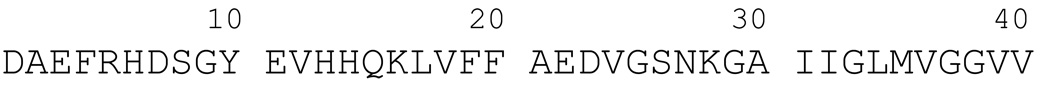Abstract
The ability of a single polypeptide sequence to grow into multiple stable amyloid fibrils sets these aggregates apart from most native globular proteins. The existence of multiple amyloid forms is the basis for strain effects in yeast prion biology, and may also contribute to variations in Alzheimer’s disease pathology. However, the structural basis for amyloid polymorphism is poorly understood. We report here five structurally distinct fibrillar aggregates of the Alzheimer’s plaque peptide Aβ(1–40), as well as a non-fibrillar aggregate induced by Zn+2. Each of these conformational forms exhibits a unique profile of physical properties, and all the fibrillar forms “breed true” in elongation reactions at a common set of growth conditions. Consistent with their defining cross-β structure, we find that in this series the amyloid fibrils containing more extensive β-sheet exhibit greater stability. At the same time, side chain packing outside of the β-sheet regions also contributes to stability, and to stability differences between polymorphic forms. Stability comparison is facilitated by the unique feature that the free energy of the monomer (equivalent to the unfolded state in a protein folding reaction) does not vary, and hence can be ignored, in the comparison of ΔG° of elongation values for each polymorphic fibril obtained at a single set of conditions.
The aggregated, β-sheet rich amyloid structure represents a stable, alternatively folded state of polypeptides. Amyloid fibrils are associated with several important neurodegenerative diseases, such as Alzheimer’s and Huntington’s diseases 1, as well as a number of peripheral diseases of organ failure 2. Amyloid fibrils can be produced in vitro from many proteins, consistent with the polymeric structure of proteins and the relationship of amyloid fibrils to synthetic polymers 3. The fundamental unit of amyloid fibrils is the cross-β structure, in which β-sheet extended chains and sheet-sheet stacking interactions are perpendicular to the fibril axis and β-sheet H-bonds are parallel to the fibril axis 4. Details of the three-dimensional structures of amyloid fibrils are still being elucidated 5; 6; 7; 8; 9; 10; 11; 12
One striking feature of amyloid fibrils that sets them apart from most globular proteins is the ability of a single polypeptide chain to grow into more than one stable structure 13. The existence of multiple protein aggregate conformations, each of which can propagate with retention of structure, has long been speculated to be the basis for strain and species barrier effects in mammalian and yeast prion biology 14; 15. Polymorphism at the electron microscopy level, for example in Aβ amyloid fibrils 16; 17, has been known for some time, but it has not been clear whether these shape differences were due merely to different modes of super-assembly of a common protofilament structure, or to more substantial internal structural differences, such as β-sheet formation and side-chain packing. Previous solid state NMR and electron microscopy analyses suggest that the folded structures of Aβ(1–40) in two polymorphic amyloid fibrils are only modestly different, while the major structural differences are in how the folded peptides pack within the fibril cross-section 9; 18. Analysis of amyloid polymorphs of other protein sequences, however, suggests the possibility of polymorphic structures differing more extensively in the details of segmental folding, H-bonding and packing within the fibril 13; 19, and this is further suggested by the different manners in which sequence-related fragments from amyloid proteins pack within “β-spine” crystal structures 6.
Polymorphism in amyloid fibrils may have profound biological consequences. It has been demonstrated that different polymorphic yeast prion fibrils generated in vitro produce different prion strain behavior when these are introduced into yeast 20; 21. Two structurally and functionally different polymorphic fibrils have been generated by exposing Aβ(1–40) to different growth conditions in vitro 22, and the identification of a third polymorphic form produced by elongation of Aβ peptides in vitro using fibrils extracted from AD brain 23 supports the idea that fibril polymorphism may contribute to variations within human diseases.
In this paper we describe the creation of five self-propagating amyloid fibril structures by subjecting wild type Aβ(1–40) to different trial growth conditions. These Aβ(1–40) polymorphic fibrils vary considerably in structural properties. In particular, we find that amyloid polymorphs exhibit significant differences in the extent and locations of stable β-sheet, as probed by the number of backbone amides highly protected from hydrogen-deuterium exchange. We find that fibril stabilities assessed by their free energies of elongation 24 correlate extremely well with these β-sheet contents, consistent with the central role of β-sheet in fibril structure. The results illustrate the facility with which some peptides produce fibril polymorphs and suggest that β-sheet content contributes significantly to fibril properties.
RESULTS
Many buffer and growth conditions have been described for producing fibrils from the Alzheimer’s disease plaque peptide Aβ(1–40) (Fig. 1). Experiments by the Lansbury group emphasized stirred conditions (for accelerating aggregation rates) and contained DMSO (for removing pre-existing aggregates) 25. Our group was able to eliminate DMSO by using volatile solvents for disaggregation, and grew amyloid in PBS without agitation to better replicate in vivo conditions 26. The Tycko group has applied various growth conditions 9; 22. Different types of Aβ aggregates have been produced at pH 2 27 and in the presence of zinc 28 or other additives 29; 30; 31; 32. In our work here, we maintained pH at 7.4 while exploring variations in temperature, agitation, and salt concentration, and the presence or absence of Zn+2 28 or concentrations of SDS 32. Overall, we explored a total of eight different conditions that produced five distinct amyloid polymorphs with unique distributions of properties, as well as one non-fibrillar, β-sheet rich morphology (Table 1).
Figure 1.
Amino acid sequence of human Aβ(1–40).
Table 1.
Fibril preparation conditions.
| Polymorph | Disaggregated Monomer? |
Temperature, °C |
Buffer | Agitation? | Additions |
|---|---|---|---|---|---|
| A | Yes | 37 | PBS | No | None |
| B | No | 24 | Phosphate | No | None |
| C | Yes | 24 | PBS | Yes | None |
| C | Yes | 24 | Phosphate | Yes | None |
| D | Yes | 4 | PBS | Yes | None |
| D | Yes | 4 | Phosphate | Yes | None |
| E | Yes | 37 | Phosphate + 500mM NaCl |
No | 0.5 mM SDS |
| Z | Yes | 37 | Tris.HCl/NaCl | No | ZnCl2 |
One of these polymorphs, the “A” form, was prepared by our standard conditions, using rigorously disaggregated monomer in a non-agitated incubation in PBS at 37 °C 26; 33; 34. The “B” form was prepared with material directly dissolved from lyophilized peptide, without disaggregation, and incubated in phosphate buffer without salt and without agitation at 24 °C 35. The “C” form was obtained using disaggregated monomer with agitation (see Methods) at 24 °C in either PBS or in phosphate buffer; both conditions gave the same polymorphic form. The “D” form was obtained using disaggregated monomer with agitation at 4 °C in either PBS or in phosphate buffer; both conditions gave the same polymorphic form. The “E” form was obtained by incubating disaggregated monomer in SDS at 37 °C without agitation. The “Z” form was obtained by incubating disaggregated monomer in the presence of Zn+2 at 37 °C without agitation.
The data supporting the identical structures of the phosphate and PBS versions of the C and D forms, such as FTIR, critical concentration, EM, and hydrogen-deuterium exchange protection, are included throughout this paper. However, for consistency and simplicity, where comparisons are being made to other amyloid polymorphs, we only use the data for the C and D forms made in PBS. Our ability to identify sibling polymorphic fibrils from Aβ(1–40) by comparing the fine details of a number of experimental parameters underscores the robust nature of the polymorphic fibril types that are the subjects of this paper.
TEM analysis
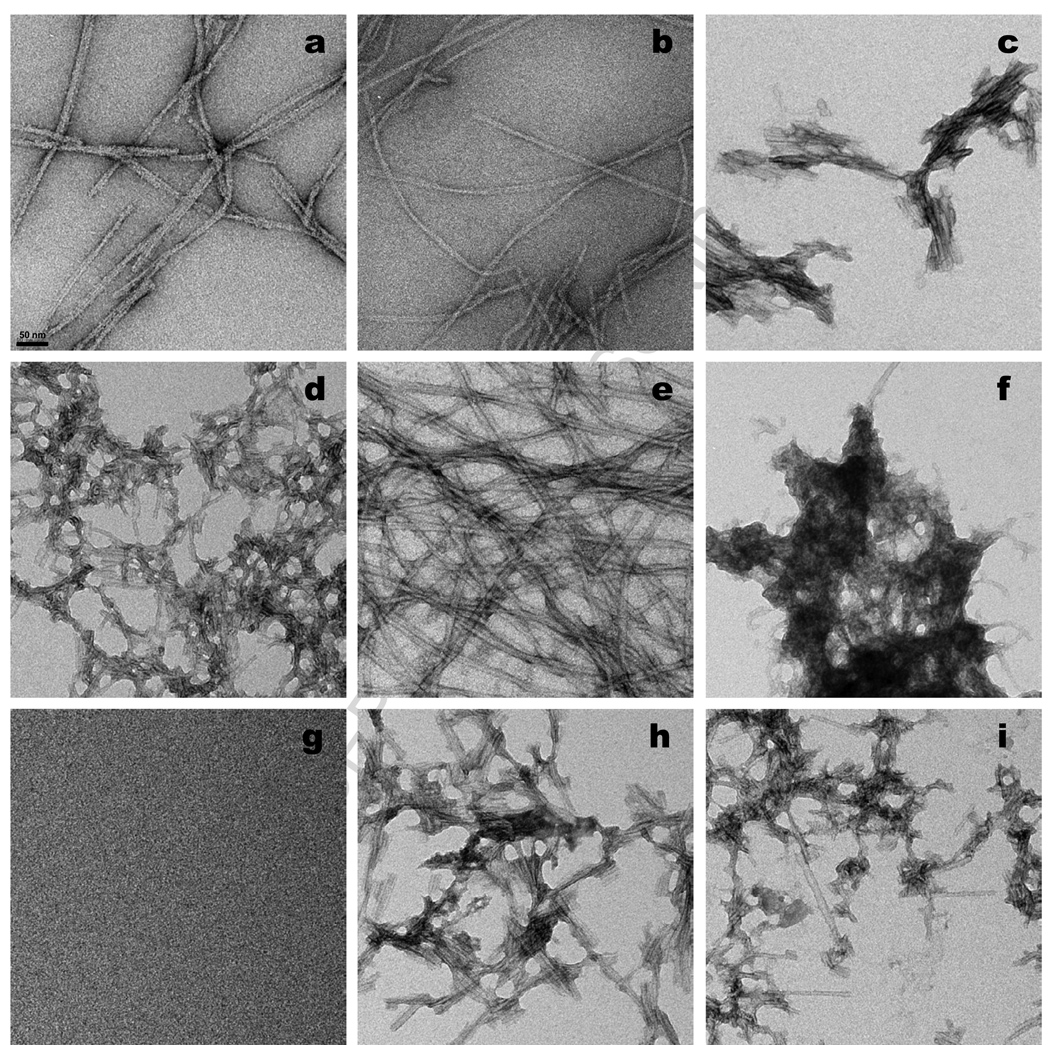
Negative staining TEM (Methods) shows that fibrils grown under quiescent conditions (A and B fibrils) tend to be long and un-branched fibrils with some aperiodic twisting (Fig. 2 a,b). In contrast, most of the aggregates grown under agitated conditions (C and D fibrils) are short, relatively straight, and appear to be composed of laterally associated filaments lacking discernable twist (Fig. 2 c,d,h,i). Fibrils formed without stirring in the presence of SDS (E fibrils) are long, unbranched, and loosely interleaved (Fig. 2 e). The aggregates grown in the presence of ZnCl2 appear to be highly clumped, short filaments (Fig. 2 f). (A mock staining of a grid exposed to ZnCl2 alone is blank (Fig. 2 g)). As discussed above, aggregates formed with agitation in PBS (Fig. 2 c,d) are morphologically similar to those formed under similar conditions in phosphate buffer (Fig. 2 h,i). While the differences in average lengths between the quiescent (Fig. 2 a,b,e) and agitated (Fig. 2 c,d,h,i) fibrils are consistent with the known sensitivity of some amyloid fibrils to shear forces 13, it is also possible that the shorter average length in agitated fibrils reflects a greater number of nuclei being formed in these reactions.
