture medium. The cells were transfected with 3 μg of vectors according to the manufacturing protocol (JetPEI, Polyplus transfection). After 24 h, stable transfected cells were selected with geneticin 500 μg/ml for 3 weeks before analysis. Membrane fractions of transfected cells were isolated as previously described for neutrophils [20].
Reconstitution of NADPH Oxidase Activity
Neutrophil membranes and cytosol for cell-free assays were prepared as described previously after cell sonication (3 times 10 s at 40 W) and centrifugation at 1,000 g for 15 min at 4°C. The supernatant was then centrifuged at 200,000 g for 1 h at 4°C to separate the cytosol (supernatant) and the crude membrane (pellet) [20]. Neutrophil membranes (30 μg) were pre-incubated with p22-phox-specific or irrelevant mAb (50 μg) for 30 min on ice. Then neutrophil cytosol (300 μg), 40 μM GTPγS and 5 mM MgCl2 were added to the membrane fraction in a final volume of 100 μl. An optimum amount of arachidonic acid (40–100 nmol) was used for activation [20]. After 10 min incubation at 25°C, the medium was transferred to a photometric cuvette. The reaction was initiated by 150 μM NADPH. Reconstituted oxidase activity was assessed by measuring the superoxide dismutase-sensitive portion of ferricytochrome c reduction recorded at 550 nm (absorption coefficient at 550 nm: 21.1 mM−1 cm−1).
Phage-Display Epitope Mapping
The 6 mAbs (17A2, 13C4, 13D4, 12E6, 16G6 and 16G7) were epitope mapped by selecting peptide sequences from the J404 nonapeptide library [22] by 3 successive rounds of affinity purification on antibodies covalently coupled to Sepharose followed by amplification on bacterial lawns on nutrient medium, as previously described [23]. After selection, high-affinity mAb-binding clones were identified after replica plating on nitrocellulose and immunoblot analysis, and then purified and sequenced. Sixteen to twenty nonapeptide sequences were obtained for the 6 mAbs selections and were aligned visually as well as by the unique alignment algorithm used by the program FINDMAP [24].
SDS-PAGE and Western Blotting
Samples were loaded on a 10% or 11% SDS-PAGE [25] and electrotransferred to nitrocellulose, as previously described [26]. Immunodetection was performed using mAbs raised against p22-phox (mAb 44.1, 17A2, 13C4, 13D4, 12E6, 16G6 and 16G7; dilution 1:1,000 to 1:5,000) or polyclonal antibody directed against the C-terminal part of p22-phox (residues 184–195; dilution 1:1,000). The immune complexes were detected with a secondary antibody combined with peroxidase. The bound peroxidase activity was detected using ECL reagents.
Slot Blot Analysis of Purified Cytochrome b558
Cytochrome b558 (10 pmol/well) purified from resting or PMA-stimulated neutrophils was directly adsorbed on nitrocellulose under vacuum. The nitrocellulose was then saturated with 1% (w/v) low-fat milk proteins and immunodetection was performed using mAb 12E6 (dilution 1:1,000). Immune complexes were detected with a secondary antibody combined with peroxidase. The bound peroxidase activity was detected using ECL reagents.
Results
Characterization of Six Novel Monoclonal Antibodies
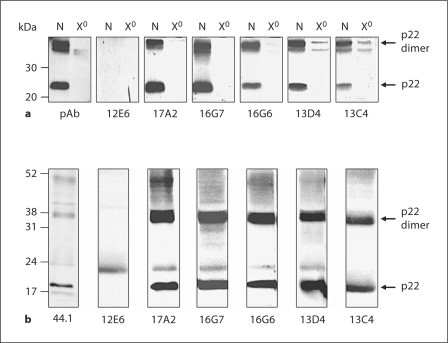
In order to further elucidate the topology of cytochrome b558, 6 novel mAbs, named 17A2, 13C4, 13D4, 12E6, 16G6 and 16G7, were obtained after mouse immunization with cytochrome b558 immunogens. The antigen consisted of a mixture of purified cytochrome b558 fractions obtained from resting and PMA-stimulated neutrophils in 0.1% (w/v) Triton X-100. These fractions had a specific activity in the range of 14–17 nmol heme b · mg protein−1. The 6 mAbs specifically recognized Triton X-100-solubilized (0.1% w/v) cytochrome b558 purified either from resting or PMA-stimulated neutrophils by ELISA, suggesting that they bound the detergent solubilized protein (data not shown). To determine the subunit specificity of the mAbs, immunoblot analyses were performed on Triton X-100-soluble extracts prepared either from control or from X910 CGD neutrophils (fig. 1a) as well as on neutrophil membrane fractions (fig. 1b). Two bands at approximately 24 and 44 kDa were recognized by 5 mAbs as well as by a control polyclonal antibody (fig. 1a). In contrast, these bands were not recognized in extracts from X910CGD neutrophils (fig. 1a), indicating that the bands corresponded to p22-phox and an aggregated dimer due to sample preparation. For mAb 12E6, no protein was detected by immunoblot, suggesting that this antibody recognized a conformational epitope on cytochrome b558 (fig. 1). Similar results were obtained with purified cytochrome b558 fractions (data not shown).
Fig. 1.
