Abstract
Nociceptive neurons innervate the skin with complex dendritic arbors that respond to pain-evoking stimuli such as harsh mechanical force or extreme temperatures. Here we describe the structure and development of a model nociceptor, the PVD neuron of C. elegans, and identify transcription factors that control morphogenesis of the PVD dendritic arbor. The two PVD neuron cell bodies occupy positions on either the right (PVDR) or left (PVDL) sides of the animal in posterior lateral locations. Imaging with a GFP reporter revealed a single axon projecting from the PVD soma to the ventral cord and an elaborate, highly-branched arbor of dendritic processes that envelop the animal with a web-like array directly beneath the skin. Dendritic branches emerge in a step-wise fashion during larval development and may use an existing network of peripheral nerve cords as guideposts for key branching decisions. Time-lapse imaging revealed that branching is highly dynamic with active extension and withdrawal and that PVD branch overlap is prevented by a contact-dependent self-avoidance, a mechanism that is also employed by sensory neurons in other organisms. With the goal of identifying genes that regulate dendritic morphogenesis, we used the mRNA tagging method to produce a gene expression profile of PVD during late larval development. This microarray experiment identified > 2,000 genes that are 1.5 X elevated relative to all larval cells. The enriched transcripts encode a wide range of proteins with potential roles in PVD function (e.g., DEG/ENaC and Trp channels) or development (e.g., UNC-5 and LIN-17/frizzled receptors). We used RNAi and genetic tests to screen 86 transcription factors from this list and identified eleven genes that specify PVD dendritic structure. These transcription factors appear to control discrete steps in PVD morphogenesis and may either promote or limit PVD branching at specific developmental stages. For example, time-lapse imaging revealed that the MEC-3 (LIM homeodomain) is required for branch initiation in early larval development whereas EGL-44 (TEAD domain) prevents ectopic PVD branching in the adult. A comparison of PVD-enriched transcripts to a microarray profile of mammalian nociceptors revealed homologous genes with potentially shared nociceptive functions. We conclude that PVD neurons display striking structural, functional and molecular similarities to nociceptive neurons from more complex organisms and can thus provide a useful model system in which to identify evolutionarily conserved determinants of nociceptor fate.
Keywords: Dendritic morphogenesis, nociceptor development, transcription factors
Introduction
Somatosensory neurons detect external stimuli such as touch and temperature. The nociceptor class of somatosensory neurons responds to noxious stimuli to trigger the sensation of pain and to evoke aversive behavior. Nociceptors typically display a complex, highly branched arbor of dendritic processes directly beneath the skin. This feature of nociceptor architecture has been widely observed in both vertebrate and invertebrate organisms and thus is likely to reflect fundamental, conserved mechanisms of development and function (Blackshaw et al., 1982; McGlone and Reilly, 2010; Tracey et al., 2003). The elaborate branching patterns that these sensory neurons display are constrained by a basic organizing principle in which dendrites with shared sensory function do not overlap and thus occupy separate spatial domains. The term “tiling” refers to the segregation of receptive fields for sensory neurons of common modality whereas “self-avoidance” describes the tendency of dendritic branches from a single neuron (i.e., sister dendrites) to avoid contact with each other. The mechanisms that govern sister dendrite avoidance are largely unknown but may depend on repulsive interactions mediated by specific cell surface components (Long et al., 2009; Matthews et al., 2007).
Different classes of sensory neurons are distinguished by the size and branching complexity of their dendritic arbors. Recent studies have shown that these differences in dendritic architecture are subject to transcriptional control (Crozatier and Vincent, 2008; Grueber et al., 2003a; Jinushi-Nakao et al., 2007; Moore et al., 2002; Parrish et al., 2006). For example, the branching complexity of Drosophila embryonic sensory neurons is defined by dose-dependent expression of the Cut homeodomain protein (Grueber et al., 2003a). In another case, the transcription factor Spineless may either promote or limit dendritic branching in different classes of sensory neurons. The context-dependent function of Spineless points to the likely role of combinatorial mechanisms that modify transcription factor output (Crews and Brenman, 2006; Kim et al., 2006). The general importance of transcriptional control in dendritic morphogenesis is underscored by a recent study in which a genome-wide RNAi screen identified > 75 transcription factors with roles in somatosensory neuron architecture (Parrish et al., 2006). It is noteworthy that homologs of many of these transcription factors are expressed in vertebrate neurons (Gao, 2007; Garel et al., 2002). Moreover, studies of mammalian neurons in culture have shown that different classes of neurons maintain their distinctive morphologies in vitro (Bartlett and Banker, 1984; Temple, 1989). Together these findings are indicative of evolutionarily conserved genetic programs that drive intrinsic pathways of neuronal differentiation (Scott and Luo, 2001). Although these genetic approaches have now shown that transcription factors control key features of dendritic morphogenesis, little is known of the downstream targets that define dendritic architecture (Parrish et al. 2007).
Studies in the nematode C. elegans have identified specific nociceptive neurons that mediate avoidance responses to mechanical force, temperature or noxious molecules(O’Hagan et al., 2005; Tobin et al., 2002; Wittenburg and Baumeister, 1999). Although this repertoire of sensory modalities parallels that of vertebrate nociceptors, C. elegans nociceptive neurons typically adopt a much simpler architecture with little or no dendritic branching (White et al., 1986). A striking exception to this difference was described in recent reports showing that the C. elegans PVD neuron displays a large and highly branched dendritic arbor directly beneath the hypodermal “skin” that envelops the worm (Halevi et al., 2002; Oren-Suissa et al., 2010; Tsalik et al., 2003)(Albeg et al., manuscript submitted). The occurrence of this elaborate subdermal array of PVD dendritic branches is also consistent with an earlier finding that PVD mediates an avoidance response to the application of harsh mechanical force to the external surface of the animal (Way and Chalfie, 1989b). Here we use live imaging studies with a bright PVD-expressed GFP reporter gene to provide a comprehensive description of PVD anatomy. We adopt a simple classification scheme for PVD dendritic branches (Oren-Suissa et al., 2010) and use time-lapse imaging to describe their emergent morphology and the developmental timing of each branching decision. We find that subsets of PVD branches fasciculate with an underlying network of peripheral nerve cords, which are likely sources of local guidance cues. Time-lapse imaging also revealed that PVD dendritic morphology is sculpted by striking examples of self-avoidance. To identify genes with potential roles in PVD differentiation or function, we utilized a cell specific microarray profiling strategy to catalog PVD genes (Roy et al., 2002; Von Stetina et al., 2007b). This approach revealed > 2,000 highly expressed genes encoding a wide array of proteins of different molecular classes. To illustrate the utility of this data set, we used RNAi knockdown or genetic mutants of 86 transcription factors from this list and identified eleven genes that control PVD dendritic architecture. Thus, this report firmly establishes the PVD neuron as a useful model for nociceptor development and provides a detailed anatomical and molecular foundation for future studies of nociceptor morphogenesis and function that exploit the simplicity and genetic utility of C. elegans biology.
RESULTS
PVD displays a net-like array of dendritic branches that envelops the animal
C. elegans contains two PVD neurons (PVDL and PVDR), one on each side of the adult animal. Both PVD neurons are generated post-embryonically during the L2 larval stage from an ectodermal precursor cell (V5). The PVD cell body is located in a posterior-lateral sensory organ (postdeirid) that also includes other V5-derived cells (Sulston and Horvitz, 1977). Reconstruction of the C. elegans nervous system from electron micrographs (EM) of serial sections suggested a relatively simple PVD architecture comprised of elongated, unbranched lateral processes projecting from anterior and posterior sides of each PVD soma and a single axon that grows downward to enter the ventral nerve cord (White et al., 1986). However, images of PVD obtained in the light microscope after immunostaining for a PVD-expressed membrane receptor (Halevi et al., 2002; Yassin et al., 2001; Yassin et al., 2002) or with a PVD-specific GFP reporter revealed a much more elaborate morphology with many additional dendritic branches(Tsalik and Hobert, 2003). Here we have used a bright PVD::GFP marker (F49H12.4::GFP) (Watson et al, 2008) (Fig 1) to reveal that PVD architecture is defined by a complex but well-ordered array of non-overlapping sister dendrites and that the creation of this structure involves a stereotypical series of branching decisions. The single PVD axon projects downward from the PVD cell body to join the ventral nerve cord. Dendritic branching, however, is much more elaborate. A 1O dendritic branch extends from the PVD cell soma along the anterior/posterior (A/P) axis at the location of the lateral nerve fascicle (Fig 1B,C). Orthogonal arrays of 2O, 3O, and 4O dendritic branches envelop the animal along the dorsal/ventral (D/V) and anterior/posterior (A/P) axes to produce a network of sensory processes. A single 20 branch can be seen as the “stem” for a “menorah-like” collection of 30 (“base”) and 40 (“candles”) branches (Fig 1B,C) (Oren-Suissa et al., 2010). A mature PVD (adult stage, see below) exhibits ~38 menorah-like structures (Supplemental Table 1). This web-like dendritic architecture is stereotypic of a wildtype PVD neuron.
Figure 1. PVD displays an elaborate dendritic arbor that envelops the animal in a net-like array.
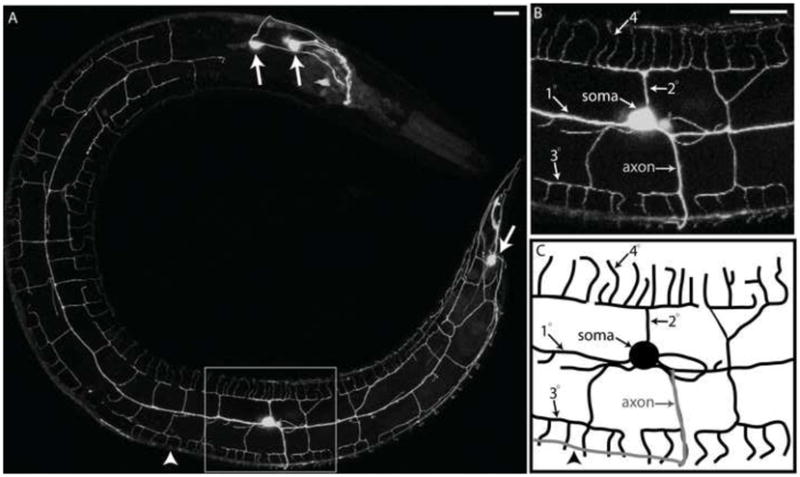
(A). Confocal image of an adult worm (anterior to left, ventral to bottom) showing the PVD::GFP marker (arrows denotes other neurons in head and tail that express GFP). Insets show more highly magnified image (B) and schematic tracing (C) of region surrounding PVD soma. Note dendritic branches (1O, 2O, 3O, and 4O) and single ventrally projecting axon (arrowhead denotes location of ventral nerve cord). Scale bar is 15 um.
