Abstract
Notch signaling is critical to animal development, and its dysregulation leads to human maladies ranging from birth defects to cancer. Although endocytosis is currently thought to promote signal activation by delivering activated Notch to endosome-localized γ secretase, the data are controversial and the mechanisms that control Notch endocytosis remain poorly defined. Here, we investigated the relationship between Notch internalization and signaling. siRNA-mediated depletion studies reveal that Notch endocytosis is clathrin-dependent and requires epsin1, AP2, and Nedd4. Moreover, we show that epsin1 interaction with Notch is ubiquitin-dependent. Contrary to the current model, we demonstrate that internalization defects lead to elevated γ secretase-mediated Notch processing and downstream signaling. These results indicate that signal activation occurs independently of Notch endocytosis and that γ secretase cleaves Notch at the plasma membrane. These observations support a model where endocytosis serves to down-regulate Notch in signal receiving cells.
Keywords: epsin1, AP2, endocytosis, Notch, Nedd4
Introduction
The highly conserved Notch signaling pathway performs critical roles in metazoan development by regulating cell proliferation, viability, cell-fate specification, and differentiation (1, 2). Additionally, the continued role of Notch in adults is exemplified by the fact that disrupting the signaling pathway can result in a variety of human malignancies ranging from leukemia to brain cancer (3–5). The canonical Notch signaling pathway is initiated when Notch binds one of several transmembrane ligands belonging to the Delta, Serrate, and Lag2 (DSL) family, which are expressed on neighboring cells (1).
Following ligand binding, a conformational change within the Notch extracellular domain is thought to occur (6, 7) that enables cleavage by either Adam17/TACE (8) or Adam10 (9). This releases the Notch ectodomain from the cell surface, which, in complex with its ligand, is subsequently internalized into the signaling cell (7, 10). The remaining, membrane-tethered, Notch extracellular truncation fragment (NEXT) undergoes cleavage by γ secretase, an intramembrane protease complex consisting of at lease four subunits: presenilin, nicastrin, APH-1, and PEN-2 (11–14). This cleavage event releases the Notch intracellular domain (NICD) from the membrane, which then translocates to the nucleus to regulate gene expression in concert with CSL and Mastermind (15).
Genetic studies in Drosophila implicate a role for endocytosis in the Notch signaling pathway, where internalization of Notch and its ligand are thought to be critical for productive signaling (10, 16–18). However, the role of Notch internalization remains controversial. Struhl and Adachi (2000) originally reported that presenilin-dependent Notch cleavage remains unaffected in flies expressing a temperature-sensitive mutant form of shibire, the gene encoding Dynamin (19). From this, the authors concluded that Notch cleavage by presenilin occurs independently of endocytosis. This conclusion is supported by two subsequent studies in C. elegans where internalization-defective LIN-12/Notch rescue the sterility and lethality of Lin-12/Notch null animals (20, 21).
By contrast, in mammalian cell culture systems, disrupting endocytosis by overexpression of dominant-negative dynamin was found to prevent γ secretase-dependent cleavage and nuclear targeting of Notch (22). This observation suggested that endocytosis promotes Notch signaling by delivering NEXT to endosome-localized γ secretase. In agreement, mosaic analysis in flies demonstrated that mutant dynamin overexpression (17) or complete loss of Dynamin or Clathrin (23) impairs Notch signaling in ovarian tissues.
Here, we re-evaluate the relationship between receptor internalization and Notch signaling. To do so, we developed a robust endocytosis assay to quantitatively measure the internalization of a NEXT mimic in mammalian cell culture. Using this assay, we find that Notch endocytosis occurs via a clathrin-dependent mechanism that requires the coordinated activities of two endocytic adaptors, AP2 and epsin1, as well as the E3 ubiquitin ligase Nedd4. We also provide evidence that γ secretase-mediated Notch cleavage and subsequent activation of downstream signaling is independent of receptor internalization. Taken together, our findings support the previously proposed model where receptor internalization serves to down-regulate the Notch signaling pathway (20).
Results
γ secretase-mediated Notch cleavage does not require endocytosis
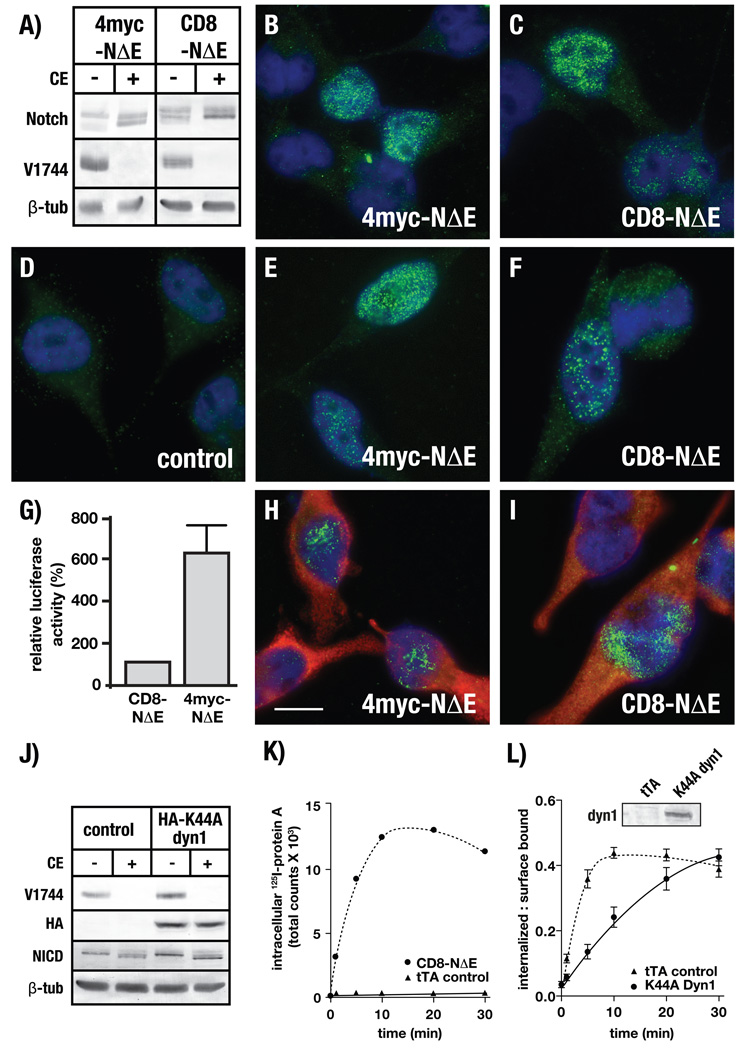
To directly test the idea that NEXT internalization is required for productive Notch signaling in mammalian systems, we first engineered a Notch chimera encoding four amino terminal copies of the myc epitope followed by 20 amino acids of the Notch extracellular domain through the carboxy terminal cytoplasmic tail (4myc-NΔE). To validate that 4myc-NΔE accurately reflects NEXT, we evaluated its γ secretase-dependent processing, targeting to the nucleus, and ability to promote gene expression. To resolve if 4myc-NΔE is an effective γ secretase substrate, we used antisera that specifically recognize γ secretase-derived NICD (V1744NICD, 24) to analyze protein lysates from 4myc-NΔE expressing cells. Immunoblot analysis reveals V1744NICD production following 4myc-NΔE overexpression (Figure 1A). The V1744NICD cleavage product is specific for γ secretase activity as its generation is inhibited by pretreating cells with a γ secretase inhibitor, Compound E (25). Consistent with V1744NICD targeting to the nucleus following γ secretase-mediated cleavage, immunolocalization analysis reveals V1744NICD localized to nuclei in 4myc-NΔE expressing cells (Figure 1B). Additionally, we find that 4myc-NΔE promotes expression of endogenous c-myc (Figure 1E), a known downstream Notch target (26). From these observations, we conclude that 4myc-NΔE faithfully mimics NEXT in its post-translational processing and ability to promote downstream changes in gene expression.
Figure 1.
