Abstract
microRNAs (miRNAs) comprise an extensive class of post-transcriptional regulatory molecules in higher eukaryotes. Intensive research in Drosophila has revealed that miRNAs control myriad developmental and physiological processes. Interestingly, several of the best-studied miRNAs impact multiple biological processes, often by regulating distinct key target genes in each setting. Here we discuss the roles of some of these pleiotropic miRNAs, and their implications for studying and interpreting the roles of miRNAs in gene regulatory networks.
Introduction
microRNAs (miRNAs) are an ancient class of genes that are abundant in higher eukaryotic genomes. Most animal miRNAs derive from primary transcripts bearing hairpin structures that are sequentially cleaved by the Drosha and Dicer RNase III enzymes to produce mature miRNAs (Ghildiyal and Zamore, 2009; Kim et al., 2009). In addition, a subclass of miRNAs known as mirtrons exploit the splicing machinery to generate their precursor hairpins prior to Dicer cleavage (Okamura et al., 2007; Ruby et al., 2007a). In either case, mature miRNAs are loaded into Argonaute complexes and guide them to target transcripts, generally leading to mRNA destabilisation, translational inhibition, or both (Liu, 2008).
Most characterized miRNA-target interactions include Watson-Crick pairing between the miRNA "seed" (positions 2–8 relative to its 5' end) and 3' untranslated regions (3' UTRs) of target mRNAs, although alternative pairing configurations and site locations have been documented (Bartel, 2009; Brennecke et al., 2005; Lai, 2002). On the basis of conserved seed matches in multigenome alignments, individual miRNAs are frequently predicted to target hundreds of transcripts (Krek et al., 2005; Lewis et al., 2005; Ruby et al., 2007b; Xie et al., 2005) (Figure 1A). Microarray and proteomic studies confirm the repression of large target cohorts by individual miRNAs (Baek et al., 2008; Lim et al., 2005; Selbach et al., 2008), although in most cases the consequence of miRNA-mediated regulation is quantitatively subtle.
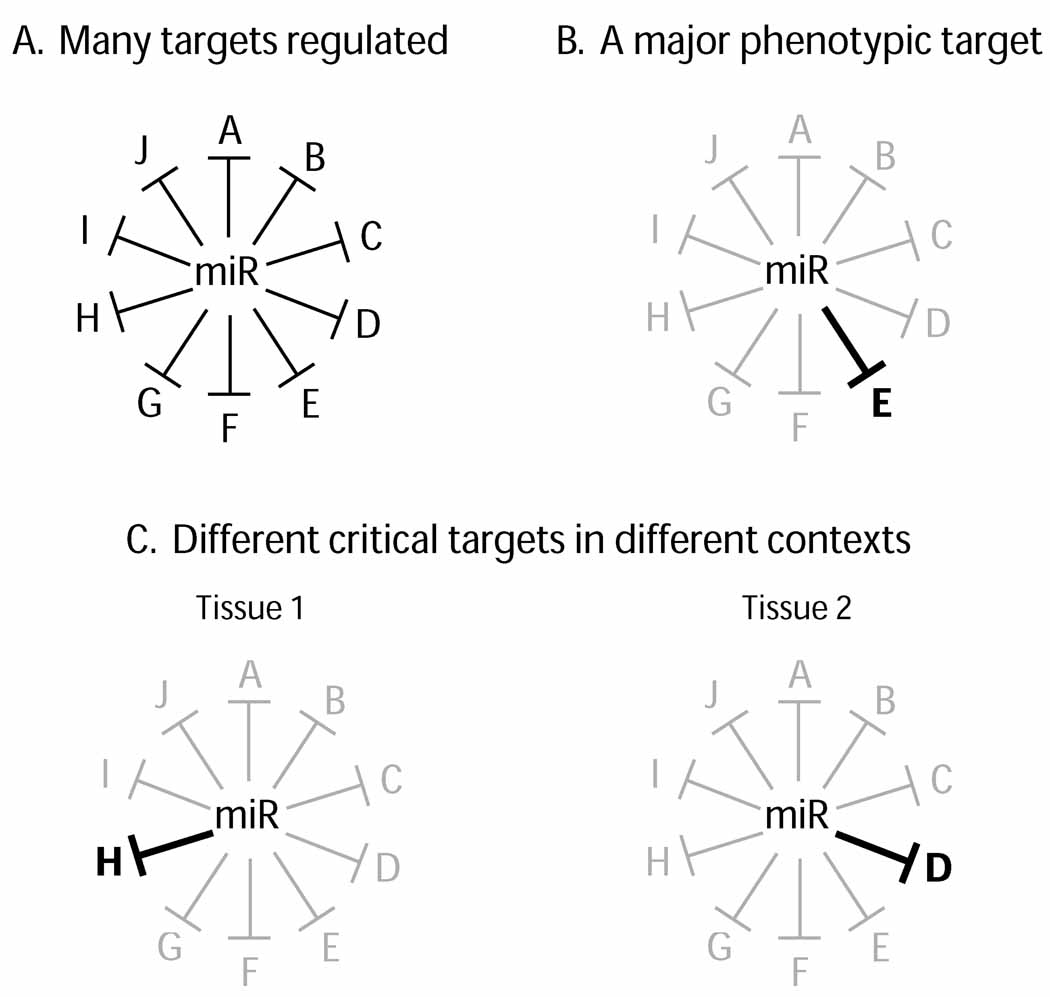
Figure 1. Modes of miRNA regulation.