Western blot analysis with 6 novel cytochrome b558 antibodies. Several neutrophil fractions were submitted to SDSPAGE followed by immunoblot identification: a Neutrophil cellular extracts (40 μg/well) from either X0 CGD patient (X0) or normal subject (N), b neutrophil crude membrane (Mb: 30 μg/well). The presence of gp91-phox was detected using the 6 novel mAbs (12E6, 17A2, 16G7, 16G6, 13D4 and 13C4) or control antibodies (mAb 44.1 or a polyclonal antibody, pAb, directed against the C-terminal peptide of p22-phox). The immune complexes were detected by ECL.
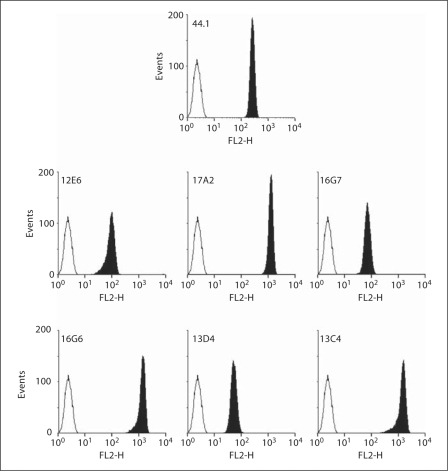
The binding of the 6 mAbs to native antigen was further investigated by flow cytometry on both intact and saponin-permeabilized neutrophils. No mAb binding was observed on intact neutrophils (data not shown), while the 6 mAbs bound strongly to permeabilized neutrophils like mAb 44.1, which is specific of a complex intracellular epitope formed by the regions 29TAGRF33 and 183PQVNPI188 on p22-phox (fig. 2). Similar results were obtained with saponin-permeabilized EBV-B lymphocytes (data not shown). These data indicated that the 5 mAbs (17A2, 13C4, 13D4, 16G6 and 16G7) recognized an intracellular epitope on p22-phox and that mAb 12E6 bound an intracellular region of cytochrome b558 that does not survive SDS-PAGE.
Fig. 2.
Flow cytometry analysis of cytochrome b558 in permeabilized human neutrophils. Purified human neutrophils (5 × 105 cells) were fixed with 1% (w/v) paraformaldehyde, permeabilized with 0.01% (w/v) saponin and labeled with specific mAbs (12E6, 17A2, 16G7, 16G6, 13D4, 13C4 or 44.1; 5 μg; black area) or irrelevant mAb (5 μg; white area) for 30 min on ice. The antibody-labeled cells were stained with PE-conjugated secondary antibody, and the fluorescence (FL2) was measured.
Determination of mAb Epitopes by Phage Display Epitope Mapping
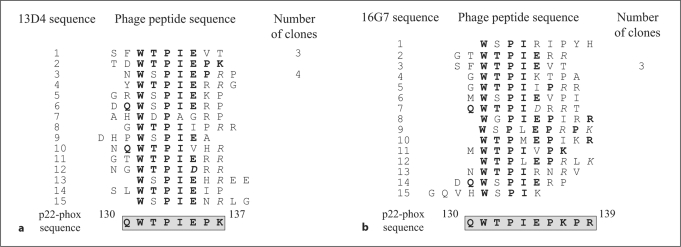
In order to localize the mAb epitope regions, each p22-phox mAb was epitope mapped by phage display analysis. After immunopurification on mAb affinity matrices, the sequences of peptides interacting with each mAb were determined. Alignment of phage peptide sequences led to the identification of a consensus peptide sequence for 3 mAbs, 13D4, 16G7 and 12E6 (fig. 3, 4), while no clear consensus sequence was obtained for mAbs 16G6, 17A2 and 13C4. The consensus sequence was then compared with the primary structure of p22-phox. The resulting epitopes for the 3 mAbs are shown in figures 3 and 4. For both mAb 13D4 and 16G7, the epitope region was similar and corresponded to the 130QWTPIEPK137 region of p22-phox (fig. 3). However, the 16G7 epitope seemed to be longer than that of 13D4 (fig. 3b, 130–139 residues).
Fig. 3.
Phage display epitope mapping for mAb 13D4 (a) and mAb 16G7 (b). After selection of phages on an mAb 13D4 (a) or mAb 16G7 (b) affinity matrix, the nucleotide sequence was determined on isolated phage clones. The corresponding peptide sequences were aligned to identify a consensus sequence, which was compared to p22-phox sequence in order to identify the epitope region. Residues of phage peptides identical to residues of p22-phox are in bold. The epitope sequence is framed.
Fig. 4.
Analysis of the mAb 12E6 interaction site. a Phage display epitope mapping for mAb 12E6 was performed as described in figure 3. The consensus sequence was compared to the primary structure of p22-phox. Residues of phage peptides identical to residues of p22-phox are in bold. The potential epitope region is framed. b Immunoprecipitation studies with mAbs 12E6, 16G7 and 16G6. Cytochrome b558 present in the membrane soluble extract (200 μg) was immunoprecipitated with specific mAbs (2 μg of mAbs 12E6, 16G7 or 16G6) or irrelevant mAb. The filtrate (F) and the immunoprecipitate (IP) were submitted to SDS-PAGE followed by immunoblot identification of p22-phox with mAb 16G7. The immune complexes were detected by ECL.