γ secretase-dependent Notch cleavage does not require endocytosis. A) 4myc-NΔE or CD8-NΔE expressing cells were pretreated with either DMSO (control) or compound E (CE) and analyzed by immunoblot for V1744NICD production. 4myc-NΔE or CD8-NΔE expressing cells were evaluated by immunolocalization for nuclear recruitment of V1744NICD (B/C, green) or induction of endogenous c-myc gene expression using pAbs against endogenous c-myc (E/F, green), control represents tTA infected cells (D). 4myc-NΔE or CD8-NΔE expressing cells were analyzed using a RBP-Jk luciferase reporter assay (G) or infected with K44A dyn1 (red) encoded adenovirus and analyzed by for V1744NICD nuclear recruitment (H/I, green) or by immunoblot to evaluate V1744NICD production levels (J). Merged images include DAPI stain (blue) to mark nuclei. K) tTA HeLa cells were infected with adenovirus encoding tTA (control) or CD8-NΔE and incubated with the mAb 51.1 to evaluate endocytosis by measuring total internalized gamma counts (see Materials and Methods). L) CD8-NΔE expressing cells were infected with tTA control or K44A dyn1 encoded adenovirus and analyzed for CD8-NΔE uptake. Immunoblot inset shows K44A dyn1 expression levels. Error bars represent ± SEM of 5 independent experiments, bar = 10 µm.
We then evaluated the impact of disrupting endocytosis on 4myc-NΔE. To do so, we first overexpressed a mutant form of the dynamin 1 GTPase (K44A dyn1), which competes with endogenous endocytic machinery to disrupt internalization (27, 28). Based on published reports (22), we anticipated that disrupting endocytosis by overexpressing K44A dyn1 would prevent 4myc-NΔE processing by γ secretase. To the contrary, we found that K44A dyn1 overexpression, to levels that significantly impair transferrin uptake (Figure S1), did not prevent γ secretase-dependent 4myc-NΔE cleavage, as evidenced by V1744NICD production, recruitment to nuclei (Figure 1H), and c-myc induction in K44A dyn1 expressing cells (Figure S2). Collectively, these observations provide evidence that K44A dyn1 expression does not prevent γ secretase-mediated NEXT cleavage.
V1744NICD production following K44A dyn1 overexpression (Figures 1H) suggests that γ secretase-mediated NEXT cleavage occurs at the plasma membrane. If this is the case, the membrane-tethered Notch cytoplasmic tail would be released into the cytosol, thus elminating our ability to robustly track 4myc-NΔE endocytosis. To test this idea, we replaced the 4myc tag with the extracellular domain of CD8 to generate a larger Notch chimera (CD8-NΔE) since increases in the size of the extracellular domain were reported to reduce presenilin-dependent Notch cleavage efficiency in Drosophilia (19). Indeed, CD8-NΔE expressing cells generate less V1744NICD relative to that produced in 4myc-NΔE expressing cells (Figure 1A). Despite the reduced rate of V1744NICD production in CD8-NΔE expressing cells, V1744NICD was still detectable within nuclei and capable of promoting endogenous c-myc expression (Figures 1C and 1F). To quantitatively evaluate differences in Notch-dependent downstream signaling between 4myc-NΔE and CD8-NΔE, we employed an RBP-Jk luciferase reporter assay and found that 4myc-NΔE signaling is ~6 fold greater than that of CD8-NΔE (Figure 1G). However, similar to that observed for 4myc-NΔE, co-expressing CD8-NΔE with K44A dyn1 did not impact V1744NICD production or nuclear targeting (Figures 1I). Although differences in the level of nuclear-localized V1744NICD can be observed between individual cells by immunolocalization, immunoblot analysis, which evaluates the entire cell population, revealed that K44A dyn1 overexpression leads to somewhat increased V1744NICD levels relative to controls (Figure 1J). The observed increases are consistent with published reports indicating elevated Notch signaling following K44A dyn1 overexpression (29).
To evaluate CD8-NΔE endocytosis, we first infected tTA HeLa cells with adenovirus encoding CD8-NΔE to maximize the number of Notch chimera-expressing cells. Cells were then incubated with 51.1, a monoclonal antibody that specifically recognizes the extracellular CD8 tag (30), to measure single-round internalization kinetics (see Materials and Methods). In contrast to 4myc-NΔE, we were able to quantitatively measure CD8-NΔE internalization, which reaches a maximum uptake within 10 min (Figure 1K). After 10 min, we observed a consistent decrease, indicating CD8-NΔE recycles back to the plasma membrane following endocytosis. By comparison, control infected tTA HeLa cells show only background levels of 51.1 uptake when treated under identical conditions, demonstrating that the observed CD8-NΔE uptake is specific.
Our results and those of others (16, 19, 29) demonstrate that overexpression of dominant-negative dynamin forms did not prevent NEXT-mediated Notch signaling. These observations support the idea that NEXT endocytosis is not required for signaling by Notch. However, it remains possible, as previously suggested (17), that NEXT uptake occurs via an endocytic pathway that is unaffected by K44A dyn1 overexpression. To test this possibility, we measured the impact of K44A dyn1 overexpression on CD8-NΔE endocytosis. We found that the CD8-NΔE internalization rate is significantly reduced when K44A dyn1 is overexpressed (Figure 1L). We also found that CD8-NΔE continued to accumulate in cells overexpressing K44A dyn1 over time. This suggests that K44A dyn1 also disrupts CD8-NΔE recycling, consistent with the known impact of mutant dyn1 overexpression on receptor recycling from early endosomes (31, 32). Given that CD8-NΔE internalization is impaired following K44A dyn1 overexpression, yet V1744NICD is still produced and targeted to nuclei, these data provide evidence that γ secretase-dependent NEXT cleavage does not require endocytosis.
Clathrin drives CD8-NΔE internalization
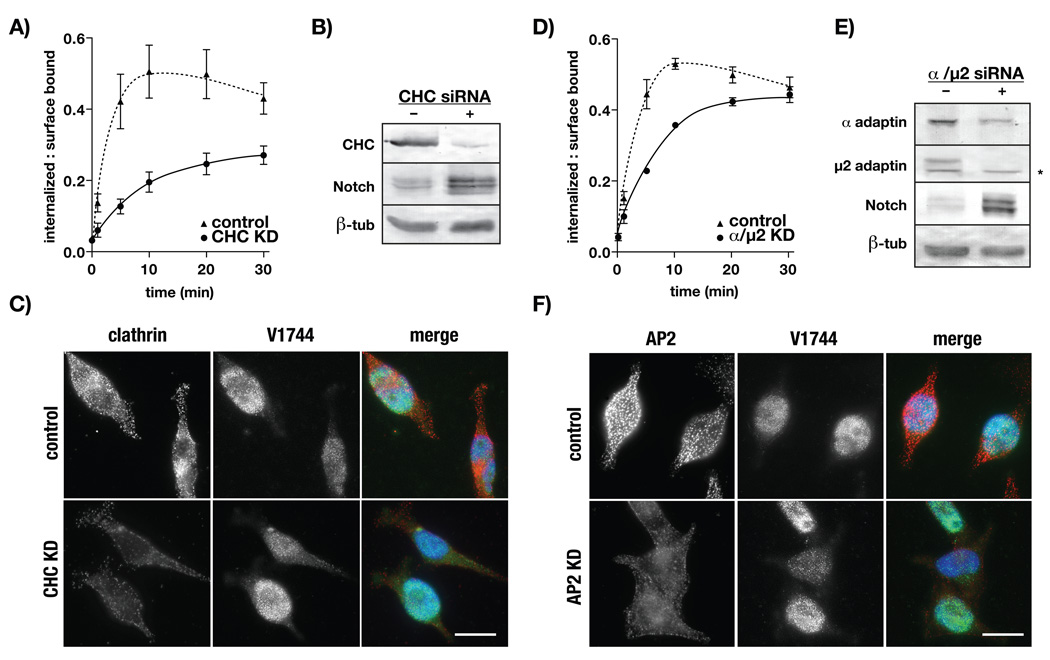
Results presented here and several published reports link Notch internalization to the clathrin-mediated endocytic pathway (33). However, the requirement for clathrin in Notch endocytosis has not been directly tested in mammalian systems. To define the mechanisms that govern Notch internalization, we tested if CD8-NΔE clearance from the plasma membrane is clathrin-dependent using an siRNA depletion strategy. Following clathrin heavy chain (CHC) depletion, we observed a marked reduction in CD8-NΔE internalization and an increase in CD8-NΔE stability relative to controls (Figures 2A and 2B). Similar to that observed following K44A dyn1 overexpression, CHC knock-down did not impact V1744NICD production or targeting to the nucleus (Figure 2C).
Figure 2.