Figure 2.
Negative stained electron micrographs of samples freshly harvested from aggregation reactions. (a) polymorph A; (b) polymorph B; (c) polymorph C made in PBS; (d) polymorph D made in PBS; (e) polymorph E; (f) polymorph Z; (g) mock staining of grid treated with ZnCl2 in PBS; (h) polymorph C made in phosphate buffer; (i) polymorph D made in phosphate buffer. Scale bar, 50 nm.
Weight-normalized ThT binding
Previously we found that amyloid fibrils grown from single point mutants of Aβ(1–40) can differ substantially in the yield of ThT fluorescence obtained per unit weight of amyloid fibril 36. Similarly, using the same fluorescence instrument and conditions, fibrils from different proteins can give significantly different weight-normalized (see Methods) ThT signals. We show here that polymorphic aggregated forms of the identical sequence - wild type Aβ(1–40) - also vary considerably in ThT fluorescence values (Table 2). The highest fluorescence yields are obtained from C and E fibrils. Although the low ThT value for the Z aggregates might be related to a more protofibril-like structure (Aβ(1–40) protofibrils yield relatively low ThT values 37), it is noteworthy that the seeding competent (and hence composed of fibrils rather than protofibrils) D aggregates (Fig 2 d,i) also exhibit a low ThT value. Comparison of fibril lengths (Fig. 2) and ThT values (Table 2) rules out any simple relationship between ThT signal and the number of fibril termini. Our data show a rough correlation of ThT fluorescence with the number of highly protected β-sheet H-bonds (Table 2), and hence are consistent with the often-stated view that ThT binds to β-sheet. While the structural basis for differences in ThT response remains to be elucidated 38, weight normalized ThT values are clearly useful for distinguishing among polymorphic forms (Table 2).
Table 2.
Properties of Aβ(1–40) polymorphic fibrils.
| Polymorphic form | A | B | Ce | De | E | Z |
|---|---|---|---|---|---|---|
| ThTa | 55 ± 10 | 35 ± 4 | 242 ± 25 200 ± 22 |
17 ± 3 28 ± 2 |
190 ± 25 | 12 ± 2 |
| Cr, µM, spontaneous growtha |
1.06±0.18 | 20.3± 2.5 | 0.22±0.06 0.94±0.32 |
1.35±0.07 1.32±0.14 |
0.13±0.03 | 1.36±0.15 |
| Cr, µM, seeded elongation, phosphate b |
1.3 | 16 | 0.23 0.66 |
0.79 1.3 |
0.28 | nd |
| Cr, µM, seeded elongation, PBS b |
1.33 | 5 | 0.23 0.25 |
0.36 0.56 |
0.13 | 1.1 |
| N-H exchanged, spontaneous growtha,c |
16.7± 1.3 | 24.2± 0.2 | 10.5±0.11 10.6±0.32 |
13.8±0.32 14.0±0.25 |
14.1±0.44 | 24.2±0.26 |
| N-H exchanged, spontaneous growtha,d |
17.1± 1.3 | 25.0± 0.2 | 11.4±0.11 11.4±0.32 |
15.2±0.32 14.7±0.25 |
13.8±0.44 | 24.6±0.26 |
| N-H exchanged, seeded elongation, phosphate b,d |
17.2 | 24.1 | 11.7 11.2 |
15.0 15.0 |
14.5 | nd |
| N-H exchanged, seeded elongation, PBS b,d |
17.2 | 20.6 | 11.9 12.0 |
15.0 15.0 |
14.5 | 17.0 |
Mean of three or more independent fibril growth experiments starting with monomeric Aβ.
Monomeric Aβ(1–40) seeded (7.5% wt/wt) with different polymorphic fibrils and incubated without shaking at 37 °C in phosphate or PBS.
Data corrected for artifactual exchange.
Data not corrected for artifactual exchange.
Top value, from polymorph grown in PBS; bottom value, from polymorph grown in phosphate.
Note that the weight normalized ThT values for both the C (agitated, 24 °C) and D (agitated, 4 °C) fibrils are quite similar (Table 2, top and bottom values within each cell) regardless of whether they were generated in PBS or phosphate.
Fourier transform infrared spectroscopy (FTIR)
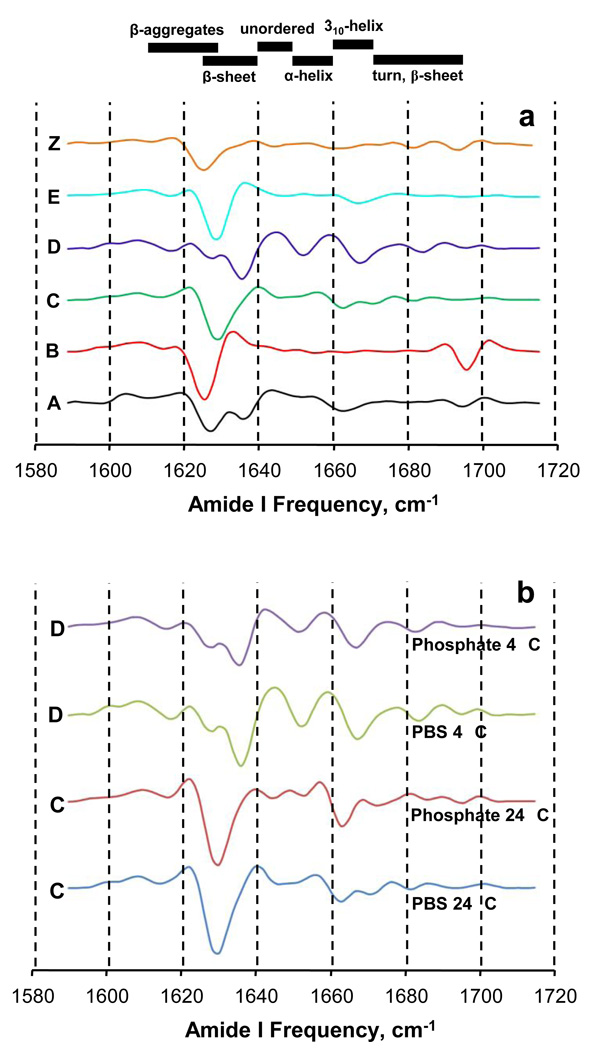
FTIR is well suited for obtaining amyloid fibril secondary structural information as the amide I band is very sensitive to β-sheet structures 39. Second derivative FTIR spectra show that all conformers, including the Z conformation grown in presence of Zn+2, exhibit intense bands in the β-sheet region (Fig. 3 a,b). Four aggregates (B, C, E and Z) display only one band (Fig. 3 a) in the major β-sheet region spanning 1610–1640 cm−1 39; this band is centered around 1625–1628 cm−1, in the range associated with higher stability H-bonds 39. In contrast, two other polymorphic aggregates (A and D) reveal a second, less stable H-bond band (~ 1634 cm−1) in addition to the higher stability H-bond band (~ 1628 cm−1). While the higher stability H-bond bands are very likely attributable to parallel β-sheet 39 known to dominate WT Aβ(1–40) amyloid structures 18, the nature of the lower stability β-sheet H-bonds is not clear. Each of the conformers exhibits one or more bands of varying intensity in the 1640–1660 cm−1 region, which is generally assigned to α-helix and random coil 39. All the polymorphs also have one or more bands in the 1660–1695 cm−1 region normally assigned to 310 helix, turns, and β-sheet conformations 39. Detailed molecular descriptions of these FTIR spectra currently are not possible, but it is clear that the FTIR spectra can be used as fingerprints to identify sets of fibrils with similar structures, as has also been reported for mammalian prion amyloids 40. For example, FTIR further supports the structural similarity of the PBS and phosphate forms of the C and D fibril morphologies (Fig. 3 b)
Figure 3.
Second derivative FTIR spectra of various aggregated polymorphic forms of Aβ(1–40). Secondary structure frequency ranges at the top of the figure are estimates from reference 39. (a) Comparison of the six polymorphic forms described herein. (b) Comparisons of the two sets of sibling fibrils for the C and D polymorphs, each prepared in either PBS or phosphate buffer.
H/D exchange mass spectrometry (HX-MS)
Although structural sequestration can inhibit hydrogen exchange, the dramatic exchange protection in amyloid, that persists even after hundreds of hours incubation 41, is probably primarily associated with very stable H-bonds involved in the structure of the amyloid β-sheet network. In fact, HX-MS with in-line pepsin digestion provides segmental exchange protection information that is in verygood agreement with the location of H-bonded β-sheet in the same fibril type as identified by other techniques 42; 43.
To obtain the HX-MS data, aggregates are suspended in D2O and deuterated buffer and incubated, and portions of the reaction mixture analyzed over time by quickly dissolving the aggregates and analyzing by MS. After correction for adventitious exchange with H2O during the workup 44, the mass change of the Aβ peptide (Fig. 4) indicates the number of backbone amide protons exchanged and protected 43; 45. Since almost all of the exchange observed into Aβ fibrils occurs within the first 24 hrs 41, we used the 24 hour time point to analyze and compare both global and segmental protection data for all the conformers.