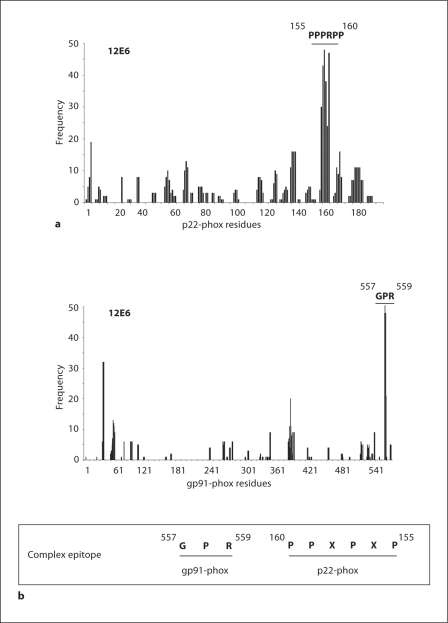
For mAb 12E6, which did not detect denatured cytochrome b558 by immunoblot, a strong consensus sequence was determined ‘GPRPPXPXP’ (fig. 4a). This consensus demonstrated a high degree of similarity to the polyproline-containing region of p22-phox, 159PRPPPNSP152, suggesting that mAb12E6 bound to p22-phox. In order to confirm that mAb 12E6 recognized p22-phox, immunoprecipitation of p22-phox with mAb12E6 was performed. An octyl-glucoside-soluble extract of neutrophil membrane was used for this assay. A similar experiment was done with mAbs 16G7 and 16G6. Results showed that mAbs 16G6 and 16G7 were able to precipitate p22-phox. In contrast, no significant immunoprecipitation was observed with mAb12E6 as compared with the control performed with a nonspecific mAb (fig. 4b). This result suggested that recognition of the epitope by mAb12E6 was altered in octyl glucoside. Three possibilities could explain such a result: (1) the polyproline region of p22-phox could possess a three-dimensional structure modified during the solubilization process; (2) the 12E6 epitope could be a complex epitope implicating several regions of the cytochrome b558; (3) the 12E6 was less effective in octyl glucoside.
To gain a more comprehensive understanding of the mAb12E6 epitope, an extensive analysis of the peptide sequences obtained by phage display was performed by using the computer program FINDMAP [24] in order to map the epitopes to discontinuous segments of the protein that are distant in the primary sequence, but are in close spatial proximity in the structure. This analysis was carried out on both gp91-phox and p22-phox sequences. Histograms presenting results confirmed a good matching of the mAb 12E6 consensus sequence on the polyproline region of p22-phox especially with the region 155PPPRPP160. Surprisingly, the analysis showed a strong match of a 3-residue segment of the consensus sequence with a short segment of the C-terminus of gp91-phox corresponding to residues 557GPR559 (fig. 5).
Fig. 5.
FINDMAP analysis for mAb 12E6. Alignment of phage sequences to discontinuous protein regions of p22phox (a) or gp91-phox (b) by using the program FINDMAP [24]. Histograms presented the frequency of phage-peptide amino acids recovered in the target protein sequence. The potential epitope is presented in frame with amino acids belonging to both gp91-phox and p22-phox.
Confirmation of mAb Epitope Regions by Using Truncated Forms of p22-Phox
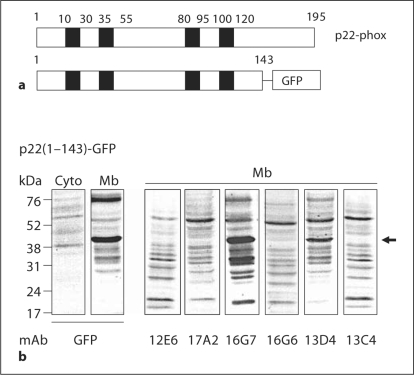
In order to get more information about the regions recognized by these mAbs, a p22-phox-truncated form corresponding to amino acids (1M-Q143) was expressed as a GFP-tagged protein in HEK293 cells (fig. 6a). To control the expression of the p22(1M-Q143)-GFP-truncated form, an immunoblot was performed on transfected HEK293 cells with an anti-GFP mAb (fig. 6b, left panel). Results showed a strong expression of the truncated form in the membrane of transfected HEK293 cells. Interestingly, only 2 p22-phox mAbs, 16G7 and 13D4, recognized the p22(1M-Q143)-truncated form confirming the epitope determined by phage display. Absence of detection of the truncated form by the 3 mAbs 17A2, 16G6 and 13C4 indicated that they bound a region localized at the C-terminal part of p22-phox (144I-V195). As observed with the full-length p22-phox, mAb 12E6 did not bind to the truncated form in Western blot analysis.
Fig. 6.
Western blot of a p22-phox-truncated form. A truncated form of p22-phox corresponded to residues 1–143 [p22(1 M-Q143)] coupled to GFP at its C-terminus (a) was expressed in HEK293E cells. After geneticin selection, cells were isolated, fractioned into membrane and cytosol fractions and analyzed for their expression of the truncated p22-phox (b). Control of expression was performed with an anti-GFP mAb on both cytosolic (50 μg protein/lane) and membrane (50 μg protein/lane) fractions. A membrane fraction (50 μg protein/lane) was analyzed by Western blot to evaluate the capacity of anti-p22-phox mAbs to bind truncated forms.
