Abstract
HIV-1 Tat protein is an important pathogenic factor in HIV-1-associated neurological diseases. One hallmark of HIV-1 infection of the central nervous system (CNS) is astrocytosis, which is characterized by elevated GFAP expression in astrocytes. We have shown that Tat activates GFAP expression in astrocytes (Zhou, et al., Mol. Cell. Neurosci. 27:296, 2004) and that GFAP is an important regulator of Tat neurotoxicity (Zou, et. al., Am. J. Pathol. 171:1293, 2007). However, the underlying mechanisms for Tat-mediated GFAP up-regulation are not understood. In the current study, we reported concurrent up-regulation of adenovirus E1a-associated 300 kDa protein p300 and GFAP in Tat-expressing human astroytoma cells and primary astrocytes. We showed that p300 was indeed induced by Tat expression and HIV-1 infection and that the induction occurred at the transcriptional level through the cis-acting elements of early growth response 1 (Egr-1) within its promoter. Using siRNA, we further showed that p300 regulated both constitutive and Tat-mediated GFAP expression. Moreover, we showed that ectopic expression of p300 potentiated Tat transactivation activity and increased proliferation of HIV-1-infected astrocytes, but had little effect on HIV-1 replication in these cells. Taken together, these results demonstrate for the first time that Tat is a positive regulator of p300 expression, which in turn regulates GFAP expression, and suggest that the Tat-Egr-1-p300-GFAP axis likely contributes to Tat neurotoxicity and predisposes astrocytes to be an HIV-1 sanctuary in the CNS.
Keywords: HIV-1 Tat, GFAP, astrocytes, p300, Egr-1, transcription activation
INTRODUCTION
HIV-1 infection of the CNS is associated with neurological disorders ranging from mild cognitive disorder to HIV-1-associated dementia The paradox between HIV-1-infected target cells, i.e., microglia/macrophages and astrocytes and HIV-1-affected cells, i.e., neurons in the CNS has led to a number of indirect mechanisms for HIV-1 neuropathogenesis. HIV-1 Tat protein is present in the CNS, it can be directly from HIV-1-infected cells in the CNS, or derived from HIV-1-infected cells outside the CNS (Banks et al. 2005; Chang et al. 1997; Chen et al. 1995). Tat can be taken up by neighboring neurons and other brain cells (Frankel and Pabo 1988; Liu et al. 2000). Thus, Tat has been proposed to be one of the most important viral elements responsible for HIV-1 neuropathogenesis. A growing body of evidence has accumulated to support this hypothesis. Firstly, direct exposure to Tat is neurotoxic, so-called acute Tat neurotoxicity, through alterations of neuronal integrity, homeostasis and survival, neuroexcitatory property, endoplasmic reticulum calcium load, and oxidative state (Aprea et al. 2006; Brailoiu et al. 2006; Caporello et al. 2006; Norman et al. 2007). Secondly, Tat possesses a chemokine-like activity; its presence results in infiltration of monocytes/macrophages and lymphocytes into the CNS, which in turn causes neurotoxicity (Benelli et al. 2000; de Paulis et al. 2000; Jones et al. 1998; Park et al. 2001). Thirdly, Tat alters neuronal gene expression through directly regulating gene expression mechanisms or indirectly eliciting intracellular signaling cascades (Liu et al. 2000; Peruzzi 2006). In addition to those direct neurotoxic effects, Tat can also affect neuronal function and survival indirectly by interacting with other brain cells such as astrocytes and brain endothelial cells (Andras et al. 2008; Chauhan et al. 2003; Kim et al. 2003b; Price et al. 2006; Toborek et al. 2003; Zhong et al. 2009; Zhou and He 2004; Zhou et al. 2004). In agreement with these findings, Tat expression in or injection into the CNS in the absence of HIV-1 infection is sufficient to cause neuropathologies similar to most of those noted in the brain of AIDS patients (Jones et al. 1998; Kim et al. 2003a). It is evident from all these studies that Tat is a major contributing factor to HIV-1 neuropathogenesis.
Glial fibrillary acidic protein (GFAP) is a type III intermediate filament protein; it is exclusively expressed in astrocytes and has been widely used as a marker for these cells [see review (Eng et al. 2000)]. GFAP expression is developmentally and pathophysiologically regulated. It begins to express as astrocytes mature, and its expression is always increased in reactive astrocytes, or astrocytosis, which is one of the main characteristics of the astrocytic reaction commonly observed in the CNS response to physical, pathological, or chemical insults (Norenburg 1997). Interestingly, both over-expression and disrupted expression of GFAP are problematic and result in neuropathologies. Transgenic mice with higher levels of GFAP expression die prematurely (Messing et al. 1998), while GFAP knockout mice exhibit some developmental defects (Gomi et al. 1995) and CNS abnormalities in the white matter, myelination and the blood-brain barrier (Liedtke et al. 1996; Pekny et al. 1998). In addition, GFAP mutations have been linked to a fatal disease of infants, juveniles, and adults called Alexander's Disease (AxD) (Brenner et al. 2001). AxD is pathologically characterized by formation of Rosenthal fibers (cytoplasmic inclusions) in astrocytes, which contain increased amounts of GFAP in association with stress proteins similar to that of GFAP transgenic mice (Eng et al. 1998). Thus, tightly regulated GFAP expression in astrocytes appears to be important for maintaining astrocyte function and CNS homeostasis. Nevertheless, the molecular mechanisms of increased GFAP expression during CNS injury are largely unknown.
Astrocytosis, or increased GFAP expression in astrocytes is one of the very few common and faithful hallmarks of HIV-1 infection of the CNS (Bell et al. 2006). We have shown that Tat expression in the absence of HIV infection activates GFAP expression and alters astrocyte function and eventually neuron survival (Kim et al. 2003a; Zhou and He 2004; Zhou et al. 2004). We have also shown that disruption of GFAP expression significantly relieves Tat-induced neurotoxicity (Zou et al. 2007). In this study, we aimed to further determine the relationship between Tat and GFAP expression and its effects on HIV-1 interaction with astrocytes.
MATERIALS AND METHODS
Cells and cell cultures
Human astrocytoma cell line U373.MG cells and human kidney epithelial cells 293T were purchased from ATCC (Manassas, VA). Tat-expressing U373.MG cells (U373-Tat) were described previously (Zhou and He 2004; Zhou et al. 2004). All these cells were maintained in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin and 50 μg/ml streptomycin in a 37°C, 5% CO2 incubator. Primary human fetal astrocytes were isolated from 18-19 weeks old human fetal brain tissue (Advanced Bioscience Resources, Inc., Alameda, CA). Briefly, human fetal brain tissue was aseptically and mechanically separated and dissociated in Ca2+/Mg2+ free Hanks balanced salt solution (HBSS) and then treated with 0.25% trypsin at 37°C for 30 min. Trypsin digestion was stopped by washing cells with Ca2+/Mg2+ free HBSS supplemented with 10% FBS, followed by low speed centrifugation to remove debris. Cells were suspended and plated at an appropriate density in a high glucose DMEM containing 10% FBS, 50 units/ ml penicillin, 50 μg/ml streptomycin and 1 mM sodium pyruvate. Culture medium was changed 24 hr following the initial plating and then every 3-4 days in order to remove non-adherent cells. The culture was maintained in a 37°C, 5% CO2 incubator. The primary astrocytes used in the experiments were from the 3rd or 4th passage of these cells.
