SUMMARY
RIG-I is a cytosolic sensor of viral RNA that plays crucial roles in the induction of type I interferons. The C-terminal domain (CTD) of RIG-I is responsible for the recognition of viral RNA with 5′ triphosphate (5′ ppp). However, the mechanism of viral RNA recognition by RIG-I is still not fully understood. Here we show that RIG-I CTD binds 5′ ppp dsRNA or ssRNA, as well as blunt-ended dsRNA, and exhibits the highest affinity for 5′ ppp dsRNA. Crystal structures of RIG-I CTD bound to 5′ ppp dsRNA with GC- and AU- rich sequences revealed that RIG-I recognizes the termini of the dsRNA and interacts with the 5′ triphosphate through extensive electrostatic interactions. Mutagenesis and RNA binding studies demonstrated that similar binding surfaces are involved in the recognition of different forms of RNA. Mutations of key residues at the RNA binding surface affected RIG-I signaling in cells.
INTRODUCTION
Viral pathogens can infect hosts from bacteria to mammals. To combat viral infection, highly sophisticated immune systems have been developed in vertebrates to sense viral infection and initiate antiviral responses. In addition to the adaptive immune responses mediated by B cells and T cells, the innate immune system maintains several families of pattern recognition receptors (PRRs) that recognize molecular motifs of viral pathogens and launch quick responses toward infection (Janeway and Medzhitov, 2002; Pichlmair and Reis e Sousa, 2007). Two major families of PRRs, Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs), play key roles in innate immune responses toward viral infection (Akira et al., 2006). TLR3, TLR7 and TLR8 sense viral RNA in the endosomes following phagocytosis of the pathogens (Kawai and Akira, 2008; Thompson and Locarnini, 2007). RIG-I-like receptors RIG-I and MDA5 detect viral RNA from replicating viruses in the cytoplasm (Pichlmair and Reis e Sousa, 2007; Yoneyama and Fujita, 2009). LGP2, a homolog of RIG-I and MDA5 lacking the N-terminal caspase recruitment domains, regulates the signaling of the RLRs (Rothenfusser et al., 2005; Satoh et al., 2010; Venkataraman et al., 2007). Stimulation of the RLRs by viral RNA induces the secretion of type I interferons (IFN-α and -β) and other proinflammatory cytokines, conferring antiviral activity to the host cells and activating acquired immune responses (Palm and Medzhitov, 2009; Pichlmair and Reis e Sousa, 2007; Stetson and Medzhitov, 2006; Yoneyama and Fujita, 2009).
RIG-I and MDA5 respond to distinct but overlapping sets of viruses, suggesting that they recognize different forms or structures of viral RNA (Kato et al., 2006; Takeuchi and Akira, 2009). The 5′ triphosphate group was initially identified as a key structural feature of viral RNA that was sensed by RIG-I and it was suggested that 5′ ppp ssRNA is the primary ligand of RIG-I (Hornung et al., 2006; Pichlmair et al., 2006; Schlee et al., 2009a). However, recent studies demonstrated that dsRNA with 5′ triphosphate are potent activators of RIG-I (Schlee et al., 2009b; Schmidt et al., 2009). In addition, several other studies demonstrated that synthetic dsRNA or short dsRNA mimetics derived from RNase digestion of poly I:C can also activate RIG-I (Kato et al., 2008; Takahasi et al., 2008, Ranjith-Kumar et al., 2009). The crystal structure of human LGP2 CTD bound to dsRNA showed that the termini of dsRNA are recognized by LGP2 (Li et al., 2009b). Consistent with this finding, blunt-ended dsRNA rather than dsRNA with 5′ or 3′ overhangs binds and stimulates the activation of RIG-I (Li et al., 2009b; Marques et al., 2006). Although blunt-ended dsRNA with 5′ triphosphate potently stimulates the activation of RIG-I, triphosphate dsRNA with 5′ overhangs does not stimulate the activation of RIG-I effectively (Schlee et al., 2009b). Since different forms of RNA, cell lines and assays were used in these studies, questions remain about the structural feature(s) of viral RNA recognized by RIG-I. Recent studies by Rehwinkel et al. demonstrated that the genomic RNA of influenza A virus and Sendai virus are the physiological agonists of RIG-I (Rehwinkel et al., 2010). It was suggested that the panhandle structure formed by the 5′ and 3′ ends of the viral RNA is likely a structural feature recognized by RIG-I (Fujita, 2009; Rehwinkel et al., 2010; Schlee et al., 2009b).
Limited proteolysis of RIG-I bound to dsRNA revealed that its CTD (residues 792–925) is responsible for RNA binding (Takahasi et al., 2008). Structures of the CTDs of RIG-I, LGP2 and MDA5 have been determined recently, revealing a novel and conserved RNA binding domain structure (Cui et al., 2008; Li et al., 2009a; Pippig et al., 2009; Takahasi et al., 2009; Takahasi et al., 2008). The structure of LGP2 CTD bound to dsRNA showed that the RNA binding surface is located on a positively charged surface made up of strands β5 to β8 and loop 5–6 (Li et al., 2009b). NMR titrations of RIG-I and MDA5 CTD showed that similar binding surfaces are involved in RNA binding by RIG-I and MDA5 (Li et al., 2009a; Takahasi et al., 2009; Takahasi et al., 2008). Mutagenesis of several positively charged residues on this surface disrupted RNA binding and signaling by RIG-I (Cui et al., 2008; Takahasi et al., 2008). Based on the results from these studies, the triphosphate binding site of RIG-I was predicted to be located at a large positively charged surface around residues Lys858 and Lys861 (Cui et al., 2008; Takahasi et al., 2008). However, how RIG-I recognizes the 5′ triphosphate of viral RNA remains to be established.
To elucidate the structural basis of 5′ ppp dsRNA recognition by RIG-I, we expressed the CTD of human RIG-I, conducted extensive binding studies using RNA with different structures, and determined the crystal structures of RIG-I CTD bound to a 14-bp GC-rich and a 12-bp AU-rich dsRNA. The structures revealed that RIG-I CTD recognizes the termini of dsRNA and interacts with the 5′ triphosphate through extensive electrostatic interactions. Mutagenesis of key residues at the binding surface affected RNA binding and signaling by RIG-I.
RESULTS
RIG-I CTD binds 5′ ppp dsRNA with high affinity
Understanding of RIG-I structure and function requires the elucidation of the chemical basis for viral RNA recognition by RIG-I. We have expressed and purified human RIG-I CTD and characterized its ligand binding properties with various forms of RNA.
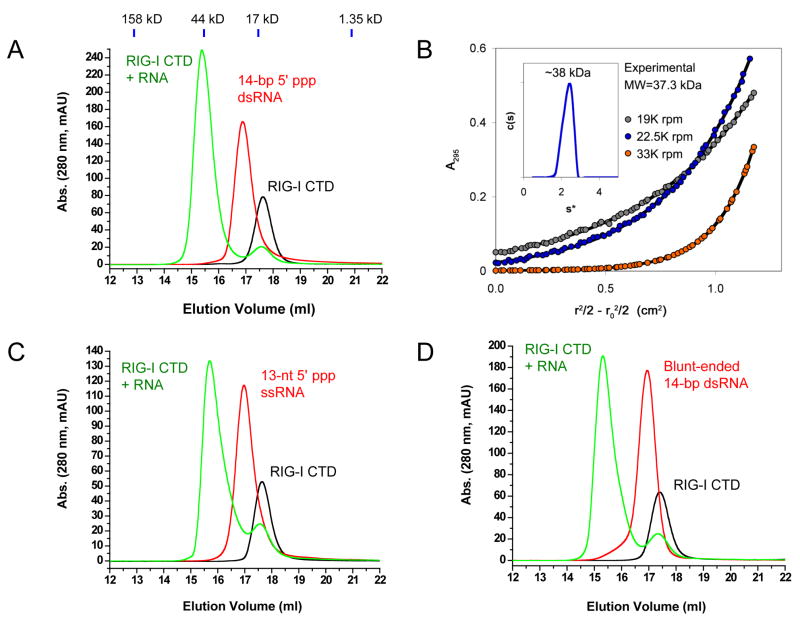
First, we studied the RNA binding of RIG-I CTD by gel filtration chromatography. A 14-bp GC-rich dsRNA, a 12-bp AU-rich dsRNA and a 13-nucleotide (nt) ssRNA were synthesized by in vitro transcription using T7 RNA polymerase (Table S1). We found that the 14-bp dsRNA with 5′ triphosphate binds RIG-I CTD and forms a stable complex (Figure 1A). Analytical ultracentrifugation (AUC) sedimentation velocity and equilibrium analyses of purified RIG-I CTD bound to the 14 GC 5′ ppp dsRNA demonstrated that the stoichiometry between the protein and the dsRNA is 2:1 (Figure 1B). The 12-bp AU-rich dsRNA also binds RIG-I CTD and forms a complex of 2:1 stoichiometry (Figure S1), indicating that RIG-I has no preference for specific bases within the RNA. Surprisingly, the 13-nt ssRNA with 5′ triphosphate also binds RIG-I CTD (Figure 1C). In addition, we found a blunt-ended dsRNA with the same sequence as the 14-bp GC-rich 5′ ppp dsRNA but lacking the 5′ triphosphate also binds RIG-I CTD and forms a stable complex with a stoichiometry of 2:1 (Figure 1D).