Effect of mAb anti-p22-Phox on the Phagocyte NADPH Oxidase Activation Process in vitro
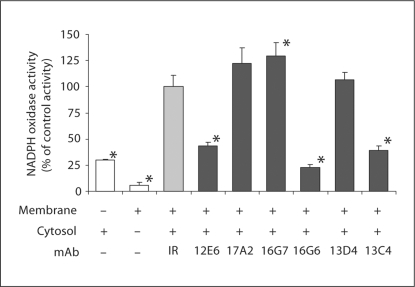
In order to identify new specific Nox2 inhibitors, the effect of mAbs on the NADPH oxidase activity reconstituted in a homologous cell-free system was analyzed. In this experiment, neutrophil membranes were pre-incubated with a 37-fold excess of mAb for 30 min on ice, before adding neutrophil cytosol and an optimal concentration of arachidonic acid (fig. 7). A control experiment was performed with an irrelevant antibody. In presence of mAbs 17A2, 16G7 or 13D4 the production of superoxide was not altered. In contrast, the addition of mAb 12E6, 16G6 or 13C4 resulted in a strong inhibition of the reconstituted NADPH oxidase activity in the range of 65–80% compared to the activity measured in presence of an irrelevant mAb. The 4 mAbs 12E6, 17A2, 16G6 and 13C4 recognized an epitope localized in the C-terminal region of p22-phox-containing residues (144I-V195). Among these 4 mAbs, only mAb 17A2 did not inhibit the reconstituted NADPH oxidase activity, suggesting that the mAb 17A2 epitope was not located in domains involved in the interaction with cytosolic regulatory factors.
Fig. 7.
Effect of the p22-phox mAbs on the NADPH oxidase activity reconstituted in a cell-free system. Neutrophil membrane fractions (30 μg) were incubated with anti-p22-phox mAbs (12E6, 17A2, 16G7, 16G6, 13D4, 13C4; 50 μg), or an irrelevant mAb (IR; 50 μg) for 30 min on ice before adding neutrophil cytosol (300 μg), 40 μM GTPγS, 5 m M MgCl2 and an optimal concentration of arachidonic acid. The superoxide anion production was measured after addition of 150 μM NADPH by the SOD-sensitive portion of the cytochrome c reduction recorded at 550 nm. Results were expressed as a percentage of the control activity obtained in the presence of an irrelevant mAb (100% corresponded to 168 ± 18 nmol min−1 mg membrane protein−1). They were presented as the average of at least 3 experiments ± SD. * p<0.05 vs. presence of an irrelevant mAb.
Immunological Analysis of the Phagocyte NADPH Oxidase Activation Process
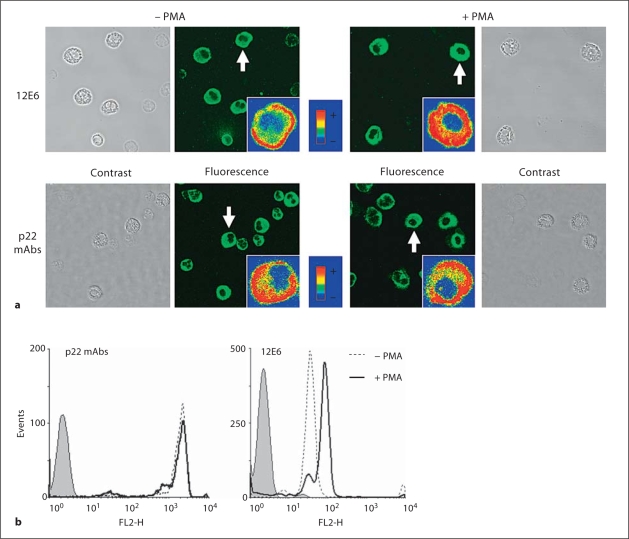
In our study, mAbs were produced against a mixture of cytochrome b558 purified from resting and PMA-stimulated neutrophils. We analyzed the binding of mAbs to permeabilized neutrophils before and after stimulation with 130 nM PMA for 10 min at 37°C by confocal microscopy (fig. 8). Control experiments were done in absence of the primary antibody or in presence of an irrelevant antibody. No significant fluorescence was detected in both cases (data not shown). Image acquisition on control, resting and PMA-stimulated cells was performed with exactly the same configuration settings of the confocal microscope in order to allow the qualitative comparison of the fluorescence intensities. Confocal microscopy done on resting cells with the 6 mAbs showed a green fluorescence inside cells (fig. 8a, left panels). After PMA stimulation, the fluorescence signal intensity was not modified in cells stained with mAbs 17A2, 16G7, 16G6, 13D4 and 13C4. In contrast, the mean fluorescence obtained with the mAb 12E6 was strongly increased leading to a saturating signal (fig. 8a, right panels). A similar increase in fluorescence was observed by flow cytometry in PMA-stimulated neutrophils incubated with mAb 12E6 (fig. 8b). These data suggested that the mAb 12E6 bound preferentially active cytochrome b558 in PMA-stimulated neutrophils. Moreover, the increased fluorescence in PMA-stimulated cells indicated that the binding of cytosolic factors to cytochrome b558 did not exclude the binding of mAb 12E6 to its epitope probably because of the epitope complexity.
Fig. 8.