Plasmids
Tat72Myc, pHIV-GFP, pHCMV-G, p300-luc and p300abcdef-luc were described elsewhere (Li et al. 2002; Yu et al. 2004; Zhou and He 2004). pCMV-β-gal was from Clontech (Mountain View, CA). pGfa2-luc3 reporter plasmid and p300.HA were gifts from Dr. M. Brenner of University of Alabama at Birmingham, Birmingham, AL and Dr. C-H Chang of Indiana University School of Medicine, Indianapolis, IN, respectively. pLTR-Luc was from Drs. R. Jeeninga and B. Berkhout through the NIH AIDS Reagent Program.
Western blotting analysis
Cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lyzed in PBS/TDS buffer (10 mM Na2HPO4, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 0.2% sodium azide, 0.004% sodium fluoride, 1 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mM sodium orthovanadate, pH 7.25) on ice for 20 min for whole cell lysate preparation. Protein concentration was determined using a Bio-Rad DC protein assay kit (Bio-Rad, Hercules, CA). Whole cell lysate was electrophoretically separated by 6% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) for p300 detection (100 μg protein) or by 10% SDS-PAGE for Egr-1, GFAP and β-actin detection (50 μg protein), and processed for blotting and ECL detection with antibodies for GFAP and β-actin antibodies (Sigma, St. Louis, MO), α-Egr-1 (Santa Cruz Biotechnologies, Santa Cruz, CA), or α-p300 (Novus Biologicals (Littleton, CO). Blots were scanned and analyzed by the NIH Image J software, the relative protein levels were compared to the loading control (Rel.).
RNA isolation and semi-quantitative reverse transcriptase (RT)-PCR
Total RNA was isolated using a Trizol Reagent kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RT-PCR was performed on a PE Thermocycler 9700 (PE Applied Biosystem, Foster City, CA) using a Titan One Tube RT-PCR kit (Boehringer Mannheim, Indianapolis, IN). The primers for p300 were 5′-ATG AAC AAC CCC AAT CCT TAT GG-3′ and 5′-CAG GAC AAT CAT GTC TTG TAC-3′ with a program of 50°C for 30 min, 94°C for 3 min, followed by 22 cycles of 94°C for 1 min, 50°C for 1 min and 68°C for 1 min and one cycle of 68°C for 7 min. The primers for GFAP were 5′-AAG CAG ATG AAG CCA CCC TG-3′ and 5′-GTC TGC ACG GGA ATG GTG AT-3′ with a program of 50°C for 30 min, 94°C for 3 min, followed by 25 cycles of 94°C for 1 min, 52°C for 1 min and 68°C for 1 min and one cycle of 68°C for 7 min. The primers for Tat were 5′-GTC GGG ATC CTA ATG GAG CCA GTA GAT CCT-3′ and 5′-TGC TTT GAT AGA GAA ACT TGA TGA GTC-3′ with a program of 50°C for 30 min, 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 50°C for 30 sec and 72°C for 45 sec and one cycle of 72°C for 5 min. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was included in the RT-PCR as an internal control with primers 5′-CTC AGT GTA GCC CAG GAT GC-3′ and 5′-ACC ACC ATG GAG AAG GCT GG-3′. The expected sizes for GFAP, Tat, p300, and GAPDH amplification products were 625, 216, 494 and 500 bp, respectively. Control reactions containing no RT were included to ensure no genomic DNA contamination in the DNA preparations.
Preparation of VSV-G-pseudotyped HIV-GFP viruses and the RT assay
293T cells were transfected with pHIV-GFP plasmid and pHCMV-G plasmids (Li et al. 2002) using the standard calcium phosphate precipitation method. Culture supernatants were collected 72 hr after transfection, clarified by brief low speed centrifugation to remove cell debris, and stored in aliquots at a −80°C freezer. Mock virus with envelope was produced in the same manner and used as a control. The virus titer was determined by the reverse transcriptase assay.
HIV infection of U373.MG cells and primary human fetal astrocytes
U373.MG and primary human fetal astrocytes were plated at an appropriate density and allowed to grow for 24 hr, followed by infection with VSV-G pseudotyped HIV-GFP viruses or mock virus at the dosage of 50K cpm/per million U373.MG cells or 80K cpm/per million primary human fetal astrocytes at 37°C for 2 hr in the presence of 8 μg/ml polybrene. The cells were then thoroughly washed with culture medium to remove unbound viruses and further cultured an additional 48 hr. The dosages of the viruses for infection were titrated for U373.MG and for human primary fetal astrocytes to ensure minimal induction of cell death and maximal infection efficiency. Bright field and GFP images were captured using a Zeiss digital camera mounted on a Zeiss Axiovert M200 fluorescence microscope (Carl Zeiss, Thornwood, NY).
siRNA and transfection
A smart pool of p300 siRNA and a scramble control siRNA were purchased from Dharmacon (Lafayette, CO). U373.MG cells were plated in a 12-well plate at a density of 6 × 104/well or in a 6-well plate at a density of 2 × 105/well and were transfected with siRNA using Lipofectamine according to the manufacturer's instructions (Invitrogen). Cells were harvested 24 hr after transfection for RNA analysis and 72 hr after transfection for protein analysis. For experiments involving both siRNA and plasmid transfection, U373.MG cells were first transfected with siRNA, cultured for 24 hr, and then transfected with plasmid DNA.
β-galactosidase and luciferase reporter gene and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays
Cells were harvested in ice-cold PBS and collected by brief centrifugation. After washes with PBS, the cells were suspended in 0.25 M Tris.HCl, pH 8.3 and subject to three cycles of freezing and thawing. The clear supernatant was saved as the cell extracts to determine the relative β-galactosidase activity. The β-galactosidase activity was then used to normalize the transfection variations among transfections for the subsequent luciferase reporter gene assay, which was done using a luciferase assay system (Promega, Madison. WI). The luciferase activity was quantitated using an Opticomp Luminometer (MGM Instruments, Hamden, CT). Cell viability was determined using the MTT assay as previously described (Zhou et al. 2004).
[3H]-thymidine incorporation assay
U373.MG cells were plated in a 96-well plate at a density of 1 × 104 cells per well and allowed to grow overnight. Then, the cells were pulse-chased with 1 μCi [3H]-thymidine (Perkin Elmer) in each well for 48 hr. Cells were then harvested by incubating with 100 μl 0.25% Trypsin/1 mM EDTA for 20 min and transferred onto a filter paper for scintillation counting.