FLP sensory neurons in the head adopt a dendritic morphology similar to PVD
We first sought to characterize the posterior and anterior reach of the PVD dendritic array. PVD processes extend posteriorly into the tail. In the head region, however, PVD terminates near the base of the pharynx (Fig 1A). We have established that this location corresponds to the posterior boundary of two bilaterally symmetric sensory neurons, FLP (L + R)(White et al., 1986). Interestingly, FLP neurons show a dendritic architecture that is strikingly similar to that of PVD with prominent menorah-like structures located along the sub-lateral nerve cords (Fig 2B). By examining animals co-expressing dsRed (FLP) and GFP (PVD) markers, we established that FLP and PVD dendritic branches rarely overlap (Fig 2C, inset). This “tiling” effect is characteristic of functionally related sensory neurons in other species and ensures efficient coverage of the receptive field (Parrish et al., 2007). The similar dendritic branching patterns and distinct receptive fields for each neuron (Fig 2D), are consistent with evidence that FLP and PVD function as nociceptive neurons (Albeg et al., manuscript submitted) (Chatzigeorgiou et al., 2010Chatzigeorgiou et al., in press)(Kaplan and Horvitz, 1993).
Figure 2. PVD dendrites tile with FLP dendritic branches in the anterior.
Lateral view of adult from left side (anterior to left, ventral to bottom). PVD::GFP (A) with FLP neuron marker, pmec-7::RFP, (B) and merged image (C) demonstrate that PVD dendritic branches (green) do not overlap with FLP (red) in the anterior (inset). Schematic showing that PVD and FLP envelop the animal with similar dendritic branching patterns (D). Scale bar is 15 um.
PVD dendrites fasciculate with pre-existing neuronal tracks
The predictable architecture of PVD processes is suggestive of distinct landmarks that determine the location of dendritic branches. To test this idea, we used a panneural reporter to mark neurons (CAN, ALA) in the lateral nerve cord. Dual color imaging of the dsRed panneural and PVD::GFP markers confirmed that PVD 10 branches are closely apposed to the lateral nerve cord as previously observed by EM reconstruction (Fig 3D, 3H, 3L) (White et al., 1986).
Figure 3. PVD branches fasciculate with motor neuron commissures and sub-lateral nerve cords.
Confocal images of PVD::GFP marker (A-D,P), panneural::dsRed (E-H,P) and merged reporters (I-L,O,P) show PVD dendritic branches, motor neuron commissures (arrow head) and sub-lateral nerve cords (arrow). PVD secondary branches lie in the same plane as motor neuron commissures as shown in rotated Z-stack from PVDR [(B,F,J)(rotated 55O on the X-axis and 45O on the y-axis)]. Rotated Z-stack of left side (ventral up) shows circumferential 4O branches [(C,G,K (rotated 80O on X-axis and 90O on Y-axis)]. PVD 3O branches fasciculate with dorsal and ventral sublateral nerve cords (D,H,L,O)(anterior left, ventral down). Schematic transverse section (M) shows PVD (L+R) (black) and fasciculation of some 2O branches (left) but not others (right) with motor neuron commissures (red). Lateral view of PVDR (N,O) showing 3O branches fasciculated with sub-lateral nerve chords (arrow). PVDR fasciculates with processes in the sub-lateral nerve cords (P, arrow) but does not contact the touch neuron, PVMR (P, arrowhead). Scale bars are 10um (A-C,E-G,I-K,O) or 15 um (D,H,L,P). See supplemental movie 1.
The panneural reporter also revealed that some PVD 2O branches fasciculate with motor neuron commissures (Fig 3A-L, supplemental movie 1). Motor neuron commissures extend around the circumference from the ventral to dorsal sides. These commissural processes are located directly beneath the hypodermis and course over the top of body muscle quadrants on dorsal and ventral sides (Fig 3E-H)(White et al., 1986). Confocal imaging indicates that PVD 2O branches are also located in this subdermal region and that a significant fraction of PVD 2O branches fasciculate with motor neuron commissures (Fig 3A–C, 3E–G, 3I–K) (Supplemental Table 1). The left and right sides contain unequal numbers of motor neuron commissures with 7 on the left and 27 on the right (White et al., 1986). This asymmetry is also reflected in the fraction of PVD secondary branches that fasciculates with motor neuron commissures which is greater on the right (43%) than on the left (14%) (Supplemental table 1). This result shows that the frequency of 2O branch fasciculation is correlated with the number of available motor neuron commissures on each side. In both cases, however, the majority of PVD secondary branches do not fasciculate with motor neuron commissures which suggests that 2O branch outgrowth may depend on separate mechanisms that either rely on the existing motor neuron commissure or extend independently. The occurrence of more PVD secondary branches on the right side vs the left is consistent with a model in which fasciculation with motor neuron commissures stabilizes 2O branches (Supplemental Table 1). We also note the both PVDL and PVDR show a greater number of dorsally projecting 2O branches than ventral 2O branches (Supplemental Table 1). PVD 3O branches are consistently positioned adjacent to sublateral nerve cords on both dorsal and ventral sides and fasciculation is extensive along the A/P axis in these locations (Fig 3D, 3H, 3L, 3M-O). The dorsal and ventral sub-lateral nerve cords are composed of processes contributed by a small number (2–5) of neurons. These co-linear nerve cords are discontinuous with specific processes exiting and new ones joining the sublaterals in the vulval region (White et al., 1986). In the posterior, PVD 3O branches on the dorsal side fasciculate with posterior-dorsal sub-lateral neurons, presumably ALN and SDQ (Fig 3P dorsal arrow). 3O branches on the ventral side fasciculate with the posterior-ventral sub-lateral nerve cord, likely comprised of PLN (Fig 3P ventral arrow), but do not fasciculate with the more dorsally located sub-lateral neuronal process of the touch neuron PLM (Fig 3P arrowhead). PVD 3O branches anterior to the vulva also fasciculate with specific sub-lateral processes. In this anterior region, 3O branches on the ventral side fasciculate with ventral sub-lateral nerve cord neurons, likely SIAV, SIBV, SMBC, SMDV, and PLN (Fig 3L). On the dorsal side, 3O branches fasciculate with the anterior-dorsal sub-lateral neuronal processes of SDQ, SIAD, SIBD, SMBD and SMDD. PVD 3O branches located on the dorsal side do not fasciculate with the dorsal sub-lateral process of the touch neuron ALM, which is located more ventrally than the dorsal sub-lateral nerve cord (Fig 3D, 3H, 3L, Supplemental figure 2). In summary, PVD 3O branches fasciculate with either the dorsal or ventral sublateral nerve cords but different individual neurons contribute to each of these process bundles in anterior vs posterior regions. Fasciculation of PVD 3O branches with the discontinuous sublateral nerve cords could be indicative of a local signal from surrounding tissues that guides independent outgrowth of both sublateral nerve processes and PVD 3O branches in this location. For example, the PVD 3O branches and sublateral nerve cords are positioned along the medial edges of the body muscle quadrants which are thus potential sources of a morphogenic cue. In an alternative model, PVD 3O branches could respond directly to fasciculation signals provided by both anterior and posterior sub-lateral processes.
PVD 4O branches are also located directly beneath the hypodermis but fasciculation with motor neuron commissures is rarely observed (Fig 3A-L) (data not shown). PVD 4O processes originate from 3O branches located at medial edge of the longitudinal bands of underlying body muscle cells to produce a series of finger-like projections that extend across the width of each body muscle quadrant. Although the significance of the close association of PVD dendritic branches with body muscle cells is unclear, recent studies showing that PVD could function as a proprioceptor suggest the intriguing possibility that this arrangement could provide a feedback mechanism of stretch-induced PVD activity that controls body posture (Albeg et al., manuscript submitted).
PVD dendritic morphology emerges from a series of orthogonal branching decisions
We used the PVD::GFP marker to visualize PVD dendritic branching during development in order to provide a detailed description of each step in PVD morphogenesis. The PVD::GFP reporter is initially detected in the mid-L2 larva immediately after the PVD cell soma appears in the postdeirid (Sulston and Horvitz, 1977). By the end of the L2 stage, the single PVD axon has projected to the ventral nerve cord (Fig 4A,B). During this period, the 1O dendritic branches emerge from the PVD cell body to join the lateral nerve cord, one extending toward the anterior and the other growing posteriorly (Fig 4A,B). The lateral and sub-lateral nerve cords with which adult PVD dendritic branches fasciculate are already in place before PVD dendritic outgrowth is initiated (Fig 4C-F). Beginning in the late L2 larva and continuing into the early L3, the 2O branches emerge at periodic intervals from both the dorsal and ventral sides of the 1O processes. In each case, 2O branches are perpendicular to the established 1O dendritic branch. 3O branches (“base of the menorah-like structure”) appear in the early L3 and extend along the sub-lateral nerve cords (Fig 4G, 4H). 3O branch outgrowth continues into the early L4 stage when 4O dendrites begin to emerge. The mature PVD dendritic arbor is established by the end of the L4 larval stage when it ultimately envelops the animal in a non-overlapping web of sensory processes (Fig 4I, 4J).
Figure 4. PVD dendritic architecture is defined by orthogonal branches.
Confocal images (left) and schematic tracings (right) of PVD in L2 larval stage (A,B), panneural (C,D) and merged panels (E,F) demonstrate that both motor neuron commissures (arrowheads) and sub-lateral nerve cords (arrow) are established before the majority of PVD dendritic branches emerge. PVD 1O branches arise in the L2 stage (B,D) followed by sequential orthogonal branching of 2O and 3O branches in L3 larval stage (G,M). A mature PVD neuron with 4O branches is largely completed by late L4 larval stage (I,J). Scale bar is 15 um.
Time-lapse imaging of PVD dendritic outgrowth reveals dynamic branching events
As described above, we deduced the order and timing of PVD dendritic branching by observing several different animals at successive stages during larval development. Our results are suggestive of an orderly progression of dendrite outgrowth along alternating orthogonal axes. Time-lapse imaging of single animals confirmed the successive outgrowth of dendritic branches but also revealed important details of how these branches are generated.