CD8-NΔE internalization is clathrin-dependent. tTa HeLa cells were transfected with scrambled (control) siRNA or siRNAs directed against clathrin heavy chain (CHC, A–C) or the α and μ2 subunits of AP2 (D–F). Cells were then analyzed for CD8-NΔE internalization (A/D), by immunoblot to score protein expression (B/E), or by immunofluorescence (C/F), as described in the Materials and Methods. Errors bars represent the SEM of 5–6 independent experiments. The asterisk (2E) indicates a non-specific band. C/F) V1744NICD (green) and clathrin or AP2 (red) were detected with epitope-specific antisera and merged images include DAPI (blue) to mark nuclei, bar = 10 µm.
Epsin and AP2 promote Notch internalization
Defects in CD8-NΔE internalization following CHC knockdown indicates that Notch endocytosis occurs via a clathrin-dependent mechanism. However, clathrin coupling to endocytic cargo requires additional endocytic accessory factors (34, 35). One such factor is the heterotetrameric adaptor protein complex AP2 (36). We tested the possibility that AP2 promotes Notch uptake by evaluating CD8-NΔE endocytosis following adaptor complex depletion. To maximize AP2 expression knockdown, we pretreated CD8-NΔE-expressing cells with siRNAs that target the α and μ2 subunits (37). We found that the rate of CD8-NΔE endocytosis is impaired in AP2-depleted cells relative to controls (Figure 2D). Moreover, like that for CHC knockdown, we observed increased CD8-NΔE stability with no defect in V1744NICD nuclear localization (Figures 2E and 2F).
While these results indicate that AP2 performs an important role in Notch endocytosis, the CD8-NΔE internalization defect observed was less severe relative to clathrin loss (Figure 2A). This raised the possibility that our conditions were not sufficient to maximally disrupt AP2 activity. To test this, we measured 125I-labeled transferrin endocytosis using the same AP2-depleted cell population as that used for CD8-NΔE uptake (Figure 2D). In agreement with published reports (37), we found that transferrin uptake was potently impaired (Figure S3). This demonstrated that our experimental conditions sufficiently disrupted AP2 activity, and suggested that additional endocytic adaptors perform more critical roles in promoting Notch endocytosis.
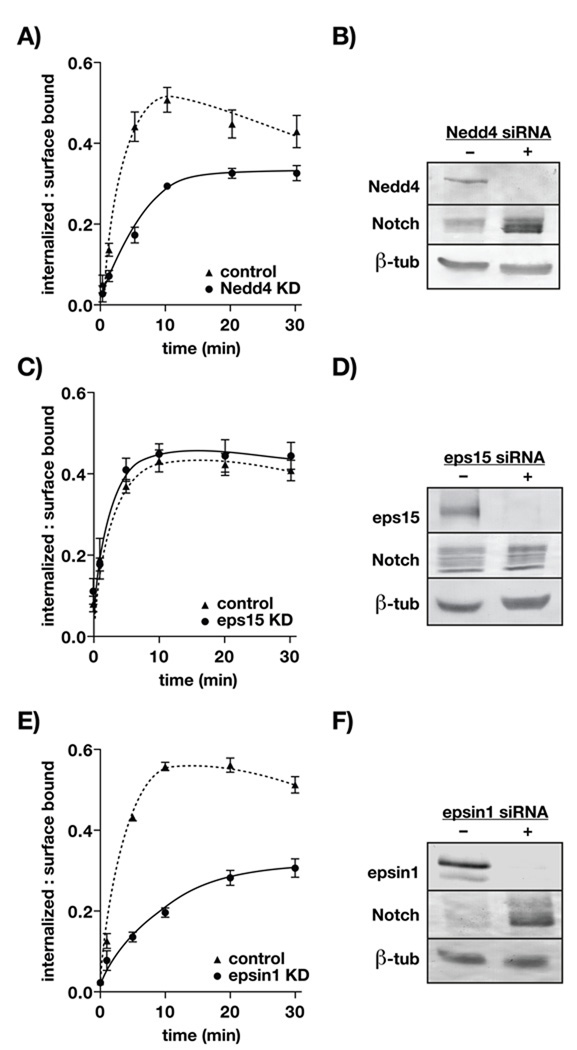
Studies in Drosophila reveal an important role for the E3 ubiquitin ligase, Nedd4, in Notch down-regulation by endocytosis (38). Although Nedd4 null mutations are embryonic lethal in flies, Sakata and colleagues found that mutant forms of Nedd4 disrupt ligand-independent Notch uptake when overexpressed and siRNA-mediated Nedd4 depletion in S2 cells increased Notch stability. Consistent with these results, we found that siRNA-mediated Nedd4 depletion in mammalian cells disrupts CD8-NΔE endocytosis and increases CD8-NΔE stability (Figures 3A and 3B). These observations reinforce the idea that ubiquitination is critical for Notch endocytosis. Moreover, these results implicate an essential role for endocytic adaptors that engage ubiquitinated cargo.
Figure 3.
Robust CD8-NΔE internalization requires Nedd4 and epsin1. A) tTa HeLa cells were transfected with control siRNA or siRNAs targeting Nedd4 (A/B), eps15 (C/D), or epsin1 (E/F). CD8-NΔE internalization was then scored and immunoblot analysis performed to validate protein expression and equal loading, as described in Figure 2. Errors bars represent ± SEM from 5–8 independent experiments.
The endocytic adaptor proteins Eps15 and epsin both encode functional ubiquitin interacting motifs (39, 40). Thus, we postulated that these adaptors might perform essential roles in Notch endocytosis. We tested this by measuring CD8-NΔE internalization following siRNA-mediated depletion of each adaptor. Despite significant expression knockdown, CD8-NΔE endocytosis was not impacted by a reduction in Eps15 (Figure 3C). By contrast, epsin1 depletion revealed a marked reduction in CD8-NΔE internalization and increased stability (Figures 3E and 3F). This suggests that epsin1 performs a key role in Notch internalization.
γ secretase-dependent NEXT cleavage occurs at the plasma membrane
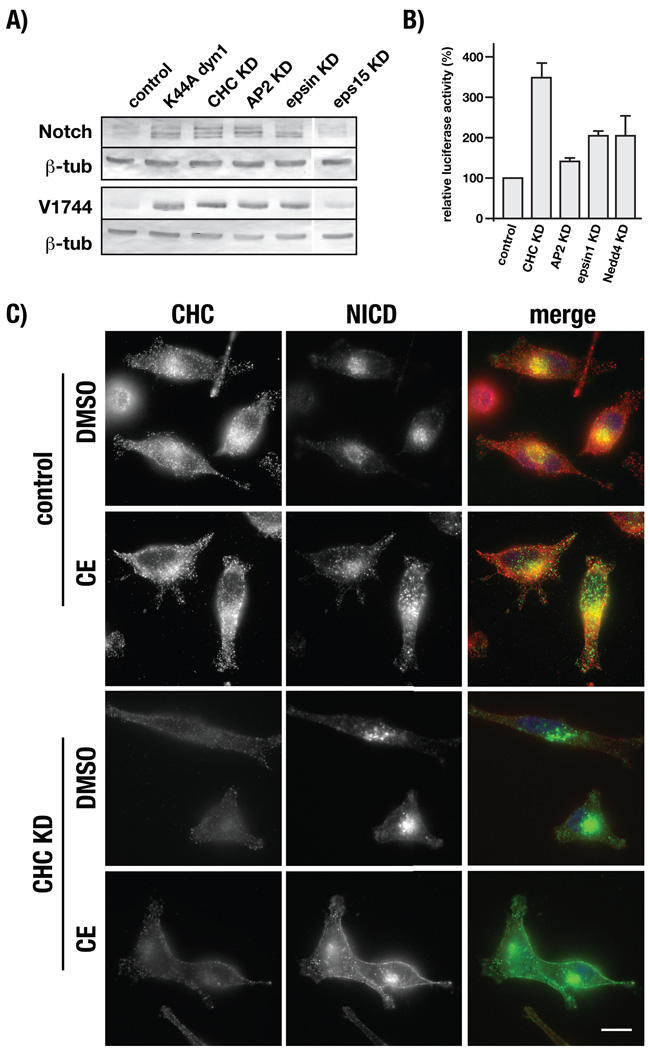
Our immunolocalization analyses indicated that V1744NICD production and nuclear targeting occurs independent of receptor endocytosis (Figures 1 and 2). However, the extent of nuclear-localized V1744NICD between cells within a given population was heterogeneous. Therefore, we used immunoblot analysis to evaluate the potential impact of impaired CD8-NΔE internalization on V1744NICD production. In each case where endocytosis is disrupted by siRNA-mediated depletion of CHC, AP2, or epsin1, we observed increased V1744NICD production relative to controls (Figure 4A). Conversely, no significant change in V1744NICD was observed in Eps15-depleted cells, which show control levels of CD8-NΔE endocytosis (Figure 3C). To ensure that increased V1744NICD production reflects increased downstream signaling, we used a RBP-Jk luciferase reporter assay to measure Notch signaling activity. Consist with our immunoblot analyses, we observed increases in luciferase activity relative to controls when CD8-NΔE internalization was disrupted by clathrin, AP2, epsin1 or Nedd4 depletion (Figure 4B). Together, these observations support the conclusion that elevated Notch signaling results from increased CD8-NΔE exposure to plasma membrane-localized γ secretase. We therefore predicted that Notch should accumulate at the plasma membrane when both γ secretase activity and CD8-NΔE internalization are disrupted. To test this idea, CD8-NΔE expressing cells were depleted of clathrin and treated with compound E. In these cells we observed a significant increase in plasma membrane-localized CD8-NΔE that was not observed in control cells (Figure 4C).