Data analysis
All experiment data were analyzed by 2-tailed student's t test. A p < 0.05 was considered to be statistically significant and marked as “*”, a p < 0.01 was considered to be statistically highly significant and marked as “**”.
RESULTS
Identification of increased p300 cDNA level in Tat-expressing astrocytoma cells and primary astrocytes
Our early studies have shown that Tat expression leads to increased levels of GFAP expression in astrocytes (Kim et al. 2003a; Zhou et al. 2004). During this study, we always noticed an additional minor RT-PCR product of about 1 kb in length that was only amplified from Tat expressing U373-Tat cells (Fig. 1A). To ensure that this product was not an artifact derived from RT-PCR or a selection bias from generation of U373-Tat stable cell lines, we isolated primary embryonic astrocytes from inducible astrocyte-specific Tat transgenic mice (Kim et al. 2003a; Zhou et al. 2004) along with primary astrocytes from wild-type C57BL/6 mice, the breeding congenic strain for Tat transgenic mice. We cultured these cells in vitro in the presence of 5 mg/ml doxycycline to induce Tat expression, which was demonstrated by RT-PCR using Tat specific primers (middle panel, Fig. 1B). There was a 1 kb RT-PCR product amplified in Tat-expressing primary murine astrocytes (upper panel, Fig. 1B). We then recovered the DNA from the agarose gel, cloned it into the TA TOPO cloning vector, and sequenced the insert. BLAST searches matched the sequence of this DNA fragment to p300 cDNA between nucleotide (nt.) 6475 and nt. 7436 (GenBank accession # NM_001429). The unintended amplification of p300 was likely due to degenerative alignment of the 5′ GFAP primer to p300 cDNA between nt. 6475 and nt. 6493 and the 3′ GFAP primer to p300 cDNA between nt. 7417 and nt. 7436.
Figure 1. Identification of p300 up-regulation in Tat-expressing U373 cells and primary astrocytes.
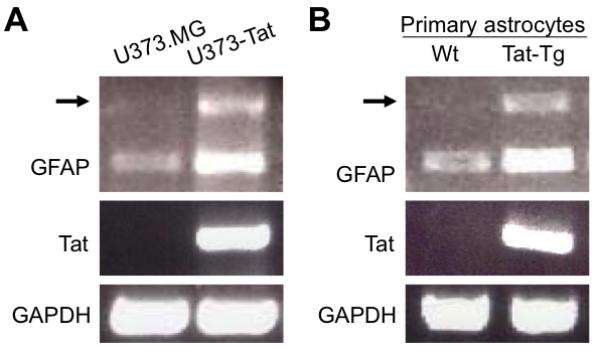
Total RNA was isolated from U373.MG and U373-Tat (A) or from primary astrocytes (B), which were prepared from wild-type C57BL/6 mice or inducible Tat transgenic mice and cultured in the presence of 5 mg/ml doxycycline for 3 days. The total RNA was subject to RT-PCR using specific GFAP and Tat primers. GAPDH was included as an equal loading control. Arrow: the unknown DNA products of about 1 kb amplified with GFAP primers from Tat-expressing cells.
Up-regulated p300 expression in Tat-expressing astrocytes and HIV-infected astrocytes
To determine whether Tat expression indeed led to p300 up-regulation, we prepared total RNA from U373.MG and U373-Tat cells and performed RT-PCR using p300-specific primers. Meanwhile, we also prepared whole cell lysates from these cells and performed Western blot analysis using an anti-p300 antibody. Compared to U373.MG cells, U373-Tat cells had significantly higher levels of p300 expression at both mRNA and protein levels (Fig. 2A). To further ascertain this phenomenon, we also transiently transfected U373.MG cells with HIV-1 Tat expression plasmid and determined p300 expression in those cells and obtained similar results (Fig. 2B). To further determine the physiological relevance of Tat-up-regulated p300 expression, we infected U373.MG cells with VSV-G-pseudotyped HIV-GFP viruses, which were use to override the HIV-1 entry restriction in astrocytes and facilitate HIV-1 infection of these cells. The infection efficiency of these viruses was estimated to be about 40% (Fig. 3A). We then compared p300 mRNA and protein levels between HIV-infected and mock infected-U373.MG cells. In agreement with the findings obtained from Tat expression, HIV-1 infection resulted in increased p300 expression at both mRNA and protein levels (Fig. 3B). Moreover, we also infected primary human fetal astrocytes with the HIV viruses and determined p300 expression. The infection efficiency of these cells was about 25%, slightly lower than that of U373.MG cells (data not shown). Nevertheless, increased p300 mRNA and protein were also noted in those cells infected with HIV (Fig. 3C). Taken together, these results demonstrate that Tat expression alone or in the context of HIV-1 infection leads to up-regulated p300 expression.
Figure 2. p300 induction in astrocytes by HIV-1 Tat expression.
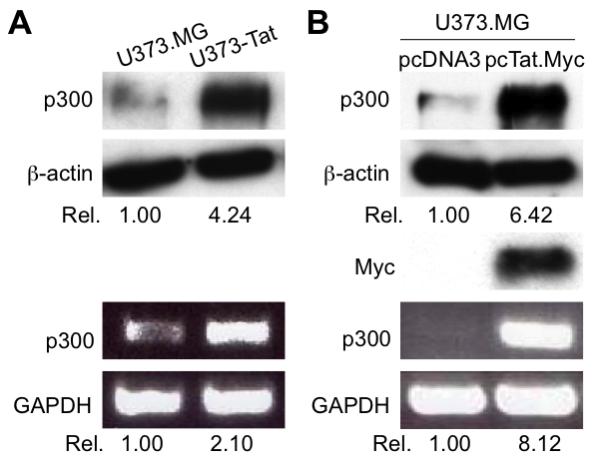
A. Induction of p300 protein and mRNA in U373-Tat cells. B. Induction of p300 protein and mRNA in U373 cells that were transiently transfected with HIV-1 Tat expressing plasmid. U373.MG cells were plated in a 6-well plate at a density of 3 × 105/well and transfected with pcTat.Myc or control pcDNA3 plasmid. Total RNA and whole cell lysates were then prepared 48 hr after transfection for RT-PCR and Western blotting analysis, respectively. GAPDH and β-actin were used as equal loading controls for RT-PCR and Western blot, respectively.
Figure 3. p300 induction in astrocytes by HIV-1 infection.