In the first instance, we noted highly dynamic outgrowth of 2O branches throughout the anterior/posterior length of the PVD 1O process. In L2 animals, 2O dendrites grow ventrally or dorsally toward sub-lateral nerve cords. Time-lapse imaging revealed that potential 2O branches are frequently initiated and then retracted. At periodic intervals, a subset of these projections appears to stabilize and reach the sub-lateral nerve cord whereas other nascent 2O branches in flanking regions are consistently withdrawn (Fig 5A, C, D, supplemental movie 2). This pattern of dynamic growth is replicated at successive stages with processes alternately extending and retracting until the final adult pattern is produced (Supplemental movie 3, 4, Supplemental figure 3).
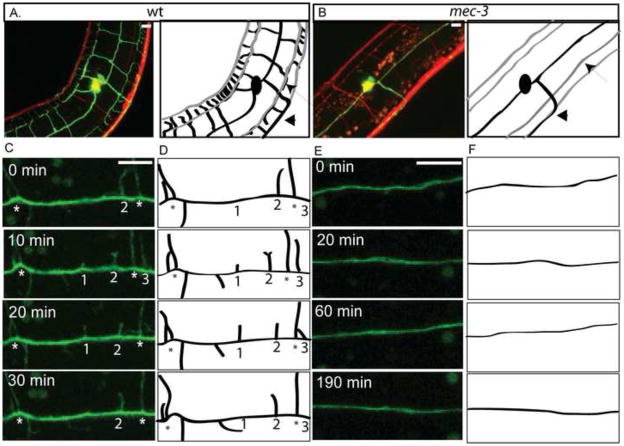
Figure 5. Dynamic initiation of PVD secondary branches is disrupted in mec-3 mutants.
Confocal images and schematic tracings of PVD::GFP (green/black) and panneural::dsRed (red/gray) (anterior left, ventral down) show that sub-lateral nerve cords (arrow) and PVD axon (arrowhead) are not altered in mec-3 mutants (B) in comparison to WT (A). Images (C) and schematics (D) from time-lapse confocal microscopy of wt L2 larval stage demonstrate dynamic PVD 2O branches (1–3) that initiate and retract in vicinity of established 2O branches (*) over 30 min period. Images (E) and schematics (F) of mec-3 mutants do not show PVD 2O branch initiation during 190 min of observation. Scale bar is 5 um. See supplemental movie 2 for wt and supplemental movie 7 for mec-3.
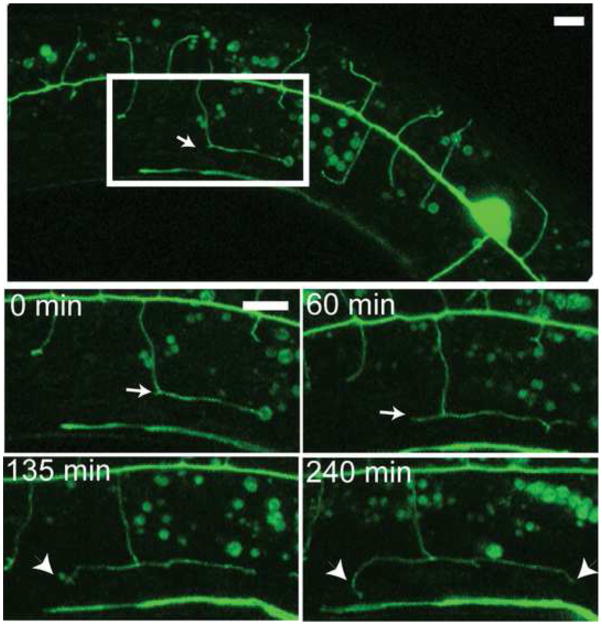
As 2O dendrites approach the sub-lateral nerve cord, they initiate 3O branch morphogenesis by turning 90O to project along the A/P axis. In each case, the initial 3O process growing in either the anterior or posterior direction is joined by a new process that sprouts at the point of turning (Fig 6, arrow) (Supplemental movie 3) to extend in the opposite direction along the A/P axis. The net result is that each 3O branch is composed of an anterior and posterior arm both emanating from a single 2O dendrite.
Figure 6. PVD dendritic branches turn 90O to establish orthogonal pattern.
Time-lapse confocal images of L3 larva depict PVDL dendritic outgrowth (anterior left, ventral down). (Top panel) PVD 2O branch makes a 90o turn (arrow) to fasciculate with sub-lateral nerve cord where it becomes a tertiary branch (inset, 0 min). A 3O branch with opposite polarity emerges from the point of turning (arrow) and grows toward the posterior (60 min). 4O branches are established by a similar mechanism (240 min) in which 4O branches at each end of the menorah-like structure (arrowheads) are generated by 90O turns. Additional, interstitial 4O branches emerge from the outer edge of the 3O branch. Scale bar is 5 um. See supplemental movie 3 for example.
The development of 4O branches proceeds via a similar mechanism with the tip of an outgrowing 3O dendrite eventually making an orthogonal turn (see below) to project along the D/V axis (Fig 6, arrowhead) (Supplemental movie 3). Additional 4O branches emerge at intervals along the length of each 3O branch (Fig 7, arrow). PVD 4O branches demonstrate dynamic growth, with branches initiating and retracting throughout the L4 larval stage (Fig 7, Supplemental movie 5). This pattern of rapid branch initiation and withdrawal is strikingly similar to that seen for 2O branch outgrowth during the L2 larval stage (Fig 5 and 7, asterisk). 4O branches terminate as they approach either the dorsal or ventral nerve cords to complete the architecture of the menorah-like structures rooted in the PVD 1O dendritic process (Fig 1).
Figure 7. PVD 4O branches exhibit dynamic growth.
Time-lapse confocal images and schematic tracings of L4 larval stage (anterior left, ventral down) illustrating dynamic outgrowth of 4O dendrites from established 3O branches. Nascent 4O branches (0 min) continue to grow throughout the L4 stage until they produce the mature menorah-like structures observed in the adult. Arrow denotes an example of a maturing 4O branch. Asterisk (*, 0 min) indicates a nascent 4O branch that ultimately retracts (60 min). Scale bar indicates 25 um. See supplemental movie 5.
Non-overlapping dendritic architecture of PVD is established by contact-dependent self-avoidance
In the mature PVD neuron, 3O branches from adjacent menorah-like structures point toward each other but do not touch (Fig 1). This feature of non-overlapping dendritic processes is a universal characteristic of sensory neurons (Corty et al., 2009; Parrish et al., 2007) and thus prompted us to consider a mechanism that could account for this outcome. Two possibilities seemed likely: either PVD 3O processes (1) stop outgrowth upon reaching a fixed, mature location or (2) continue growing until contact with the tip of a neighboring 3O branch induces withdrawal. We used time-lapse imaging to distinguish between these models. At the sub-lateral nerve cords, adjacent 3O branches initially grow toward each other along the A/P axis. In fact, the tips of adjacent 3O branches are frequently observed in closer proximity during larval development than in the adult (Fig 8, 0 min, supplemental movie 6). Upon contact, these sister 3O dendrites characteristically stop outgrowth and withdraw (Fig 8, 30 min) (Supplemental movie 6). Following retraction, 3O dendrites remain separate and the gap between them is preserved in the mature PVD architecture. Our results are thus consistent with the second mechanism in which the final length of each 3O branch is limited by contact with an adjacent sister dendrite. In fact, this phenomenon of self-avoidance was also observed for other transient dendritic extensions in which dendrites rapidly withdrew upon contact with each other or with previously established PVD branches (supplemental figure 3, supplemental movies 3 & 4). We therefore conclude that contact-dependent self-avoidance is likely to contribute to overall non-overlapping dendritic architecture of PVD.
Figure 8. PVD tertiary branches demonstrate contact-dependent self-avoidance.
Time-lapse confocal images of L3 larval stage (anterior left, ventral down) PVD 3O branches growing toward each other (0–27.5 min, arrow indicates gap between branches), achieving contact (30 min, arrowhead) and then retracting (32.5–35 min) to leave intervening space (arrow). This spacing is preserved in the adult PVD dendritic network. Scale bar is 5 um. See supplemental movie 6.
A gene expression profile of PVD nociceptive neurons
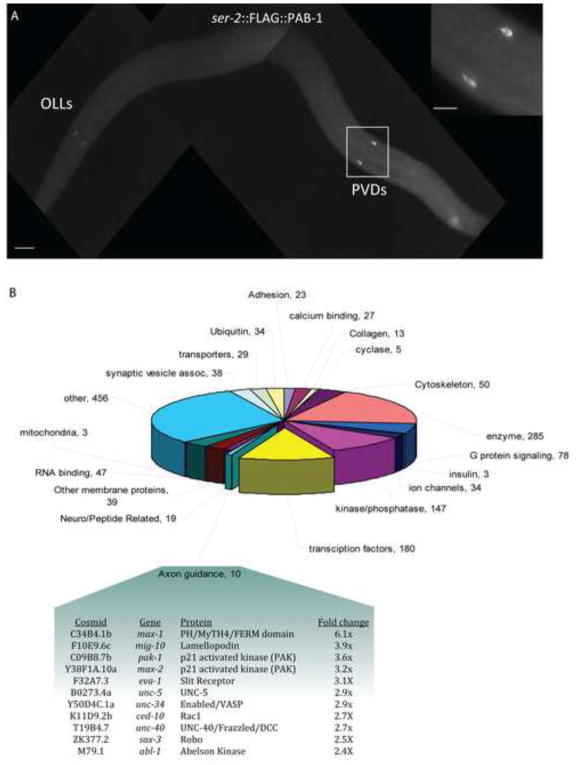
Having defined the detailed architecture and development of the PVD sensory network, we next generated a gene expression profile of PVD in order to identify transcripts with possible roles in PVD morphogenesis and function. For this purpose, we employed the mRNA tagging strategy in which an epitope-tagged poly-A-binding protein (FLAG::PAB-1) is used to co-immunoprecipiate cell-specific transcripts (Roy et al., 2002; Von Stetina et al., 2007b). Immunostaining with anti-FLAG confirmed specific expression in PVD and in OLL neurons (Fig 9A) as predicted for the ser2prom3 promoter used to construct our PVD mRNA tagging line (Tsalik and Hobert, 2003). Three independent samples were obtained from synchronized populations of early L4 larvae and applied to the Affymetric Gene Chip Array. PVD-enriched transcripts were detected by comparing these results to a reference microarray data set obtained from all larval cells of an age-matched wildtype sample also prepared in triplicate. Statistical analysis revealed 2,213 transcripts that are enriched (≥ 1.5 X) (Supplemental File 1) and 1,009 depleted (≤ 0.66 X) (Supplemental File 6) in the PVD/OLL profile vs all L3/L4 larval cells at a False Discovery Rate (FDR) ≤ 1 % (See Methods). We also identified a larger group of 4,977 “Expressed Genes” or “EGs” (see Methods) that are reliably detected by the PVD/OLL microarray profile but which may also be expressed at comparable levels in other cell types (Von Stetina et al., 2007b) (Supplemental File 2). Of genes previously described as expressed in PVD, 14/32 (~44%) are included in the enriched transcripts and 20/32 (~62%) are EGs. A smaller fraction of known OLL genes are detected with 8/43 (19%) enriched and 15/43 (35%) detected as EGs (Supplemental File 3). Previously noted PVD genes including the nicotinic acetylcholine receptor subunits (nAChRs) deg-3 (18x) and des-2 (17x), p21-activated kinase/PAK, max-2 (3.1X) and the homeodomain transcription factors, mec-3 (4.9x) and unc-86 (2.6x) are especially prominent (Lucanic et al., 2006; Way and Chalfie, 1989b; Yassin et al., 2001). As an additional test of the specificity of the microarray profile, we scored in vivo expression of 18 promoter::GFP fusions for representative genes from the list of enriched transcripts (Supplemental Table 2, Supplemental figure 4). Of the GFP reporters tested, 44% (8/18) are detected in PVD and 55% (10/18) are expressed in OLL with a total of 78% (14/18) expressed in either PVD or OLL (Supplemental Table 2). For example, the promoter-GFP fusion for EGL-3, a proprotein convertase that functions to process neuropeptide precursors (Kass et al., 2001), is highly expressed in PVD where it outlines the dendritic network with strong GFP staining (Supplemental figure 4). egl-3::GFP is also expressed in the OLL neurons in the head (data not shown). Expression of the egl-3 GFP reporter in the intestine and in many additional neurons underscores the sensitivity of the microarray analysis to differential expression. The broad range of enrichment (1.7x – 9.4X) of the PVD or OLL-expressed GFP reporters in this list provides a representative sample of transcripts with differing levels of expression. Together, these GFP reporter data validate the prediction that a majority of transcripts in this microarray data set are in fact expressed in either PVD or OLL neurons in vivo. Although a significant fraction of transcripts in this data set may be derived from OLL and not PVD, on the basis of these validation experiments we estimate that approximately half of the genes in our enriched and EG data sets are expressed in PVD. Therefore, this list provides a useful compendium of candidate genes to test for potential roles in PVD morphogenesis and function.