Immunological analysis of cytochrome b558 activation ex vivo. a Detection of p22-phox in resting and PMA-stimulated neutrophils by confocal microscopy. Neutrophils were incubated with PMA (130 nM) or DMSO for 10 min at 37°C. Resting and stimulated cells (2 × 105 cells) were then fixed, permeabilized and labeled with anti-p22-phox mAbs (5 μg) for 1 h at room temperature as described in Materials and Methods. An Alexa Fluor 488 secondary antibody was used to detect the mAb binding. Inserts show typical cells (indicated by a white arrow) in false colors for the sake of comparison of fluorescence levels. Phase contrast, differential interference contrast and fluorescence images were recorded. b Flow cytometry analysis of cytochrome b558 in resting and PMA-stimulated neutrophils. Purified human neutrophils (5 × 105 cells) were stimulated (plain line) or not (dotted line) with 130 nM PMA for 10 min at 37°C, washed, fixed with 1% (w/v) paraformaldehyde, permeabilized with 0.01% (w/v) saponin and labeled with specific mAbs, 12E6 mAb or other p22-phox mAbs (17A2, 16G7, 16G6, 13D4 or 13C4; 5 μg; dotted and plain lines) or irrelevant mAb (5 μg; grey area) for 30 min on ice. The antibodylabeled cells were stained with PE-conjugated secondary antibody, and the fluorescence (FL2) was measured.
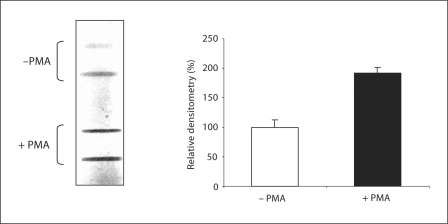
In order to confirm the conformational binding of mAb 12E6 to active cytochrome b558, an in vitro slot-blot experiment was performed on native cytochrome b558 purified either from resting or from stimulated neutrophils. In a previous work, we showed that cytochrome b558 purified from PMA-stimulated cells displayed the capacity to form a constitutively active complex in the presence of cytosolic regulatory factors while cytochrome b558 purified from resting cells did not, indicating that cytochrome b558 was pre-activated upon PMA stimulation, and that this conformation change was retained during the whole purification process [6]. In the present work, results clearly showed a strongly increased binding of mAb 12E6 to the same amount (10 pmol) of cytochrome b558 purified from PMA-stimulated neutrophils as compared to that observed with cytochrome b558 isolated from resting cells (fig. 9, left panel). The slot-blot densitometry analysis indicated that the interaction between mAb 12E6 and cytochrome b558 was doubled after stimulation with PMA (fig. 9, right panel).
Fig. 9.
Slot-blot analysis of mAb 12E6 binding to active and inactive cytochrome b558 conformations. Duplicate samples of cytochrome b558 (10 pmol) purified from resting (– PMA) or PMA-stimulated (+ PMA) neutrophils were adsorbed to the nitrocellulose under vacuum conditions. The membrane was then incubated with mAb 12E6. The immune complexes were detected by ECL. Histograms showed the relative densitometry analysis of the slot-blot (100% corresponded to the densitometry obtained with cytochrome b558 purified from resting cells). Results were expressed as the mean of densitometry values obtained after analysis of duplicate bands ± SD.
Discussion
The present work describes 6 novel mAbs directed against an intracellular domain of p22-phox, the small subunit of the phagocyte cytochrome b558. As no structural data are available for cytochrome b558, mAbs are essential tools to get novel information about p22-phox membrane topology and regions involved in NADPH oxidase activation.
In this study, the 6 mAbs recognized purified native cytochrome b558 in 0.1% (w/v) Triton X-100 by ELISA, while only 5 mAbs (mAbs 17A2, 16G7, 16G6, 13D4 and 13C4) were able to detect denatured p22-phox in membrane or in solubilized membrane fractions by immunoblotting. The inability of the 6 mAbs to bind to intact neutrophils favors the intracellular localization of the epitopes, even if we could not completely rule out that the mAbs bind to extracellular regions that are masked on the cell surface until perturbation by permeabilization.
Phage display epitope mapping strongly suggested that mAbs 13D4 and 16G7 interact with the (130Q-K137) and (130Q-R139) regions of p22-phox, respectively.
For mAbs 17A2, 16G6 and 13C4, phage display results were not clear. The lack of recognition of the p22(1M-Q143)-truncated form by the 3 mAbs was consistent with detection of a cytoplasmic domain of p22-phox as shown by flow cytometry on permeabilized cells. Future refinements of FINDMAP with different substitution matrices and enhanced matching algorithms such as EPIMAP [27] may aid in the analysis of the clues to the structure of complex epitopes. Together, these results suggest that the target-region for mAbs 17A2, 16G6 and 13C4 is located in the C-terminal part of p22-phox that includes residues (144I-V195).