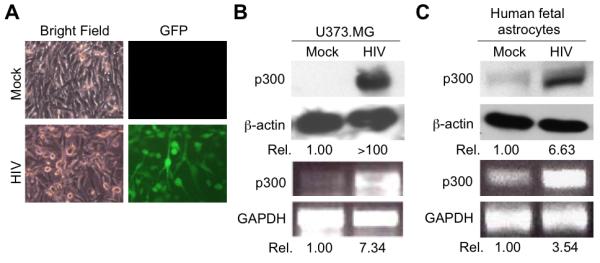
U373.MG cells were infected with 50K cpm RTase equivalent VSV-G-pseudotyped HIV-GFP viruses. HIV-1 infection was monitored by GFP expression and estimated to be about 40% (A). HIV-GFP viruses containing no envelope (mock) were included as a control. Total RNA and whole cell lysates were prepared from these infected cells 48 hr after infection and subject to RT-PCR and Western blotting analysis for p300 expression (B). C. Up-regulated p300 expression in HIV-1-infected primary human fetal astrocytes. Primary human fetal astrocytes were infected as stated above except for that 80K cpm RTase equivalent VSV-G-pseudotyped HIV-GFP viruses were used and analyzed for p300 expression as above.
Transactivation of p300 promoter by HIV-1 Tat and its requirement for transcription factor early growth response-1 (Egr-1)
Up-regulation of p300 mRNA by Tat expression raises the possibility that Tat activates p300 expression at the transcriptional level. To address this possibility, we transfected U373.MG cells with a luciferase reporter gene under the control of p300 promoter (Yu et al. 2004) along with increasing amounts pcTat.Myc expression plasmid and determined the direct effect of Tat on p300 promoter activity. Tat expression activated the p300 promoter-driven luciferase reporter gene expression in a direct and dose-dependent manner (Fig. 4A). Since early growth resonse-1 is a major transcriptional regulator of p300 gene transcription and expression and there are a total of four potential Egr-1 DNA binding motifs within the p300 promoter (Yu et al. 2004), we then performed co-transfections of the Egr-1 motifs-deleted p300 promoter with HIV-1 Tat expressing plasmid to investigate a possible role of Egr-1 in Tat-activated p300 expression. We found that deletion of Egr-1 DNA binding motifs completely abolished the activation activity of Tat on the p300 promoter. These results suggest that Egr-1 was required for Tat activation of p300 promoter and prompted us to further determine the relationship between Tat expression and Egr-1 expression in astrocytes. To this end, we performed Western blot analysis for Egr-1 expression in Tat-expressing astrocytes using an anti-Egr-1 antibody. Compared to U373.MG cells, U373-Tat cells had a much higher level of Egr-1 (Fig. 4B). Taken together, these findings indicate that Tat activates p300 expression in astrocytes and involves the cis-acting elements of Egr-1 within the p300 promoter.
Figure 4. Transactivation activity of HIV-1 Tat on the p300 promoter.
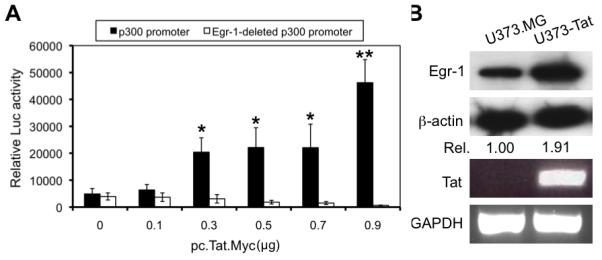
A. Transactivation of the p300 promoter by HIV-1 Tat and its requirement for Egr-1. U373.MG cells were transfected with p300 promoter-driven luciferase reporter gene (p300 promoter, closed bar) or a mutated p300 promoter-driven reporter gene that lacks all four Egr-1 DNA binding sites (Egr-1-deleted p300 promoter, open bar) and increasing amounts of pcTat.Myc plasmid. Cell lysates were prepared 48 hr after transfection for the Luc activity assay. pCMV-βGal was included in transfections to normalize the transfection variations. All samples were compared to the one that was not transfected with pcTat.Myc for statistical analysis. B. Up-regulated expression of Egr-1 protein in U373-Tat cells. Total RNA and whole cell lysates were prepared from U373.MG and U373-Tat cells and subject to RT-PCR and Western blotting analysis, respectively. GAPDH and β-actin were used as equal loading controls for RT-PCR and Western blotting analysis, respectively.
Regulation of constitutive and Tat-activated GFAP expression by p300
Having shown that HIV-1 Tat expression leads to GFAP activation (Kim et al. 2003a; Zhou and He 2004; Zhou et al. 2004), we then investigated the relationship between p300 and GFAP expression. To this end, we first determined the effect of p300 over-expression on GFAP expression. We transfected U373.MG cells with the p300 expression plasmid and analyzed GFAP expression by Western blotting. We found a significantly elevated level of GFAP in p300-transfected cells (Fig. 5A). Next, we determined the effect of p300 knockdown on GFAP expression. We took advantage of the siRNA strategy and transfected U373.MG cells with p300 siRNA. p300 siRNA was very effective in knocking down constitutive p300 mRNA as well as p300 protein expression, while the control siRNA had little effect (upper panels, Fig. 5B). In parallel, p300 knockdown resulted in a significantly lower level of GFAP expression (lower panel, Fig. 5B). In addition, we performed similar experiments in U373.Tat cells. p300 knockdown also effectively suppressed GFAP expression in Tat-expressing astrocytes (Fig. 5C). These results indicate that p300 not only plays an important regulatory role in constitutive GFAP expression, but also directly participates in Tat-mediated GFAP up-regulation.
Figure 5. Relationship between p300 and GFAP expression.
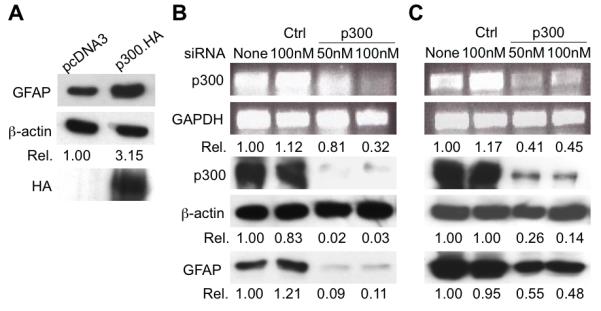
A. GFAP up-regulation in astrocytes by p300 over-expression. U373.MG cells were transfected with p300.HA expressing plasmid or control pcDNA3. Whole cell lysates were prepared 48 hr after transfection and subject to Western blotting analysis for GFAP, p300 and β-actin expression. B & C. GFAP down-modulation in U373.MG (B) and U373-Tat cells (C) by p300 knockdown. U373.MG and U373-Tat cells were transfected with p300 siRNA or control siRNA at the indicated concentrations. Total RNA and whole cell lysates were prepared after transfection for RT-PCR and Western blotting analysis, respectively. GAPDH and β-actin were used as equal loading controls for RT-PCR and Western blotting analysis, respectively.