Figure 9. Expression profile reveals transcripts for PVD/OLL-enriched gene families.
(A) Anti-FLAG immunostaining of L4 larval stage animal shows specific ser-2 prom3B::FLAG::PAB-1 expression in PVD (L and R) (box, inset) and OLL (L and R). Scale bar is 25 um. (B) Genes (with Wormbase annotation) encoding transcripts with elevated expression (1.5x) in the PVD/OLL microarray data set organized according to protein families or functional groups. Numbers denote genes in each group. (Table Inset) Enrichment of axon guidance proteins, including multiple UNC-6/Netrin pathway transcripts, enriched in the PVD/OLL microarray.
A cutoff of ≥ 2.5 X enrichment provides a shorter list of 1,022 candidate PVD/OLL-specific genes (Supplemental File 1). This treatment results in significantly less overlap with enriched genes from profiles of embryonic muscle (Fox et al., 2007) or larval intestinal cells (Pauli et al., 2005) while retaining a robust representation of transcripts from a microarray data set obtained from the entire C. elegans nervous system (Watson et al., 2008) (Supplemental figure 8). This more stringent list offers the advantage of including fewer false positives but it also excludes transcripts with known PVD functions (see below).
Gene families represented in the enriched PVD/OLL profile
Transcripts for a wide range of protein families are included in the enriched PVD/OLL data set (Fig 9B) Supplemental File 1) and may be suggestive of specific functions. The coordinate occurrence of genes encoding known components of the UNC-6/Netrin axon guidance pathway is particularly striking, for example (Fig 9B). UNC-6/Netrin functions as an exogenous cue for cell migration, axon guidance, neuronal asymmetry and synaptogenesis (Adler et al., 2006; Colon-Ramos et al., 2007; Hedgecock et al., 1990; Poon et al., 2008; Wadsworth et al., 1996). The UNC-6/Netrin receptors, UNC-5 and UNC-40/DCC and several additional components with cytoplasmic roles in the UNC-6/Netrin signaling pathway are also highly enriched. For example, UNC-6/Netrin signaling is proposed to induce changes in cytoskeletal structure by promoting CED-10/Rac1-dependent localization of MIG-10/Lamellipodin in axon growth cones (Quinn et al., 2008); both ced-10 and mig-10 are elevated in the PVD/OLL profile. We have recently shown that mutants for several of these components including unc-6/netrin, unc-5, unc-40/DCC, unc-34/Ena, and mig-10/lamellopodin show similar defects in PVD morphology that are indicative of roles in a canonical UNC-6/Netrin signaling pathway that controls the elaboration of the PVD dendritic arbor (C.J. Smith, J.D. Watson, D.M. Miller, unpublished data). We also note enrichment of other classes of transcripts with potential roles in morphogenesis including Wnt receptors (lin-18, cam-1, lin-17, cfz-2) and receptors (eva-1, sax-3/Robo) for the axon guidance cue, SLT-1/Slit (Supplemental File 1)(Eisenmann, 2005; Fujisawa et al., 2007).
In addition to identifying genes that govern PVD morphogenesis, the microarray profile has also detected strong candidates for roles in PVD nociceptive function. For example, members of the DEG/ENaC family of cation channel subunit proteins have been implicated in pain sensation induced by either mechanical stimuli or low pH (O’Hagan et al., 2005; Wemmie et al., 2006). The C. elegans genome encodes 28 predicted DEG/ENaC proteins (Goodman and Schwarz, 2003). Two of these, MEC-4 and MEC-10, are expressed in the “light touch” mechanosensory neurons (AVM, PVM, PLML, PLMR, ALML, ALMR) where they evoke an aversive response to physical contact (O’Hagan et al., 2005; Suzuki et al., 2003). mec-10 expression in PVD has been previously reported (Huang and Chalfie, 1994) and the mec-10 transcript is enriched in the PVD/OLL profile.
Three additional DEG/ENaC genes (del-1, asic-1, F25D1.4), but not mec-4, are also elevated (Supplemental file 1). Recent work has shown that one of these DEG/ENaC channel proteins, F25D1.4/DEGT-1, is required along with MEC-10 to mediate a PVD-dependent response to strong mechanical stimuli. Co-localization of DEGT-1 and MEC-10 to punctate structures in PVD dendrites could be indicative of joint function in a mechanoreceptor complex (Chatzifgeorgiou et al, in press). The absence of other components of the touch neuron mechanosensory apparatus, including the stomatins mec-2 and unc-24 (Zhang et al., 2004), and accessory proteins, mec-14 and mec-18 (Gu et al., 1996), in the PVD/OLL-enriched profile (Supplemental File 1), suggests that the harsh touch response in PVD may depend on a largely separate set of mechanosensory factors that are not utilized in the touch neurons (Way and Chalfie, 1989b). For example, the stomatins, sto-4 and sto-5, are strongly elevated in the PVD/OLL profile (Supplemental File 1) but have not been previously implicated in touch neuron function. We also note enrichment of a C. elegans homolog (B0416.1) of a human protein, Tmc1, associated with hereditary deafness (Kurima et al., 2002). The role of Tmc1 in mechanosensitive cells in the human ear is poorly understood and thus studies of B0416.1 function in C. elegans could be useful for elucidating a cellular mechanism for Tmc1 in auditory function.
Other candidates for nociceptive function in PVD include members of the TRP family of cation channels. TRP proteins have been linked to a wide array of sensory modalities including mechanosensation, thermosensation, osmosensation and olfaction (Kahn-Kirby and Bargmann, 2006; Xiao and Xu, 2009). The C. elegans genome includes > 20 genes predicted to encode TRP-type ion channel proteins (Goodman and Schwarz, 2003; Kahn-Kirby and Bargmann, 2006; Xiao and Xu, 2009). Transcripts for four of these TRP genes, trp-2, gtl-1, gon-2, and pkd-2 are enriched in the PVD/OLL data set (Supplemental file 1). Each of these TRP channels has been shown to function in C. elegans in specific roles ranging from male mating behavior (pkd-2) (Barr et al., 2001) to adaptation to nicotine exposure (trp-2) (Feng et al., 2006). osm-9 which has been previously shown to function in the ciliated ASH neuron in chemotransduction and mechanosensation is not enriched (Colbert et al., 1997). It will be interesting to determine if these PVD-expressed TRP genes confer additional sensory modalities such as the response to temperature that is characteristic of sensory neurons in other species with similarly elaborate topical networks of dendritic processes (Tracey et al., 2003). This idea is consistent with the recent discovery that homologs of three of these C. elegans TRP channels, trp-2, gtl-1, and gon-2, are also prominently represented in a gene expression profile of mammalian sensory neurons that mediate pain pathways for mechanical and thermal stimuli (Abrahamsen et al., 2008). A significant fraction of additional classes of ion channels and receptor proteins from the mammalian nociceptor data set are also represented in the enriched PVD/OLL profile (Supplemental file 5).
Mechanical stimulation of PVD evokes avoidance behavior in which the animal moves away from the point of contact (Way and Chalfie, 1989b). PVD inputs to the motor circuit command interneurons PVC and AVA in the ventral nerve cord could mediate this response (White et al., 1986) although the role of these connections has not been directly tested. We note that several neuropeptide-encoding transcripts are enriched in PVD and could also modulate motor circuit behavior (Supplemental Table 3). Indeed, the preproconvertase, egl-3, shows strong expression in PVD and has been previously shown to mediate the response to body touch (Supplemental File 1, Supplemental figure 4) (Kass et al., 2001).