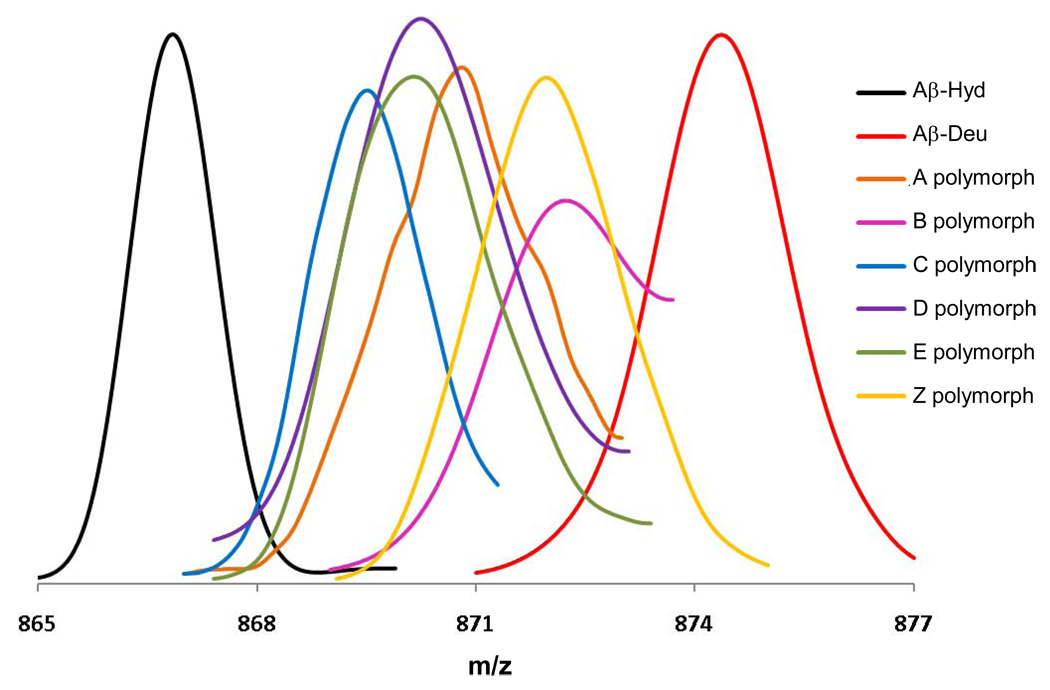
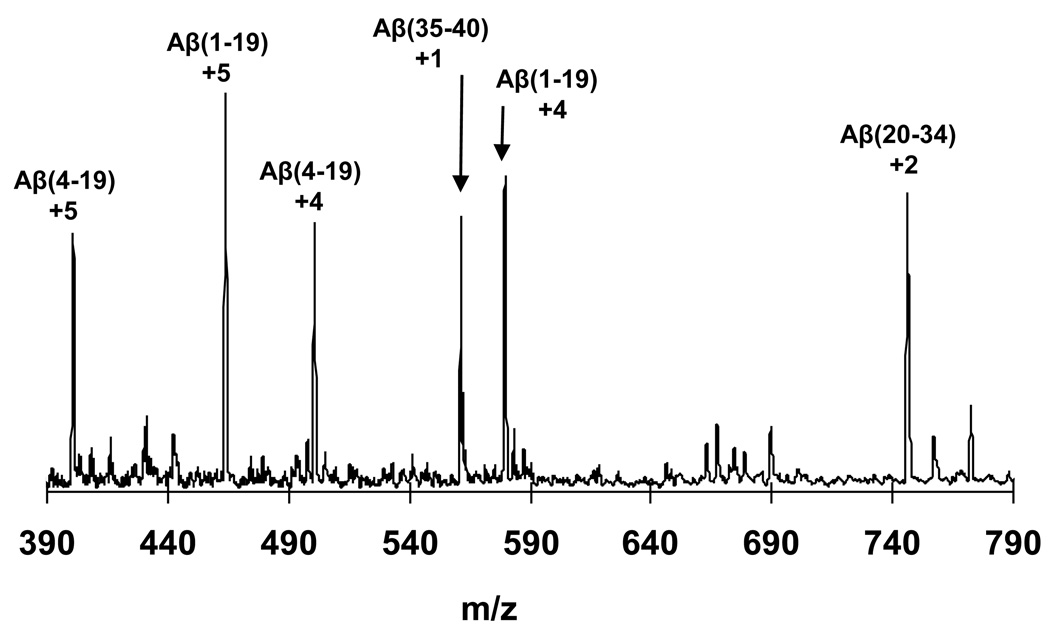
Figure 4.
Electrospray mass spectrograms of the +5 charge state of the polymorphic Aβ(1–40) aggregate preparations described in this paper. “Aβ-Hyd” and “Aβ-Deu” are the Aβ(1–40) A polymorph fibrils grown in either H2O or D2O. Polymorphic fibrils (and the Zn induced aggregate “Z”) were grown in H2O, harvested, and subjected to D2O exchange for 24 hrs. All were analyzed by in-line H/D exchange as described, in which fibrils are dissolved in-line and streamed into the MS. Small peaks appearing at the “fully exchanged” m/z value at the “Aβ-Deu” position, which appear to be due to equilibrium exchange of deuterated monomeric Aβ(1–40) into fibrils during the exchange incubation 41, are not shown.
Most of the fibrils grown under different conditions exhibit different degrees of protection (first data column, Table 3). In agreement with our earlier studies 44, about 22 of the 39 backbone amide hydrogens of Aβ(1–40) are highly protected in the A form of Aβ(1–40) fibrils (Table 3). In contrast, C fibrils each have about 28 protected amide protons, suggesting a significantly expanded β-sheet network. The B conformation has the least number of protected amide protons, similar to that of the Z aggregate and only slightly more than the 12 protected amide protons previously characterized in Aβ(1–40) protofibrils 37. D and E fibrils both exhibit about 25 protected amide hydrogens, intermediate between the A and C fibrils (Table 3). There is excellent agreement in the amounts of protection in the PBS and phosphate versions of C fibrils and of D fibrils (Table 3).
Table 3.
Segmental hydrogen-deuterium exchange analysis of polymorphic fibrils
| Polymorphic form |
Number of protected backbone amide protons, 24 hrsa,b, In intact Aβ and pepsin fragments (total exchangeable protons) |
||||
|---|---|---|---|---|---|
| 1–40 (39) | 1–19 (18) | 4–19 (15) | 20–34 (14) | 35–40 (5) | |
| A | 22.3±1.3 | 7.56±0.7 | 7.24±0.7 | 9.73±0.8 | 1.1±0.1 |
| B | 14.8±0.2 | 5.3±0.6 | 5.7±0.6 | 7.1±0.7 | 1.1±0.06 |
| Cc | 28.5±0.11 28.4±0.32 |
11.9±0.17 12.1±0.32 |
8.9±0.25 9.2±0.5 |
12.1±0.06 12.2±0.17 |
1.3±0.14 1.3±0.3 |
| Dc | 25.2±0.2 25.0±0.25 |
9.8±0.45 9.1±0.6 |
7.0±0.46 6.4±0.4 |
10.91±0.09 11.1±0.13 |
1.3±0.17 1.9±0.14 |
| E | 24.9±0.44 | 8.73±0.45 | 6.0±0.3 | 10.2±0.35 | 2.0±0.3 |
| Z | 14.8±0.26 | 5.25±0.19 | 4.95±0.5 | 6.20±0.22 | 1.2±0.18 |
| Ad | 21.8 | 7.6 | 8.4 | 9.7 | 1.1 |
Data corrected for artifactual exchange.
From three or more independent determinations.
Top value, from polymorph grown in PBS; bottom value, from polymorph grown in phosphate.
Data from analysis with a triaxial probe 42.
To probe within these global levels of protection, we used in-line pepsin digestion to cleave the Aβ(1–40) monomers freshly released by dissolution of deuterium exchanged fibrils (see Methods). Pepsin cleaves Aβ(1–40) into 4 fragments corresponding to sequence positions 1–19, 4–19, 20–34 and 35–40 42. Previously our group determined the segmental protection for the A form of Aβ(1–40) fibrils 42 using in-line pepsin digestion and a triaxial probe, and found protected protons distributed over all of the pepsin fragments, with the sum total of protected protons from the fragment analysis agreeing very well with the global protection value. Thus, in the previously published data 42 shown in the last row of Table 3, summing the numbers of protected protons for the 1–19, 20–34 and 35–40 fragments of Aβ(18.4), and adding 2 protons for the amides destroyed by pepsin, gives a value of 20.4 – very close to the 21.8 value obtained for the entire Aβmolecule (residues 19–20 and 34–35 have generally been assigned to be in β-sheet and thus are expected to also be protected). In this paper we describe a new, simplified T-tube protocol (see Methods), in which we obtained efficient cleavage of Aβ(1–40) into fragments (Fig. 5) with exchange protection in very good agreement with the previous results (compare two entries for the A form of fibrils, Table 3). We therefore analyzed all the Aβ(1–40) aggregates by this new, simplified procedure.
Figure 5.
Mass spectrogram resulting from dissolution of a suspension of A type Aβ(1–40) H-fibrils with pepsin in aqueous formic acid in the mixing stream of a T-tube front end of the mass spectrometer, showing the major charge state species of the resulting pepsin digestion fragments.
We found that the distributions of protected amide hydrogens vary significantly among the various aggregates we analyzed (Table 3). We found that the C-terminal 35–40 fragment is not well protected in any of these aggregates: of the five backbone amide protons in this segment, two are protected in the E form, while only one is protected in the other aggregates. Although the short N-terminal Aβ fragment generated by pepsin is not isolated, it is possible to obtain protection information for residues 1–4 by comparing the protection in the 1–19 fragment to that observed for the 4–19 fragment. For the two native buffer quiescent fibrils and for the zinc aggregate (A, B and Z), the levels of protection within the 1–19 and 4–19 fragments are identical, within error, suggesting that the amide protons of residues 2, 3 and 4 are not protected in these aggregates. This is as expected for Aβ fibrils, whose N-termini are accessible to proteases 46 and to antibodies 47, and have been shown by other methods 35; 48 to be relatively disordered in fibril structure. In contrast, however, the C, D and E fibrils all exhibit three additional protected amide protons in the 1–19 fragment compared with the 4–19 fragment, consistent with the amide protons of residues 2, 3 and 4 being involved in highly protective – presumably H-bonded – structure (Table 3).
More variation exists in the levels of protection of the central fragments. For the 4–19 pepsin fragment, levels of protection of about 5, 6, 7, or 9 protons are observed, depending on the fibril. For the 14 backbone amide protons of the 20–34 fragment, levels of protection of about 6, 7, 10, 11, or 12 protons are observed. These data strongly support the existence of significant differences in the major implicated β-sheet regions in the different polymorphic fibrils.
As with other analytical methods described here, the “sibling” fibrils formed by agitation at 24 °C in either phosphate buffer or PBS (C fibrils), and those formed by agitation at 4 °C in these two buffers (D fibrils), give very good agreement in their segmental HX-MS protection (Table 3).