Figure 4.
γ-secretase-dependent CD8-NΔE cleavage occurs at the plasma membrane. A) tTa HeLa cells were transfected with control siRNA or siRNAs targeting clathrin heavy chain (CHC), AP2 (α and μ2 subunits), epsin1, or Eps15. Cells were then infected with CD8-NΔE encoded adenovirus and V1744NICD production was evaluated by immunoblot. B) Notch signaling was evaluated using a RBP-Jk luciferase reporter assay in control cells or those depleted of CHC, AP2, epsin1, or Nedd4. C) CHC-dependent cells expressing CD8-NΔE were pretreated with either DMSO or compound E (CE) and analyzed by immunolocalization for clathrin (red) or the cytoplasmic Notch tail (green). Merged images include DAPI stain (blue) to mark nuclei (C). Error bars represent ± SEM of 4 independent experiments, bar = 10 µm.
Consistent with published reports (19), our observations indicate that 4myc-NΔE cleavage by γ secretase is significantly more efficient than that of CD8-NΔE (Figure 1G). Moreover, we found that endocytosis defects did not impair γ secretase-dependent 4myc-NΔE or CD8-NΔE cleavage (Figures 1H and 1I). These observations likely explain our inability to consistently measure 4myc-NΔE endocytosis using our current protocol. For example, our current method requires two pre-binding steps at 4°C to allow antibody and 125I-protein A binding and to prevent their premature endocytosis into cells expressing 4myc-NΔE or CD8-NΔE. We postulate that γ secretase remains active during these pre-binding steps, it efficiently cleaves 4myc-NΔE, and eliminates our ability to quantitatively track 4myc-NΔE uptake. To test this idea, 4myc-NΔE expressing cells were incubated with pre-warmed media containing 9E10 for 10 min before fixation. Immunolocalization analysis reveals significant antibody binding and uptake after 10 minutes, demonstrating 4myc-NΔE is expressed at the cell surface and capable of internalization (Figure S4). From these observations, we suspect that γ secretase cleaves 4myc-NΔE during the 4°C pre-incubation step and prevents us from quantitatively measuring 4myc-NΔE uptake. This limitation is circumvented by using the CD8-NΔE chimera since its cleavage by γ secretase is less efficient (Figures 1A and 1G).
However, the possibility remained that 4myc-NΔE and CD8-NΔE might be transported differently within cells and thus account for the differences in their signaling capacity (Figure 1G). To address this possibility, we tested 4myc-NΔE signaling following clathrin depletion. Consistent with that observed for CD8-NΔE, we find that siRNA-mediated clathrin depletion leads to increased signaling by 4myc-NΔE relative to siRNA controls (Figure S4B). Pretreating cells with CE abolishes signaling indicating that the signaling is γ secretase-dependent. From this we conclude that 4myc-NΔE and CD8-NΔE employ the same internalization mechanism.
The increases in V1744NICD production and signaling when endocytosis is disrupted, combined with Notch accumulation at the plasma membrane following clathrin depletion and γ secretase inhibition (Figure 4), leads us to conclude that γ secretase is active at the plasma membrane.
Clathrin is essential at multiple Notch transport steps
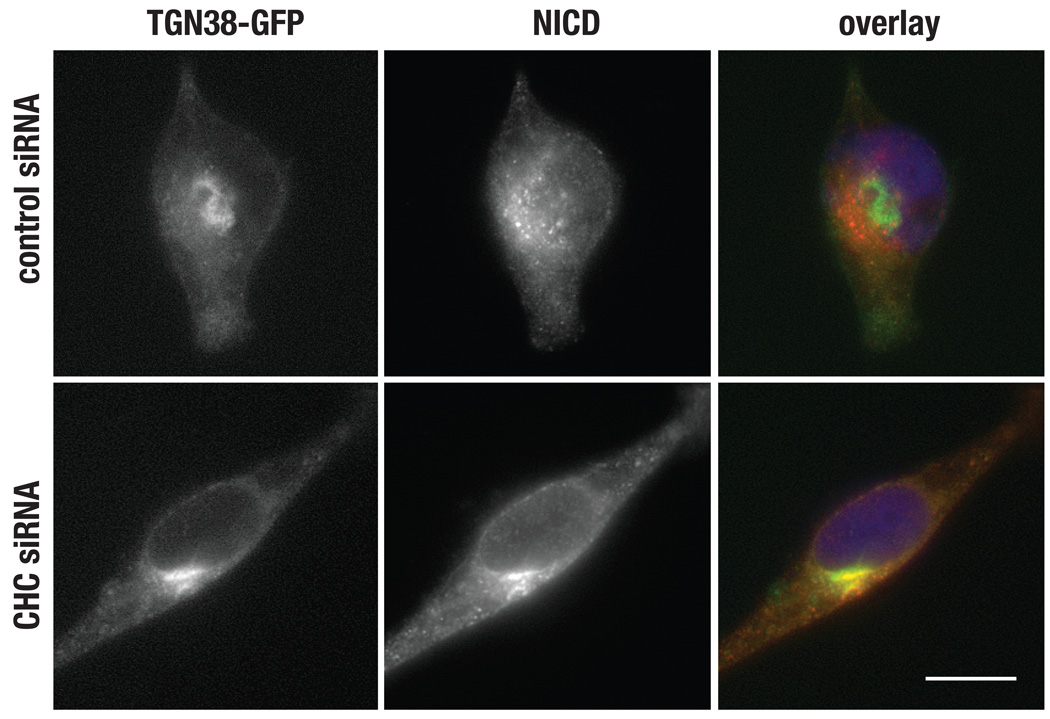
Immunolocalization analysis of CD8-NΔE following clathrin depletion revealed accumulation within a perinuclear compartment (Figure 4C). In addition endocytosis, clathrin also functions at the trans Golgi network (TGN) and endosome to facilitate receptor transport within the cell. The perinuclear accumulation of CD8-NΔE suggested the possibility that clathrin might also function in Notch transport from the TGN. To test this, we transfected CD8-NΔE expressing cells with a TGN marker, TGN38-GFP (41), and evaluated Notch localization. In control cells, we find little colocalization between CD8-NΔE and TGN38-GFP. By contrast, a marked increase in co-localization is observed when clathrin is depleted (Figure 5). Despite impaired transport from the TGN, CD8-NΔE is still observed in association with cytoplasmic vesicles when clathrin expression is reduced. We interpret this to mean that efficient Notch transport from the TGN requires clathrin. Athough, the TGN transport defect that results in clathrin-depleted cells can be overcome by protein overexpression.
Figure 5.
Clathrin functions in Notch transport from the TGN. tTA HeLa cells were treated with control or CHC-specific siRNA for 50 hr. Cells were then infected with adenovirus encoding CD8-NΔE and transfected with plasmid encoding TGN38-GFP. After an additional 16 hr incubation, cells were then fixed and processed for immunolocalization. NICD was detected using polyclonal antisera directed against the Notch cytoplasmic tail. Overlays indicate colocalization of NICD (red) with TGN38-GFP (green) in yellow. Blue marks nuclei., bar = 10 µm.