Transcriptional regulation of GFAP by p300
p300 is a multifunctional transcription co-activator with inherent histone acetyltransferase (HAT) activity. It regulates a number of cellular genes through its direct binding to other transcription factors or through its intrinsic HAT activity. Thus, we then investigated whether p300 regulation of GFAP expression also occurred at the transcription level. We transfected U373.MG or U373-Tat cells with a luciferase reporter gene under the control of the GFAP promoter (pGfa2-luc3) (Su et al. 2004) along with increasing amounts of the p300 expression plasmid and determined the luciferase activity. p300 expression activated the GFAP promoter in U373.MG and U373-Tat cells (Fig. 6A). The more pronounced activation of the GFAP promoter-driven reporter gene obtained in U373-Tat cells likely results from p300 activation by Tat and the subsequent additive effects. To further ascertain the role of p300 in Tat-mediated GFAP activation, we also transfected U373.MG cells with p300 siRNA, followed by the pGfa2-luc3 plasmid and pcTat.Myc. As previously shown (Zhou et al. 2004), Tat expression activated the GFAP promoter-driven luciferase reporter gene expression (Fig. 6B). In agreement with previous findings (Fig. 5C), p300 knockdown abolished Tat-mediated activation of the GFAP promoter-driven luciferase reporter gene expression (Fig. 6B). These results offer additional evidence to support the notion that p300 is a transcription regulator of both constitutive and Tat-induced GFAP expression.
Figure 6. Involvement of p300 in constitutive and Tat-mediated GFAP expression.
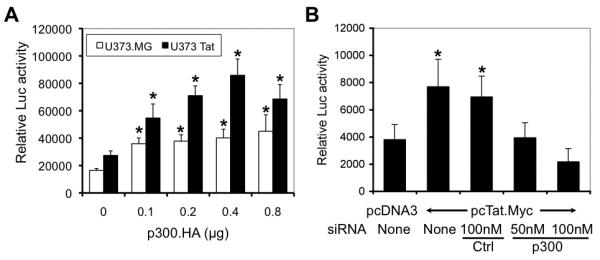
A. Transactivation of the GFAP promoter by p300. U373.MG (open bar) and U373-Tat (closed bar) cells were transfected with the GFAP promoter-driven luciferase reporter gene plasmid pGfa2-luc3 and increasing amounts of p300 expressing plasmid. Cell lysates were prepared 48 hr after transfection for the luciferase activity assay. pCMV-βGal was included in the transfection to normalize transfection variations. All samples were compared to the one that was not transfected with p300.HA for statistical analysis. B. Attenuation of Tat-mediated GFAP activation by p300 knockdown. U373.MG cells were transfected with p300 siRNA or control siRNA, cultured for 24 hr, and then transfected again with pGfa2-luc3 and pcTat.Myc. Cell lysates were prepared 48 hr after the 2nd transfection for the luciferase activity assay. pCMV-βGal was included in transfections to normalize the transfection variations. All samples were compared to the one that was transfected only with pcDNA3 for statistical analysis.
p300, HIV-1 gene expression and replication in astrocytes, and astrocyte proliferation
There are multiple blocks that contribute to the non-productive replication of HIV-1 in astrocytes. To determine whether Tat activation of p300 could play a role in the restricted HIV-1 restriction in these cells, we first focused on possible roles of p300 in HIV-1 gene transcription. To test this, we transfected U373.MG cells with a luciferase reporter gene under the control of the HIV-1 LTR promoter along with pcTat.Myc and increasing amounts of the p300 expression plasmid and determined the luciferase activity. p300 expression potentiated Tat transactivation activity on the HIV-1 LTR promoter (Fig. 7A). We next determined the effect of p300 on HIV-1 replication. We transfected U373.MG cells with increasing amounts of the p300 expression plasmid followed by infection with VSV-G-pseudotyped HIV-GFP viruses, and then monitored HIV-1 replication. Surprisingly, p300 exhibited somewhat inhibitory effects on HIV-1 replication in these cells, although the inhibition was not statistically significant (Fig. 7B). Meanwhile, we performed the MTT assay to determine whether p300 expression would alter the proliferation of HIV-infected astrocytes. We found that p300 expression increased the proliferation of HIV-infected astrocytes in a dose-dependent manner (Fig. 7C). Similar results were obtained using the [3H]-thymidine incorporation assay (7D).
Figure 7. Effects of p300 on HIV-1 gene expression and replication in astrocytes and on survival of HIV-1-infected astrocytes.
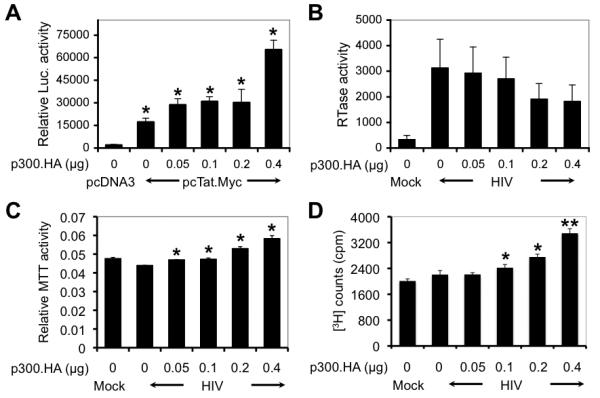
A. Activation of the HIV-1 LTR promoter by p300. U373.MG cells were transfected with the HIV-1 LTR promoter-driven luciferase reporter gene plasmid pHIV LTR-Luc, pcTat.Myc, and increasing amounts of p300 as indicated. Cell lysates were prepared 48 hr after transfection for the luciferase activity assay. pCMV-βGal was included in transfections to normalize the transfection variations. All samples were compared to the one that was transfected only with pcDNA3 for statistical analysis. B. No significant effects of p300 on HIV-1 replication. U373.MG cells were transfected with increasing amounts of p300 expressing plasmid as indicated, cultured for 24 hr, and then infected with VSV-G-pseudotyped HIV-GFP viruses. Cell culture supernatants were collected 48 hr after infection for the RT activity assay. Mock infection was included as a control. C & D. Increased proliferation of HIV-1-infected astrocytes by p300. U373.MG cells were transfected and infected as stated above and then subject to the MTT assay 48 hr after infection (C), or pulse-chased with [3H]-thymidine for 48 hr for scintillation counting (D). All samples were compared to the one that was not transfected with p300.HA but infected with HIV-1 for statistical analysis (B-D).