Recent studies based on deletion mutagenesis experiments showed that N- and C-terminal regions of p22-phox contained specific determinants critical for cytochrome b558 maturation and NADPH oxidase activation [28]. In particular, it was shown that deletion in the N-terminal (amino acids 6W-R90) or the C-terminal portion (amino acids 131W-V195) leads to a defect in Nox2 maturation and thus in NADPH oxidase activity [28]. Interestingly, deletion of the polyproline-containing region (142P-V195) did not influence cytochrome b558 maturation but resulted in the absence of NADPH oxidase activity [28]. These results suggest that the amino acids (131W-R141) are essential for Nox2 stabilization and maturation (fig. 10). The mAb NS2 that was earlier described to interact with the (131W-R139) sequence of p22-phox was unable to immunoprecipitate detergent-solubilized cytochrome b558[13]. In our study, mAbs 13D4 and 16G7 that recognize a similar region of p22-phox (130Q-K137 for mAb 13D4 and 130Q-R139 for mAb 16G7) were shown to immunoprecipitate detergent-solubilized cytochrome b558, suggesting exclusive properties of both mAbs 13D4 and 16G7 in comparison to mAb NS2. In regard to the importance of the target epitope, these mAbs might be useful for cellular analysis of cytochrome b558 maturation by evaluating their effect on p22-phox/gp91-phox heterodimer formation. As previously described for mAb NS2 [13], both mAbs 13D4 and 16G7 did not influence the NADPH oxidase activity, suggesting that the region (130Q-R139) of p22-phox is not involved in enzyme activation or activity.
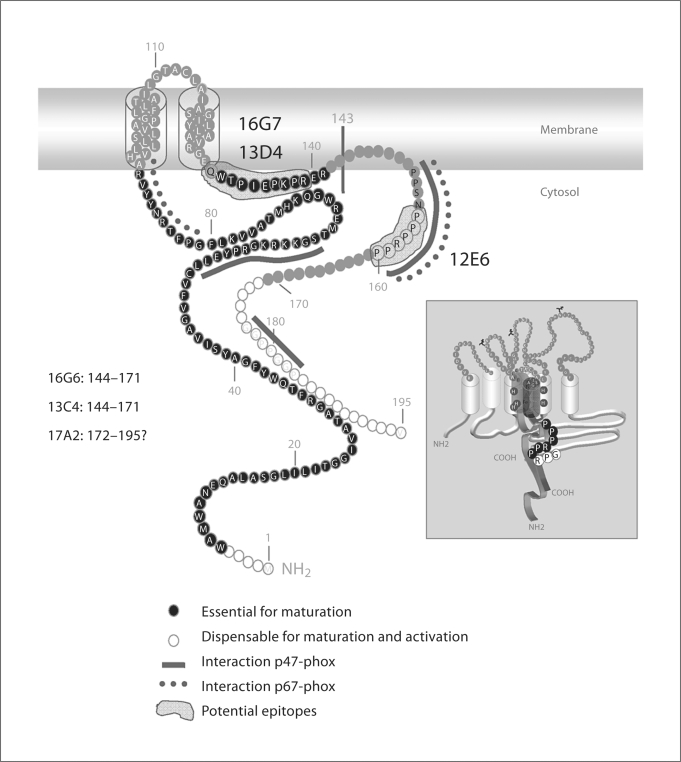
Fig. 10.
Topological model of p22-phox. The model highlights 2 transmembrane domains, regions containing amino acids shown to be essential or dispensable for cytochrome b558 maturation and activation [27] as well as regions involved in interactions with p47-phox and p67-phox. Epitope regions for novel mAbs resulting from data combining phage display analysis and p22-phox truncated forms investigation are depicted in this model. The insert shows potential epitope for mAb 12E6 shared by p22-phox (155PPPRPP160) and gp91-phox (557GPR559).
MAb 12E6 displays interesting properties. It is unable to bind denatured cytochrome b558, arguing in favor of a conformational target epitope easily lost or masked by mild detergent. Moreover, data obtained by confocal microscopy and flow cytometry demonstrate that mAb 12E6 binds activated neutrophils more strongly than resting cells. This effect is also observed in vitro on cytochrome b558 purified from PMA-stimulated cells, suggesting a conformational epitope which is unmasked during NADPH oxidase activation. Phage display analysis performed with mAb 12E6 indicates a potential antigen-determinant in the region (152P-P159) of p22-phox, the region containing a polyproline domain required for the interaction with p47-phox and p67-phox upon activation (fig. 10) [14,15]. The polyproline region is probably inaccessible in the cytochrome b558 resting state in order to avoid uncontrolled NADPH oxidase activation. Upon cytochrome b558 activation, the p22-phox polyproline region may change its conformation leading to a better accessibility of cytosolic factors to the domain. This hypothesis was confirmed by the inhibitory effect of mAb 12E6 on the NADPH oxidase activity reconstituted in vitro in the presence of neutrophil membrane and cytosol. Moreover, the analysis of the phage peptide sequences using FINDMAP revealed a potential epitope shared between p22-phox (155P-P160 residues) and the C-terminal part of gp91-phox (557G-R559 residues). Although such an assignment is highly speculative and requires confirmation, such complexity could explain the epitope lability in SDS and the nonionic detergent octyl glucoside. If indeed the epitope is shared by gp91-phox and p22-phox, mAb 12E6 will be useful to study interactions between cytochrome b558 subunits during protein maturation and enzyme activation processes.