Figure 1.
RIG-I CTD binds RNA with or without a 5′ triphosphate. (A) Binding study of RIG-I CTD with a 14-bp GC-rich 5′ ppp dsRNA by gel filtration chromatography. RIG-I CTD is shown by the black chromatogram. The RNA is shown by the red chromatogram. The mixture of the protein and the RNA is shown by the green chromatogram. Elution volumes of four protein standards are shown above the chromatograms. (B) Stoichiometry between RIG-I CTD and the 14-bp GC-rich 5′ ppp dsRNA in a purified protein RNA complex determined by sedimentation velocity (inset) and equilibrium. The experimental molecular weight values from velocity (~38 kDa) and equilibrium (37,339 Da) data were consistent only with the formation of a 2:1 complex. (C) Binding study of RIG-I CTD with a 13-nt 5′ ppp ssRNA by gel filtration chromatography. (D) Binding study of RIG-I CTD with a 14-bp blunt-ended dsRNA without a 5′ triphosphate.
Second, we tested whether the 5′ or 3′ end overhanging regions of 5′ ppp dsRNA affect RNA binding to RIG-I CTD. We found that triphosphate dsRNA with either 3′ or 5′ overhanging nucleotides retained binding to RIG-I CTD (Table S1, Figure S1). However, treatment of these dsRNA with shrimp alkaline phosphatase (SAP) that removes the 5′ triphosphate groups dramatically reduced their binding with RIG-I CTD (Figure S1), demonstrating the 5′ triphosphate increases the affinity of the dsRNA for RIG-I. Consistent with this observation, treatment of the12-bp 5′ ppp dsRNA without 3′ or 5′ overhangs using SAP also reduced its binding for RIG-I CTD (Figure S1).
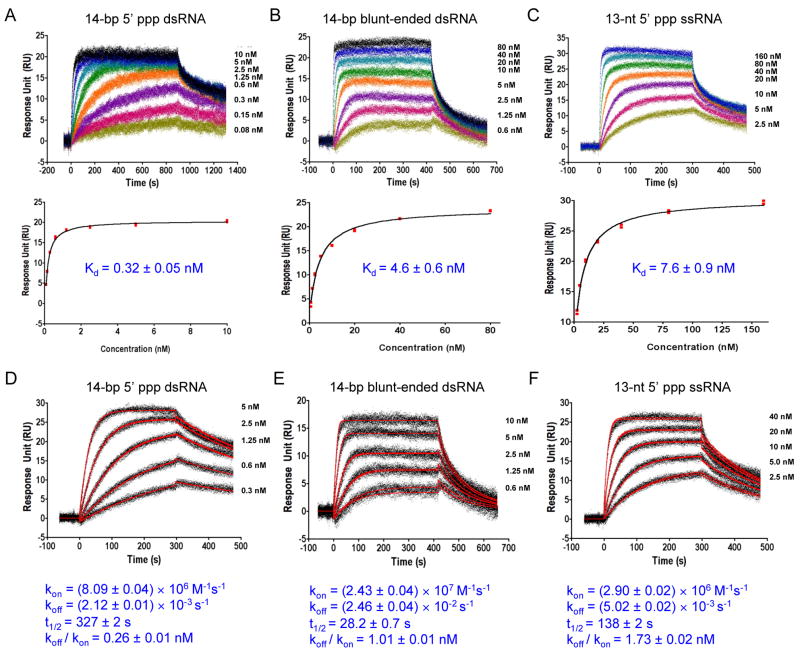
The affinity and kinetic properties of ligand binding to immunoreceptors determine the strength and duration of the signals initiated by these receptors. To determine the affinities and kinetic properties of RNA binding by RIG-I CTD, we expressed and purified GST-RIG-I CTD and conducted RNA binding studies by surface plasmon resonance (SPR). Equilibrium binding studies showed that the affinity of RIG-I CTD for the 14-bp 5′ ppp dsRNA is 0.32 ± 0.05 nM (Figure 2A). The affinity of RIG-I CTD for the 14-bp dsRNA without 5′ triphosphate is 4.6 ± 0.6 nM (Figure 2B), about 15 times lower than the affinity for the 5′ ppp dsRNA. The 13-nt 5′ ppp ssRNA binds RIG-I CTD with an affinity of 7.6 ± 0.9 nM (Figure 2C). The calculated dissociation constants (KD,calc = koff/kon) derived from the kinetic binding studies are similar to those derived from the equilibrium binding studies (Figure 2). Although RIG-I CTD interacts with RNA primarily through electrostatic interactions, binding of the 14-bp 5′ ppp dsRNA by RIG-I CTD shows slow association rate and slow dissociation rate (Figure 2D), suggesting conformational change of the protein is likely needed for RNA binding. It is possible that other types of interactions such as hydrogen bonding and hydrophobic interactions may also contribute to RNA binding. In contrast, the binding of blunt-ended dsRNA lacking a 5′ triphosphate shows faster association and dissociation rates (Figure 2E). Binding of the 13-nt 5′ ppp ssRNA shows kinetic properties similar to that of the 5′ ppp dsRNA (Figure 2F). These results indicate the 5′ triphosphate plays an important role in determining the kinetic properties of RNA binding by RIG-I CTD. The higher affinity of the 5′ ppp dsRNA and the longer half-life (t1/2 = 327 ± 2 s) of its complex with RIG-I suggest that dsRNA with 5′ triphosphates should be more potent stimulator of RIG-I.
Figure 2.
RIG-I CTD binds 5′ triphosphate dsRNA with high affinity. (A) Equilibrium binding study of RIG-CTD with the 14-bp GC-rich 5′ ppp dsRNA by surface plasmon resonance (SPR). The dissociation constant (Kd) was derived by fitting of the equilibrium binding data to a one site binding model (lower panels). (B) Equilibrium binding study of RIG-I CTD with the 14-bp blunt-ended dsRNA. (C) Equilibrium binding study of RIG-I CTD with a 13-nt 5′ ppp ssRNA. (D) Kinetic binding studies of RIG-CTD with the 14-bp GC-rich 5′ ppp dsRNA. The association and dissociation rate constants (kon and koff) were derived from global fitting of the binding data to a 1:1 binding model (red curves). Half – life (t1/2) for each of the RNA: RIG-I CTD complex was calculated from the koff. The calculated dissociation constant (KD, calc) was derived by division of the dissociation rate constant (koff) by the association rate constant (kon). (E) Kinetic binding studies of RIG-I CTD with the 14-bp blunt-ended dsRNA. (F) Kinetic binding studies of RIG-I CTD with the 13-nt 5′ ppp ssRNA.
RIG-CTD recognizes the termini of 5′ triphosphate dsRNA
To elucidate the structural basis of 5′ triphosphate dsRNA recognition by RIG-I, we determined the crystal structures of RIG-I CTD (residues 803 to 923) bound to a 14-bp GC-rich dsRNA (with self complementary sequence of 5′ pppGGCGCGCGCGCGCC 3′) and a 12-bp AU-rich dsRNA (with self complementary sequence of 5′ pppAUAUAUAUAUAU 3′). The structures were determined by molecular replacement and refined at 2.6 and 2.15 Å resolution, respectively. Crystals of the 12-bp AU-rich dsRNA complex are merohedrally twinned. The structure was refined against the twinned data using CNS (Brunger et al., 1998). Statistics of data collection and structural refinement are listed in Table 1.
Table 1.