Self Propagation and condition-matched critical concentrations
Since Aβ(1–40) fibril formation is the result of non-covalent assembly of monomers, in principle the assembly reaction should decay toward a position of dynamic equilibrium at which monomer association with fibrils and monomer dissociation from fibrils are balanced, generating an equilibrium concentration of monomer – the critical concentration (Cr). This has been demonstrated for the A form of Aβ(1–40) fibrils, wherein it was shown that the Cr is related to the ΔG°elong for Aβ(1–40) fibrils 24. These ΔG°elong values for A form fibrils grown from point mutants of Aβ(1–40) allow quantitative assessment of the mutational effects on fibril stability and hence offer hints on fibril structure 3; 49. In the context of this paper, it becomes very interesting to use these Cr values for different polymorphic fibrils formed by wild type Aβ(1–40).
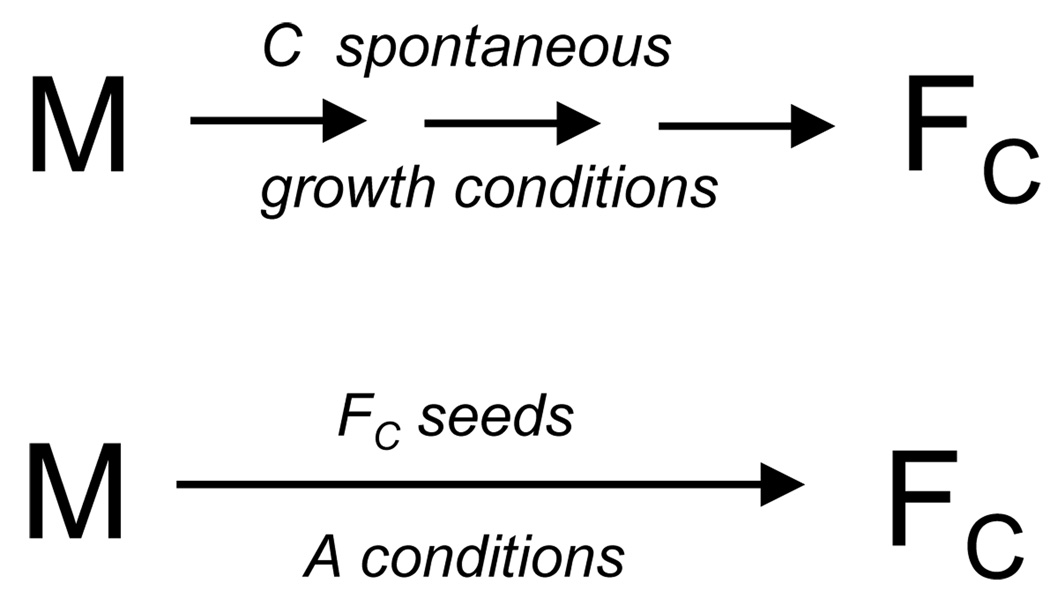
To a first approximation, we might simply determine the residual monomer concentration at equilibrium for each of the unique growth conditions required for producing the different polymorphic fibrils. However, just as is well-known to be the case for globular protein stability, we must assume that fibril stability will depend on environmental factors like temperature and buffer, i.e., the kinds of parameters that we are varying to produce the different polymorphs. For this reason, it becomes important to determine the Cr values of each polymorph under identical conditions of buffer, temperature, etc. This is possible in theory because of the ability of fibrils to propagate with fidelity of structure in a seeded elongation reaction. Thus, if fibril structure is strictly conserved when the fibril is propagated by seeded elongation under a different set of conditions, then a monomer-fibril equilibrium can be established for a common set of conditions for all fibrils, allowing direct stability comparisons. This is illustrated in the schematic in Figure 6.
Figure 6.
Schematic summary of seeding experiments. In this example, monomeric Aβ(1–40) “M” is incubated under the solution conditions (agitation, PBS, 24 °C) that generate the type C polymorph. The type C fibrils (“FC”) are isolated, then used to seed a solution of monomer incubated under the normal conditions (PBS, 37 °C, no agitation) for obtaining the type A polymorph. If fibril structure is, as expected, controlled by the structure of the seed and not by the incubation conditions, the product fibrils should also be of the C type. Since the elongation equilibrium is achieved in this case under A conditions, however, the Cr and derived ΔG°elong should reflect this.
We seeded reactions of freshly disaggregated monomer, in both PBS and phosphate buffer, with 7.5 % w/w of seeds from each of the fibrils isolated from different growth conditions, and incubated each of these reactions at 37 °C under quiescent conditions until the reactions reached equilibrium. To confirm that the seeding reactions took place with fidelity of fibril conformation, we analyzed all product fibrils by HX-MS, and found – with two exceptions - very good agreement when comparing properties of seed aggregates and product aggregates (Table 2), consistent with the expected abilities of these polymorphic forms to propagate with retention of conformation.
One of the exceptions occurred when using Z aggregates as seeds. At the conditions under which A fibrils are normally grown (including the absence of Zn+2), the reaction of monomers plus Z aggregates led to A fibrils (Table 2). This is most likely due to spontaneous initiation of A fibril formation within the monomer pool, which might occur, for example, if the Z aggregates are themselves not competent as elongation seeds.
In the other exception, B fibrils were found to seed elongation with retention of configuration under one set of conditions and not the other. In phosphate buffer at 37 °C without agitation, B fibrils replicated their polymorphic form in the fibril elongation product (Table 2). In contrast, however, in PBS buffer at 37 °C without agitation, B fibril seeds produced a fibrillar product (B*) that exhibits four additional protected amide protons and a four-fold reduced Cr (Table 2). There are a number of possible explanations for these observations: (a) Fibril conformation in this case itself may be unstable and sensitive to [NaCl] leading to a conformational change within the fibril on shifting to PBS conditions. (b) The elongation face of B fibrils may be somewhat promiscuous, being capable of seeding the growth of different fibril structures depending on solution conditions. In such an instance, it is possible for competing elongation reactions to produce a mixture of polymorphic fibrils, controlled by relative elongation rates 50. (c) Seeding efficiency of B under the PBS conditions may be so poor that spontaneous nucleation of A fibrils by the monomer becomes competitive, as with the Z aggregates above, leading to results reflecting a mixture of types. It remains to be seen whether B* constitutes a sixth polymorphic form. Uncovering a molecular explanation for this result will require additional studies.
With this demonstration of fidelity of propagation for most of the fibril polymorphs, the Cr values of the fibrils produced by seeded elongation can now be rigorously compared, since the fibril growth was in each case conducted under exactly the same conditions. The results for phosphate−37 °C-quiescent conditions indicate fibril stabilities, as judged by elongation thermodynamics, that vary over about 70-fold (Table 2). The most stable fibrils are the C, D and E forms, and the least stable is the B form. Surprisingly, the Cr values turned out to not change appreciably when comparing the two different sets of elongation conditions (phosphate buffer vs. PBS, Table 2). Thus, the presence or absence of 0.15 M NaCl has only a modest effect on the stability of the propagated fibrils.
Correlation of fibril stability and β-sheet content
It is well-known that the dominant structural motif in amyloid fibrils, in fact their defining motif, is a form of β-sheet. Hence the structural interactions underlying β-sheet structure – the H-bonds, the intra-sheet side-chain packing, the sheet-sheet side chain packing 51 – would be expected to make a major contribution to fibril stability. At the same time, while β-sheet is clearly a dominant structure of the protofilament units of the fibril, packing interactions between protofilaments must also play a stabilizing role. Little is known, however, about the nature of these interactions. In addition, it is well known that even for the simple peptide Aβ not all portions of the peptide chain of the amyloid component protein are involved in H-bonded β-sheet 46, and how these non-β components pack within the fibril may also contribute significantly to fibril stability. Thus, it becomes of interest to determine the extent to which β-sheet energetics dominate fibril stability. With our data on these polymorphic Aβfibrils, in which no allowances have to be made for energetic contributions of mutations, we are in a position to address this question.
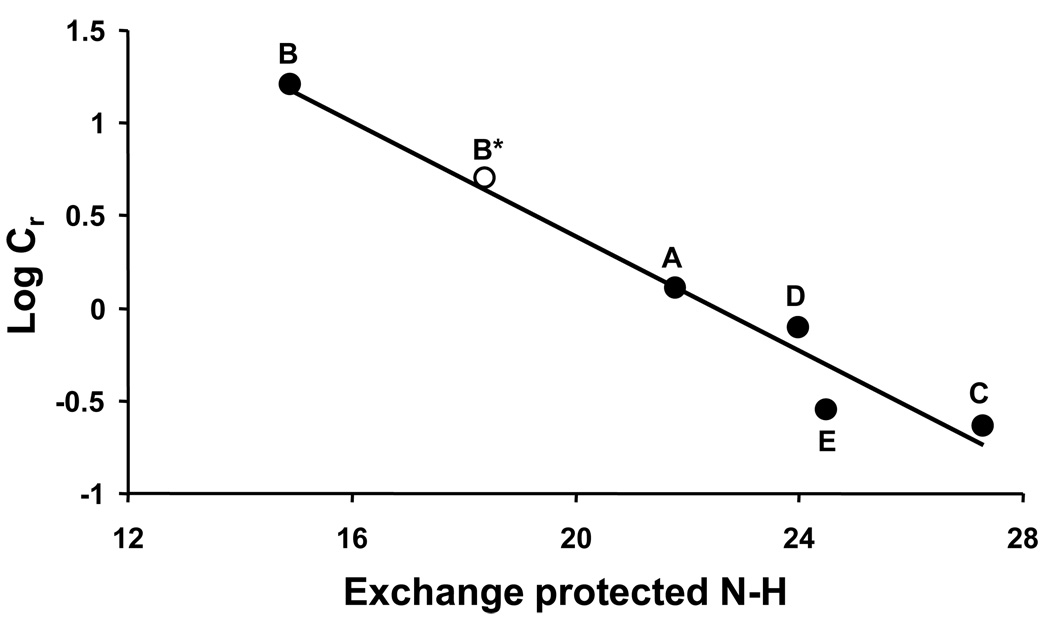
In fact, we find an excellent correlation between the number of highly protected backbone amide protons – presumed to be in β-sheet - and fibril stability as assessed by the elongation Cr values (Fig. 7). Good correlations were obtained whether the Cr values were obtained in the original growth conditions for each fibril (Table 2), or were observed in what we believe to be more rigorous experiments where fibril polymorphs are propagated from the original seeds into a set of common solution conditions (Fig. 7). Figure 7 shows the correlation of Cr and HX data from the fibrils obtained by seeding disaggregated monomeric Aβ(1–40) in phosphate buffer with the various fibril polymorphs described above. After this data was fit to a line, the additional data point was added that represents the possible additional fibril polymorph B* obtained by propagating B fibrils in PBS buffer (see above). Gratifyingly, this data point falls very close to the fitted line. Overall, the data strongly suggests that the net total of strong H-bonds, that resist hydrogen exchange even after one day incubation in D2O, contribute greatly to fibril stability as assessed by the position of the elongation equilibrium.
Figure 7.