DISCUSSION
p300 is a large nuclear molecule and evolutionarily conserved. It is a multifunctional transcriptional co-activator with inherent histone acetyltransferase (HAT) activity, mediates various cross-talks between different signaling pathways and participates in cellular functions including DNA repair, cell growth, differentiation, and apoptosis (Blobel 2002). These biological functions are primarily regulated via its HAT activity through phosphorylation and interactions with other cellular proteins (Kalkhoven 2004). Compared to its well-characterized biological functions, only a very few studies have reported on its transcriptional regulation. Transcription factor early growth response 1 (Egr-1) has been shown to regulate p300 transcription through the Egr-1 DNA-binding cis-acting elements within the p300 promoter, intracellular signaling, or viral infection or expression of viral protein (Cai et al. 2006; Nair et al. 2006; Saegusa et al. 2008; Yu et al. 2004). Meanwhile, Tat is known to alter various intracellular signaling pathways (Liu et al. 2002; Peruzzi 2006). In the current study, we showed that Tat expression in or HIV-1 infection of astrocytes led to p300 transactivation (Fig. 1-3) and that Egr-1 was directly involved in this process (Fig. 4). Although the underlying mechanisms for Tat-induced Egr-1 or p300 expression remain to be determined, cross-talk among various intracellular signaling pathways is likely involved. Tat-mediated p300 up-regulation appeared to be specific to astrocytes, as the opposite effect was noted in other cell types [our unpublished data and (de la Fuente et al. 2002)]. Taken together, the current study provides further evidence to support the notion that transcription is indeed a regulatory mechanism for p300 expression and activity. In addition, these findings from the current study suggest that Tat likely exerts its plethora of effects on the host through gene expression involving more general and upstream transcription factors like Egr-1 and p300 (Fan et al.).
The initial observation of concurrent induction of p300 and GFAP by Tat prompted us to further examine the relationship between p300 and GFAP expression. Our results demonstrated that ectopic expression of p300 resulted in increased levels of GFAP expression, whereas p300 knockdown led to decreased levels of GFAP expression (Fig. 5). We further showed similar effects using a GFAP promoter-driven reporter gene system and astrocytes stably expressing Tat protein (Fig. 5 and Fig. 6). These results suggest that p300 is important for both constitutive and Tat-induced GFAP expression. There are several response elements in the upstream region of the GFAP promoter, including nuclear factor (NF)-1, NF-κB, estrogen response element, glucocorticoid response element, activating protein-1 binding site, and 12-O-tetradecanoylphorbol 13-acetate response element (Laping et al. 1994). Accordingly, the main regulators of GFAP expression are hormones such as thyroid hormone and glucocorticoids, growth factors, and inflammatory cytokines (Gomes et al. 1999). On the other hand, p300 regulates gene expression mainly through the HAT activity (chromatin remodeling) and/or binding to basal transcription factors and other transcription activators or other cellular proteins (Xu et al. 1999). Thus, GFAP is probably one of the target genes of p300 in astrocytes. Interestingly, complex formation of p300 with STAT3 and Smads has indeed been shown to be essential for differentiation of neuronal progenitors into astrocytes, as assessed by GFAP expression (Nakashima et al. 1999; Yanagisawa et al. 2001).
Quite a few studies have documented Tat interaction with p300 and its importance in Tat-mediated transactivation of the HIV-1 LTR promoter. p300 acetylates Tat at Lys50 and Lys51 and enhances Tat recycling from the TAR complex to activate transcription elongation of the HIV-1 LTR promoter (Bres et al. 2002; Ott et al. 2004). Meanwhile, Tat binds to and recruits p300 and other proteins to the transcription machinery within the integrated HIV-1 LTR promoter, which subsequently leads to transactivation of the LTR promoter (Marzio et al. 1998; Wong et al. 2005). Consistent with these findings, our results showed that p300 potentiated Tat-mediated transactivation of the HIV-1 LTR promoter in astrocytes (Fig. 7A). Interestingly, in the context of HIV-1 infection, p300 expression appeared to inhibit, albeit in an insignificant manner, HIV gene expression (Fig. 7B). This discrepancy between the LTR-driven reporter gene assay and the HIV gene expression assay may be due to the state of the LTR, i.e., unintegrated LTR in the reporter gene assay vs. integrated LTR in the gene expression assay, or p300 interaction with other HIV gene products that differ between these two assay systems. Nevertheless, p300 expression significantly promoted proliferation of HIV-infected astrocytes (Fig. 7C), suggesting that p300 up-regulation by Tat may contribute to virus persistence and latency in the CNS following initial HIV infection. It is clear that p300 up-regulation in astrocytes by Tat, as shown in this study, adds another layer of complexity to the axis of Tat-p300 interaction and its role in HIV gene expression and pathogenesis.
In summary, these results reveal a cascade of transcription networks in astrocytes that is initiated by Tat and manifested by GFAP up-regulation and suggest two additional pathways for astrocyte-mediated Tat neurotoxicity: one is GFAP accumulation that subsequently alters astrocyte function, the other is enhanced survival of HIV-infected astrocytes that leads to the establishment of an HIV-1 sanctuary in the CNS.
Acknowledgments
This work was supported in part by the grants R01NS039804 and R01NS065785 from the National Institutes of Health.
REFERENCES
- Andras IE, Rha G, Huang W, Eum S, Couraud PO, Romero IA, Hennig B, Toborek M. Simvastatin protects against amyloid beta and HIV-1 Tat-induced promoter activities of inflammatory genes in brain endothelial cells. Mol Pharmacol. 2008;73(5):1424–33. doi: 10.1124/mol.107.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea S, Del Valle L, Mameli G, Sawaya BE, Khalili K, Peruzzi F. Tubulin-mediated binding of human immunodeficiency virus-1 Tat to the cytoskeleton causes proteasomal-dependent degradation of microtubule-associated protein 2 and neuronal damage. J Neurosci. 2006;26(15):4054–62. doi: 10.1523/JNEUROSCI.0603-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks WA, Robinson SM, Nath A. Permeability of the blood-brain barrier to HIV-1 Tat. Exp Neurol. 2005;193(1):218–27. doi: 10.1016/j.expneurol.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Bell JE, Anthony IC, Simmonds P. Impact of HIV on regional & cellular organisation of the brain. Curr HIV Res. 2006;4(3):249–57. doi: 10.2174/157016206777709401. [DOI] [PubMed] [Google Scholar]