For mAbs 17A2, 16G6 and 13C4, the epitope region was not clearly determined. However, immunoblots performed on the p22(1M-Q143)-truncated form suggested that their target-epitope was located in the cytosolic C-terminal 52 residues (144I-V195) of p22-phox. Deletion mutagenesis performed on p22-phox previously showed that the N-terminal residues (1M-5E) and the C-terminal residues (172A-V195) are not necessary for cytochrome b558 maturation and NADPH oxidase activity (fig. 10) [28]. Pre-incubation of neutrophil membranes with either mAb 16G6 or mAb 13C4 leads to a complete inhibition of the reconstituted NADPH oxidase activity, indicating that they interact with a region important for NADPH oxidase activation, located between amino acids 144I and 171E. In contrast, mAb 17A2 has no effect on the NADPH oxidase activity reconstituted in vitro, suggesting a potential interaction site in the C-terminal part of p22-phox dispensable for activation (after the polyproline motif) (161A-V195).
In conclusion, among the 6 new p22-phox mAbs described in this study, 2 (13D4 and 16G7) recognize a region required for cytochrome b558 maturation (130Q-R139). Three mAbs, 16G6, 13C4 and 17A2, bind an epitope localized in the C-terminal region (144IV195) but they display specific properties: mAbs 16G6 and 13C4, but not mAb 17A2, interact with a region involved in NADPH oxidase activation. According to data resulting from deletion mutagenesis performed on p22-phox [28], the C-terminal part of p22-phox (172A-V195) is not necessary for enzyme activity, suggesting that epitopes for mAbs 16G6 and 13C4 are located in the (144I-E171) part of p22-phox. The last mAb (12E6) recognizes a conformational epitope localized partially in the polyproline region of p22-phox. This mAb is unique as this is the first one described to detect this p22-phox domain. Our results lend support to the speculative view that this region may be part of a complex epitope shared between gp91-phox and p22-phox that may be produced during the activation process. However, there is as yet no confirming evidence to support this structural interpretation.
As p22-phox is an essential partner for novel Nox (Nox1, Nox3 and Nox4), these mAbs represent a valuable set of probes to investigate maturation and activation processes of NADPH oxidases.
Acknowledgements
We thank Dr. J. C. Renversez (DBI, CHU de Grenoble) for isotyping the monoclonal antibodies.
This work was supported by grants from the Ministère de l'Enseignement supérieur de la Recherche et Technologie, Paris, the UFR de Médecine, Université Joseph Fourier, Grenoble, the Région Rhône Alpes, programme Emergence 2003, the Groupement des Entreprises Françaises dans la lutte contre le Cancer, délégation de Grenoble, the Fondation pour la Recherche Médicale, Isère, the Délégation Régionale de la Recherche Clinique, CHU Grenoble and the French association La Ligue Nationale contre le Cancer. A.J.J. is supported by United States Public Health Service grants R56AI22735 and 5RO1AI26711.
References
- 1.Vignais PV. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell Mol Life Sci. 2002;59:1428–1459. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nauseef W. Nox enzymes in immune cells. Semin Immunopathol. 2008;30:195–208. doi: 10.1007/s00281-008-0117-4. [DOI] [PubMed] [Google Scholar]
- 3.Cross AR, Segal AW. The NADPH oxidase of professional phagocytes-prototype of the NOX electron transport chain systems. Biochim Biophys Acta. 2004;1657:1–22. doi: 10.1016/j.bbabio.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bokoch GM, Zhao T. Regulation of the phagocyte NADPH oxidase by Rac GTPase. Antioxid Redox Signal. 2006;8:1533–1548. doi: 10.1089/ars.2006.8.1533. [DOI] [PubMed] [Google Scholar]
- 5.Paclet MH, Coleman AW, Vergnaud S, Morel F. P67-phox-mediated NADPH oxidase assembly: imaging of cytochrome b558 liposomes by atomic force microscopy. Biochemistry. 2000;39:9302–9310. doi: 10.1021/bi000483j. [DOI] [PubMed] [Google Scholar]
- 6.Paclet MH, Berthier S, Kuhn L, Garin J, Morel F. Regulation of phagocyte NADPH oxidase activity: identification of two cytochrome b558 activation states. FASEB J. 2007;21:1244–1255. doi: 10.1096/fj.06-6852com. [DOI] [PubMed] [Google Scholar]
- 7.Heyworth PG, Cross AR, Curnutte JT. Chronic granulomatous disease. Curr Opin Immunol. 2003;15:578–584. doi: 10.1016/s0952-7915(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 8.Morel F. Molecular aspects of chronic granulomatous disease. Bull Acad Natl Med. 2007;191:377–390. [PubMed] [Google Scholar]
- 9.Cross AR, Segal AW. The NADPH oxidase of professional phagocytes-prototype of the NOX electron transport chain systems. Biochim Biophys Acta. 2004;1657:1–22. doi: 10.1016/j.bbabio.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLeo FR, Burritt JB, Yu L, Jesaitis AJ, Dinauer MC, Nauseef WM. Processing and maturation of flavocytochrome b558 include incorporation of heme as a prerequisite for heterodimer assembly. J Biol Chem. 2000;275:13986–13993. doi: 10.1074/jbc.275.18.13986. [DOI] [PubMed] [Google Scholar]