Data collection and refinement statistics
| Data Collection | RIG-I CTD:14-bp dsRNA | RIG-I CTD:12-bp dsRNA |
| Space group | P212121 | P65 |
| Cell dimensions | ||
| a, b, c (Å) | 41.49, 96.87, 103.40 | 82.90, 82.90, 111.16 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50-2.60 (2.69-2.60)a | 50.00-2.15(2.23-2.15)a |
| Rsym (%) | 8.6 (58.7) | 7.1(45.7) |
| I/σI | 27.4(3.2) | 36.0(4.4) |
| Completeness (%) | 98.7(96.3) | 93.5(90.4) |
| Redundancy | 4.5(4.4) | 6.5(6.3) |
| Refinement | ||
| Resolution (Å) | 50-2.60 | 50-2.15 |
| No. Reflections | 12530 | 20357 |
| Rwork/Rfree | 22.1/28.5 | 18.7/22.7 |
| No. atoms | ||
| Protein | 1998 | 1998 |
| RNA | 620 | 522 |
| Ion | 2 | 2 |
| Water | 54 | 0 |
| B-factor | ||
| Protein | 45.9 | 37.3 |
| RNA | 69.5 | 70.7 |
| Ligand/ion | 45.0 | 35.1 |
| Water | 49.0 | NA |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.013 | 0.009 |
| Bond angles (°) | 1.69 | 1.54 |
Diffraction data were collected using one single crystal for each structure. Values in the parentheses are for the highest-resolution shell.
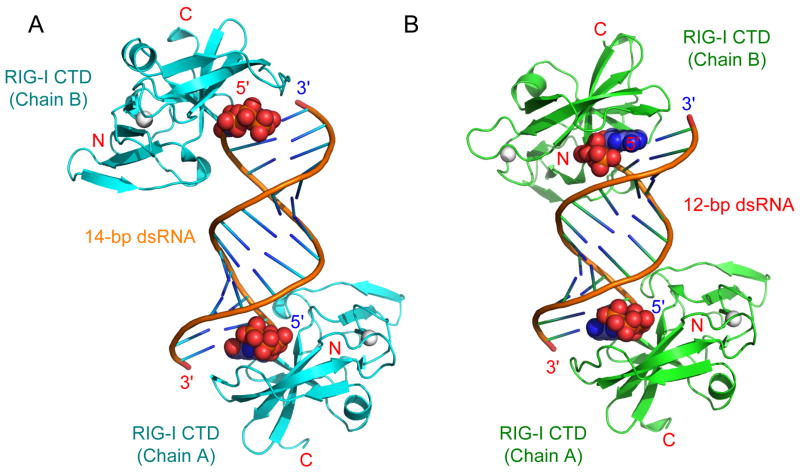
Consistent with the stoichiometry determined by AUC, the crystallographic asymmetric units for the two complex crystals contain a 2:1 complex between RIG-I CTD and the dsRNA (Figure 3A, B). The complexes exhibit pseudo two-fold symmetry along the two-fold axis of the palindromic dsRNA. The orientations of the dsRNA relative to the RIG-I CTD in the two complexes are similar (Figure 3). The 5′ triphosphates of the dsRNA are well defined at the four binding sites in the two complexes and interact with the protein in similar ways (Figure S2). Structures of four RIG-I CTDs in the two complexes are similar; the root mean square deviations (r.m.s.d) between the Cα atoms of RIG-I CTD within each complex or between the two complexes are only 0.3 to 0.5 Å. Discussions of the complex structure below are based on the structure of RIG-I CTD (Chain A) in the 14-bp GC-rich dsRNA complex due to better-defined electron density map at this interface (Figure S2) and more complete diffraction data used in the structural refinement (Table 1).
Figure 3.
Structures of RIG-I CTD bound to 5′ triphosphate double-stranded RNA. (A) Crystal structure of RIG-I CTD bound to the 14-bp GC-rich 5′ ppp dsRNA. The nucleotides at the 5′ ends of the dsRNA containing the triphosphate are shown as space filling models. The zinc ions bound to RIG-I CTD are shown as gray spheres. (B) Structure of the RIG-I CTD bound to the 12-bp AU-rich 5′ ppp dsRNA. The orientations of RIG-I CTD (Chain A) relative to the dsRNA are the same in the two structures.
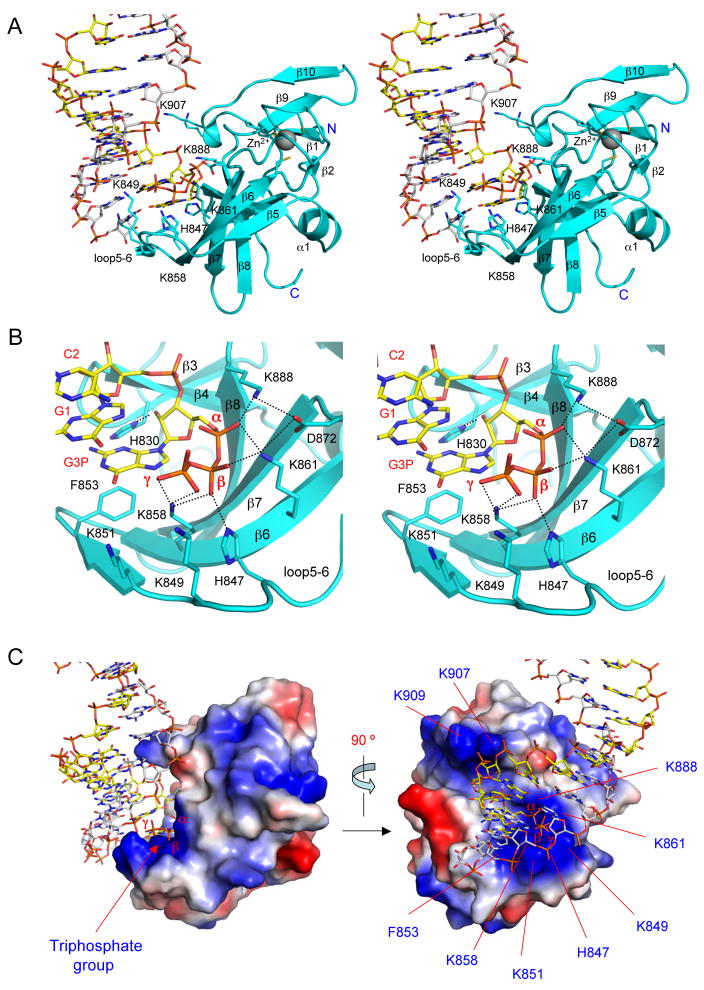
As predicted from our previous structural studies of the LGP2 CTD:dsRNA complex (Li et al., 2009b), RIG-I CTD recognizes the termini of the 5′ ppp dsRNA (Figure 3). The dsRNA in the complex adopts a typical A-form double helical structure. The RNA binding surface of RIG-I CTD is located at a large saddle-shaped surface defined by the hairpin containing strands β3 and β4, the loop connecting strands β5 and β6, the β-sheet containing strands β5 to β8, the loop connecting strands β8 and β9, and the loop connecting strand β10 to the C-terminus (Figure 4A). RIG-I CTD interacts with the dsRNA primarily through extensive electrostatic interactions with the 5′ triphosphate and the phosphate backbone (Figure 4A, B). The exposed base pairs at the termini of the dsRNA interact with RIG-I CTD through hydrophobic interactions (Figure 4B). The RNA-binding surface of RIG-I CTD exhibits a high degree of shape and charge complementarity to the blunt-ends of 5′ ppp dsRNA (Figure 4C). The total buried surface area at the RIG-CTD:dsRNA interface is approximately 1,200 Å2, with primary contributions from the first four nucleotides at the 5′ end of the dsRNA (buried surface area of ~900 Å2). The last three nucleotides at the 3′ end of the complementary strand only make minor contributions to dsRNA binding (buried surface area of ~300 Å2). The average shape correlation statistic (Sc) is 0.62 for the four dsRNA protein interfaces; the Sc value is a measure of the degree to which two contacting surfaces are geometrically matched, with an Sc value of 1.0 indicating a perfect fit (Lawrence and Colman, 1993).
Figure 4.
Structural basis of 5′ triphosphate dsRNA recognition by RIG-I CTD. (A) Stereo close up of the interface between RIG-I CTD (Chain A) and the terminus of the 14-bp 5′ ppp dsRNA. RIG-I CTD is shown as cyan ribbons and the dsRNA is shown as stick models. Key residues of RIG-I CTD involved in RNA binding are shown as stick models. (B) Stereo close up of the interactions between RIG-I CTD and the 5′ triphosphate of the 14-bp GC-rich dsRNA. (C) Surface representation of RIG-I CTD bound to 5′ ppp dsRNA showing the shape and charge complementarity between RIG-I CTD and the terminus of the 5′ ppp dsRNA. Positively charged surfaces are colored blue and negatively charged surfaces are red. The orientation of RIG-I CTD relative to the dsRNA on the left panel is similar to the orientation of the protein in panel A.