Ubiquitination enhances Notch-epsin1 interaction
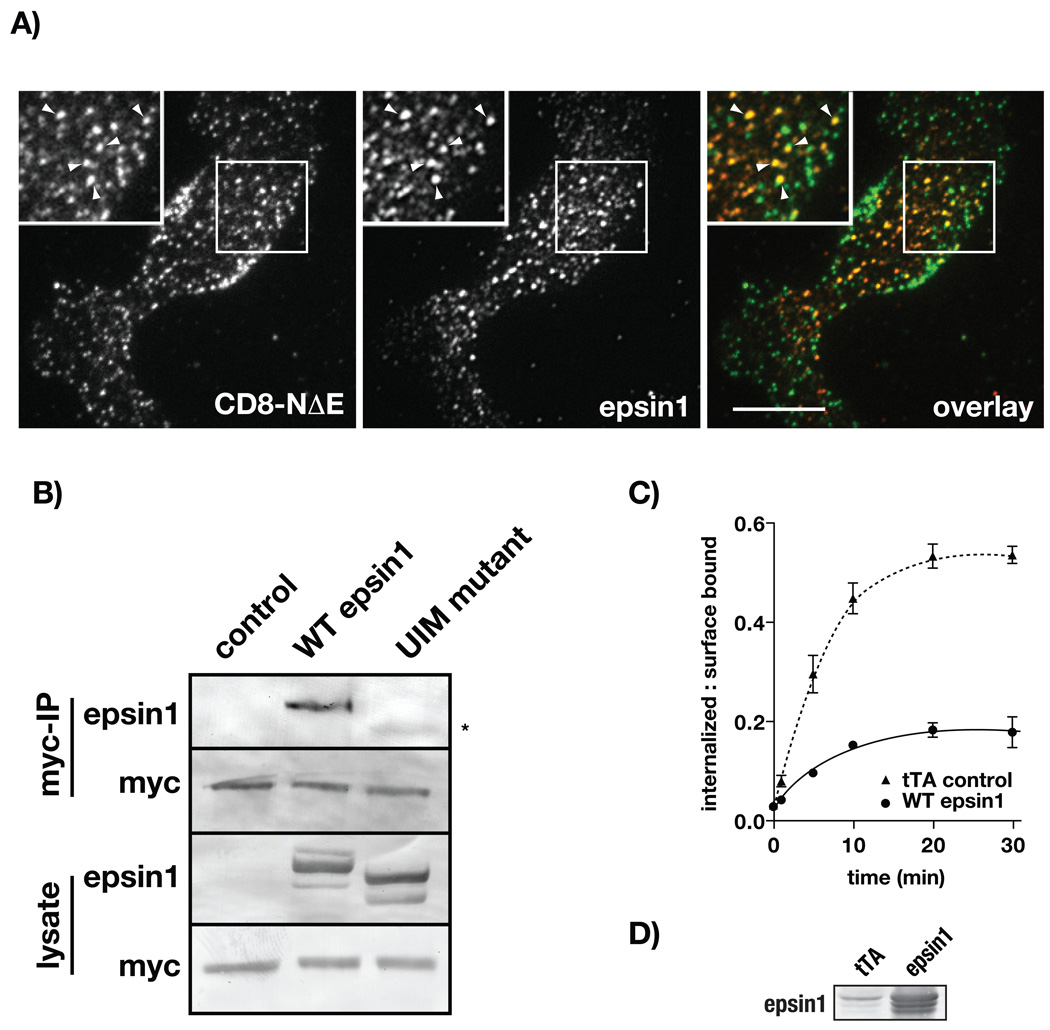
Internalization data presented here indicate an important role of Nedd4 and epsin1 in promoting CD8-NΔE endocytosis (Figure 3). Nedd4 is known to ubiquitinate Notch (38) and epsin1 is thought to couple receptors to the internalization machinery by engaging ubiquitinated cargo (34). Given this, we postulated that ubiquitination might regulate epsin1 activity in coordinating Notch uptake. To test this idea, we first performed immunolocalization analysis to ask if endogenous epsin1 colocalized with CD8-NΔE at the plasma membrane. Using total internal reflection fluorescence we observe significant colocalization between endogenous epsin1 and CD8-NΔE at the cell surface (Figure 6A). To extend our analysis, we next evaluated a potential interaction between epsin1 and Notch by co-immunoprecipitation. To do so, we co-expressed WT epsin1 with either CD8-NΔE or 4myc-NΔE in tTA HeLa cells. Notch chimeras were then immunoprecipitated from cell lysates and analyzed for WT epsin1. We found that WT epsin1 was detected by immunoblot from cells co-expressing recombinant WT epsin1 and either CD8-NΔE (not shown) or 4myc-NΔE (Figure 6B).
Figure 6.
Ubiquitination promotes epsin1 binding to Notch. A) CD8-NΔE expressing tTA HeLa cells were analyzed by TIRF microscopy using using antibodies against the extracellular CD8 epitope (green) and pAb against endogenous epsin (red). Boxed regions indicate areas of higher magnification with arrowheads marking colocalization examples. B) 4myc-NΔE expressing tTA HeLa cells were transfected with either WT epsin1 or UIM mutant epsin1 (Materials and Methods). The mAb 9E10 was then used to immunoprecipitate 4myc-NΔE from cell lysates, which were then analyzed for epsin1 binding by immunoblot. The asterisk indicates a minor interaction between 4myc-NΔE and UIM mutant epsin1. C) CD8-NΔE expressing cells were transfected with WT epsin1 or infected with tTA expressing adenovirus (control) and analyzed for CD8-NΔE internalization. Epsin1 expression levels were evaluated by immunoblot. Errors bars represent ± SEM of 3 independent experiments, bar = 10 µm.
We next tested if the epsin1 interaction with the Notch cytoplasmic tail was ubiquitin-dependent by coimmunoprecipitation analysis of 4myc-NΔE with an epsin1 ubiquitin-binding mutant, previously reported incapable of binding ubiquitinated proteins (42). Immunoblot analysis revealed significantly less mutant epsin1 coimmunoprecipiation with 4myc-NΔE than that observed for WT epsin1 (Figure 6B). This result suggests that epsin1 recruitment to the Notch cytoplasmic tail is enhanced by receptor ubiquitination. To evaluate epsin1 mechanism of action, we next wanted to test if the epsin1 ubiquitin-binding mutant could rescue the CD8-NΔE internalization defect that results from endogenous epsin1 depletion. However, this was not possible since WT epsin1 overexpression was found to impair CD8-NΔE internalization (Figure 6C). Similar to published reports, this dominant-negative effect likely reflects competition with other endocytic factors including clathrin and AP2, both of which are known to stably bind epsins (43). Consistently, we also found that overexpression of other endocytic factors like AAK1 (WT or kinase dead, 44) or the T102A Numb1 phosphomutant (45), both of which impair clathrin-mediated endocytosis by competing with core endocytic machinery, disrupt CD8-NΔE uptake (Figure S5). Taken together, these results support a conclusion that ubiquitination is critical to promoting epsin1-mediated Notch endocytosis.
Discussion
The observations we report here provide additional mechanistic insight into Notch internalization and signaling. Our results argue that clathrin-mediated receptor internalization downregulates Notch signaling. This interpretation reinforces the conclusions of previous published work (19–21) and is supported by three main results in this study. First, we demonstrate that when the CD8-NΔE internalization rate is impaired by siRNA-mediated depletion of multiple endocytic components (i.e. clathrin, AP2, or epsin1), Notch signaling continues. Likewise, K44A dyn1 overexpression disrupts endocytosis but did not reduce Notch signaling. Second, our results show that disrupting internalization, which prolongs CD8-NΔE transit time at the plasma membrane, leads to elevated 1744NICD production and increases in downstream Notch signaling. Third, Notch accumulates at the plasma membrane when receptor uptake and γ secretase activity are inhibited.
NEXT cleavage and signal activation occurs at the plasma membrane
Endocytosis is currently modeled as a requirement for NEXT cleavage by endosome-localized γ secretase (2, 46). In support of this model, mosaic analysis of the developing Drosophila ovary demonstrates that follicle cells, which overexpress a dominant-negative Dynamin or those that lack Clathrin or Dynamin, are unable to activate the Notch signaling pathway (17, 23). Likewise, K44A dyn1 overexpression in mammalian cells was reported to limit γ secretase cleavage of a truncated form of Notch that mimics NEXT (22).
Based on our observations, we favor the originally proposed model in which γ secretase-mediated NEXT cleavage occurs at the plasma membrane (19). Although processing within the endosome cannot be completely excluded. Our quantitative analyses reveal that Notch signaling is independent of endocytosis and instead proportional to the level of γ secretase activity at the plasma membrane. The model for plasma membrane-linked cleavage is supported by data from previous studies in both genetic and cell-based systems which show productive Notch signaling despite receptor internalization defects.