Correlation of fibril stability with β-sheet content. Cr values were obtained from seeded elongation fibril reactions conducted in phosphate buffer, where seeding was with different fibril polymorphs, as shown in a general way in Figure 6 (Table 1). Log Cr (●) is plotted against corrected HX-MS values of each fibril product. Slope = − 0.1548, R2 = 0.9598. The open circle (○) represents the data for the product (“B*”) of elongation in PBS at 37 °C of Aβ(1–40) monomers seeded with B fibrils. This data point was not used in the linear fit.
Sensitivity of polymorphic elongation stability to Ala mutation
Previously we used the elongation Cr values coupled with point mutagenesis within the Aβ(1–40) peptide to map elements of structure within the A fibril polymorphic type 3; 36; 49; 52; 53; 54. Very broadly, Pro replacements were used to explore chain configuration effects 52, Cys replacements were used to explore solvent accessibility of side chains 36, as well as side chain-side chain contact 53, and Ala replacements were used to explore packing interactions 54. Although it was not possible to conduct all of these scanning exercises on all of the polymorphs as part of this work, we report here the results of an Ala mutational scan of fibril elongation stability for the C fibrils produced by agitation in PBS buffer at 24 °C. Purified mutant Aβ(1–40) peptides were disaggregated and incubated under the standard C polymorph growth conditions to initiate fibril growth, which was allowed to continue to an equilibrium position at which the residual monomer concentration no longer changed with time 24. These Cr values were then used to calculate a free energy of elongation 24.
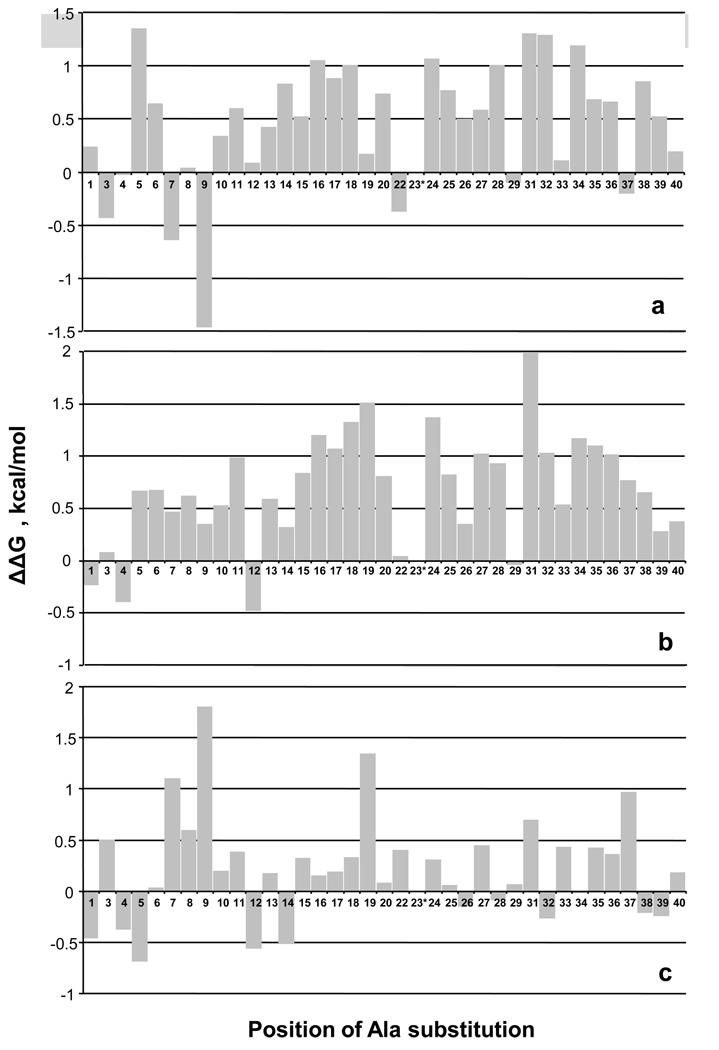
Figure 8a summarizes the results by showing ΔΔG°elong for type C fibril growth for each mutation, while Figure 8b shows data previously acquired for the A polymorph 54. These values are calculated by subtracting the ΔG° of elongation for wild type from the ΔG°elong of the mutant. Positive values indicate that the Ala mutation has a net destabilizing effect on the thermodynamics of the repetitive elongation reaction that underlies fibril stability. The data show that, as expected, reducing the size of most of the hydrophobic side chains of Aβ reduces the packing stabilization of the fibril, and this is true for both the A (Fig. 8b) and C (Fig. 8a) polymorphs. Since in many cases the lost packing is probably largely due to the role of the amino acid side chain in β-sheet structure, it is not surprising that the implicated packing interactions are dominantly located in the C-terminal 2/3 of the molecule that has been implicated to contain the bulk of the H-bonded structure (Table 3) 35; 48. Some residues implicated in packing interactions in this data are not necessarily involved in β-structure, however, and might be involved in other types of packing interactions, including, for example, side chain packing within and between monomer units in the protofibril, and protofilament-protofilament interactions within the fibril. As previously discussed 54, mutational effects on free energies of fibril elongation necessarily derive from effects on both monomer and fibril free energies. However, patterns in free energy effects of mutation are broadly consistent with other structural probes such as HX and ssNMR 54, suggesting that effects on monomer free energies generally do not dominate. This might especially be true in comparisons of two ΔΔG°elong values involving the same mutated monomer, such as shown in Figure 8c.
Figure 8.
Impact of Ala mutations on Aβ(1–40) amyloid fibril stability. Monomeric Aβ(1–40) Ala point mutants were subjected to spontaneous growth conditions and the Cr values assessed after ThT signal development reached a plateau. ΔG°elong values were calculated from the Cr values 24. Bar graphs represent pairwise comparisons, with positive ΔΔG values indicating lower stability. (a) ΔG°WT − ΔG°Ala for fibrils grown under C conditions; positive values indicate the Ala mutation decreases fibril stability. (b) ΔG°WT − ΔG°Ala for fibrils grown under A conditions; positive values indicate the Ala mutation decreases fibril stability. Data from reference 54. (c) ΔG °A − ΔG°C for each Ala point mutant. Positive values indicate that the Ala mutation is more destabilizing of A fibrils than of C fibrils, or, in other words, the Ala mutation is better accommodated into C fibrils than into A fibrils.
Ala mutations of Gly residues will also be expected to play roles in fibril stabilization, if they either allow or disfavor the formation of chain conformations required for propagation of fibril structure. Wild type Aβ (1–40) has Gly residues at positions 9, 25, 29, 33, 37 and 38. The data in Figure 8a show that Gly->Ala mutations are well tolerated by the type C fibril polymorph at positions 29, 33 and 37. In contrast, this replacement is modestly destabilizing of the monomer – fibril equilibrium at positions 25 and 38. Previously we showed that residue 25 is part of a sequence that in the type A polymorph is likely to be in a non-β segment with some significant geometric requirements 54, and this might explain the destabilization by a Gly-> Ala mutation in both the A and C polymorphs. The modestly destabilizing effect of Gly->Ala at position 38, which is unlikely to be in strongly H-bonded β-sheet (Table 3), is less clear. The most interesting Gly->Ala mutation is at position 9, where for the C fibril polymorph the mutation is unique among all the Ala mutations in being highly stabilizing (Fig. 8a). At a minimum, this result indicates that the φ,ψ angles of residue 9 in the fibril structure must be reasonably consistent with the allowed φ,ψ space for Ala. If such configurations are not highly populated within the monomer ensemble, restriction of the φ,ψ space in the Ala mutant would be stabilizing compared to the wild type sequence with Gly at position 9. In addition, the small Ala side chain may contribute to some packed structure within the fibril that is not possible when position 9 is occupied by Gly.
To more easily visualize the differences between these two fibril forming conditions, the ΔΔG° for the C polymorph (Fig. 8a) was subtracted from the ΔΔG° for the A polymorph (Fig. 8b) to generate a ΔΔG° directly relating the two polymorphs to each other (Fig. 8c). Thus, for example, the Gly->Ala mutation at position 9 is relatively destabilizing for the A polymorph vs. the C polymorph. The small values for many residues in Figure 8c suggests that the environments of many of the Aβ (1–40) amino acids may be very similar between these two polymorphic forms. Against this backdrop of very modest effects, several residues stand out for which Ala replacement is more stabilizing in the C polymorph than in the A polymorph. One of these is Phe19, and the other is the cluster of Asp7, Ser8 and Gly9. Although the physical explanation of these effects is not clear, it is interesting that our HX data implicates the presence of some strongly protected H-bonding - and hence a significant degree of order - in the Aβ N-terminal segment in the C polymorph (Table 3). It would therefore not be surprising if Ala replacement of small, hydrophilic amino acids in this region might have a small relatively positive effect on fibril stability. While much more work would be required to elucidate the structural differences between the A and C polymorphic fibrils, the Ala scanning results provide supporting evidence that there are discrete structural differences between these polymorphs that extend well beyond gross fibril morphology.
DISCUSSION
In recent years, attention in the Alzheimer’s disease field has shifted from a focus on mature amyloid fibrils and plaques to consideration of potential roles for non-fibrillar Aβ aggregation intermediates in disease mechanisms 55. Nonetheless, detailed pathological surveys distinguishing different types of Aβ deposits show a strong correlation of disease severity with details of advancing amyloid plaque pathology in AD brains 56. Furthermore, a recent report of a new familial AD mutation can be interpreted as being consistent with a dominant role for mature fibrils 57. Much remains to be learned about the relative importance of the various states of Aβ peptides in the molecular mechanism of AD. In this paper, we address structure-function relationships among mature Aβ(1–40) fibrils, focusing on the nature and consequences of a unique aspect of amyloid, its ability to exist in multiple polymorphic states.
In spite of their molecular evolution into sequences that tend to form condensed monomeric states, proteins - as polymeric molecules – also have an intrinsic tendency to form condensed polymeric states 3. Like other condensed polymers, these protein aggregates can exhibit polymorphism 3; 13. It has been shown that Aβ(1–40) can grow into a large number of amyloid fibrils with different gross morphologies in the EM 16; 17, and in principle it seems possible that fibril polymorphism at the EM level might derive from different quaternary arrangements of a single, common folding unit of the monomer 9. At the same time, the generic amyloid folding motif not only can tolerate single or multiple conservative mutations in the monomeric building block 58, but can also clearly accommodate within the folded polypeptide subunit much more substantial, mutationally forced structural differences of a kind that are normally thought to be essentially immutable in evolved globular protein structures, like H-bond arrangements, chain alignments, and sheet-sheet packing interactions 3. This suggests that fibril polymorphisms might sometimes derive from even substantially different folding patterns within the monomeric subunit. The experiments described here show that polymorphism in Aβ(1–40) fibrils extends to details of H-bonding, which can differ significantly between polymorphs (Table 3). The precise residue locations of each backbone amide H-bond, and how these locations differ between polymorphs, are yet to be worked out. Furthermore, the locations and roles of the individual H-bonds characteristic of these polymorphic structures – within monomers, between monomers within a single spine of a filament, or providing lateral stabilization between spines, filaments or fibrils – cannot be deduced with the HX technique. In addition to H-bonding, comparative alanine scanning analysis suggests that fibril polymorphs can differ significantly in how various residue side chains are packed within the folded structure of the fibril (Fig. 8).