The MEC-3 homeodomain transcription factor is required for the initiation of PVD 2O branch outgrowth
Extensive genetic studies have documented the key roles of transcription factors in sensory neuron morphogenesis (Moore et al., 2002; Parrish et al., 2007; Parrish et al., 2006) (Grueber et al., 2003a; Kim et al., 2006). As a first step toward identifying specific transcription factors that control PVD morphogenesis, we compiled a list of 112 transcription factor-encoding genes that are ≥ 2X enriched in the PVD/OLL data set (Table 1) (Supplemental File 4). This list includes a diverse array of transcription factor families with the largest groups represented by nuclear hormone receptors and homedomain proteins. One of the most highly enriched (~5x) homeodomain proteins, MEC-3, is the only transcription factor in our data set that has been previously shown to affect PVD morphology. The complex PVD dendritic arbor is largely absent in mec-3 mutants imaged with the ser2prom3::GFP reporter gene (Tsalik et al., 2002). We have confirmed this result with our PVD::GFP marker which shows that the single PVD axon and 1O dendritic process appear normal in mec-3(e1338) but higher order branches (2O, 3O, 4O) are missing (Fig 5B). To explore the potential origin of the mec-3 PVD branching defect, we initially used the pan neural RFP marker to visualize other neuronal processes that PVD dendrites contact during outgrowth. These images showed that the lateral nerve cord (fasciculates with PVD 1O branch), motor neuron commissures (fasciculate with subset of PVD 2O branches) and dorsal and ventral sublateral nerve cords (fasciculate with PVD 3O branches) are intact in the mec-3(e1338) mutant (Fig 5A,B). These results are consistent with a cell-autonomous role for mec-3 in promoting PVD dendritic branching. As reported above, in wild type animals, branches are actively extended and retracted along the length of the 1O dendrite throughout the L2 and L3 larval stages. A subset of these nascent 2O branches is stabilized and contacts the sublateral nerve cords whereas others emerging from nearby regions during this time eventually collapse (Fig 5C, D, supplemental movie 2). We therefore hypothesized that mec-3 mutants are either (1) unable to stabilize 2O branches or (2) are defective in 2O branch initiation. We used time lapse imaging to distinguish between these alternative models. These experiments showed that the PVD 1O dendritic process is remarkably quiescent in mec-3 mutants with virtually no saltatory branching even during extended periods (e.g., 10 hours) of observation (Fig 5E, F, Supplemental movie 7). This result supports the hypothesis that MEC-3 is required for the initiation of PVD 2O dendrite outgrowth.
Table 1. Transcription factor families.
112 Transcription factors that are ≥ 2X enriched in the PVD/OLL profile versus all larval cells. Transcription factors are grouped according to shared homology of function. General transcription refers to factors with broad roles in transcription. Other includes sequences with weak homology to transcription factor motifs. (Established by BLAST searches at NCBI)
| Family/Domain | # in PVD data set |
|---|---|
| Nuclear Hormone receptors | 27 |
| Homeobox | 16 |
| General transcription | 12 |
| Basic region leucine zipper transcription factor | 10 |
| Zinc finger | 6 |
| SMAD | 4 |
| HMG | 4 |
| Forkhead | 4 |
| GATA-4/5/6 transcription factors | 4 |
| bHLH | 4 |
| ETS | 2 |
| Aryl-hydrocarbon receptor | 1 |
| Atrophin-like protein | 1 |
| CDK9 kinase-activating protein cyclin T | 1 |
| CREB/ATF | 1 |
| doublesex/MAB-3 domain | 1 |
| E2F-like protein | 1 |
| LIM domain | 1 |
| MADS box | 1 |
| Mlx interactors and related transcription factors | 1 |
| NGF1-A binding protein domain | 1 |
| Nuclear factor erythroid 2-realted factor 2 | 1 |
| PAX and HOX domains | 1 |
| TBX2 and related T-box transcription factors | 1 |
| TEAD family | 1 |
| Other | 5 |
A targeted RNAi screen of transcription factors genes reveals regulators of PVD dendritic morphogenesis
We used RNAi to test other ≥ 2X enriched transcription factors in our microarray data set for roles in PVD morphogenesis (Supplemental File 4). We selected late L4 larvae (F1 progeny of RNAi-treated parents) for screening with the idea that the appearance of the final structure could reveal transcription factors with roles at any stage of PVD morphogenesis. RNAi with empty vector served as a negative control and consistently resulted in a wildtype PVD dendritic architecture (Fig 10A,B). mec-3 RNAi-treated animals displayed fewer dendritic branches after RNAi treatment (Fig 10E, F). As expected, the Mec-3 RNAi phenotype was less severe than that of mec-3 mutants in which the 1O process shows virtually no branching activity (Fig 5B). Of the 86 transcription factors screened via RNAi, nine resulted in PVD defects. In most cases, corresponding genetic mutants were examined to confirm the RNAi defect. Two additional transcription factor determinants of PVD morphology (ahr-1 and egl-46) that did not produce RNAi phenotypes were detected in genetic mutants for a total of eleven transcription factor genes including mec-3 that regulate some aspect of PVD differentiation or morphogenesis (Table 2) (Supplemental file 4).
Figure 10. Transcription factors enriched in PVD expression profile control dendritic morphogenesis.

Confocal images (left) and schematics (right) of RNAi-treated animals expressing PVD::GFP marker (anterior left, ventral down). (A,B) Empty vector (EV)-treated negative control. Positive control, mec-3 RNAi (E-F), results in reduced 2O and 3O branches. lin-39 RNAi-treated animals (C-D) do not show PVD neurons (open circle indicates location of wt PVD cell body, arrow points to tail neuron that also expresses PVD::GFP marker). Mutants egl-46(gk692) shows fewer 2O branches (G,H) and ahr-1(ju145) displays increased numbers of 2O branches (I,J) (Supplemental File 4). Proposed temporal order of transcription factor function during PVD morphogenesis (M). (Table 2)
Table 2.
Transcription factors that regulate PVD morphology.
| Gene | Family/Domain | enrichment | Orthologues | RNAi | mutant | phenotype |
|---|---|---|---|---|---|---|
| lin-39 | HOX homeodomain | 4.4 | Scr (DM)/Hox-B5 (HS) | + | ND | no PVD |
| zag-1 | homeodomain | 3.4 | Afh-1 (DM) | + | + | posterior defects, PVD duplicated on right side |
| unc-86 | POU homeodomain | 2.6 | Acj6 (DM)/ Brn3a (MM) | + | + | unbranched 10 dendrite |
| dpl-1 | E2F-like protein | 2.8 | Dp (DM) | + | ND | misguided 10 branch |
| mec-3 | LIM homeodomain | 4.9 | Lim-1 (DM)/Lhx5 (HS) | + | + | unbranched 10 dendrite |
| unc-30 | homeodomain | 2.1 | Pitx-1 (DM)/Pit1 (HS) | + | + | fewer 20 branches |
| ahr-1 | aryl-hydrocarbon receptor | 3 | Spineless/SS (DM) | − | + | increased number of 20 branches |
| egl-46 | Zinc Finger | 3.6 | nerfin 2A (DM) | − | + | fewer 20 branches |
| atf-2 | bZip Superfamily | 2.3 | + | ND | overlapping 30 branches | |
| thoc-2 | General Transcription | 2.0 | Tho2 (DM) | + | ND | overlapping 30 branches |
| egl-44 | TEAD | 3.9 | Sd(DM)/TEF-3 (MM) | + | + | ectopic branching |
DM=D. melanogaster, MM= M. musculus, HS = H. sapiens
lin-39, which encodes a conserved member of the HOX family of homedomain proteins, appears to have an early role as PVD is either not detected or shows an unbranched architecture in lin-39 RNAi-treated animals (Fig 10C). The Lin-39 PVD-defective phenotype is consistent with the established role of LIN-39 in the specification of cell fates in the mid body region of C. elegans (Clark et al., 1993). RNAi knock down of unc-86 (POU homeodomain) phenocopies the mec-3 mutant with an unbranched 1O dendrite. The unc-86 mutant, however, is more severely affected; the PVD soma and axonal projection to the ventral cord are normal but the 1O dendritic process fails to emerge (data not shown). This result indicates that UNC-86 is required for initiating dendritic outgrowth and is consistent with an earlier report that unc-86 activates mec-3 expression in PVD (Way and Chalfie, 1989) (Table 2).
The ZAG-1 transcription factor (homeodomain) (Clark and Chiu, 2003) displays a unique mutant phenotype in which two apparent PVDR neurons are consistently observed on the right side of the animal whereas PVDL on the left side is not duplicated. The striking asymmetry of the Zag-1 defect offers an explanation for a previous report of incompletely penetrant duplication of PVD in zag-1 mutants (Wacker et al. 2003). Other transcription factors detected in our screen appear to affect successive steps in the placement or elaboration of dendritic branches (Fig 10M). To determine the developmental role of these transcription factors, we noted the larval stage at which the RNAi phenotype initially appeared. Misplacement of the 1O branch in dpl-1 RNAi-treated animals is observed during the L2 stage, which suggests that DPL-1 (E2F-like protein) functions early in PVD morphogenesis to regulate targets that guide initial outgrowth along the lateral nerve cord. Other transcription factors appear to define the overall number of 2O branches with unc-30 (Pitx homeodomain)(Jin et al., 1994) and egl-46 (Zn finger/Nerfin) (Wu et al., 2001) mutants showing fewer 2O branches (Fig 10G,H) (Supplemental figure 5) and ahr-1 animals displaying an increased number of 2O dendrites (Fig 10I, J) (Supplemental figure 5). The role of ahr-1 (aryl-hydrocarbon receptor) in this case is intriguing because its Drosophila homolog, Spineless (SS), also controls the complexity of sensory neuron dendritic branching (Kim et al., 2006). The importance of transcription to later stages of PVD morphogenesis is revealed by 3O branch defects in atf-2 (bZip superfamily) mutant and in thoc-2 (general transcription) deficient animals. In both cases, 3O branches are elongated and frequently overlap (Fig 10K, L). The apparent failure of the contact-dependent self-avoidance mechanism indicates that atf-2 and thoc-2 may control downstream genes that mediate this characteristic feature of 3O branch morphogenesis. thoc-2 RNAi-treated animals also frequently show other PVD defects including misplaced 1O processes and a general failure to elaborate dendritic branches anterior to the PVD soma. Lastly, one of the transcription factor mutants detected in our screen, egl-44 (TEAD domain) (Wu et al., 2001) did not demonstrate any obvious PVD dendritic defects during early development but showed extensive ectopic branching in the adult stage (Supplemental figure 6). Thus, egl-44 must normally act to limit excessive branching at later stages of development. We note that many of the ectopic branches observed in egl-44 mutants appear to overlap which could mean that egl-44 also regulates target genes that function in the self-avoidance mechanism that maintains the discrete sensory field for each dendritic branch in the PVD arbor. In the future, it will be important to confirm the cell autonomous roles of these transcription factors in PVD morphogenesis. For example, UNC-30 is highly expressed in GABAergic motor neurons where it is required for outgrowth of commissures to the dorsal side of the animal (Jin et al., 1994; McIntire et al., 1992). Thus, in this case, the loss of PVD 2O branches in unc-30 mutants could derive from either a necessary function in PVD or from indirect elimination of motor neuron commissures that normally guide 2O branch outgrowth.