- 11.Ambasta RK, Kumar P, Griendling KK, Schmidt HHHW, Busse R, Brandes RP. Direct interaction of the novel nox proteins with p22phox is required for the formation of a functionnally active NADPH oxidase. J Biol Chem. 2004;279:45935–45941. doi: 10.1074/jbc.M406486200. [DOI] [PubMed] [Google Scholar]
- 12.Ueno N, Takeya R, Miyano K, Kikuchi H, Sumimoto H. The NADPH oxidase Nox3 constitutively produces superoxide in a p22phox-dependent manner: its regulation by oxidase organizers and activators. J Biol Chem. 2005;280:23328–23339. doi: 10.1074/jbc.M414548200. [DOI] [PubMed] [Google Scholar]
- 13.Taylor RM, Burritt JB, Baniulis D, Foubert TR, Lord CI, Dinauer MC, Parkos CA, Jesaitis AJ. Site-specific inhibitors of NADPH oxidase activity and structural probes of flavocytochrome b: characterization of six monoclonal antibodies to the p22-phox subunit. J Immunol. 2004;173:7349–7357. doi: 10.4049/jimmunol.173.12.7349. [DOI] [PubMed] [Google Scholar]
- 14.Sumimoto H, Kage Y, Nunoi H, Sasaki H, Nose T, Fukumaki Y, Ohno M, Minakami S, Takeshige K. Role of Src homology 3 domains in assembly and activation of the phagocyte NADPH oxidase. Proc Natl Acad Sci USA. 1994;91:5345–5349. doi: 10.1073/pnas.91.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahan I, Issaeva I, Gorzalczany Y, Sigal N, Hirshberg M, Pick E. Mapping of functional domains in the p22-phox subunit of flavocytochrome b558 participating in the assembly of the NADPH oxidase complex by ‘peptide walking’. J Biol Chem. 2002;277:8421–8432. doi: 10.1074/jbc.M109778200. [DOI] [PubMed] [Google Scholar]
- 16.Berthier S, Paclet MH, Lerouge S, Roux F, Vergnaud S, Coleman AW, Morel F. Changing the conformation state of cytochrome b558 initiates NADPH oxidase activation: MRP8/MRP14 regulation. J Biol Chem. 2003;278:25499–25508. doi: 10.1074/jbc.M209755200. [DOI] [PubMed] [Google Scholar]
- 17.Campion Y, Paclet MH, Jesaitis AJ, Marques B, Grichine A, Berthier S, Lenormand JL, Lardy B, Stasia MJ, Morel F. New insights into the membrane topology of the phagocyte NADPH oxidase: characterization of an anti-gp91-phox conformational monoclonal antibody. Biochimie. 2007;89:1145–1158. doi: 10.1016/j.biochi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Batot G, Martel C, Capdeville N, Wientjes F, Morel F. Characterization of neutrophils NADPH oxidase activity reconstituted in a cell-free assay using specific monoclonal antibodies raised against cytochrome b558. Eur J Biochem. 1995;234:208–215. doi: 10.1111/j.1432-1033.1995.208_c.x. [DOI] [PubMed] [Google Scholar]
- 19.Paclet MH, Henderson LM, Campion Y, Morel F, Dagher MC. Localization of Nox2 N-terminus using polyclonal antipeptide antibodies. Biochem J. 2004;382:981–986. doi: 10.1042/BJ20040954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paclet MH, Coleman AW, Burritt J, Morel F. NADPH oxidase of Epstein-Barr-virus immortalized B lymphocytes: effect of cytochrome b558 glycosylation. Eur J Biochem. 2001;268:5197–5208. doi: 10.1046/j.0014-2956.2001.02455.x. [DOI] [PubMed] [Google Scholar]
- 21.Vergnaud S, Paclet MH, El Benna J, Pocidalo MA, Morel F. Complementation of NADPH oxidase in p67-phox-deficient CGD patients p67-phox/p40-phox interaction. Eur J Biochem. 2000;267:1059–1067. doi: 10.1046/j.1432-1327.2000.01097.x. [DOI] [PubMed] [Google Scholar]
- 22.Burritt JB, Bond CW, Doss KW, Jesaitis AJ. Filamentous phage display of oligopeptide libraries. Anal Biochem. 1996;238:1–13. doi: 10.1006/abio.1996.0241. [DOI] [PubMed] [Google Scholar]
- 23.Burritt JB, Quinn MT, Jutila MA, Bond CW, Jesaitis AJ. Topological mapping of neutrophils cytochrome b epitopes with phage-display libraries. J Biol Chem. 1995;270:16974–16980. doi: 10.1074/jbc.270.28.16974. [DOI] [PubMed] [Google Scholar]
- 24.Mumey BM, Bailey BW, Kirkpatrick B, Jesaitis AJ, Angel T, Dratz EA. A new method for mapping discontinuous antibody epitopes to reveal structural features of proteins. J Comput Biol. 2003;10:555–567. doi: 10.1089/10665270360688183. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mumey BM, Ohler N, Angel T, Jesaitis AJ, Dratz EA. Filtering epitope alignments to improve protein surface prediction; in Lecture Notes in Computer Science. Frontiers of High Performance Computing and Networking – ISPA. Berlin/Heidelberg: Springer; 2006. pp. 648–657. [Google Scholar]
- 28.Zhu Y, Marchal CC, Casbon AJ, Stull N, Lohneysen KV, Knaus UG, Jesaitis AJ, McCormick S, Nauseef WM, Dinauer MC. Deletion mutagenesis of p22-phox subunit of flavocytochrome b558: identification of regions critical for gp91-phox maturation and NADPH oxidase activity. J Biol Chem. 2006;281:30336–30346. doi: 10.1074/jbc.M607191200. [DOI] [PubMed] [Google Scholar]