We prepared Aβ(1–40) aggregates under eight different sets of conditions, and derived six different aggregated forms. Five of these structures are self-propagating amyloid fibrils, and the sixth is a non-fibrillar structure that nonetheless has many attributes of Aβ(1–40) protofibrils, including a relatively low content of strongly protected H-bonds, a low ThT response, and an inability to seed elongation from low concentrations of monomers 37; 59. Since we obtained five structurally distinct fibril types after testing only eight sets of growth conditions, it seems quite likely that additional polymorphic forms of Aβ(1–40). fibrils exist.
With two exceptions 5; 60, most models of Aβ(1–40) fibril structure include an N-terminus that is not involved in stable structure. Data reported here for all fibrils prepared with agitation show clearly that the first 3–4 residues of Aβ, at a minimum, are involved in stable structure – quite possibly in β-sheet. This result has several biological implications: (a) Recently, de Fede et al. described a novel familial Alzheimer’s disease mutation, Ala2Val 57. Patients homozygous for this mutation experience onset of AD at an early age, and homogeneous solutions of the mutated peptide aggregate in vitro more rapidly than the WT sequence. It is possible that the replacement of Ala with the β-branched amino acid Val might tip the energetic balance to favor amyloid β-sheet formation that now includes the N-terminus. Structural studies must be conducted to confirm this. (b) There is continued guarded optimism for immune therapy approaches to AD therapy 61. The immunodominant epitope of Aβ is at residues 3–7, and antibodies to this segment are able to bind to the Aβ monomer as well as various aggregated species, thus theoretically enabling all proposed mechanisms of immunoprotection against AD 47. However, if a major therapeutic mechanism is binding to Aβ plaques and recruitment of a cell-mediated immune response 61, then plaques containing fibrils in which most of the Aβ N-termini are involved in exchange-protected structure might be much less capable of binding antibodies against this major linear epitope. This would be especially significant in a passive immune therapy approach in which only specific monoclonal antibodies directed against the immuno-dominant 3–7 segment were administered 62.
The role of the C-terminus of Aβ(1–40) in fibril structure has received significant attention. We are confident of the low level of strong H-bonded structure in the Aβ C-termini of these polymorphs revealed in this study. First, NMR analysis of exchange into A fibrils with single residue sensitivity shows a similar lack of protection in these residues 63. Second, relative insensitivity of residues 36–40 to proline mutagenesis in A fibrils 3; 49; 52 supports the absence of structurally critical β-sheet in the C-terminus. Third, results of electron paramagnetic resonance spectroscopy on spin-labeled Aβ (1–40) fibrils are inconsistent with a highly structured C-terminus 48. Fourth, in agreement with HX-NMR analysis 5, we recently found highly protective structure in the C-terminus of amyloid fibrils formed from the 1–42 variant of Aβ using the same HX-MS methods as utilized in this paper (D. Kaleta, S. Chemuru, R. Kodali, K. D. Cook and R. Wetzel, Ms. in preparation); it therefore seems unlikely that the low protection we observe in Aβ(1–40) fibrils is an artifact of our methods. Fifth, relatively poor HX protection in the C-terminus of Aβ(1–40) fibrils has been independently reported by another group 60. There are at least two possible explanations for the differences in secondary structural models for Aβ(1–40) fibrils based on solid state NMR data 9; 18 and HX-MS data: (a) There may be real structural differences due to differences in fibril preparation protocols. (b) There may be relatively weak and/or transient H-bonds in the C-terminus of fibrillar Aβ(1–40) that are stable in lyophilized ssNMR samples but are easily broken in solution phase HX-MS experiments.
Since β-sheet is the dominant – in fact, defining – structural characteristic of amyloid fibrils, it would not be surprising to find that fibril stability is influenced by varying extents of β-sheet. In fact, we find an excellent correlation, among the polymorphic fibrils reported here, between elongation Cr values and the number of strongly protected backbone amide hydrogens (Fig. 7). This correlation must be due, not simply to H-bond content, but more broadly to H-bonds plus the well-known, attendant stabilizing components of β-sheet structure such as packing between strands and sheets 51; 64. The role of side chain packing in amyloid stabilization is clearly evident in results from unbiased mutational analysis of Aβ(1–42) fibril formation 58 and from alanine scanning of Aβ(1–40) fibril stability 54 (Fig. 8).
The existence of strain effects in both yeast 20; 21 and mammalian 65 prion phenomena is often interpreted as a reflection of the high fidelity of fibril elongation, even when the amyloid fibril templates are polymorphic assembled states of the same polypeptide sequence. Our data reported here illustrate the robustness of this effect within the universe of Aβ(1–40) fibril polymorphs. We show that, regardless of the growth conditions used to prepare the fibrils, when these fibrils are isolated and used to seed new growth at a common set of conditions, the fibril progeny in most cases reflect the structural properties of the seed, not the growth conditions. Where fibrils do not elongate with fidelity, it appears to be due to their poor stability, suggesting that fibril seeds that tend to completely disintegrate (as opposed to fracture) under the growth conditions may not produce structurally identical progeny.
Mutational analysis of protein folding reactions, including amyloid formation, generally suffer from the caveat that mutations can potentially affect the free energies of both the folded and unfolded states, so that mutation-related differences in ΔΔG° of folding reactions cannot be unequivocally attributed to differences between free energies of folded states. Interestingly, the energetic comparison of polymorphic condensed states from a single, common peptide sequence, at a common set of elongation conditions, does not suffer from this caveat. Since each elongation reaction leading to a characteristic Cr and ΔG° is conducted at exactly the same solution conditions, the monomer ensemble in each reaction must possess exactly the same free energy (Figure 9). Thus, the differences in elongation thermodynamics reported here (Table 2) can be attributed only to free energy differences between the different polymorphic fibril structures. Our results show that in an ideal case when sequence variation is not involved, the extent of β-sheet formation correlates very well with fibril stability (Fig. 7). The Weissman group showed that, in comparing two yeast prions, increased stability against shear-induced fragmentation correlated with a large increase in the extent of β-sheet in the fibril core 19. Our results extend these observations to a series of fibril polymorphs and to a more quantitative measure of fibril stability, and show a stepwise impact for each additional β-sheet H-bond. In spite of the above results, a dominant role for β-sheet in controlling fibril stability does not seem to be universal; previously we found that for some proline insertion mutants of Aβ(1–40), fibril products can be destabilized even while exhibiting increases in the number of protective H-bonds 52. These Pro mutant data are more difficult to interpret, however, due to possible mutational effects on the free energy of the monomer ensemble, as discussed above.
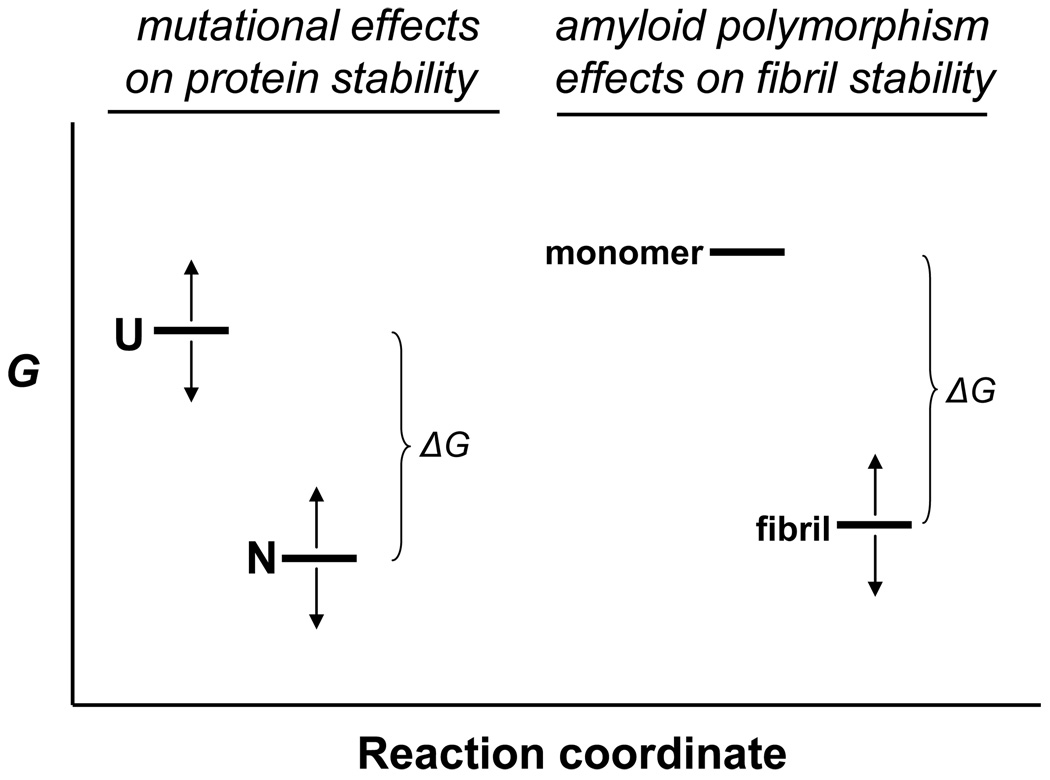
Figure 9.
Interpretation of free energy changes in protein transformations. In mutational analyses of protein folding reactions, mutations can modify the free energy (G) of both the unfolded state (U) and the folded, native state (N), as indicated by the arrows. This means that the physically measurable parameter, ΔG° - the free energy change on folding - cannot be rigorously ascribed to a change in the free energy of the folded state. In contrast, in the comparison of free energy changes of fibril elongation for different polymorphic fibrils from the same amino acid sequence, as determined in seeded elongation of these polymorphs under identical solution conditions, the difference in measured ΔG° values between polymorphic fibrils must be identical to the difference in free energies between the fibrils themselves, since the G of the monomer does not change under the standard elongation conditions used.
Our results on the molecular basis of amyloid polymorphism are consistent with the suggestion that alternative packing schemes for peptide segments, as visualized in microcrystals of amyloidogenic peptides, might serve as the basis for amyloid polymorphism 6. Interestingly, one polymorphic crystalline form of the peptide MVGGVV - derived from the Aβ C-terminus - does not exhibit packing interactions involving the terminal valines 6 in spite of the general strong tendency of β-branched amino acids like Val to exist in β-sheet structure 66. This is consistent with our finding of a lack of strong H-bonded β-sheet in the Aβ(1–40) C-terminus in these amyloid polymorphs (Table 3).