For example, acute disruption of endocytosis in Drosophila did not impact presenilin-dependent NEXT processing or subsequent downstream signaling when flies expressing a temperature-sensitive Shibire allele were shifted to a non-permissive temperature (19). In support of this observation, Notch was found to accumulate at apical membranes in cells expressing mutant forms of nicastrin or presenilin (47, 48). The idea that NEXT processing occurs at the plasma membrane is reinforced by results in C. elegans where mutations in the Lin12/Notch cytoplasmic tail, which disrupt receptor internalization, remain functional for signaling and rescue the lethality caused by loss of endogenous Lin-12/Notch (20, 21).
Consistently, results obtained in both flies and worms are supported by findings in mammalian cell culture where Notch signaling is unimpaired or even elevated in cells overexpressing dominant-negative K44A dyn1 (29, 49). Moreover, Tarassishin and colleagues reported that treating HEK293 cells with a membrane-impermeable γ secretase inhibitor, MRL631, significantly disrupted Notch signaling mediated by a NEXT mimic, while γ secretase-mediated cleavage of amyloid precursor protein within the endosome remained unperturbed (50).
Our observations support these findings and, collectively, they provide strong evidence for a model in which γ secretase initiates Notch signaling by cleaving NEXT at the plasma membrane.
Endosomal Notch activation
How do we reconcile our observations with those demonstrating a requirement for NEXT internalization in activating the Notch signaling pathway? In mammalian cells, Gupta-Rossi et al failed to detect nuclear accumulation of NICD by immunocytochemistry following overexpression of mutant forms of dynamin or Eps15 (22). However, in this study, changes in the kinetics of V1744NICD production were never specifically evaluated. Moreover, it is possible that endosomal transport of the truncated Notch form employed, which lacks nearly half the cytoplasmic tail, may not reflect the behavior of wild-type NEXT. By contrast, Vaccari and colleagues clearly demonstrate defective Notch signaling in Drosophila follicle cells overexpressing a mutant Shibire allele (17). Similarly, a more recent study demonstrates defects in Notch signaling in folicle cells lacking Clathrin or Dynamin (23). Given the well characterized requirement for clathrin and dynamin in endocytosis, the simple interpretation of these results is that Notch internalization is essential for signaling. However, it is possible that loss of dynamin and/or clathrin function, in addition to disrupting endocytosis, may also impact Notch transport throughout the cell. Indeed, we find that Notch accumulates at the TGN in clathrin-depleted cells. This indicates that clathrin functions at multiple Notch transport steps. Moreover, it suggests the possibility that failure to observe Notch signaling in fly ovarian cells, which completely lack Clathrin or Dynamin, might result from limited endogenous Notch delivery to the plasma membrane. Indeed, immunolocalization analysis reveals less Notch targeting to the plasma membrane in fly ovarian cells that lack either dynamin or clathrin relative to controls (23). By contrast, cells lacking AP2, the archetypal endocytic adaptor, reveal strong Notch accumulation at the plasma membrane, yet Notch signaling remains unaffected (23). This latter observation is consistent with our findings that efficient Notch endocytosis is AP2-dependent and that signaling continues when receptor uptake is impaired.
Clathrin-dependent endocytosis down-regulates Notch
Our findings and those of others support a model in which endocytosis serves to down-regulate, not activate, the Notch signaling pathway (20, 21, 29, 49, 50). We demonstrate that Notch internalization occurs via a clathrin-dependent mechanism. Consistent with this, we discovered that robust internalization requires AP2. However, the requirement for AP2 during Notch internalization appears to be less than that observed for clathrin. We suggest that AP2 performs an auxillary role in promoting Notch uptake and that receptor internalization is more heavily reliant on other endocytic factors. In contrast, we discovered that epsin1 depletion disrupts Notch endocytosis to a similar extent as that observed following clathrin loss. Given the recently described redundancy of epsin1 and epsin2 in mice (51), our observation suggests that the epsin2 isoform in HeLa cells is not sufficient to compensate for epsin1 loss. In general agreement, genome-wide expression analyses indicate that HeLa cells may express as much as 7-fold more epsin1 than epsin2 (52).
Although the molecular details by which epsin1 coordinates Notch internalization remain to be resolved, two key observations implicate receptor ubiquitination as a critical step in promoting Notch endocytosis. 1) Mammalian Nedd4 depletion impairs Notch endocytosis. This observation agrees well with similar results in Drosophila S2 cells where Notch becomes trapped at the plasma membrane following siRNA-mediated Nedd4 knockdown (38). 2) The epsin1 ubiquitin-binding mutant shows impaired association with Notch. Given that epsin1 is required for Notch endocytosis, these observations provide new information on how post-translational modification mediates the trafficking of Notch. Thus, we propose that Nedd4-mediated receptor ubiquitination promotes epsin1 interaction with Notch. Epsin1 could then deliver Notch to clathrin-coated pits via its interaction with other components of the endocytic machinery (i.e. AP2 and/or clathrin, 53) to promote receptor uptake into the cell. In future studies, it will be important to further test this mechanism and to elucidate the regulatory pathways that control Notch receptor ubiquitination.
Materials and Methods
Reagents
The mAb E7, 12CA5 and 9E10 were used to identify β-tubulin, HA- and myc-tagged protein, respectively. The mouse hybridoma 51.1 (HB-230) was obtained from the ATCC and rabbit antisera against CD8-α (H-160) was obtained from Santa Cruz Biotechnology. Mouse antisera against μ2 (611350) and mAb hudy1 (05–319) were obtained from BD Transduction Laboratories and Upstate, respectively. Polyclonal rabbit antisera against myc (600-401-381) was obtained from Rockland Immunochemicals. Antibodies against eps15 and epsin1 were a generous gift from Dr. Sandra Schmid (The Scripps Research Institute, CA), and the CD8-LDLR receptor construct, which was used as a backbone for generation of CD8 Notch fusion constructs was a generous gift of Dr. Margaret Robinson (University of Cambridge, UK). The mAbs TD.1 and AP.6 were used to detect clathrin and α-adaptin, respectively. Rabbit antisera against Notch (ab27526) was obtained from Abcam and was a generous gift from Drs. Neetu Gupta-Rossi and Alain Israël (Insitiut Pasteur, France). Nedd4 (2740) and Notch V1744 (2421 and 4147) antisera was purchased from Cell Signaling Technology. AAK1 antisera was previously described (44). 125I-Protein A and 125I-transferrin was purchased from Perkin Elmer (NEX146 and NEX212, respectively). γ secretase inhibitor XXI (Compound E) was purchased from Calbiochem (565790).
Constructs
Wildtype and T102A Numb1 adenovirus was used as previously described (45). WT and K74A AAK1 adenovirus (amino acids 1–863) were used as previously described (44). HA-K44A dynamin1 adenovirus was a gift from the Schmid lab. Rat WT epsin1 and UIM mutant [deletion of second UIM and two point mutations (D234A D235A) in the third UIM] in pcDNA-3myc were a generous gift from Dr. Pietro De Camilli (Yale University School of Medicine, CT). Epsin1 constructs were subcloned into the EcoRI and XhoI sites of pcDNA3.1(+). NΔE Notch (aa 1704–2532) in pCeMM-CTAP(SG)-6W was a generous gift from Dr. Alain Israel (Insitiut Pasteur, France). TGN38-GFP plasmid was a generous gift from Dr. George Banting (University of Bristol, UK).