- Benelli R, Barbero A, Ferrini S, Scapini P, Cassatella M, Bussolino F, Tacchetti C, Noonan DM, Albini A. Human immunodeficiency virus transactivator protein (Tat) stimulates chemotaxis, calcium mobilization, and activation of human polymorphonuclear leukocytes: implications for Tat-mediated pathogenesis. J Infect Dis. 2000;182(6):1643–51. doi: 10.1086/317597. [DOI] [PubMed] [Google Scholar]
- Blobel GA. CBP and p300: versatile coregulators with important roles in hematopoietic gene expression. J Leukoc Biol. 2002;71(4):545–56. [PubMed] [Google Scholar]
- Brailoiu E, Brailoiu GC, Mameli G, Dolei A, Sawaya BE, Dun NJ. Acute exposure to ethanol potentiates human immunodeficiency virus type 1 Tat-induced Ca(2+) overload and neuronal death in cultured rat cortical neurons. J Neurovirol. 2006;12(1):17–24. doi: 10.1080/13550280500516427. [DOI] [PubMed] [Google Scholar]
- Brenner M, Johnson AB, Boespflug-Tanguy O, Rodriguez D, Goldman JE, Messing A. Mutations in GFAP, encoding glial fibrillary acidic protein, are associated with Alexander disease. Nat Genet. 2001;27(1):117–20. doi: 10.1038/83679. [DOI] [PubMed] [Google Scholar]
- Bres V, Kiernan R, Emiliani S, Benkirane M. Tat acetyl-acceptor lysines are important for human immunodeficiency virus type-1 replication. J Biol Chem. 2002;277(25):22215–21. doi: 10.1074/jbc.M201895200. [DOI] [PubMed] [Google Scholar]
- Cai Y, Liu Y, Zhang X. Induction of transcription factor Egr-1 gene expression in astrocytoma cells by Murine coronavirus infection. Virology. 2006;355(2):152–63. doi: 10.1016/j.virol.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporello E, Nath A, Slevin J, Galey D, Hamilton G, Williams L, Steiner JP, Haughey NJ. The immunophilin ligand GPI1046 protects neurons from the lethal effects of the HIV-1 proteins gp120 and Tat by modulating endoplasmic reticulum calcium load. J Neurochem. 2006;98(1):146–55. doi: 10.1111/j.1471-4159.2006.03863.x. [DOI] [PubMed] [Google Scholar]
- Chang HC, Samaniego F, Nair BC, Buonaguro L, Ensoli B. HIV-1 Tat protein exits from cells via a leaderless secretory pathway and binds to extracellular matrix-associated heparan sulfate proteoglycans through its basic region. Aids. 1997;11(12):1421–31. doi: 10.1097/00002030-199712000-00006. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Turchan J, Pocernich C, Bruce-Keller A, Roth S, Butterfield DA, Major EO, Nath A. Intracellular human immunodeficiency virus Tat expression in astrocytes promotes astrocyte survival but induces potent neurotoxicity at distant sites via axonal transport. J Biol Chem. 2003;278(15):13512–9. doi: 10.1074/jbc.M209381200. [DOI] [PubMed] [Google Scholar]
- Chen LL, Frankel AD, Harder JL, Fawell S, Barsoum J, Pepinsky B. Increased cellular uptake of the human immunodeficiency virus-1 Tat protein after modification with biotin. Anal Biochem. 1995;227(1):168–75. doi: 10.1006/abio.1995.1267. [DOI] [PubMed] [Google Scholar]
- de la Fuente C, Santiago F, Deng L, Eadie C, Zilberman I, Kehn K, Maddukuri A, Baylor S, Wu K, Lee CG. Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals. BMC Biochem. 2002;3:14. doi: 10.1186/1471-2091-3-14. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paulis A, De Palma R, Di Gioia L, Carfora M, Prevete N, Tosi G, Accolla RS, Marone G. Tat protein is an HIV-1-encoded beta-chemokine homolog that promotes migration and up-regulates CCR3 expression on human Fc epsilon RI+ cells. J Immunol. 2000;165(12):7171–9. doi: 10.4049/jimmunol.165.12.7171. [DOI] [PubMed] [Google Scholar]
- Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP-thirty-one years (1969-2000) Neurochem Res. 2000;25(9-10):1439–51. doi: 10.1023/a:1007677003387. [DOI] [PubMed] [Google Scholar]
- Eng LF, Lee YL, Kwan H, Brenner M, Messing A. Astrocytes cultured from transgenic mice carrying the added human glial fibrillary acidic protein gene contain Rosenthal fibers. J Neurosci Res. 1998;53(3):353–60. doi: 10.1002/(SICI)1097-4547(19980801)53:3<353::AID-JNR9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Fan Y, Zou W, Green LA, Kim BO, He JJ. Activation of Egr-1 Expression in Astrocytes by HIV-1 Tat: New Insights into Astrocyte-Mediated Tat Neurotoxicity. J Neuroimmune Pharmacol. doi: 10.1007/s11481-010-9217-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55(6):1189–93. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- Gomes FC, Paulin D, Moura Neto V. Glial fibrillary acidic protein (GFAP): modulation by growth factors and its implication in astrocyte differentiation. Braz J Med Biol Res. 1999;32(5):619–31. doi: 10.1590/s0100-879x1999000500016. [DOI] [PubMed] [Google Scholar]
- Gomi H, Yokoyama T, Fujimoto K, Ikeda T, Katoh A, Itoh T, Itohara S. Mice devoid of the glial fibrillary acidic protein develop normally and are susceptible to scrapie prions. Neuron. 1995;14(1):29–41. doi: 10.1016/0896-6273(95)90238-4. [DOI] [PubMed] [Google Scholar]
- Jones M, Olafson K, Del Bigio MR, Peeling J, Nath A. Intraventricular injection of human immunodeficiency virus type 1 (HIV-1) tat protein causes inflammation, gliosis, apoptosis, and ventricular enlargement. J Neuropathol Exp Neurol. 1998;57(6):563–70. doi: 10.1097/00005072-199806000-00004. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68(6):1145–55. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Kim BO, Liu Y, Ruan Y, Xu ZC, Schantz L, He JJ. Neuropathologies in Transgenic Mice Expressing Human Immunodeficiency Virus Type 1 Tat Protein under the Regulation of the Astrocyte-Specific Glial Fibrillary Acidic Protein Promoter and Doxycycline. Am J Pathol. 2003a;162(5):1693–707. doi: 10.1016/S0002-9440(10)64304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TA, Avraham HK, Koh YH, Jiang S, Park IW, Avraham S. HIV-1 Tat-mediated apoptosis in human brain microvascular endothelial cells. J Immunol. 2003b;170(5):2629–37. doi: 10.4049/jimmunol.170.5.2629. [DOI] [PubMed] [Google Scholar]
- Laping NJ, Teter B, Nichols NR, Rozovsky I, Finch CE. Glial fibrillary acidic protein: regulation by hormones, cytokines, and growth factors. Brain Pathol. 1994;4(3):259–75. doi: 10.1111/j.1750-3639.1994.tb00841.x. [DOI] [PubMed] [Google Scholar]