While the results presented here are confined to fibrils composed of the Aβ(1–40) sequence, the current view is that longer versions of Aβ, such as Aβ(1–42), are more important to AD disease pathology. It is not clear to what extent our results might be extended to the possibilities of polymorphism in Aβ(1–42). Two groups have reported slightly different HX protection patterns for Aβ(1–42) fibrils 5; 67, whose differences may well be due to polymorphisms due to the different growth conditions employed, or to the fact that in one case 5 the Aβ contained an oxidized Met 35. Interestingly, both structures found strong protection of the Aβ C-terminus, in contrast to the Aβ(1–40) structures described here but consistent with our own unpublished analysis of Aβ(1–42) fibrils (D. Kaleta, S. Chemuru, R. Kodali, K. D. Cook and R. Wetzel, Ms. in preparation). Collectively these results suggest that the highly related Aβ(1–40) and Aβ(1–42) peptides grow into fibrils exhibiting different fibril secondary structures, an expectation borne out by direct experiment 60. Further studies will be required to rigorously establish the existence of structural polymorphism among Aβ(1–42) amyloid fibrils.
It remains an open question as to whether Aβ pathogenicity in AD is attributable to properties of oligomeric and protofibrillar assembly intermediates 68, to the mature fibrils 69, or to both. The amyloid fibril form, in any case, appears to be the toxic entity in cases of cerebral microvascular amyloid deposition, which is associated with stroke and may also be a factor in AD 70. The possibility that fibril structural polymorphism may play a role in Aβpathology in the AD brain 23, with possible implications to disease heterogeneity, is a fascinating subject warranting experimental attention.
METHODS AND MATERIALS
Materials
Wild-type Aβ(1–40) was obtained via large-scale custom synthesis from the Keck Biotechnology Center at Yale University. ACS grade hexafluoroisopropanol, acetonitrile and formic acid were from Acros Organics, and trifluoroacetic acid was from Pierce. Procine pepsin was from Sigma-Aldrich.
Fibril growth
For most aggregation reactions, Aβ was disaggregated by our standard protocol 26. In one case (polymorph B), no prior disaggregation was carried out. All reaction buffers were at pH 7.4 and contained 0.05% sodium azide. Reactions were conducted in either PBS or 10 mM phosphate, pH 7.4, at either 4 °C, 24 °C, or 37 °C, with or without agitation by stirring. Stirring was done on a magnetic stir plate (Thermolyne) with stir setting at “6”, using a small “flea” stir bar, to generate a discernable vortex that nonetheless did not reach to the stir bar.
For the non-disaggregated reaction, peptide powder was added directly to 2mM NaOH, sonicated in a bath sonicator for 5 minutes, and buffer added to make a 250 µM solution of peptide in 10 mM phosphate, pH 7.4. For the SDS reaction, disaggregated Aβ was adjusted to 0.5 mM SDS in 500 mM NaCl in 10 mM phosphate, pH 7.4. For the zinc reaction, disaggregated peptide was dissolved in 20 mM Tris.HCl plus 150 mM NaCl at pH 7.4, its concentration determined 26, and an equimolar amount of ZnCl2 was added.
Aliquots were periodically removed from reactions and analyzed by the thioflavin (ThT) reaction 26; 71. After the ThT signal reached a plateau, reactions were incubated for one additional day, then analyzed for the molar concentration of monomeric Aβ left in the reaction as described previously 26. Knowing the amount of monomer at the beginning and end of the reaction it is possible to calculate the weight concentration of aggregates at the time when the final ThT determination is made. This allows the ThT to be normalized for the weight of fibrils in the cuvette. Values reported here as “weight normalized” refer to the ThT values associated with 1.25 µg fibrils in 400 µl PBS buffer containing 15 µM ThT in a 4 mm × 4 mm cuvette with excitation at 445 nm (5 nM slit width) and emission at 489 nm (10 nm slit width) using a Perkin Elmer LS50B spectrofluorimeter.
Electron Microscopy
Aliquots of reaction mixtures were taken at the end of reaction and directly (without further manipulations) analyzed by electron microscopy. A 5 µl aliquot of each reaction mixture was placed on freshly glow-discharged carbon-coated grids (Electron Microscopy Sciences, Hatfield), adsorbed for two mins, and the excess sample wicked away with filter paper. The sample grid was washed with deionized water, stained with 1% uranyl acetate for 15 secs, and blotted. Grids were imaged using Tecnai T12 microscope (FEI Co., Hillsboro, Oregon) operating at 120 kV and 30,000× magnification and equipped with an Ultrascan 1000 CCD camera (Gatan, Pleasanton, CA) with post-column magnification of 1.4 ×.
Fourier transform infrared spectroscopy
Aβ aggregates were harvested by centrifugation at 14,000 rpm on a bench top centrifuge for 30 mins and the pellet was washed 3 times with PBS to remove the traces of TFA and the other solutes. Pellets were re-suspended in PBS at around 10 mg/ml concentration and spectra were acquired by placing the aggregate suspension between two polished CaF2 windows using a BioCell module (BioTools, Inc) on an ABB Bomem FTIR instrument. A total of 400 scans were collected with 4 cm−1 resolution at RT and averaged for each sample. Spectra were corrected for residual buffer absorption by subtracting the buffer alone spectrum interactively until a flat baseline was obtained between 1700–1800 cm−1. Second-derivative spectra for the amide I region were calculated from the primary spectrum using PROTA software (BioTools, Inc.).
Seeding experiments
Fibrils were isolated from spontaneous growth reactions by centrifugation at 100,000 rpm for 30 mins, and washed with one or more cycles of centrifugation followed by resuspension of the pellet in the eventual elongation buffer (PBS or 10 mM sodium phosphate, pH 7.4). Fibrils were then sonicated (except for B fibrils) and the fibril weight concentrations determined by dissolving an aliquot of the fibril suspension in formic acid followed by the HPLC assay 26. Aβ(1–40) monomer was disaggregated, dissolved in PBS or 10 mM phosphate, and sonicated fibrils of the different polymorphic seeds added (7.5% w/w). Reactions were carried out at 37°C without agitation in either PBS or 10 mM sodium phosphate. Once the ThT curve reached a plateau, the reaction was judged to have reached equilibrium, and the residual monomer concentration, the characteristic Cr for these conditions, was determined. Fibrils were harvested for further studies.
From the Cr value, the equilibrium constant for the elongation reaction was calculated, and, from this, the associated free energy change 24; 26. All ΔG° values are based on the conventional, 1 M, standard states for non-gaseous solutes involved in equilibrium processes.
H/D exchange mass spectrometry
Global and segmental hydrogen-deuterium exchange (HX) was analyzed using an Agilent 1100 series single quadruple electrospray ionization mass spectrometer. Global analysis was as described previously45. Segmental analysis was by a modification (see below) of a previously described on-line pepsinolysis approach 42. Most data were corrected for artifactual exchange during sample workup with the aid of fully deuterated fibril control samples, as described 42; 44. While correction is necessary to obtain the best precision, it is not necessary for many comparisons of similar samples, and we therefore did not routinely carry out data correction. Fibril suspensions in D2O and the processing solvent (1% formic acid in 1:1 acetonitrile / H2O) were infused at 1 µl/min and 9 µl/min, respectively, into two ports of a T union (Valco instruments, Houston) and into the mass spectrometer, and measured in positive ion mode.
Previously our group described a modification of the above method which allowed determination of exchange properties of Aβ segments, by doing on-line proteolysis with pepsin in the fibril solubilization and analysis stream 42. In these previous studies, it was necessary to utilize a triaxial probe so that acetonitrile – which inhibits pepsin activity but aids MS detection of Aβ fragments - could be added late in the stream, after pepsin digestion in aqueous formic acid had occurred. More recently, we found that acetonitrile is not required for obtaining good peptide sensitivity in our Agilent MS instrument, and were thus able to use a simpler T-tube approach. Thus, fibrils in D-TRIS buffer (1µl/min) and pepsin (0.5 mg/ml in 0.5% formic acid; 9 µl/min) were infused into the T-union and directed into the mass spectrometer. All MS data from the pepsin fragmentation experiments were corrected for artifactual exchange 42; 44.