The overall structure of RIG-I CTD in the complex is similar to the structure of RIG-I CTD in isolation (r.m.s.d of ~ 0.7 Å). The largest structural difference between free and RNA bound RIG-I CTD occurs at the loop between strands β5 and β6 (loop 5–6) from His847 to Ser855 (Figure S3). The average deviation for the Cα atoms of these eight residues is 1.67 Å in the two structures. As reflected in the nine structures of RIG-I CTD in the crystallography asymmetric unit, this loop is highly flexible in the structures of RIG-I CTD in isolation (Cui et al., 2008). The structure of RIG-I CTD in solution also revealed that this loop is flexible (Takahasi et al., 2008). However, the four structures of the RIG-I CTD in the two complexes are highly similar, suggesting that dsRNA binding stabilizes the specific conformation of loop 5–6.
Structural basis of 5′ ppp dsRNA recognition by RIG-I CTD
As predicted from previous mutagenesis studies (Cui et al., 2008; Takahasi et al., 2008), the 5′ triphosphate-binding site of RIG-I is located at a large patch of positively charged surface around residues Lys858 and Lys861. Four positively charged residues interact with the 5′ triphosphate through electrostatic interactions (Figure 4A, B; Table S2). Lys888 interacts with the α-phosphate group through electrostatic interaction (Figure 4A, B). The distance between the side chain amine group of Lys888 and the α-phosphate is only 2.4 Å. The side chain amine group of Lys861 is ~2.7 Å away from the α- and β-phosphates, interacting simultaneously with these two phosphate groups (Figure 4A, B; Table S2). Proper positioning of the side chains of Lys888 and Lys861 to interact with the α- and β-phosphate is facilitated by Asp872, which interacts with the side chains of these two residues through electrostatic interactions (Figure 4B). The β-phosphate also interacts with the side chains of Lys858 and His847 simultaneously (Figure 4B). In addition, Lys858 also interacts with the γ-phosphate that is about 5 Å away from the side chain amine groups of Lys849 and Lys851 (Figure 4B). These two positively charged residues may interact with the triphosphate through weak electrostatic interaction or solvent-mediated hydrogen bonds.
Although the electrostatic interactions between RIG-I CTD and the 5′ triphosphate play dominant roles in dsRNA binding, RIG-I CTD interacts extensively with other parts of the dsRNA (Table S2). The exposed base pair at each terminus of the dsRNA interacts with Phe853 through hydrophobic interaction; and the guanine base at the 5′ end of the RNA stacks against the phenyl side chain of Phe853 (Figure 4B). The side chain of His830 forms a hydrogen bond with the 2′ hydroxyl group of nucleotide G1 (Figure 4B). This hydroxyl group also forms two solvent-mediated hydrogen bonds with the backbone amine of Tyr831 and the carbonyl group of Ile887 (Table S2). The phosphate group of G1 forms two solvent-mediated hydrogen bonds with Ser906 and Trp908 (Table S2). The third nucleotide C2 from the 5′ end forms a solvent mediated hydrogen bond with the backbone amine of Trp908 (Table S2). In addition, the phosphate group of G3 interacts with the side chain of Lys907 through electrostatic interactions (Figure 4A). However, the RNA does not interact with the nearby Lys909 directly. The side chains of Lys849 and Lys851 are in close proximity to the phosphate of C10 and G11 near the 3′ end of the RNA (Figure 4A). However, the electron density map for the side chains of these two residues is poorly defined, reflecting their flexibility and limited contribution to dsRNA binding. The side chain hydroxyl of Ser854 forms a hydrogen bond with the 2′ hydroxyl at the 3′ terminal nucleotide (C13) of the complementary strand (Table S2). Apart from the hydrophobic interactions between the terminal bases and Phe853, no other specific interaction between RIG-I CTD and the bases was observed in the two complex structures, demonstrating RIG-I CTD binds RNA in a sequence-independent manner.
RIG-I and LGP2 bind dsRNA with and without 5′ triphosphate in different ways
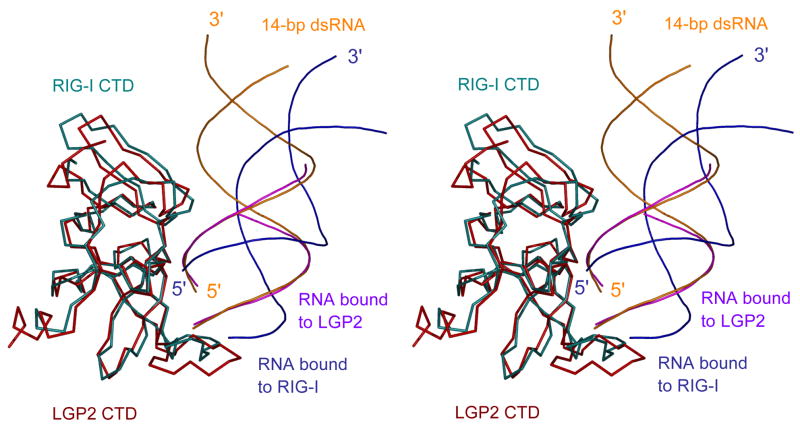
The crystal structures of RIG-I and LGP2 CTD bound to dsRNA with and without 5′ triphosphate respectively allowed us to elucidate how the RLRs recognize these two different forms of dsRNA. To facilitate this comparison, we superimposed a14-bp dsRNA onto the structure of LGP2 CTD bound to an 8-bp dsRNA (Li et al., 2009b). Superposition of LGP2 CTD onto the structure of RIG-I CTD bound to the 14-bp GC-rich dsRNA revealed that the orientations of the dsRNA in the two complexes are dramatically different (Figure 5). Although both proteins recognize the terminus of the dsRNA and similar binding surfaces are involved in RNA binding, the 14-bp 5′ ppp dsRNA was rotated by approximately 30 degrees away from RIG-I CTD compared to the 14-bp dsRNA in the LGP2 CTD:dsRNA complex model (Figure 5). As a result, RIG-I CTD has less extensive interactions with the 5′ ppp dsRNA compared to the interactions between the blunt-ended dsRNA and LGP2 CTD (Li et al., 2009b). The total buried surface area at the LGP2 CTD:dsRNA interface (~1,540 Å2) is about 30% larger than that of the RIG-I CTD:dsRNA interface. The interface between LGP2 CTD and the blunt-ended dsRNA also exhibits a higher degree of shape complementarity (Sc = 0.75). Although the interaction between LGP2 CTD and dsRNA is more extensive, the affinity of LGP2 CTD for a 10-bp blunt-ended dsRNA without triphosphate is about 100 nM (Li et al., 2009b). In addition, triphosphorylated dsRNA exhibits a slightly lower affinity for LGP2 CTD (Li et al., 2009b). Furthermore, mutations that disrupted RNA binding by LGP2 do not affect its ability to regulate RIG-I signaling (Li et al., 2009b). These results suggest that LGP2 does not regulate RIG-I signaling through direct competition with RIG-I for ligand binding, indicating that the RIG-I-RNA interaction is different from that of LGP2-RNA interaction.
Figure 5.
Distinct binding by RIG-I and LGP2 CTDs to dsRNA with and without 5′ triphosphate. Stereo representation of the structure of RIG-I CTD bound to the 14-bp GC-rich 5′ ppp dsRNA superimposed on the structure of LGP2 CTD bound to an 8-bp dsRNA without 5′ triphosphate. The 14-bp 5′ ppp dsRNA bound to RIG-I CTD is shown as blue ribbons. The 8-bp dsRNA bound to LGP2 CTD is colored magenta. A 14-bp dsRNA (orange) is superimposed on the 8-bp dsRNA in the LGP2 CTD:dsRNA complex structure to show the different orientations of the dsRNA relative the proteins in the two complexes.
Since the 5′ triphosphate has to be positioned precisely into the triphosphate-binding site, this structural requirement determines the specific orientation of the 5′ ppp dsRNA relative to RIG-I CTD. As the triphosphate makes primary contributions to 5′ ppp dsRNA binding by RIG-I CTD, the binding affinity of blunt-ended dsRNA without triphosphate should be much lower than that of the 5′ ppp dsRNA; the buried surface area between a dsRNA without 5′ triphosphate and RIG-I CTD will be reduced to ~1,050 Å2 if the RNA still binds RIG-I in the same orientation. Interestingly, SPR binding studies show that RIG-I CTD binds the 14-bp dsRNA without the 5′ triphosphate at nanomolar affinity. It is possible that RIG-I CTD binds blunt-ended dsRNA lacking a triphosphate in a similar way as LGP2 CTD to increase the binding interactions. Since dsRNA with or without triphosphate both activate RIG-I in vivo, we propose that the two forms of RNA may trigger the activation of RIG-I through different mechanisms.