Other genes with roles in PVD morphogenesis
Having identified a specific subset of transcription factors in the PVD/OLL data set with roles in dendritic morphogenesis, we next tested selected members of other protein classes for similar functions. A total of 15 additional genes with roles in PVD morphogenesis were detected by either RNAi or with genetic mutants (Supplemental figure 7, Supplemental Table 4). An important role for GTPase dependent signaling pathways is suggested by the finding that mutations in three different guanine nucleotide exchange factors (GEFs) (CllD9.1, uig-1 and sos-1) result in PVD defects. These results suggest that a systematic mutant or RNAi analysis of genes in the microarray profile should reveal additional components of pathways that direct PVD dendritic morphogenesis.
Discussion
Nociceptive function depends on elaborate networks of dendritic processes adjacent to the skin (McGlone and Spence, 2010). The complexity of this architecture and the general inaccessibility of sensory neurons to real time studies of morphogenesis have hindered the elucidation of cell biological mechanisms that govern dendritic branching. Here we describe a model nociceptor, the PVD neuron in C. elegans, that displays a complex but highly ordered sensory arbor and show that the generation of this network can be readily studied by dynamic imaging methods. This approach has revealed the step-wise emergence of PVD branches during development and identified external landmarks that correspond to key branch points. Our observations suggest that the final pattern of PVD branches also depends on an intrinsic mechanism of error correction in which sister dendrites avoid contact with each other. To identify genes with potential roles in dendritic morphogenesis, we generated a cell-specific expression profile that includes >2,000 PVD-enriched transcripts. Selected genes in this list were ablated by RNAi or in mutants to identify eleven transcription factor proteins and representative members of other functional protein classes with a range of specific roles in PVD morphogenesis.
The PVD dendritic arbor is generated by a series of defined branching decisions
Our observations show that the PVD dendritic arbor arises from a series of ordered branching decisions that correspond to specific stages of larval development. Dendritic outgrowth is initiated in late L2 larvae and continues throughout the L3 and L4 stages until the mature PVD morphology is achieved in the adult. Because PVD and its dendritic arbor are located near the surface, all of these branching events are readily observed in a live animal and can be catalogued by time-lapse imaging. A comparison of PVD morphology to the structure of the C. elegans nervous system showed that specific PVD dendrites are closely apposed to external nerve fascicles. These interactions are extensive and involve the 1O PVD processes which extend along the lateral nerve cord, a subset of 2O dendrites which fasciculate with motor neuron commissures and 3O branches that are in contact with sublateral nerves throughout their length. An interesting question to address in the future is whether these nerve cords serve as landmarks for PVD branching and outgrowth by providing local guidance cues.
The cell biological mechanisms that drive PVD branching are unknown but could involve different components for separate branching events. This idea derives from the distinct spatial environments occupied by each of the branches and from the characteristic manner in which each arises. 1O branches grow out from opposite sides of the PVD soma and project either anteriorly or posteriorly along the lateral nerve cord. In contrast, 2O branches emerge at interstitial locations along the length of the 1O process and grow in either the dorsal or ventral directions. The orthogonal switch in the geometry of these branching patterns is suggestive of a temporal change in either the intrinsic polarity of dendritic outgrowth and/or the responsiveness to external cues. The potential existence of diverse dendritic branching mechanisms is also suggested by the observation from time lapse imaging of two distinct modes of 3O branch outgrowth. A 3O process is initially generated as a 2O branch that turns at the sublateral nerve to extend in either the anterior or posterior direction. An additional branch then sprouts from the point of turning to grow in the opposite A/P direction and thus form the other arm of each 3O dendrite (Fig 6). Although it seems likely that the turning and branching events may be triggered by a common signal, perhaps provided by the sublateral nerve cord, these cell biological responses are distinct and thus could employ subsets of unique components.
The overall shape and extent of the PVD sensory arbor may also depend on negative cues that constrain dendritic growth. We note, for example, that PVD processes do not extend into the head region occupied by the FLP neurons. FLP and PVD display dendritic arbors with similar branching patterns (Fig 2) and both mediate nociceptive responses to mechanical force (Kaplan and Horvitz, 1993; Way and Chalfie, 1989b) (Chatzifgeorgiou et al, in press). The “tiling” pattern of dendritic arborization that PVD and FLP display in which sensory neurons of a given functional class occupy discrete topical domains is widely observed and may depend on mutual inhibition by outgrowing dendrites from adjacent neurons (Corty et al., 2009). In Drosophila, the Ig protein, Turtle mediates homophilic interactions that maintain separate sensory fields for neighboring R7 photoreceptors (Ferguson et al., 2009). An unknown negative cue also mediates this behavior in C. elegans and Drosophila in a shared signaling pathway involving the conserved components Furry/sax-1 and Tricorner/sax-2 (Emoto et al., 2004; Emoto et al., 2006; Gallegos and Bargmann, 2004). We note that both Furry/sax-1 and Tricorner/sax-2 are enriched in the PVD microarray data set (Supplemental File 1) and are thus candidates for regulators of PVD dendritic outgrowth.
Additional evidence of negative regulation of dendritic outgrowth derived from our time lapse imaging results showing that PVD dendritic branches are actively repelled by contact with each other (Fig 8, Supplemental figure 3, Supplemental movies 3, 4 & 6). This phenomenon of self-avoidance is commonly employed by sensory neurons and serves to prevent overlapping coverage of a given receptive field by sister dendrites from the same neuron (Grueber et al., 2003b; Sugimura et al., 2004). Studies in Drosophila have shown that the cell surface Ig superfamily proteins, Dscam and Turtle, mediate dendritic self-avoidance (Matthews et al., 2007) (Long et al., 2009). Neither of these proteins is encoded by the C. elegans genome, however, and thus alternative repulsive cues are likely utilized in PVD. These effectors of PVD dendritic self-avoidance could be potentially detected by genetic or RNAi ablation of candidate cell surface receptors (Fig 9) that are enriched in the PVD microarray data set (Supplemental File 1). For example, a recent report (Oren-Suissa et al., 2010) has shown that the enriched (5X) cell surface fusogen, EFF-1, is required for a global mechanism of error correction that limits the outgrowth and stability of ectopic PVD dendritic branches. The transcription factors, thoc-2 and aft-2, which appear to prevent overgrowth of adjacent 3O branches (Fig 10, Table 2) are strong candidates for regulators of additional downstream components that mediate dendritic self-avoidance.
Having considered patterning mechanisms that involve extracellular cues or contact-dependent interactions among sister dendrites, we also suggest the possibility of internal cytoplasmic mechanisms for limiting dendritic outgrowth. Over half of the 2O branches and most of the 4O branches do not fasciculate with external nerve cords and thus are unlikely to follow specific paths defined by previously established external structures (Supplemental Table 1). The regular spacing of 2O and 4O branches (Supplemental figure 1) could be indicative, however, of negative signals from established PVD processes that prevent the formation of additional stable branches in flanking regions. Mutual contact-dependent withdrawal of adjacent branches that deviate from parallel outgrowth could also contribute to this final pattern as seen for the comb cell in the leech (Baker and Macagno, 2007).
Transcription factors regulate specific steps in PVD dendritic morphogenesis
RNAi and genetic ablation of transcription factors identified in the PVD microarray profile detected eleven genes with roles in PVD dendritic morphogenesis (Table 2). For two of these transcription factors, UNC-86 (POU homeodomain) and MEC-3 (LIM homeodomain), our results confirm earlier findings of PVD expression and necessary roles in PVD differentiation and function (Tsalik et al., 2003; Way and Chalfie, 1989a). The PVD phenotypes of unc-86 and mec-3 mutants are consistent with a model in which unc-86 acts first to promote 1O branch outgrowth followed by mec-3 which then initiates 2O branching. The apparently sequential roles of unc-86 and mec-3 in PVD morphogenesis parallel their functions in the differentiation of the mechanosensory or touch neurons. In both cell types, unc-86 is required for mec-3 expression (Way and Chalfie, 1989b). In the touch neurons, UNC-86 also functions with MEC-3 in a heterodimeric complex to co-regulate shared targets genes(Duggan et al., 1998; Xue et al., 1993). The roles of unc-86 and mec-3 in PVD vs the touch neurons are also likely to differ. Only four of eleven canonical “mec” genes that mec-3 regulates in the touch neurons to mediate mechanosensitive function (Way and Chalfie, 1989b; Zhang et al., 2002), mec-3, mec-10, mec-12 and mec-17, are detected in the enriched PVD/OLL data set (Supplemental file 1) (Supplemental Table 5). Moreover, mec-3 promotes dendritic branching in PVD (Tsalik et al., 2003) but clearly does not activate a comparable pathway in the touch neurons which normally adopt a simple, bipolar morphology (White et al., 1986). This difference in the morphogenic roles of mec-3 in distinct sets of C. elegans sensory neurons is also observed for the Spineless transcription factor in Drosophila which may either promote or inhibit dendritic branching in separate sensory neuron types (Kim et al., 2006). These disparate outcomes are proposed to result from combinatorial interactions with other classes of transcription factors (Crews and Brenman, 2006). The key role of transcriptional control of dendritic branching is strikingly evident from the results of a genome wide RNAi screen in Drosophila that uncovered > 75 transcription factors that govern sensory neuron morphogenesis (Parrish et al., 2009). Our more limited RNAi screen revealed eleven transcription factors with morphogenic roles in PVD; experiments with mutants which typically display more penetrant phenotypes than RNAi knockdown are likely to detect additional transcription factors with necessary roles in PVD differentiation. The transcription factors that we have uncovered appear to act at different stages of PVD morphogenesis. This finding suggests that the generation of the PVD dendritic array is tightly regulated by an intricate genetic program and thus that the discovery of transcription factor targets in these pathways would provide a critical link between the regulation of gene expression and cell biological processes that control dendritic morphology. For example, the mechanisms that drive dendritic branch initiation are poorly understood. Our studies indicate that the MEC-3 transcription factor is required for the initiation of PVD branching and thus is likely to control target genes with direct roles in this morphogenic event. The mRNA tagging method is well-suited to this problem and could be used to compare PVD microarray profiles of mutant (e.g., mec-3) vs wildtype to uncover these key downstream effector genes (Von Stetina et al., 2007b).