ACKNOWLEDGMENTS
Funding support was provided from NIH R01 AG018416 and Investigator Initiated Research Grant 08–91990 from the Alzheimer’s Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Martin JB. Molecular basis of the neurodegenerative disorders. N Engl J Med. 1999;340:1970–1980. doi: 10.1056/NEJM199906243402507. [published erratum appears in N Engl J Med 1999 Oct 28;341(18):1407] [DOI] [PubMed] [Google Scholar]
- 2.Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349:583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 3.Wetzel R, Shivaprasad S, Williams AD. Plasticity of amyloid fibrils. Biochemistry. 2007;46:1–10. doi: 10.1021/bi0620959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serpell LC, Fraser PE, Sunde M. X-ray fiber diffraction of amyloid fibrils. Meth Enzymol. 1999;309:526–536. doi: 10.1016/s0076-6879(99)09036-9. [DOI] [PubMed] [Google Scholar]
- 5.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, Schubert D, Riek R. 3D structure of Alzheimer's amyloid-beta(1–42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 7.van der Wel PC, Lewandowski JR, Griffin RG. Solid-state NMR study of amyloid nanocrystals and fibrils formed by the peptide GNNQQNY from yeast prion protein Sup35p. J Am Chem Soc. 2007;129:5117–5130. doi: 10.1021/ja068633m. [DOI] [PubMed] [Google Scholar]
- 8.Sachse C, Fandrich M, Grigorieff N. Paired beta-sheet structure of an Abeta(1–40) amyloid fibril revealed by electron microscopy. Proc Natl Acad Sci U S A. 2008;105:7462–7466. doi: 10.1073/pnas.0712290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paravastua AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimers beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 11.Caspar DL. Inconvenient facts about pathological amyloid fibrils. Proc Natl Acad Sci U S A. 2009;106:20555–20556. doi: 10.1073/pnas.0910978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC. The common architecture of cross-beta amyloid. J Mol Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 15.Lansbury PT, Jr, Caughey B. The chemistry of scrapie infection: implications of the 'ice 9' metaphor. Chem Biol. 1995;2:1–5. doi: 10.1016/1074-5521(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 16.Goldsbury CS, Wirtz S, Muller SA, Sunderji S, Wicki P, Aebi U, Frey P. Studies on the in vitro assembly of a beta 1–40: implications for the search for a beta fibril formation inhibitors. J Struct Biol. 2000;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 17.Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fandrich M. Abeta(1–40) fibril polymorphism implies diverse interaction patterns in amyloid fibrils. J Mol Biol. 2009;386:869–877. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 21.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 22.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 23.Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci U S A. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R. Thermodynamics of abeta(1–40) amyloid fibril elongation. Biochemistry. 2005;44:12709–12718. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- 25.Evans KC, Berger EP, Cho CG, Weisgraber KH, Lansbury PT., Jr Apolipoprotein E is a kinetic but not a thermodynamic inhibitor of amyloid formation: implications for the pathogenesis and treatment of Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92:763–767. doi: 10.1073/pnas.92.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Nuallain B, Thakur AK, Williams AD, Bhattacharyya AM, Chen S, Thiagarajan G, Wetzel R. Kinetics and thermodynamics of amyloid assembly using a high-performance liquid chromatography-based sedimentation assay. Methods Enzymol. 2006;413:34–74. doi: 10.1016/S0076-6879(06)13003-7. [DOI] [PubMed] [Google Scholar]
- 27.Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. On the nucleation and growth of amyloid beta-protein fibrils: detection of nuclei and quantitation of rate constants. Proc Natl Acad Sci U S A. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noy D, Solomonov I, Sinkevich O, Arad T, Kjaer K, Sagi I. Zinc-amyloid beta interactions on a millisecond time-scale stabilize non-fibrillar Alzheimer-related species. J Am Chem Soc. 2008;130:1376–1383. doi: 10.1021/ja076282l. [DOI] [PubMed] [Google Scholar]
- 29.Yang DS, Yip CM, Huang TH, Chakrabartty A, Fraser PE. Manipulating the amyloid-beta aggregation pathway with chemical chaperones. J Biol Chem. 1999;274:32970–32974. doi: 10.1074/jbc.274.46.32970. [DOI] [PubMed] [Google Scholar]
- 30.McLaurin J, Franklin T, Zhang X, Deng J, Fraser PE. Interactions of Alzheimer amyloid-beta peptides with glycosaminoglycans effects on fibril nucleation and growth. Eur J Biochem. 1999;266:1101–1110. doi: 10.1046/j.1432-1327.1999.00957.x. [DOI] [PubMed] [Google Scholar]
- 31.Nichols MR, Moss MA, Reed DK, Cratic-McDaniel S, Hoh JH, Rosenberry TL. Amyloid-beta protofibrils differ from amyloid-beta aggregates induced in dilute hexafluoroisopropanol in stability and morphology. J Biol Chem. 2005;280:2471–2480. doi: 10.1074/jbc.M410553200. [DOI] [PubMed] [Google Scholar]
- 32.Yagi H, Ban T, Morigaki K, Naiki H, Goto Y. Visualization and classification of amyloid beta supramolecular assemblies. Biochemistry. 2007;46:15009–15017. doi: 10.1021/bi701842n. [DOI] [PubMed] [Google Scholar]
- 33.Wood SJ, Maleeff B, Hart T, Wetzel R. Physical, morphological and functional differences between pH 5.8 and 7.4 aggregates of the Alzheimer's peptide Aβ. J. Mol. Biol. 1996;256:870–877. doi: 10.1006/jmbi.1996.0133. [DOI] [PubMed] [Google Scholar]
- 34.O'Nuallain B, Williams AD, Westermark P, Wetzel R. Seeding specificity in amyloid growth induced by heterologous fibrils. J Biol Chem. 2004;279:17490–17499. doi: 10.1074/jbc.M311300200. [DOI] [PubMed] [Google Scholar]
- 35.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer's beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shivaprasad S, Wetzel R. Scanning cysteine mutagenesis analysis of Abeta-(1–40) amyloid fibrils. J Biol Chem. 2006;281:993–1000. doi: 10.1074/jbc.M505091200. [DOI] [PubMed] [Google Scholar]
- 37.Williams AD, Sega M, Chen M, Kheterpal I, Geva M, Berthelier V, Kaleta DT, Cook KD, Wetzel R. Structural properties of Abeta protofibrils stabilized by a small molecule. Proc Natl Acad Sci U S A. 2005;102:7115–7120. doi: 10.1073/pnas.0408582102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson M, Mantsch HH. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit Rev Biochem Mol Biol. 1995;30:95–120. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- 40.Jones EM, Surewicz WK. Fibril conformation as the basis of species- and strain-dependent seeding specificity of Mammalian prion amyloids. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 41.Kheterpal I, Zhou S, Cook KD, Wetzel R. Abeta amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc Natl Acad Sci U S A. 2000;97:13597–13601. doi: 10.1073/pnas.250288897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kheterpal I, Chen M, Cook KD, Wetzel R. Structural differences in Aβ amyloid protofibrils and fibrils mapped by hydrogen exchange--mass spectrometry with on-line proteolytic fragmentation. J Mol Biol. 2006;361:785–795. doi: 10.1016/j.jmb.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 43.Kheterpal I, Wetzel R. Hydrogen/deuterium exchange mass spectrometry--a window into amyloid structure. Acc Chem Res. 2006;39:584–593. doi: 10.1021/ar050057w. [DOI] [PubMed] [Google Scholar]
- 44.Kheterpal I, Wetzel R, Cook K. Enhanced correction methods for hydrogen exchange-mass spectrometry studies of amyloid fibrils. Protein Sci. 2003;12:635–643. doi: 10.1110/ps.0225703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kheterpal I, Cook KD, Wetzel R. Hydrogen/deuterium exchange mass spectrometry analysis of protein aggregates. Methods Enzymol. 2006;413:140–166. doi: 10.1016/S0076-6879(06)13008-6. [DOI] [PubMed] [Google Scholar]
- 46.Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Structural features of the Aβ amyloid fibril elucidated by limited proteolysis. Biochem. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 47.Gardberg AS, Dice LT, Ou S, Rich RL, Helmbrecht E, Ko J, Wetzel R, Myszka DG, Patterson PH, Dealwis C. Molecular basis for passive immunotherapy of Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:15659–15664. doi: 10.1073/pnas.0705888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torok M, Milton S, Kayed R, Wu P, McIntire T, Glabe CG, Langen R. Structural and Dynamic Features of Alzheimer's Abeta Peptide in Amyloid Fibrils Studied by Site-directed Spin Labeling. J Biol Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 49.Wetzel R. Kinetics and thermodynamics of amyloid fibril assembly. Acc Chem Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi K, Takahashi S, Kawai T, Naiki H, Goto Y. Seeding-dependent propagation and maturation of amyloid fibril conformation. J Mol Biol. 2005;352:952–960. doi: 10.1016/j.jmb.2005.07.061. [DOI] [PubMed] [Google Scholar]
- 51.Cohen FE, Sternberg MJ, Taylor WR. Analysis of the tertiary structure of protein beta-sheet sandwiches. J Mol Biol. 1981;148:253–272. doi: 10.1016/0022-2836(81)90538-6. [DOI] [PubMed] [Google Scholar]
- 52.Williams AD, Portelius E, Kheterpal I, Guo JT, Cook KD, Xu Y, Wetzel R. Mapping Aβ amyloid fibril secondary structure using scanning proline mutagenesis. J Mol Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Shivaprasad S, Wetzel R. An intersheet packing interaction in Aβ fibrils mapped by disulfide cross-linking. Biochemistry. 2004;43:15310–15317. doi: 10.1021/bi048019s. [DOI] [PubMed] [Google Scholar]
- 54.Williams AD, Shivaprasad S, Wetzel R. Alanine scanning mutagenesis of Aβ(1–40) amyloid fibril stability. J Mol Biol. 2006;357:1283–1294. doi: 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 55.Irvine GB, El-Agnaf OM, Shankar GM, Walsh DM. Protein aggregation in the brain: the molecular basis for Alzheimer's and Parkinson's diseases. Mol Med. 2008;14:451–464. doi: 10.2119/2007-00100.Irvine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thal DR, Rub U, Schultz C, Sassin I, Ghebremedhin E, Del Tredici K, Braak E, Braak H. Sequence of Abeta-protein deposition in the human medial temporal lobe. J Neuropathol Exp Neurol. 2000;59:733–748. doi: 10.1093/jnen/59.8.733. [DOI] [PubMed] [Google Scholar]
- 57.Di Fede G, Catania M, Morbin M, Rossi G, Suardi S, Mazzoleni G, Merlin M, Giovagnoli AR, Prioni S, Erbetta A, Falcone C, Gobbi M, Colombo L, Bastone A, Beeg M, Manzoni C, Francescucci B, Spagnoli A, Cantu L, Del Favero E, Levy E, Salmona M, Tagliavini F. A Recessive Mutation in the APP Gene with Dominant-Negative Effect on Amyloidogenesis. Science. 2009;323:1473–1477. doi: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim W, Hecht MH. Generic hydrophobic residues are sufficient to promote aggregation of the Alzheimer's Abeta42 peptide. Proc Natl Acad Sci U S A. 2006;103:15824–15829. doi: 10.1073/pnas.0605629103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kheterpal I, Lashuel HA, Hartley DM, Walz T, Lansbury PT, Jr, Wetzel R. Aβ protofibrils possess a stable core structure resistant to hydrogen exchange. Biochemistry. 2003;42:14092–14098. doi: 10.1021/bi0357816. [DOI] [PubMed] [Google Scholar]
- 60.Olofsson A, Lindhagen-Persson M, Sauer-Eriksson AE, Ohman A. Amide solvent protection analysis demonstrates that amyloid-beta(1–40) and amyloid-beta(1–42) form different fibrillar structures under identical conditions. Biochem J. 2007;404:63–70. doi: 10.1042/BJ20061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brody DL, Holtzman DM. Active and passive immunotherapy for neurodegenerative disorders. Annu Rev Neurosci. 2008;31:175–193. doi: 10.1146/annurev.neuro.31.060407.125529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jicha GA. Is passive immunization for Alzheimer's disease 'alive and well' or 'dead and buried'? Expert Opinion on Biological Therapy. 2009;9:481–491. doi: 10.1517/14712590902828285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whittemore NA, Mishra R, Kheterpal I, Williams AD, Wetzel R, Serpersu EH. Hydrogen-deuterium (H/D) exchange mapping of Aβ1–40 amyloid fibril secondary structure using NMR spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 64.Merkel JS, Sturtevant JM, Regan L. Sidechain interactions in parallel beta sheets: the energetics of cross-strand pairings. Structure Fold Des. 1999;7:1333–1343. doi: 10.1016/s0969-2126(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 65.Colby DW, Giles K, Legname G, Wille H, Baskakov IV, Dearmond SJ, Prusiner SB. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci U S A. 2009;106:20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Proteins: Structures and Molecular Properties. New York: W. H. Freeman; 1984. p. 235. [Google Scholar]
- 67.Olofsson A, Sauer-Eriksson AE, Ohman A. The solvent protection of alzheimer amyloid-beta-(1–42) fibrils as determined by solution NMR spectroscopy. J Biol Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- 68.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 69.Wogulis M, Wright S, Cunningham D, Chilcote T, Powell K, Rydel RE. Nucleation-dependent polymerization is an essential component of amyloid-mediated neuronal cell death. J Neurosci. 2005;25:1071–1080. doi: 10.1523/JNEUROSCI.2381-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith EE, Greenberg SM. Beta-amyloid, blood vessels, and brain function. Stroke. 2009;40:2601–2606. doi: 10.1161/STROKEAHA.108.536839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.LeVine H. Quantification of β-sheet amyloid fibril structures with thioflavin T. Meth Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]