Similar binding surfaces of RIG-I CTD mediate its interactions with different forms of RNA
Based on the structures of RIG-I CTD bound to 5′ ppp dsRNA, we expressed and purified twelve mutant proteins of RIG-I CTD and studied their binding properties with a 5′ ppp dsRNA, a blunt-ended dsRNA, and a 5′ ppp ssRNA by electrophoretic mobility shift assay (EMSA). Ten of these twelve mutants were also analyzed for RNA binding by gel filtration chromatography.
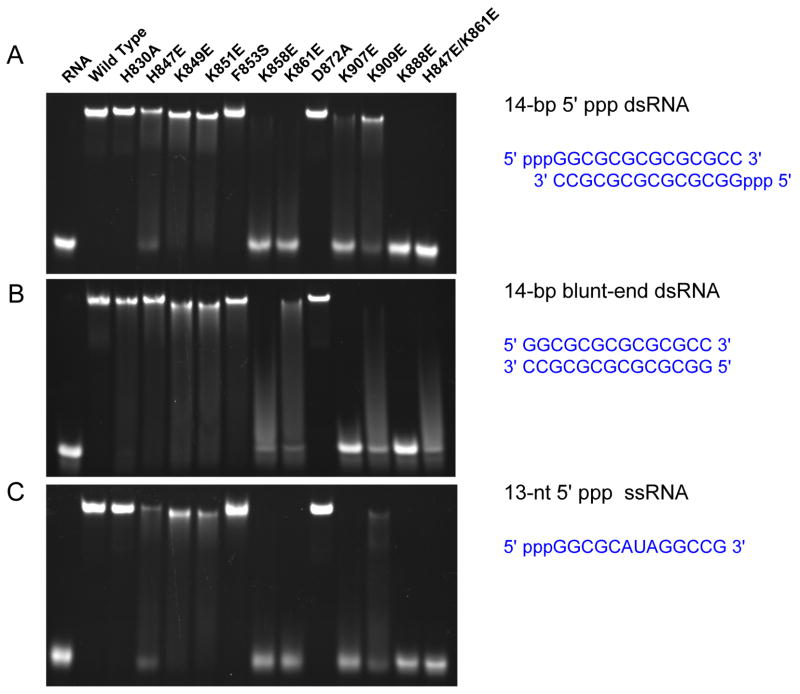
First, mutation of each of the three positively charged residues, Lys858, Lys861 and Lys888 that are involved in direct contact with the 5′ triphosphate to a negatively charged glutamate almost abolished 5′ ppp dsRNA and ssRNA binding in the gel shift assays (Figure 6). Consistent with these results, no binding for 5′ ppp ssRNA and dsRNA were observed for these mutants by gel filtration chromatography (Figure S4). Similarly, mutation of Lys888 to glutamate also abolished blunt-end dsRNA binding (Figure 6). However, substitutions of either Lys858 or Lys861 by glutamate dramatically reduced but did not abolish blunt-ended dsRNA binding (Figure 6 and Figure S4). Substitution of His847 that interacts directly with the β-phosphate with a glutamate significantly reduced the binding of 5′ ppp dsRNA and ssRNA, but only slightly reduced the binding of blunt-ended dsRNA (Figure 6 and Figure S4). Consistent with these results, mutations of both His847 and Lys861 to glutamate disrupted binding of 5′ ppp dsRNA or ssRNA and nearly abolished blunt-ended dsRNA binding (Figure 6). Second, we analyzed the effects of mutations of four other positively charged residues Lys849, Lys851, Lys907, and Lys909 on RNA binding. Mutation of Lys907 that interacts with the phosphodiester backbone of the dsRNA to glutamate dramatically reduced binding for all three forms of RNA (Figure 6), demonstrating the critical role of this residue in recognizing the RNA backbone. Gel filtration chromatography confirmed that mutant K907E failed to bind all three forms of RNA (Figure S4). In contrast, substitution of the nearby Lys909 with a glutamate reduced the binding of 5′ ppp dsRNA but almost abolished the binding of blunt-ended dsRNA and 5′ ppp ssRNA (Figure 6 and Figure S4). Mutations of either Lys849 or Lys851 to glutamate reduced binding for all three forms of RNA, but did not abolish RNA binding (Figure 6 and Figure S4). Third, we analyzed the mutations of His830, Phe853 and Asp872 that interact with the dsRNA through hydrogen bonding, hydrophobic interactions or facilitate the binding of the triphosphate group. Replacement of Asp872 with an alanine did not affect RNA binding (Figure 6 and Figure S4). Mutation of His830 to alanine did not affect 5′ ppp dsRNA or ssRNA binding, but reduced blunt-ended dsRNA binding (Figure S4). Although gel shift assays showed that substitution of Phe853 by serine did not affect RNA binding significantly (Figure 6), gel filtration chromatography showed that this mutation dramatically reduced triphosphorylated RNA binding and nearly abolished blunt-ended dsRNA binding (Figure S4).
Figure 6.
Similar binding surfaces of RIG-I CTD mediate the interactions with different forms of RNA. (A) Binding studies of wild type and mutants of RIG-I CTD for the 14-bp 5′ ppp dsRNA by electrophoretic mobility shift assay (EMSA). (B) Binding studies of RIG-I CTD mutants with a 14-bp blunt-ended dsRNA lacking 5′ triphosphate. (C) Binding studies of RIG-I CTD mutants with a 13-nt 5′ ppp ssRNA.
These mutagenesis and binding studies demonstrated that the binding surface observed in the complex structures mediates RNA binding by RIG-I CTD in solution. It is also important to note that while similar binding surfaces are involved in contacting blunt-ended dsRNA as well as dsRNA and ssRNA with 5′ triphosphates, the fact that mutations in the CTD can differentially affect binding of the three forms of RNA illustrate distinct recognition of these RNA ligands by RIG-I.
Mutations of key residues at the RNA binding surface affect RIG-I signaling
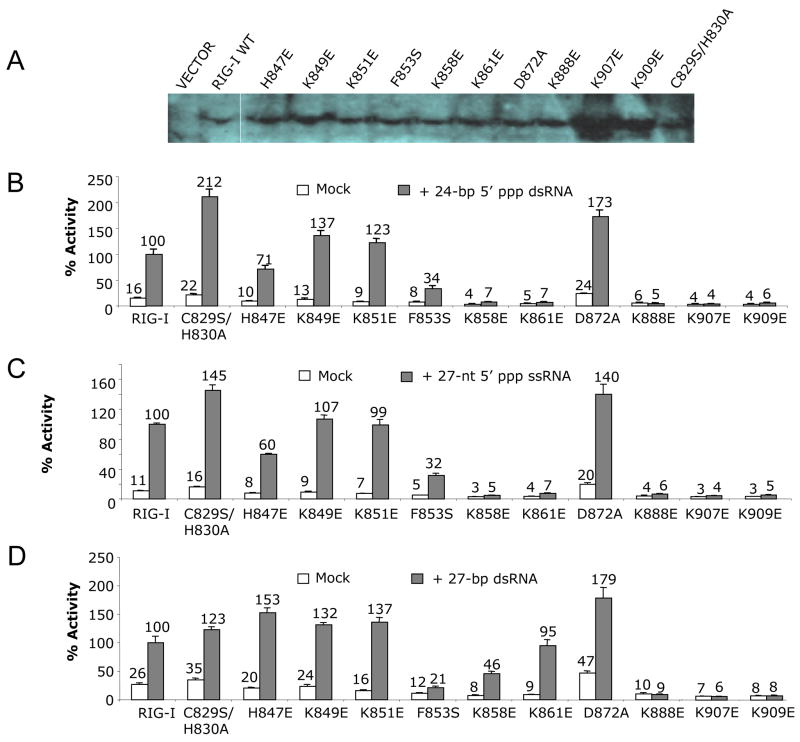
The twelve mutations tested for RNA binding were built into constructs of full-length RIG-I to assess how these mutations affect RIG-I signaling in transiently-transfected HEK293T cells. The cells were first transfected to express the RIG-I mutants along with the IFN-β luciferase reporter plasmids. Expression of wild type RIG-I and various mutants in the transfected cells was confirmed by Western blot (Figure 7A). After 24 hours, different RNA ligands were transfected into the cells to analyze their abilities to activate RIG-I dependent signaling. A 24-bp 5′ ppp dsRNA, a 27-nt 5′ ppp ssRNA and a 27-bp blunt-ended dsRNA were tested in these assays. The relative activities of triphosphorylated RNAs are similar, while the blunt-ended dsRNA is slightly less active (Figure S5).
Figure 7.