In addition to sharing morphological similarities with nociceptors in other organisms, PVD may also utilize common sets of genes for differentiation and function. At least two of the transcription factors uncovered in our RNAi screen for PVD morphogenic defects, unc-86/Brn3a/acj6 and ahr-1/Spineless, are also known to govern sensory neuron dendritic morphogenesis in other species (Supplemental figure 5) (Ichikawa et al., 2002; Kim et al., 2006; Komiyama and Luo, 2007). In addition, a significant fraction of ion channel components known to be expressed in mammalian nociceptors are also detected in the PVD microarray data set (Abrahamsen et al., 2008). These shared proteins include members of the TRP family of ion channels with established roles in mechanosensation and nociception (Supplemental file 5). The striking contact-dependent mechanisms of error correction that we have documented for the PVD neuron in C. elegans are likely to be universally employed by sensory neurons in other species that characteristically establish non-overlapping dendritic fields (Grueber et al., 2003b; Sugimura et al., 2003). Taken together, these results indicate that the C. elegans PVD neuron affords an attractive model for defining fundamental mechanisms of nociceptor differentiation and function. This work provides a detailed structural, developmental, and molecular foundation for these studies.
Methods
Nematode Strains and Genetics
The wild-type C. elegans Bristol strain N2 was used for all experiments and cultured as previously described (Brenner, 1974). Also used in this study were mutants: CZ2485 ahr-1 (ju145), FX00321 ceh-38 (tm321), FX00237 ceh-48 (tm237), MT2246 egl-43 (n1079), MT2247 egl-44 (n1080), MT2243 egl-46 (n1076), GR1373 eri-1 (mg366), VC349 lim-9 (gk210), CB1338 mec-3 (e1338), CB845 unc-30 (e191), CB1416 unc-86 (e1416), RB774 zfp-1 (ok554), VH4 zag-1 (rh315); rhIs4. The following transgenic strains were used: NC1733 (otIs173, F25B3.3::dsred; wdIs52, F49H12.4::gfp + unc-119), NC1686 (wdIs51, F49H12.4::GFP + unc119), NC1687 (wdIs52, F49H12.4::gfp + unc119), NC1841 (wdIs52, F49H12.4::gfp; rwIs1, pmec-7::RFP), NC1908 (wdEx240, myo-3::dsRed; wdIs52, F49H12.4::gfp)
GFP reporter strains for transcripts enriched in the PVD/OLL data set were obtained from the British Columbia C. elegans Gene Expression Consortium and are listed in Supplemental Table 2. Some of the nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). All studies in this work used C. elegans hermaphrodites.
Confocal Microscopy
Nematodes were immobilized with 15 mM levamisole on a 2% agarose pad in M9 buffer. Images were obtained in a Leica TCS SP5 confocal microscope. Z-stacks were collected with either 40× (1 um/step) or 63× (0.75 um/step) objectives; single plane projections were generated with Leica Application Suite Advanced Fluorescence software.
Time-Lapse Imaging
Nematodes were immobilized with a 15 mM levamisole/0.05% tricaine mix on a 2% agarose pad, all of which was diluted with M9 buffer. Slides were sealed with 1:1 vasoline/paraplast tissue embedding medium (Gabel et al., 2008). For each time point, the 40× or 63× objective was used to collect a Z-stack (0.75 um/step) spanning the focal depth of the PVD neuron and its dendritic branches. Dendritic branch outgrowth at each time point was evaluated from a Z-projection. Larval stages were identified from morphological features: L2 (postdeirid) (Sulston and Horvitz, 1977); L3, L4, and young adult (vulval development) (Inoue et al., 2005). At least three independent movies verified each example of dynamic dendritic growth described in this report.
PVD expression profiling
The 1.6 kb ser-2prom3B promoter fragment was amplified from genomic DNA using the primers: ser-2prom3-sal-1 (5′-CGAAACGCTGTCGACTTCAACTGTAGGCG-3′) and ser-2prom3-p2b (5′-GGTACCGTTGTGATGTCACAAAAATATGCC-3′) adding a KpnI site to the 3′ end (Tsalik et al., 2003). The resultant PCR product was cloned into pCR2.1-TOPO to generate the plasmid pWCS5 (Invitrogen). pWCS5 and the 3X::FLAG::PAB-1 plasmid pSV15 were digested with BamHI and KpnI and ligated to generate the ser-2prom3B::3XFLAG::PAB-1 mRNA-tagging construct pWCS8 (Von Stetina et al., 2007a). The transgenic line, NC221, was obtained by co-bombardment of pWCS8 with the co-selectable marker unc-119(+) minigene plasmid (MM051) (Maduro and Pilgrim, 1995). PVD and OLL expression of 3X FLAG was confirmed by immunostaining. PVD/OLL transcripts were obtained from synchronized early L4 larvae by the mRNA tagging strategy (Von Stetina et al., 2007b). A reference RNA sample was obtained from total L3-L4 larval cells. Three independent biological replicates were generated for both the PVD/OLL and reference samples. RNA (25ng) amplified by the WT-Pico method (Watson et al., 2008), labeled and hybridized to the Affymetrix Gene Chip array. Data sets were normalized by RMA and transcripts showing relative PVD enrichment (≥ 1.5 X) vs the reference sample were identified by SAM analysis (False Discovery Rate, FDR < 1%) as described (Fox et al., 2005). Expressed Genes (EGs) were estimated as previously described (Von Stetina et al., 2007b).
RNAi screen for PVD morphological defects
eri-1 (mg366); wdIs52 animals were used for RNAi transcription factor screening. Bacterial clones from an RNAi library (Kamath and Ahringer, 2003) were grown overnight at 37C. 200 ul of overnight culture was seeded to β-lactose NGM-lite plates (Golden and O’Connell, 2007). The plates were incubated at room temperature for 3 days for induction of dsRNA expression. L4 larval stage hermaphrodites were picked to the RNAi plates and grown at 21C until the F1 progeny were at the L4 larval stage. F1 progeny at the L4 larval stage were mounted on slides as above and viewed in a Zeiss Axiovert 200M microscope. ≥20 hermaphrodites were screened for each RNAi clone. A clone that disrupted PVD morphology in > 1 animal in each of three independent screenings was considered a positive hit. RNAi clones with effects on PVD morphology were confirmed by DNA sequencing. Mutants for specific transcription factors were crossed into the PVD::GFP strain NC1687 and examined as adults for PVD defects (Table 2). Mutant alleles of dpl-1 (sterile), aft-2, and thoc-2 (sterile/lethal) were not examined.
Supplementary Material
Acknowledgments
We thank Oliver Hobert for helpful advice on the manuscript; Sylvia Lee for the mec-7::RFP transgenic line; Chieh Chang and Sarah Kucenas for help with time-lapse imaging; Braden Boone, John Mote and Shawn Levy of the Vanderbilt Functional Genomics Shared Resource (FGSR) for help with microarray experiments; Shenglong Wang, Nurith Kurn and Joe Don Heath of NuGEN Technologies for help with RNA amplification; members of the Miller lab for technical advice and for comments on the manuscript. Some of the strains used in this work were provided by the C. elegans Genetic Center which is supported by NIH NCRR. This work was supported by U. S.-Israel Binational Science Foundation Grant 2005036 (MT), NIH R01 NS26115 (DMM), R21 NS06882 (DMM), NIH F31 NS49743 (JDW) and by NIH grants to Vanderbilt University: P30 CA68485, P60 DK20593, P30 DK58404, HD15052, P30 EY08126 and PO1 HL6744.
Footnotes
ACCESSION NUMBERS
All microarray data have been submitted to GEO and can be reviewed with this link: http://www.ncbi.nlm.nih.gov.proxy.library.vanderbilt.edu/geo/query/acc.cgi?token=nrafpuyuogwgqve&acc=GSE21162
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP, Nassar MA, Dickenson AH, Wood JN. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 2008;321:702–705. doi: 10.1126/science.1156916. [DOI] [PubMed] [Google Scholar]
- Adler CE, Fetter RD, Bargmann CI. UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat Neurosci. 2006;9:511–518. doi: 10.1038/nn1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MW, Macagno ER. In vivo imaging of growth cone and filopodial dynamics: evidence for contact-mediated retraction of filopodia leading to the tiling of sibling processes. J Comp Neurol. 2007;500:850–862. doi: 10.1002/cne.21228. [DOI] [PubMed] [Google Scholar]
- Barr MM, DeModena J, Braun D, Nguyen CQ, Hall DH, Sternberg PW. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol. 2001;11:1341–1346. doi: 10.1016/s0960-9822(01)00423-7. [DOI] [PubMed] [Google Scholar]
- Bartlett WP, Banker GA. An electron microscopic study of the development of axons and dendrites by hippocampal neurons in culture. I. Cells which develop without intercellular contacts. J Neurosci. 1984;4:1944–1953. doi: 10.1523/JNEUROSCI.04-08-01944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw SE, Nicholls JG, Parnas I. Physiological responses, receptive fields and terminal arborizations of nociceptive cells in the leech. J Physiol. 1982;326:251–260. doi: 10.1113/jphysiol.1982.sp014189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Chisholm AD, Horvitz HR. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell. 1993;74:43–55. doi: 10.1016/0092-8674(93)90293-y. [DOI] [PubMed] [Google Scholar]
- Clark SG, Chiu C. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development. 2003;130:3781–3794. doi: 10.1242/dev.00571. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Ramos DA, Margeta MA, Shen K. Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science. 2007;318:103–106. doi: 10.1126/science.1143762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development. 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews ST, Brenman JE. Spineless provides a little backbone for dendritic morphogenesis. Genes Dev. 2006;20:2773–2778. doi: 10.1101/gad.1487706. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Vincent A. Control of multidendritic neuron differentiation in Drosophila: the role of Collier. Dev Biol. 2008;315:232–242. doi: 10.1016/j.ydbio.2007.12.030. [DOI] [PubMed] [Google Scholar]
- Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM. Wnt signaling. WormBook : the online review of C elegans biology. 2005:1–17. doi: 10.1895/wormbook.1.7.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Feng Z, Li W, Ward A, Piggott BJ, Larkspur ER, Sternberg PW, Xu XZ. A C. elegans model of nicotine-dependent behavior: regulation by TRP-family channels. Cell. 2006;127:621–633. doi: 10.1016/j.cell.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson K, Long H, Cameron S, Chang WT, Rao Y. The conserved Ig superfamily member Turtle mediates axonal tiling in Drosophila. J Neurosci. 2009;29:14151–14159. doi: 10.1523/JNEUROSCI.2497-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R, Von Stetina S, Barlow S, Shaffer C, Olszewski K, Moore J, Dupuy D, Vidal M, Miller D. A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics. 2005;6:42. doi: 10.1186/1471-2164-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R, Watson J, Stetina SV, Mcdermott J, Brodigan T, Fukushige T, Krause M, Miller D. The embryonic muscle transcriptome of Caenorhabditis elegans. Genome Biology. 2007;8:R188. doi: 10.1186/gb-2007-8-9-r188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa K, Wrana JL, Culotti JG. The slit receptor EVA-1 coactivates a SAX-3/Robo mediated guidance signal in C. elegans. Science. 2007;317:1934–1938. doi: 10.1126/science.1144874. [DOI] [PubMed] [Google Scholar]