4myc-NΔE and CD8-NΔE Chimera construction
Notch signal sequence with 4 myc tags was PCR amplified from NΔE Notch in pCeMM-CTAP(SG)-6W to incorporate a 5’ StuI site and a 3’ EcoRI site. CD8α signal sequence and extracellular domain was PCR amplified to incorporate a 5’ StuI site and a 3’ EcoRI site, preceeding the transmembrane domain of CD8. Both the 4myc and CD8α tags were then subcloned into pAdtet7. NΔE Notch (aa1704–2532) was amplified from NΔE Notch in pCeMM-CTAP(SG)-6W by PCR to incorporate EcoRI sites. NΔE Notch was ligated into the EcoRI site at either the 3’ end of the 4myc tags or extracellular domain coding sequence of CD8α chain in pAdtet7 creating a 4myc-NΔE or CD8-NΔE Notch Chimeras. Both chimeras in pADtet7 were then used for adenovirus production as previously described (54).
siRNA-mediated depletion
siRNA depletions were performed essentially as described (37). In short, two siRNA trasfections were performed, one on day 1 and another on day 2. After 50 hr incubation, cells were infected with adenovirus encoding CD8-NΔE and processed for endocytosis 10–12 hrs later. The extent of protein expression knockdown was evaluated by immunoblot using the appropriate antibody. For AP2, α siRNA target sequence was AAGAGCAUGUGCACGCUGGCCA and the μ2 target sequence was CAGCAGTCACCAAGCAGAATGTCAA. The clathrin heavy chain target sequence was UAAUCCAAUUCGAAGACCAAU. The epsin1 siRNA target sequence was GGAAGACGCCGGAGUCAUU. The Nedd4 siRNA target sequence was GUCCGUCGCTAAUUAUGCAUU. Eps15 siRNA target sequence was AAACGGAGCUACAGAUUAU. AP2 siRNAs, clathrin heavy chain, epsin1, and eps15 siRNAs were obtained from Invitrogen. Nedd4 and Silencer Negative control #1 siRNA were obtained from Ambion.
Coimmunoprecipitation
5×106 tTA HeLa cells were infected with 4myc-NΔE adenovirus alone or transfected with WT epsin1 or UIM mutant epsin1 in pCDNA3.1. Transfections were perfomed using Lipofectamine LTX (Invitrogen) according to manufacturer protocols. Cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 400 mM NaCl) supplemented with 1% NP40 and 1X protease inhibitor cocktail (Sigma-Aldrich). Cell lysates were diluted 1:1 in 50 mM Tris-HCl, pH 8, and then incubated for 1.5 h at RT with 9E10 mAb that had been prebound to protein G–sepharose (Calbiochem). Matrices were washed, and bead-bound proteins were then processed for immunoblot analysis.
Immunolocalization
tTA HeLa cells grown on coverslips and infected with adenovirus encoding CD8-NΔE or 4myc-NΔE. Cells were fixed with ice-cold acetone for 5 min and extracted with methanol. Cells were then washed with PBS containing 0.1% Triton-X 100, incubated with primary antibody for 1 hr at RT. Cells were washed and incubated for 1 hr at RT with the appropriate secondary antibody conjugated to either Alexa Fluor® 488 or Alexa Fluor® 555 (Invitrogen). Samples were then visualized by epifluorescence or TIRF microscopy using a Zeiss Axio Observer Z.1. Images were then imported, cropped, and assembled into panels using Photoshop CS4 and Illustrator CS4 (Adobe Systems, Inc.).
Endocytosis assays
tTA HeLA cells were infected with adenovirus encoding CD8-NΔE for 18 hr alone, or co-infected with adenoviruses encoding the indicated dominant-negative proteins. For siRNA depletion studies, cells were infected with CD8-NΔE adenovirus 60 hr after initial siRNA transfection. Cells were then transferred to 1.5 mL tubes, incubated at 4°C for 45 min in DMEM containing 10% fetal bovine serum supplemented with the CD8 mAb (51.1). Cells were then washed and incubated at 4°C for 45 min in DMEM containing 0.5% BSA and 125I-labeled protein A (1:200 diluted; Perkin Elmer). Tubes were moved in batch to a 37°C water bath to allow endocytosis for indicated time points and internalization was stopped by moving the cells back to 4°C. Surface-bound ligand was acid stripped (0.2 M Acetic Acid, 0.5 M NaCl), cells were pelleted, and internalized CD8-NΔE was measured by γ counting. Each time point is expressed as a percentage of total surface-bound γ counts. For internalization of 125I-transferrin, the experiment was performed 72 hr following knockdown. On the day of the experiment, cells were resuspended in PBS4+ (1× PBS, 1 mM MgCl2, 1 mM CaCl2, 5 mM glucose, 0.2% BSA) containing 0.5 µCi 125I-labeled transferrin for 45 min at 4°C. Unbound 125I-labeled transferrin was removed by washing and cells were gently pelleted by centrifugation. Cells were then resuspended in DMEM/10% FBS and transferrin internalization was evaluated as described above.
Notch signaling assay
Signaling was evaluated using a dual-luciferase RBP-Jκ reporter assay (SA Biosciences) and assayed according to manufacturers published protocols (Promega). In each case, RBP-Jκ-promoted firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency and minor differences in cell number. In all experiments, non-inducible background levels of firefly luciferase activity was less that 0.1%.
Supplementary Material
K44A dynamin 1 overexpression disrupts transferrin endocytosis. tTA HeLa cells were infected with adenovirus-encoding tTA or HA-tagged K44A dyn1. Cells were incubated with FITC-labeled transferrin (5 µg/mL, green) for 8 min at 37°C before fixation. K44A dyn1 expression was detected with the mAb hudy1 (red). Merged images include DAPI (blue) to mark cell nuclei, bar = 10 µm.
4myc-NΔE-induced expression of endogenous c-myc is not disrupted by dominant-negative dynamin overexpression. 4myc-NΔE expressing tTA HeLa cells were infected with adenovirus encoding tTA (control) or K44A dyn1. Endogenous c-myc expression (green, nuclear stain) was detected with FITC-tagged 9E10, which also recognizes endosome-localized 4myc-NΔE in the perinuclear compartment (arrowhead). Merged images include DAPI (blue) to mark cell nuclei, bar = 10 µm.
AP2 depletion potently disrupts transferrin uptake. To verify sufficient AP2 depletion conditions, control and AP2-depleted cell populations from Figure 2D were used to evaluate 125I-labeled transferrin internalization. Cells were incubated with 125I-labeled transferrin at 4°C. Unbound transferrin was removed and the rest of the experiment was performed as described for the CD8-NΔE internalization assay. Errors bars represent ± SEM from 5 independent experiments.
4myc-NΔE is expressed at the cell surface and can be internalized. A) 4myc-NΔE expressing tTA HeLa cells, grown on coverslips, were transferred to media containing 9E10 for 10 min at 37°C. Cells were then fixed and probed for 9E10 using a fluorochrome-tagged secondary to mark 4myc-NΔE (green) and for the Notch cytoplasmic tail (red). Merged image includes DAPI (blue) to mark cell nuclei. Nuclear localized Notch reflects V1744NICD produced by γ-secretase-mediated cleavage of 4myc-NΔE. B) tTA HeLa cells were treated with control siRNA or CHC siRNA. After 50 hr incubation, cells were then infected with adenovirus encoding 4myc-NΔE and incubated in the in the presence or absence of CE to inhibit γ-secretase activity. After an additional 16 hr incubation, Notch signaling was evaluated using a RBP-Jk luciferase reporter assay as described in the Materials and Methods. bar = 10 µm
Competition with core endocytic machinery impairs CD8-NΔE uptake. CD8-NΔE expressing cells were infected with adenovirus-encoding tTA (control), WT Numb1-myc or T102A Numb1-myc (A/B) or WT AAK1 or K74A AAK1 (C/D). CD8-NΔE endocytosis was evaluated as described (see Materials and Methods). Error bars represent ± SEM for 4 independent experiments. Protein overexpression levels were evaluated by immunoblot using epitope specific antisera.
Acknowledgements
We acknowledge members of the Conner laboratory for helpful discussion and technical support. In particular, we acknowledge Cosmo Saunders for help developing and characterizing Notch constructs. We also thank Drs. Gary Struhl, Thomas Hays, David Greenstein, Neetu Gupta-Rossi and Alain Israël for helpful discussions and critical feedback. This work was supported in part by a National Institute of Health Grant (GM085029) to SDC and a Developmental Biology Training grant (2T32-HD007480-11A1) to EBS.