Mutations of key residues at the RNA binding surface affect RIG-I signaling. (A) Western blot showing the expression of wild type and mutants of full-length RIG-I in transfected cells. (B) IFN-β luciferase assay showing the signaling of wild type and mutants of RIG-I in HEK 293T cells stimulated with a 24-bp 5′ ppp dsRNA. (C) IFN-β luciferase assay showing the signaling of wild type and mutants of RIG-I in cells stimulated with a 27-nt 5′ ppp ssRNA. (D) IFN-β luciferase assay showing the signaling of wild type and mutants of RIG-I in cells stimulated with a 27-bp blunt-ended dsRNA without the 5′ triphosphate.
First, we analyzed signaling of mutants modified at residues that play critical roles in the recognition of the 5′ triphosphate group. Since the Cα atom of Lys888 is only 3.8 Å away from the G1 phosphate and its side chain amine group interacts with the α-phosphate of the 5′ triphosphate, substitution of Lys888 by glutamate disrupted RNA binding by RIG-I CTD and abolished RIG-I signaling by 5′ ppp dsRNA or ssRNA and blunt-ended dsRNA (Figure 7). Although mutation of Lys861 to glutamate did not affect signaling by blunt-ended dsRNA, it did disrupt signaling by 5′ ppp dsRNA and ssRNA (Figure 7). Similarly, mutation of Lys858 to glutamate reduced signaling by blunt-ended dsRNA by about 50% and abolished signaling by 5′ ppp dsRNA and ssRNA (Figure 7). Substitution of His847 by a glutamate did not affect blunt-ended dsRNA signaling, but it reduced signals from both 5′ ppp dsRNA and ssRNA dramatically (Figure 7). Second, substitutions of Lys907 and Lys909 by glutamates disrupted signaling for all three forms of RNA (Figure 7). In contrast, mutations of either Lys851 or 849 to glutamate did not affect signaling of blunt-end dsRNA, 5′ ppp dsRNA or ssRNA (Figure 7). Third, we tested signaling of mutants of RIG-I at residues His830, Phe853 and Asp872 that have slightly reduced RNA binding. We found that mutations C829S/H830A or D872A had no effect on the signaling of RIG-I for any of the three forms of RNA tested (Figure 7). Consistent with these results, a Cys829 to serine substitution in RIG-I CTD did not affect RNA binding in vitro (Figure S4). The slightly higher activity of these two mutants compared to the wild type protein is likely due to their higher expression level or better stability in the cells. However, replacement of the hydrophobic Phe853 by a hydrophilic serine reduced signaling of 5′ ppp dsRNA or ssRNA by about 70% and reduced signaling by blunt-ended dsRNA to background level (Figure 7), demonstrating that Phe853 has a role in RNA sensing by RIG-I. These results are consistent with RNA binding properties of RIG-I CTD mutants, demonstrating that RNA binding by RIG-I CTD is essential for RIG-I signaling and RNA with or without triphosphate can activate RIG-I in vivo.
DISCUSSION
RNA binding studies showed that RIG-I CTD binds 5′ ppp dsRNA or ssRNA and blunt-ended dsRNA, with the highest affinity for 5′ ppp dsRNA. IFN-β luciferase assays demonstrated that all three forms of RNA could stimulate RIG-I signaling. The structures of RIG-I CTD bound to 5′ ppp dsRNA showed that RIG-I recognizes the termini of RNA and interacts with the 5′ triphosphate as well as the backbone phosphodiesters of the RNA. Mutagenesis and RNA binding studies demonstrated that similar binding surfaces of RIG-I CTD are involved in the binding of various forms of RNA by RIG-I. These results provide a structural framework to understand the RNA binding properties and functions of RIG-I in antiviral immune responses.
The complex structures showed that RIG-I CTD interacts primarily with the 5′ four nucleotides of the dsRNA. The complementary RNA strand only makes limited contributions to RNA binding (Figure 4, Table S2). These observations explain how RIG-I CTD can bind both dsRNA and ssRNA with triphosphate.
The amino acid residues of RIG-I involved in RNA binding can be divided into three groups. The first group of residues includes Lys888, Lys858, Lys861, and His847 that recognize the 5′ triphosphate of dsRNA. The second group of residues includes Lys907 and Lys909 that interact with the phosphate backbone of the dsRNA. The third group of residues interacts with the exposed bases at the termini of the RNA or interacts with the backbone of the RNA by hydrogen bonds. The first two groups of residues contribute to RNA binding through multiple electrostatic interactions, while the third group of residues makes additional contributions to RNA binding through hydrogen bonds and hydrophobic interactions. All these residues contribute to the binding of RNA with 5′ triphosphate while the last two groups of residues contribute primarily to the binding of dsRNA without 5′ triphosphate. Importantly, these structural features explain how RIG-I can recognize such a wide range of RNA ligands including 5′ ppp dsRNA and ssRNA as well as dsRNA without 5′ triphosphate.
The reason why ssRNA without triphosphate failed to bind RIG-I CTD is most likely due to the loss of the critical electrostatic interactions mediated by the triphosphate group. However, dsRNA without triphosphate still binds RIG-I CTD at high affinity, this is due to the additional interactions between the complementary strand and the protein (P. Li, unpublished data).
Since the α- and β-phosphates make major contributions to the recognition of the 5′ triphosphate by RIG-I, the complex structures also explain why dsRNA with one or two 5′-terminal phosphates can serve as ligands for RIG-I (Figure 4B). Previous studies showed that blunt-ended 5′ ppp dsRNA are potent activators of RIG-I, while 5′ ppp dsRNA with 5′ overhangs still stimulated RIG-I but 5′ ppp dsRNA with 3′ overhangs failed to stimulate RIG-I (Schlee et al., 2009b). However, our RNA binding studies showed that all three forms of dsRNA bind RIG-I CTD efficiently (Figure S1). Although the complex structures indicated that dsRNA with 3′ overhangs can still bind RIG-I effectively, the reason why 5′ ppp dsRNA with 3′ overhangs failed to stimulate RIG-I is most likely that these RNA molecules are unwound by RIG-I (Takahasi et al., 2008). The structures suggest that triphosphate dsRNA with 5′ overhangs should exhibit significantly reduced affinity for RIG-I just like 5′ ppp ssRNA, explaining why this kind of RNA is less effective in stimulating the activation of RIG-I (Schlee et al., 2009b).
Our binding studies showed that RIG-I CTD binds the GC- rich and AU-rich RNA with similar affinities. In addition, the two complex structures showed that RIG-I binds the two kinds of RNA in almost the same way with no specific interactions between the protein and the bases (Figure 3). These results indicate that RIG-I CTD is not responsible for the recognition of specific RNA sequences such as the polyuridylates or polyadenylates sequences observed in the 3′ non-translated regions of hepatitis C virus genome (Saito et al., 2008). The studies by Chiu et al. showed that poly (dA-dT) DNA transfected in cells can activate RIG-I due to transcription of these DNA sequences by RNA polymerase III, while GC-rich DNA failed to activate RIG-I when transfected into cells (Chiu et al., 2009). This is not likely due to the different sequences of DNA but most likely due to the inefficient transcription of GC rich DNA by RNA polymerase III that need the poly (dA-dT) sequence to initiate transcription. In summary, the ability of RIG-I to recognize a broad range of RNA structures without sequence specificity allows the receptor to sense the infection of a wide spectrum of viruses.
The crystal structures of RIG-I and LGP2 CTD in isolation and in complex with dsRNA are now available (Cui et al., 2008; Li et al., 2009b; Pippig et al., 2009). The 1.45 Å resolution structure of MDA5 CTD has also been determined (Li et al., 2009a). These structures revealed that the CTD of the RLRs are highly conserved and similar binding surfaces are involved in RNA binding by all the three proteins (Figure S3). The structures of RIG-I CTD and LGP2 CTD bound to dsRNA with and without 5′ triphosphate demonstrated that the RLRs recognize the termini rather than the ribose-phosphate backbone of various forms of RNA (Figure 5). However, the three proteins exhibit different ligand binding properties and play different roles in antiviral immune responses. Comparisons of the structures of RIG-I CTD in complex with triphosphorylated dsRNA and LGP2 CTD in complex with blunt-ended dsRNA revealed that the orientations of the RNA are significantly different (Figure 5). This suggests that the triphosphorylated and blunt-ended dsRNA interact with RIG-I differently and probably induce different conformations of full-length RIG-I upon binding. This is further supported by the differential binding of triphosphorylated dsRNA and blunt-ended dsRNA by RIG-I CTD mutants (Figure 6) and the abilities of RIG-I mutants to activate luciferase reporters in cells stimulated with different forms of RNA (Figure 7).