- Gabel CV, Antonie F, Chuang CF, Samuel AD, Chang C. Distinct cellular and molecular mechanisms mediate initial axon development and adult-stage axon regeneration in C. elegans. Development. 2008;135:1129–1136. doi: 10.1242/dev.013995. [DOI] [PubMed] [Google Scholar]
- Gallegos ME, Bargmann CI. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 2004;44:239–249. doi: 10.1016/j.neuron.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Gao FB. Molecular and cellular mechanisms of dendritic morphogenesis. Curr Opin Neurobiol. 2007;17:525–532. doi: 10.1016/j.conb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Yun K, Grosschedl R, Rubenstein JL. The early topography of thalamocortical projections is shifted in Ebf1 and Dlx1/2 mutant mice. Development. 2002;129:5621–5634. doi: 10.1242/dev.00166. [DOI] [PubMed] [Google Scholar]
- Golden A, O’Connell KF. Silence is golden: combining RNAi and live cell imaging to study cell cycle regulatory genes during Caenorhabditis elegans development. Methods. 2007;41:190–197. doi: 10.1016/j.ymeth.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Goodman MB, Schwarz EM. Transducing touch in Caenorhabditis elegans. Annu Rev Physiol. 2003;65:429–452. doi: 10.1146/annurev.physiol.65.092101.142659. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003a;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of Distinct Classes of Drosophila Sensory Neurons Show Different Capacities for Homotypic Repulsion. Current Biology. 2003b;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. %U http://www.sciencedirect.com.proxy.library.vanderbilt.edu/science/article/B616VRT-648CW619W613-M/612/49946c10163ae49647dfb49946f49956bf49940fcdaba. [DOI] [PubMed]
- Gu G, Caldwell GA, Chalfie M. Genetic interactions affecting touch sensitivity in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1996;93:6577–6582. doi: 10.1073/pnas.93.13.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevi S, McKay J, Palfreyman M, Yassin L, Eshel M, Jorgensen E, Treinin M. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. Embo J. 2002;21:1012–1020. doi: 10.1093/emboj/21.5.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E, Culotti J, Hall D. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]
- Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- Ichikawa H, Mo Z, Xiang M, Sugimoto T. Effect of Brn-3a deficiency on nociceptors and low-threshold mechanoreceptors in the trigeminal ganglion. Brain Res Mol Brain Res. 2002;104:240–245. doi: 10.1016/s0169-328x(02)00355-8. [DOI] [PubMed] [Google Scholar]
- Inoue T, Wang M, Ririe T, Fernandes J, Sternberg P. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc Natl Acad Sci U S A. 2005;102:4972–4977. doi: 10.1073/pnas.0408122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Hoskins R, Horvitz HR. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature. 1994;372:780–783. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- Kahn-Kirby AH, Bargmann CI. TRP channels in C. elegans. Annu Rev Physiol. 2006;68:719–736. doi: 10.1146/annurev.physiol.68.040204.100715. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kaplan JM, Horvitz HR. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1993;90:2227–2231. doi: 10.1073/pnas.90.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass J, Jacob TC, Kim P, Kaplan JM. The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J Neurosci. 2001;21:9265–9272. doi: 10.1523/JNEUROSCI.21-23-09265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MD, Jan LY, Jan YN. The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev. 2006;20:2806–2819. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Luo L. Intrinsic control of precise dendritic targeting by an ensemble of transcription factors. Curr Biol. 2007;17:278–285. doi: 10.1016/j.cub.2006.11.067. [DOI] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- Long H, Ou Y, Rao Y, van Meyel DJ. Dendrite branching and self-avoidance are controlled by Turtle, a conserved IgSF protein in Drosophila. Development. 2009;136:3475–3484. doi: 10.1242/dev.040220. [DOI] [PubMed] [Google Scholar]
- Lucanic M, Kiley M, Ashcroft N, L’Etoile N, Cheng HJ. The Caenorhabditis elegans P21-activated kinases are differentially required for UNC-6/netrin-mediated commissural motor axon guidance. Development. 2006;133:4549–4559. doi: 10.1242/dev.02648. [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- McGlone F, Reilly D. The cutaneous sensory system. Neurosci Biobehav Rev. 2010;34:148–159. doi: 10.1016/j.neubiorev.2009.08.004. [DOI] [PubMed] [Google Scholar]
- McGlone F, Spence C. The cutaneous senses: touch, temperature, pain/itch, and pleasure. Neurosci Biobehav Rev. 2010;34:145–147. doi: 10.1016/j.neubiorev.2009.08.008. [DOI] [PubMed] [Google Scholar]
- McIntire SL, Garriga G, White J, Jacobson D, Horvitz HR. Genes necessary for directed axonal elongation or fasciculation in C. elegans. Neuron. 1992;8:307–322. doi: 10.1016/0896-6273(92)90297-q. [DOI] [PubMed] [Google Scholar]
- Moore AW, Jan LY, Jan YN. hamlet, a binary genetic switch between single- and multiple- dendrite neuron morphology. Science. 2002;297:1355–1358. doi: 10.1126/science.1072387. [DOI] [PubMed] [Google Scholar]
- O’Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- Oren-Suissa M, Hall DH, Treinin M, Shemer G, Podbilewicz B. The Fusogen EFF-1 Controls Sculpting of Mechanosensory Dendrites. Science. 2010 doi: 10.1126/science.1189095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron. 2009;63:788–802. doi: 10.1016/j.neuron.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 2005 doi: 10.1242/dev.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon VY, Klassen MP, Shen K. UNC-6/netrin and its receptor UNC-5 locally exclude presynaptic components from dendrites. Nature. 2008 doi: 10.1038/nature07291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn CC, Pfeil DS, Wadsworth WG. CED-10/Rac1 mediates axon guidance by regulating the asymmetric distribution of MIG-10/lamellipodin. Curr Biol. 2008;18:808–813. doi: 10.1016/j.cub.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Sugimura K, Satoh D, Estes P, Crews S, Uemura T. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron. 2004;43:809–822. doi: 10.1016/j.neuron.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Sugimura K, Yamamoto M, Niwa R, Satoh D, Goto S, Taniguchi M, Hayashi S, Uemura T. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci. 2003;23:3752–3760. doi: 10.1523/JNEUROSCI.23-09-03752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kerr R, Bianchi L, Frokjaer-Jensen C, Slone D, Xue J, Gerstbrein B, Driscoll M, Schafer WR. In vivo imaging of C. elegans mechanosensory neurons demonstrates a specific role for the MEC-4 channel in the process of gentle touch sensation. Neuron. 2003;39:1005–1017. doi: 10.1016/j.neuron.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Temple S. Division and differentiation of isolated CNS blast cells in microculture. Nature. 1989;340:471–473. doi: 10.1038/340471a0. [DOI] [PubMed] [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, Barstead R, Maricq A, Bargmann C. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron. 2002;35:307–318. doi: 10.1016/s0896-6273(02)00757-2. [DOI] [PubMed] [Google Scholar]
- Tracey WD, Jr, Wilson RI, Laurent G, Benzer S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- Tsalik EL, Hobert O. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J Neurobiol. 2003;56:178–197. doi: 10.1002/neu.10245. [DOI] [PubMed] [Google Scholar]
- Tsalik EL, Niacaris T, Wenick AS, Pau K, Avery L, Hobert O. LIM homeobox gene-dependent expression of biogenic amine receptors in restricted regions of the C. elegans nervous system. Dev Biol. 2003;263:81–102. doi: 10.1016/s0012-1606(03)00447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina S, Fox R, Watkins K, Starich T, Shaw J, Miller D. UNC-4 represses CEH-12/HB9 to specify synaptic inputs to VA motor neurons in C. elegans. Genes Dev. 2007a;21:332–346. doi: 10.1101/gad.1502107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina S, Watson J, Fox R, Olszewski K, Spencer W, Roy P, Miller D. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 2007b;8:R135. doi: 10.1186/gb-2007-8-7-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth WG, Bhatt H, Hedgecock EM. Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron. 1996;16:35–46. doi: 10.1016/s0896-6273(00)80021-5. [DOI] [PubMed] [Google Scholar]
- Watson JD, Wang S, Von Stetina SE, Spencer WC, Levy S, Dexheimer PJ, Kurn N, Heath JD, Miller DM., 3rd Complementary RNA amplification methods enhance microarray identification of transcripts expressed in the C. elegans nervous system. BMC Genomics. 2008;9:84. doi: 10.1186/1471-2164-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JC, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 1989a;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes and Develop. 1989b;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- Wemmie JA, Price MP, Welsh MJ. Acid-sensing ion channels: advances, questions and therapeutic opportunities. Trends Neurosci. 2006;29:578–586. doi: 10.1016/j.tins.2006.06.014. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London. 1986;B314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wittenburg N, Baumeister R. Thermal avoidance in Caenorhabditis elegans: an approach to the study of nociception. Proc Natl Acad Sci U S A. 1999;96:10477–10482. doi: 10.1073/pnas.96.18.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Duggan A, Chalfie M. Inhibition of touch cell fate by egl-44 and egl-46 in C. elegans. Genes Dev. 2001;15:789–802. doi: 10.1101/gad.857401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Xu XZ. Function and regulation of TRP family channels in C. elegans. Pflugers Arch. 2009;458:851–860. doi: 10.1007/s00424-009-0678-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Tu Y, Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science. 1993;261:1324–1328. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]
- Yassin L, Gillo B, Kahan T, Halevi S, Eshel M, Treinin M. Characterization of the deg-3/des-2 receptor: a nicotinic acetylcholine receptor that mutates to cause neuronal degeneration. Mol Cell Neurosci. 2001;17:589–599. doi: 10.1006/mcne.2000.0944. [DOI] [PubMed] [Google Scholar]
- Yassin L, Samson AO, Halevi S, Eshel M, Treinin M. Mutations in the extracellular domain and in the membrane-spanning domains interfere with nicotinic acetylcholine receptor maturation. Biochemistry. 2002;41:12329–12335. doi: 10.1021/bi020193y. [DOI] [PubMed] [Google Scholar]
- Zhang S, Arnadottir J, Keller C, Caldwell GA, Yao CA, Chalfie M. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr Biol. 2004;14:1888–1896. doi: 10.1016/j.cub.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.