- Li J, Liu Y, Park IW, He JJ. Expression of exogenous Sam68, the 68-kilodalton SRC-associated protein in mitosis, is able to alleviate impaired Rev function in astrocytes. J Virol. 2002;76(9):4526–35. doi: 10.1128/JVI.76.9.4526-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke W, Edelmann W, Bieri PL, Chiu FC, Cowan NJ, Kucherlapati R, Raine CS. GFAP is necessary for the integrity of CNS white matter architecture and long-term maintenance of myelination. Neuron. 1996;17(4):607–15. doi: 10.1016/s0896-6273(00)80194-4. [DOI] [PubMed] [Google Scholar]
- Liu X, Jana M, Dasgupta S, Koka S, He J, Wood C, Pahan K. Human immunodeficiency virus type 1 (HIV-1) tat induces nitric-oxide synthase in human astroglia. J Biol Chem. 2002;277(42):39312–9. doi: 10.1074/jbc.M205107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Jones M, Hingtgen CM, Bu G, Laribee N, Tanzi RE, Moir RD, Nath A, He JJ. Uptake of HIV-1 tat protein mediated by low-density lipoprotein receptor-related protein disrupts the neuronal metabolic balance of the receptor ligands. Nat Med. 2000;6(12):1380–7. doi: 10.1038/82199. [DOI] [PubMed] [Google Scholar]
- Marzio G, Tyagi M, Gutierrez MI, Giacca M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A. 1998;95(23):13519–24. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing A, Head MW, Galles K, Galbreath EJ, Goldman JE, Brenner M. Fatal encephalopathy with astrocyte inclusions in GFAP transgenic mice. Am J Pathol. 1998;152(2):391–8. [PMC free article] [PubMed] [Google Scholar]
- Nair AM, Michael B, Datta A, Fernandez S, Lairmore MD. Calcium-dependent enhancement of transcription of p300 by human T-lymphotropic type 1 p12I. Virology. 2006;353(2):247–57. doi: 10.1016/j.virol.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284(5413):479–82. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Norenburg M. Astrocytes pathophysiology in disorders of the central nervous system. In: Schipper H, editor. Astrocytes: in Brain Aging and Neurodegeneration. Landes Bioscience; Austin: 1997. [Google Scholar]
- Norman JP, Perry SW, Kasischke KA, Volsky DJ, Gelbard HA. HIV-1 trans activator of transcription protein elicits mitochondrial hyperpolarization and respiratory deficit, with dysregulation of complex IV and nicotinamide adenine dinucleotide homeostasis in cortical neurons. J Immunol. 2007;178(2):869–76. doi: 10.4049/jimmunol.178.2.869. [DOI] [PubMed] [Google Scholar]
- Ott M, Dorr A, Hetzer-Egger C, Kaehlcke K, Schnolzer M, Henklein P, Cole P, Zhou MM, Verdin E. Tat acetylation: a regulatory switch between early and late phases in HIV transcription elongation. Novartis Found Symp. 2004;259:182–93. discussion 193-6, 223-5. [PubMed] [Google Scholar]
- Park IW, Wang JF, Groopman JE. HIV-1 Tat promotes monocyte chemoattractant protein-1 secretion followed by transmigration of monocytes. Blood. 2001;97(2):352–8. doi: 10.1182/blood.v97.2.352. [DOI] [PubMed] [Google Scholar]
- Pekny M, Eliasson C, Chien CL, Kindblom LG, Liem R, Hamberger A, Betsholtz C. GFAP-deficient astrocytes are capable of stellation in vitro when cocultured with neurons and exhibit a reduced amount of intermediate filaments and an increased cell saturation density. Exp Cell Res. 1998;239(2):332–43. doi: 10.1006/excr.1997.3922. [DOI] [PubMed] [Google Scholar]
- Peruzzi F. The multiple functions of HIV-1 Tat: proliferation versus apoptosis. Front Biosci. 2006;11:708–17. doi: 10.2741/1829. [DOI] [PubMed] [Google Scholar]
- Price TO, Uras F, Banks WA, Ercal N. A novel antioxidant N-acetylcysteine amide prevents gp120- and Tat-induced oxidative stress in brain endothelial cells. Exp Neurol. 2006;201(1):193–202. doi: 10.1016/j.expneurol.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Saegusa M, Hashimura M, Kuwata T, Hamano M, Watanabe J, Kawaguchi M, Okayasu I. Transcription factor Egr1 acts as an upstream regulator of beta-catenin signalling through up-regulation of TCF4 and p300 expression during trans-differentiation of endometrial carcinoma cells. J Pathol. 2008;216(4):521–32. doi: 10.1002/path.2404. [DOI] [PubMed] [Google Scholar]
- Su M, Hu H, Lee Y, d'Azzo A, Messing A, Brenner M. Expression specificity of GFAP transgenes. Neurochem Res. 2004;29(11):2075–93. doi: 10.1007/s11064-004-6881-1. [DOI] [PubMed] [Google Scholar]
- Toborek M, Lee YW, Pu H, Malecki A, Flora G, Garrido R, Hennig B, Bauer HC, Nath A. HIV-Tat protein induces oxidative and inflammatory pathways in brain endothelium. J Neurochem. 2003;84(1):169–79. doi: 10.1046/j.1471-4159.2003.01543.x. [DOI] [PubMed] [Google Scholar]
- Wong K, Sharma A, Awasthi S, Matlock EF, Rogers L, Van Lint C, Skiest DJ, Burns DK, Harrod R. HIV-1 Tat interactions with p300 and PCAF transcriptional coactivators inhibit histone acetylation and neurotrophin signaling through CREB. J Biol Chem. 2005;280(10):9390–9. doi: 10.1074/jbc.M408643200. [DOI] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9(2):140–7. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Nakashima K, Takizawa T, Ochiai W, Arakawa H, Taga T. Signaling crosstalk underlying synergistic induction of astrocyte differentiation by BMPs and IL-6 family of cytokines. FEBS Lett. 2001;489(2-3):139–43. doi: 10.1016/s0014-5793(01)02095-6. [DOI] [PubMed] [Google Scholar]
- Yu J, de Belle I, Liang H, Adamson ED. Coactivating factors p300 and CBP are transcriptionally crossregulated by Egr1 in prostate cells, leading to divergent responses. Mol Cell. 2004;15(1):83–94. doi: 10.1016/j.molcel.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Hennig B, Toborek M. Intact lipid rafts regulate HIV-1 Tat protein-induced activation of the Rho signaling and upregulation of P-glycoprotein in brain endothelial cells. J Cereb Blood Flow Metab. 2009 doi: 10.1038/jcbfm.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BY, He JJ. Proliferation inhibition of astrocytes, neurons, and non-glial cells by HIV-1 Tat protein. Neuroscience Letters. 2004;359:155–158. doi: 10.1016/j.neulet.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Zhou BY, Liu Y, Kim B, Xiao Y, He JJ. Astrocyte activation and dysfunction and neuron death by HIV-1 Tat expression in astrocytes. Mol Cell Neurosci. 2004;27(3):296–305. doi: 10.1016/j.mcn.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Zou W, Kim BO, Zhou BY, Liu Y, Messing A, He JJ. Protection against human immunodeficiency virus type 1 Tat neurotoxicity by Ginkgo biloba extract EGb 761 involving glial fibrillary acidic protein. Am J Pathol. 2007;171(6):1923–35. doi: 10.2353/ajpath.2007.070333. [DOI] [PMC free article] [PubMed] [Google Scholar]