Comparisons of the ligand binding surfaces of the RLR CTDs showed that the surface electrostatic potential of the three proteins are dramatically different (Figure S3). For the four positively-charged residues in RIG-I CTD that are responsible for the triphosphate recognition, only Lys888 is conserved in the sequences of LGP2 and MDA5 CTDs. Consistent with this observation, both MDA5 and LGP2 CTDs bind 5′ ppp dsRNA with lower affinities compared to blunt-ended dsRNA (Li et al., 2009a; Li et al., 2009b). It is evident that RIG-I is the only RLR that is responsible for the sensing of 5′ triphosphate RNA. Since RIG-I can also be activated by dsRNA without 5′ triphosphate, which is also recognized by MDA5, this could explain why RIG-I and MDA5 sense distinct but overlapping sets of viruses.
A number of factors may affect the results of RIG-I activation assays in vivo. The structure and purity of RNA ligands, their binding affinities for RIG-I, their effective concentrations in cells, and their stabilities will all affect the signal readout. Previous studies used various cell lines and different assays to demonstrate that various RNA ligands can activate signal transduction through RIG-I (Schlee et al., 2009a). Our studies demonstrated that dsRNA with or without 5′ triphosphate and ssRNA with 5′ triphosphate are the three major families of RNA that activate RIG-I. Furthermore, blunt-ended dsRNA with or without 5′ triphosphate are more effective agonists for RIG-I signaling than dsRNA with overhangs (Li et al., 2009b; Schlee et al., 2009a; Schlee et al., 2009b; Takahasi et al., 2008). Consistent with these findings, all these three families of RNA bind RIG-I CTD and blunt-ended dsRNA with 5′ triphosphate binds RIG-I CTD at a significantly higher affinity (Figures 1 and 2). Results from the IFN-β luciferase reporter assays confirmed that all these three families of RNA stimulate RIG-I signaling in cells (Figure 7). In contrast, two recent studies showed that only dsRNA with 5′ triphosphate activates RIG-I in human monocytes (Schlee et al., 2009b; Schmidt et al., 2009). It is likely that the different assay techniques or the difference in the effective concentration of ligands in the transfected cells account for these discrepancies. Since the affinities of blunt-ended dsRNA and 5′ ppp ssRNA are about 15-fold lower than that of blunt-ended 5′ ppp dsRNA, significantly higher concentration of blunt-ended dsRNA is needed for the activation of RIG-I. A number of other reports, including one of our previous studies, also demonstrated that dsRNA without 5′ triphosphate can activate RIG-I (Li et al., 2009b; Takahasi et al., 2008). In addition, it has been shown that RNase digestion of dsRNA mimetic poly I:C converted it from a ligand of MDA5 into a ligand for RIG-I (Kato et al., 2008). Moreover, the digestion of self-RNA by antiviral endonuclease RNase L generates small RNA products that initiate interferon production (Malathi et al., 2007). It is likely that higher concentration of ligands generated by RNase digestion triggers the activation of RIG-I.
Structural studies of RIG-I and LGP2 CTD bound to dsRNA provided important insight into the structural basis of viral RNA recognition by the RLRs. However, the mechanism of how RNA binding triggers the activation of RIG-I or MDA5 remains to be established. Although electron microscopy studies of RIG-I and LGP2 in isolation and in complex with dsRNA provided some information about how these proteins might bind RNA (Murali et al., 2008; Ranjith-Kumar et al., 2009), the detailed mechanism of how RNA binding activates the RLRs awaits the determination of the high resolution structures of the full-length proteins bound to RNA.
EXPERIMENTAL PROCEDURES
Purification of RIG-I CTD and its complexes with 5′ ppp dsRNA
The cDNA encoding the human RIG-I CTD (residues 802 to 925) was cloned into bacterial expression vector pET22b(+) (Novagen). Residue Cys829 was mutated to serine to prevent dimerization of the protein. The cloned DNA sequence was confirmed by plasmid sequencing. RIG-CTD was expressed in E. coli strain BL21(DE3) by induction at OD600 = 0.6–0.8 with 0.4 mM isopropyl-β-D-thiogalactoside (IPTG) overnight at 15°C. The protein was purified by nickel affinity chromatography on a His-Select nickel affinity resin (Sigma-Aldrich) column. The protein was further purified by gel filtration chromatography on a Superdex75 (1.6 × 60) column (GE Healthcare) eluted with a buffer containing 20 mM Tris, 150 mM NaCl, at pH 7.50 (buffer A). The two dsRNA constructs used to form complexes with RIG-I CTD were generated by in vitro transcription using T7 RNA polymerase and purified by gel filtration chromatography on a Superdex75 (1.6 × 60) column. RIG-I CTD was mixed with the 14-bp or the 12-bp dsRNA at a molar ratio of about 3:1 and the 2:1 (RIG-I CTD:dsRNA) complexes were purified by gel filtration chromatography on a Superdex75 (1.6 × 60) column eluted with buffer A. After purification, 5 mM DTT was added to the complexes and the samples were concentrated for crystallization.
RNA binding studies by gel filtration chromatography
Different forms of RNA were generated either by chemical synthesis or by in vitro transcription using T7 RNA polymerase. All in vitro transcribed RNA were purified by gel filtration chromatography and analyzed by denaturing polyacrylamide gel electrophoresis. Each of the purified triphosphate RNA showed a single band with the expected length on the denaturing gels. Sequences of the RNAs used in this study are shown in Table S1. Double-stranded RNA was generated by heating the ssRNA at 95 °C for 5 minutes and cooling gradually to room temperature. For the binding studies, each of the RNA samples (at ~ 100 μM) was mixed with equal volume of RIG-I CTD (at ~200 μM) and 100 μl of samples were injected over a Superdex200 (10/300 GL) column (GE healthcare) eluted with buffer A. The column was calibrated with a set of protein standards of different sizes (Bio-Rad) to allow the estimation of the sizes of the protein and RNA complexes.
Analytical ultracentrifugation
To determine the stoichiometry of RIG-I CTD binding to the 14 bp 5′ ppp dsRNA, purified RIG-I CTD:14 bp dsRNA complex was analyzed by AUC. For sedimentation velocity experiments, 400μl samples in 20 mM Tris-HCl buffer at pH 7.5, 150 mM NaCl, 10 mM β-mercaptoethanol were centrifuged overnight at 48,000 rpm at 10 °C in a Beckman XL-I using absorbance optics at 304 nm. The data were analyzed by the program Sedfit using the c(s) and c(M) models to determine differential sedimentation coefficient and apparent mass distributions, respectively. The complex sedimented as a single peak with a sedimentation coefficient of 2.72 S and an estimated molecular mass of 38 kDa, similar to that expected for a 2:1 complex (38.5 kDa). To confirm the stoichiometry, the complex was analyzed in a sedimentation equilibrium experiment. Briefly, 100 μL of samples of the complex at concentrations of 15, 50, and 150 μM were spun at 19,000, 22,500, and 33,000 rpm at 10 °C until equilibrium was reached, and were scanned at 295 and 306 nm. The data of the sedimentation equilibrium experiments were analyzed as previously described (Li et al., 2009b). Global analysis of nine data sets revealed that the data could be described by a single species with a reduced buoyant molecular weight of 1.883 (at 19,000 rpm). To convert to experimental molecular weight, the partial specific volume must be known; for a protein and RNA complex this value corresponds to the weight average of each component in the sedimenting species. The partial specific volume for RIG-I CTD alone was calculated to be 0.7258 mL/g at 10 °C using Sednterp. The partial specific volume for the 14-bp dsRNA was calculated to be 0.5688 mL/g using the NucProt Calculator server (http://www.molmovdb.org/cgi-bin/psv.cgi) and corrected to 10 °C in Sednterp. Both 2:1 and 2:2 complexes were considered, with weight-average partial specific volumes of 0.6956 or 0.6716 mL/g, respectively. The molecular weight values calculated from the reduced buoyant molecular weight and appropriate partial specific volumes were only self-consistent in the case of a 2:1 complex, yielding an experimental molecular weight of 37,339 Da (95% confidence interval: 33,882 – 40,796 Da).
RNA binding studies by surface plasmon resonance
Binding interactions between RIG-CTD and three different forms of RNA were analyzed by SPR at 25°C in HBS-EP+ (10 mM HEPES pH = 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% v/v P-20 surfactant) buffer on a Biacore T100 system (GE Healthcare). The goat anti-GST mAb (Biacore AB) at 30 μg/ml in 10 mM sodium acetate (pH = 5.0) was immobilized on a CM5 sensor chip (Biacore) by standard amine coupling chemistry. Immobilization of 13,000 response units (RU) of the antibody resulted in optimal responses in subsequent analyses. GST-RIG-CTD (at 5 μg/ml) was captured over the immobilized anti-GST mAb surface at a flow rate of 10 μl/min for 120 seconds to reach 1,200 RU response. For equilibrium binding studies, dilution series of different forms of RNA were injected in randomized duplicate runs at flow rates of 30, 50, and 50 μl/min for 10, 7, and 5 min for the 14-bp 5′ ppp dsRNA, the 14-bp blunt-ended dsRNA, and the 13-nt 5′ ppp ssRNA, respectively, followed by a 5 min dissociation phase. Concentrations for the three forms of RNA in the dilution series were 0.08 to 10 nM, 0.6 to 80 nM, and 2.5 to160 nM, respectively.