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tien AC, Rajan A, Bellen HJ. A Notch updated. J Cell Biol. 2009;184(5):621–629. doi: 10.1083/jcb.200811141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lasky JL, Wu H. Notch signaling, brain development, and human disease. Pediatr Res. 2005;5 Pt 2(57):104R–109R. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- 4.Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer Biol Ther. 2002;5(1):466–476. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- 5.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 6.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 7.Nichols JT, Miyamoto A, Olsen SL, D’Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 9.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. The metalloprotease ADAM10 is required for notch1 S2 cleavage. J Biol Chem. 2009 doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 11.Steiner H, Fluhrer R, Haass C. Intramembrane proteolysis by gamma-secretase. J Biol Chem. 2008;283:29627–29631. doi: 10.1074/jbc.R800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci U S A. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 14.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 15.Kitagawa M, Oyama T, Kawashima T, Yedvobnick B, Kumar A, Matsuno K, Harigaya K. A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol Cell Biol. 2001;21:4337–4346. doi: 10.1128/MCB.21.13.4337-4346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seugnet L, Simpson P, Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev Biol. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- 17.Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol. 2008;180:755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- 19.Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 20.Shaye DD, Greenwald I. Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature. 2002;420:686–690. doi: 10.1038/nature01234. [DOI] [PubMed] [Google Scholar]
- 21.Shaye DD, Greenwald I. LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development. 2005;132:5081–5092. doi: 10.1242/dev.02076. [DOI] [PubMed] [Google Scholar]
- 22.Gupta-Rossi N, Six E, LeBail O, Logeat F, Chastagner P, Olry A, Israel A, Brou C. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J Cell Biol. 2004;166:73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Windler SL, Bilder D. Endocytic internalization routes required for delta/notch signaling. Curr Biol. 2010;20:538–543. doi: 10.1016/j.cub.2010.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 25.Kornilova AY, Das C, Wolfe MS. Differential effects of inhibitors on the gamma-secretase complex. Mechanistic implications. J Biol Chem. 2003;278:16470–16473. doi: 10.1074/jbc.C300019200. [DOI] [PubMed] [Google Scholar]
- 26.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tagami S, Okochi M, Yanagida K, et al. Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol Cell Biol. 2008;28:165–176. doi: 10.1128/MCB.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin PJ, Ledbetter JA, Clark EA, Beatty PG, Hansen JA. Epitope mapping of the human surface suppressor/cytotoxic T cell molecule Tp32. J Immunol. 1984;132:759–765. [PubMed] [Google Scholar]
- 31.van Dam EM, Ten Broeke T, Jansen K, Spijkers P, Stoorvogel W. Endocytosed transferrin receptors recycle via distinct dynamin and phosphatidylinositol 3-kinase-dependent pathways. J Biol Chem. 2002;277:48876–48883. doi: 10.1074/jbc.M206271200. [DOI] [PubMed] [Google Scholar]
- 32.van Dam EM, Stoorvogel W. Dynamin-dependent transferrin receptor recycling by endosome-derived clathrin-coated vesicles. Mol Biol Cell. 2002;13:169–182. doi: 10.1091/mbc.01-07-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;8(132):1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- 34.Maldonado-Baez L, Wendland B. Endocytic adaptors: recruiters, coordinators and regulators. Trends Cell Biol. 2006;16(10):505–513. doi: 10.1016/j.tcb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- 36.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 37.Motley A, Bright NA, Seaman MN, Robinson MS. Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 39.Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- 40.Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 41.Girotti M, Banting G. TGN38-green fluorescent protein hybrid proteins expressed in stably transfected eukaryotic cells provide a tool for the real-time, in vivo study of membrane traffic pathways and suggest a possible role for ratTGN38. J Cell Sci. 1996;109:2915–2926. doi: 10.1242/jcs.109.12.2915. [DOI] [PubMed] [Google Scholar]
- 42.Kazazic M, Bertelsen V, Pedersen KW, Vuong TT, Grandal MV, Rodland MS, Traub LM, Stang E, Madshus IH. Epsin 1 is involved in recruitment of ubiquitinated EGF receptors into clathrin-coated pits. Traffic. 2009;10:235–245. doi: 10.1111/j.1600-0854.2008.00858.x. [DOI] [PubMed] [Google Scholar]
- 43.Rosenthal JA, Chen H, Slepnev VI, Pellegrini L, Salcini AE, Di Fiore PP, De Camilli P. The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J Biol Chem. 1999;274:33959–33965. doi: 10.1074/jbc.274.48.33959. [DOI] [PubMed] [Google Scholar]
- 44.Conner SD, Schmid SL. Differential requirements for AP-2 in clathrin-mediated endocytosis. J Cell Biol. 2003;162:773–780. doi: 10.1083/jcb.200304069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorensen EB, Conner SD. AAK1 regulates Numb function at an early step in clathrin-mediated endocytosis. Traffic. 2008;10:1791–1800. doi: 10.1111/j.1600-0854.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 46.Fortini ME, Bilder D. Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 2009 doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Schier H, St Johnston D. Drosophila nicastrin is essential for the intramembranous cleavage of notch. Dev Cell. 2002;2:79–89. doi: 10.1016/s1534-5807(01)00109-5. [DOI] [PubMed] [Google Scholar]
- 48.Guo Y, Livne-Bar I, Zhou L, Boulianne GL. Drosophila presenilin is required for neuronal differentiation and affects notch subcellular localization and signaling. J Neurosci. 1999;19:8435–8442. doi: 10.1523/JNEUROSCI.19-19-08435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaether C, Schmitt S, Willem M, Haass C. Amyloid precursor protein and Notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic. 2006;7:408–415. doi: 10.1111/j.1600-0854.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- 50.Tarassishin L, Yin YI, Bassit B, Li YM. Processing of Notch and amyloid precursor protein by gamma-secretase is spatially distinct. Proc Natl Acad Sci U S A. 2004;101:17050–17055. doi: 10.1073/pnas.0408007101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, Ko G, Zatti A, Di Giacomo G, Liu L, Raiteri E, Perucco E, Collesi C, Min W, Zeiss C, De Camilli P, Cremona O. Embryonic arrest at midgestation and disruption of Notch signaling produced by the absence of both epsin 1 and epsin 2 in mice. Proc Natl Acad Sci U S A. 2009;106:13838–13843. doi: 10.1073/pnas.0907008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 54.Damke H, Gossen M, Freundlieb S, Bujard H, Schmid SL. Tightly regulated and inducible expression of dominant interfering dynamin mutant in stably transformed HeLa cells. Methods Enzymol. 1995;257:209–220. doi: 10.1016/s0076-6879(95)57026-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
K44A dynamin 1 overexpression disrupts transferrin endocytosis. tTA HeLa cells were infected with adenovirus-encoding tTA or HA-tagged K44A dyn1. Cells were incubated with FITC-labeled transferrin (5 µg/mL, green) for 8 min at 37°C before fixation. K44A dyn1 expression was detected with the mAb hudy1 (red). Merged images include DAPI (blue) to mark cell nuclei, bar = 10 µm.
4myc-NΔE-induced expression of endogenous c-myc is not disrupted by dominant-negative dynamin overexpression. 4myc-NΔE expressing tTA HeLa cells were infected with adenovirus encoding tTA (control) or K44A dyn1. Endogenous c-myc expression (green, nuclear stain) was detected with FITC-tagged 9E10, which also recognizes endosome-localized 4myc-NΔE in the perinuclear compartment (arrowhead). Merged images include DAPI (blue) to mark cell nuclei, bar = 10 µm.
AP2 depletion potently disrupts transferrin uptake. To verify sufficient AP2 depletion conditions, control and AP2-depleted cell populations from Figure 2D were used to evaluate 125I-labeled transferrin internalization. Cells were incubated with 125I-labeled transferrin at 4°C. Unbound transferrin was removed and the rest of the experiment was performed as described for the CD8-NΔE internalization assay. Errors bars represent ± SEM from 5 independent experiments.
4myc-NΔE is expressed at the cell surface and can be internalized. A) 4myc-NΔE expressing tTA HeLa cells, grown on coverslips, were transferred to media containing 9E10 for 10 min at 37°C. Cells were then fixed and probed for 9E10 using a fluorochrome-tagged secondary to mark 4myc-NΔE (green) and for the Notch cytoplasmic tail (red). Merged image includes DAPI (blue) to mark cell nuclei. Nuclear localized Notch reflects V1744NICD produced by γ-secretase-mediated cleavage of 4myc-NΔE. B) tTA HeLa cells were treated with control siRNA or CHC siRNA. After 50 hr incubation, cells were then infected with adenovirus encoding 4myc-NΔE and incubated in the in the presence or absence of CE to inhibit γ-secretase activity. After an additional 16 hr incubation, Notch signaling was evaluated using a RBP-Jk luciferase reporter assay as described in the Materials and Methods. bar = 10 µm
Competition with core endocytic machinery impairs CD8-NΔE uptake. CD8-NΔE expressing cells were infected with adenovirus-encoding tTA (control), WT Numb1-myc or T102A Numb1-myc (A/B) or WT AAK1 or K74A AAK1 (C/D). CD8-NΔE endocytosis was evaluated as described (see Materials and Methods). Error bars represent ± SEM for 4 independent experiments. Protein overexpression levels were evaluated by immunoblot using epitope specific antisera.