To study the kinetics of RNA binding by RIG-CTD, dilution series of the three different forms of RNA were injected in randomized duplicate runs at a flow rate of 50 μl/min for 5 or 7 min, followed by a 3 or 4 min dissociation phase. Concentrations for the three forms of RNA in the dilution series were 0.3 to 5 nM, 0.6 to 10 nM, and 2.5 to 40 nM, respectively. Regeneration of the sensor chip was achieved by injection of 10 mM glycine (at pH 2.2) at a flow rate of 20 μl/min for 210 sec followed by HBS-EP+ buffer stabilization for 5 min. Sensorgrams obtained from the SPR measurements were analyzed by the double-subtraction method described by Myszka (Myszka, 1999). The signal from the reference flow cell was subtracted from the analyte binding response obtained from the flow cell with captured ligand. Buffer reference responses were then averaged from multiple injections. The averaged buffer reference response was then subtracted from analyte binding responses and the final double-referenced data were analyzed with BIAevaluation 3.0 (GE Healthcare). For the equilibrium binding data, steady-state binding levels of the analytes were plotted against analyte concentration, from which the equilibrium binding constant was estimated. For the kinetic binding data, a one to one binding model was used to globally fit the data to derive the association and dissociation rate constants.
Crystallization, data collection, and structural determination
The complex of RIG-I CTD with the 14-bp GC-rich dsRNA was concentrated to about 30 mg/ml and crystallized in 18% PEG 3,350 in 0.1 M Bis-Tris buffer (pH 5.5) containing 200 mM (NH4)2SO4 at 4 °C. Large single crystals grew in about one week. The complex of RIG-I CTD with the 12-bp AU-rich dsRNA was crystallized under similar conditions in 0.1 M Bis-Tris buffer at pH 6.5. The crystals were transferred stepwise into a cryoprotectant containing 30% PEG 400 and flash frozen in liquid nitrogen. Diffraction data were collected using a Rigaku RAXIS IV++ detector mounted on a Rigaku Micromax-007HF generator. The diffraction data were processed with the HKL2000 package (Otwinowski and Minor, 1997).
The complex of the 14-bp GC-rich dsRNA and RIG-I CTD crystallizes in orthorhombic space group P212121 with cell dimensions: a = 41.49 Å, b = 96.87 Å, c = 103.40 Å. The crystallographic asymmetric unit (ASU) contains one 2:1 (RIG-CTD:dsRNA) complex. The 12-bp AU-rich dsRNA complex crystallizes in hexagonal space group P65 with cell dimensions: a = b = 82.90 Å, c = 111.16 Å, γ = 120 °. There is one 2:1 (RIG-I CTD:dsRNA) complex in the ASU. The crystal is hemihedrally twinned with a twin fraction of 0.478 with twinning operators h, -h-k, −l. The twining fraction was determined using CNS (Brunger et al., 1998).
Structures of the RIG-I CTD in complex with the 14-bp and 12-bp dsRNA were determined by molecular replacement with MOLREP in the CCP4 suite (CCP4, 1994) using the crystal structure of RIG-I CTD in isolation as the search model (PDB code: 2QFB, chain A). The structural models were rebuilt using O. After several rounds of rebuilding and refinement with CNS, electron density for the dsRNA became apparent. A 14 bp and a 12 bp dsRNA were manually docked into the electron density map and rebuilt with O (Jones and kjeldgaard, 1997). The structures were refined by several rounds of positional, simulated annealing and individual B-factor refinement using CNS followed by manual remodeling. The structure of RIG-I CTD bound to the 12-bp AU-rich dsRNA was refined in CNS against the twinned data. A total of 20,357 reflections, which correspond to 86% of a complete dataset, were used in the structural refinement. Only 5.5% of the diffraction data (1,309 reflections) were rejected due to twinning. Non-crystallographic symmetry (NCS) restraints were not applied during the refinement due to the different crystal packing environment of the RIG-I CTDs in the two complexes.
Mutagenesis of RIG-I CTD and RNA binding studies
Mutants of RIG-I CTD were generated using the Quikchange mutagenesis kit (Stratagene). Sequences of the mutants were confirmed by plasmid DNA sequencing. The mutant proteins were expressed and purified as described above for the wild type protein. For binding studies by EMSA, each type of RNA was mixed with excess RIG-I CTD at a molar ratio of RNA to protein of 1:3 at a final concentration of 1.6 μM: 5 μM. The mixtures were separated on 12% non-denaturing polyacrylamide gel in 1× Tris-boric acid buffer without EDTA. The gels were stained with ethidium bromide and visualized using Gel Doc XR (Bio-Rad). Binding studies for the mutants with the three different forms of RNA were also carried out by gel filtration chromatography. Each type of RNA was mixed with excess RIG-I CTD at a molar ratio of RNA to protein of 1:3 (final concentration of about 12 μM:36 μM) and 100 μl of samples were analyzed on a Superdex200 (10/300 GL) column.
Mutagenesis of RIG-I and IFN-β reporter gene assays
Mutations of full length RIG-I were performed using pUNO-hRIG-I (Invivogen) as template. Expression of wild type and mutants of full-length RIG-I in the transfected cells were confirmed by Western blot using anti-RIG-I antibody (C-15, sc-48929, Santa Cruz biotechnology). The luciferase assays used actively growing HEK 293T cells plated in CoStar White 96-well plates at 4.4 × 104 cells per well. When the cells were ~80% confluent, they were transfected with a mixture of Lipofectamine 2000 reagent (Invitrogen) and constant amounts of the reporter plasmids IFN-β luc (30 ng per transfection, gift from Dr. Rongtuan Lin, Lady Davis Institute for Medical Research, Montreal, QC, Canada.), phRL-TK (5 ng, Promega), and plasmids of wild type and mutants of human RIG-I pUNO-hRIG-I (0.5 ng, Invivogen). The cells were incubated for 24 h to allow expression from the plasmids. Different forms of RNA were then transfected into the cells at a final concentration of 0.05 μM. After 16 h incubation, the cells were analyzed using the Dual-Glo Luciferase Assay System reagents (Promega), quantifying luminescence with the BioTek Synergy2 Plate Reader. The ratios of firefly luciferase over Renilla luciferase were determined and the ratio obtained with WT-RIG-I induced with RNA was considered as 100% activity. The RNAs used in the assays were purified either by preparative denaturing polyacrylamide gel electrophoresis or by gel filtration chromatography.
Supplementary Material
Acknowledgments
This research is supported in part by grants from the National Institute of Allergy and Infectious Diseases (grant AI087741 to P. Li and grant AI073335 to C. Kao) and the Robert Welch Foundation (Grant A-1687 to P. Li).
Footnotes
Accession codes: The atomic coordinates and structure factors of RIG-I CTD bound to the 14-bp and the 12-bp 5′ ppp dsRNA have been deposited with the RCSB Protein Data Bank under the accession codes: 3LRN and 3LRR.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Fujita T. A nonself RNA pattern: tri-p to panhandle. Immunity. 2009;31:4–5. doi: 10.1016/j.immuni.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Jones TA, kjeldgaard M. Electron-density map interpretation. Methods in Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Li X, Lu C, Stewart M, Xu H, Strong RK, Igumenova T, Li P. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys. 2009a;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Li X, Ranjith-Kumar CT, Brooks MT, Bharmaiah S, Herr AB, Kao C, Li P. The RIG-I like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009b;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- Murali A, Li X, Ranjith-Kumar CT, Bhardwaj K, Holzenburg A, Li P, Kao CC. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J Biol Chem. 2008;283:15825–15833. doi: 10.1074/jbc.M800542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data by collected in oscillation mode. Method in Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar CT, Murali A, Dong W, Srisathiyanarayanan D, Vaughan R, Ortiz-Alacantara J, Bhardwaj K, Li X, Li P, Kao CC. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2009;284:1155–1165. doi: 10.1074/jbc.M806219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis ESC. RIG-I Detects Viral Genomic RNA during Negative-Strand RNA Virus Infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, Hartmann G. Approaching the RNA ligand for RIG-I? Immunol Rev. 2009a;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009b;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, Inagaki F. Solution structures of cytosolic RNA sensors MDA5 and LGP2 C-terminal domains: Identification of the RNA recognition loop in RIG-I like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol. 2007;85:435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.