(A) Individual miRNAs often regulate many targets, each of which may contribute subtly to the overall biological function of the miRNA. (B) A miRNA may repress a large network of targets, but the regulation of a specific target may underlie the major phenotypically visible role of the miRNA. Note that this does not necessarily mean that it is quantitatively the most strongly repressed target of the miRNA; depending on the function of the target, a relatively modest change in activity might elicit a mutant phenotype. (C) A miRNA may function in multiple spatial or temporal contexts, and may have different critical targets in each setting.
Amongst the small population of transcripts bearing few predicted miRNA binding sites, gene ontology analysis reveals enrichment in basic cell metabolic processes such as DNA, RNA and protein synthesis (Farh et al., 2005; Stark et al., 2005). It may be generally undesirable for the activity of such "antitargets" to be hindered by miRNAs (Bartel and Chen, 2004), and these transcript categories are marked by shorter 3' UTRs and lowered density of predicted miRNA binding sites (Stark et al., 2005). On the other hand, as most transcripts are proposed to be miRNA targets (Friedman et al., 2009), most biological processes are plausibly under miRNA control in some form or fashion. By extension, one may infer that miRNA dysfunction has substantial possibilities to underlie human disease and cancer, and there is indeed a rapidly growing literature to support this notion (Croce, 2009; O'Connell et al., 2010; Williams et al., 2009).
Nevertheless, the all-encompassing nature of the proposed miRNA regulatory network has presented clear challenges for understanding the endogenous influence and importance of miRNAs. One rationalization of the large numbers of targets has been that miRNAs act as robustness factors for gene networks and gene expression patterns. For example, several cell- or tissue-specific miRNAs exhibit strong statistical enrichment for targets that are present at low levels, or are undetectable, within their expression domains (Farh et al., 2005; Sood et al., 2006; Stark et al., 2005). Largely mutually exclusive patterns of miRNA and targets led to the notion that miRNAs refine and/or reinforce gene expression patterns imposed by transcriptional regulation, perhaps to prevent spurious gene activity or to clear away earlier programs activated in progenitor cells. In this scenario, the role of miRNAs is secondary to transcriptional programs, and in select cases, comparison with transcriptional reporters suggested that miRNA activity was not the cause per se of non-overlapping miRNA and target accumulation (Stark et al., 2005).
On the other hand, genetic studies have also shown that certain miRNAs play critical instructive roles (Lee et al., 1993; Reinhart et al., 2000; Wightman et al., 1993), indicating that miRNAs are not relegated to cleanup or fine-tuning roles. It is also relevant to note that while heterozygosity or duplication of most genes is well-tolerated (Lindsley et al., 1972), minor changes in specific genes can be of substantial consequence to organismal phenotype. Taken together, these facts suggest that the deregulation of certain targets, amongst large cohorts of target genes, might be primarily responsible for observed phenotypes. Indeed, while genome-wide computational and experimental methodologies clearly reveal vast networks of miRNA:target interactions, there are now many documented cases for which the loss of individual miRNA-targeting events can largely account for phenotypes of miRNA gene mutants (Bushati and Cohen, 2007; Flynt and Lai, 2008) (Figure 1B).
Resolving these genomic and genetic viewpoints to more fully understand the biological functions of miRNAs requires additional research, particularly from in vivo contexts. Drosophila melanogaster is one of the most intensively studied model organisms, and has ~150 miRNA genes that are predicted to target >30% of its mRNAs via conserved binding sites (Ruby et al., 2007b). Previous reviews have covered many aspects of miRNA biology in this organism (Bushati and Cohen, 2007; Flynt and Lai, 2008; Smibert and Lai, 2008), and we do not intend this discussion to be inclusive of all the Drosophila literature. Instead, we focus on lessons and principles that emerge from in-depth analyses of four miRNAs: miR-7, miR-8, miR-9a and bantam. In particular, this work has led to a growing appreciation that (1) certain miRNA targets are of particular importance for organismal phenotype (Figure 1B), and that (2) individual miRNAs can have pleiotropic functions involving the regulation of different key targets in different developmental or physiological contexts (Figure 1C).
miR-7: Drosophilid and species-specific roles for an ancient animal miRNA
The mature miRNA produced by the mir-7 gene is completely conserved from annelids to mammals, indicating that it is an ancient miRNA (Lagos-Quintana et al., 2001; Prochnik et al., 2007). In fact, conserved putative regulatory sites corresponding to miR-7 binding sites were identified over a dozen years ago amongst a cohort of Drosophila genes that control neurogenesis (Lai and Posakony, 1997, 1998; Leviten et al., 1997). These include two large arrays of Notch target genes located in the Bearded-Complex (Brd-C) and the Enhancer of split-Complex [E(spl)-C], encoding multiple Bearded-family and basic helix-loop-helix (bHLH) repressor proteins. Indeed, ectopic activation of miR-7 in the animal yields specific phenotypes that phenocopy a reduction of Notch signaling during wing margin specification and peripheral nervous system development (Lai et al., 2005; Stark et al., 2003), consistent with the notion that these represent substantial in vivo targets of miR-7.
miR-7 and eye development
Initial studies of a Drosophila mir-7 deletion allele (that also removes portions of host and neighboring genes) revealed a reciprocal negative feedback loop between miR-7 and the transcription factor Yan during eye development (Li and Carthew, 2005) (Figure 2A). Yan is a neural repressor that is predominantly expressed in the undifferentiated portion of the retina, and it inhibits the transcription of mir-7 directly. Conversely, miR-7 is activated in specified photoreceptors, and directly represses yan via its 3’ UTR (Li and Carthew, 2005). These regulatory interactions help to establish their mutually exclusive domains of activity.
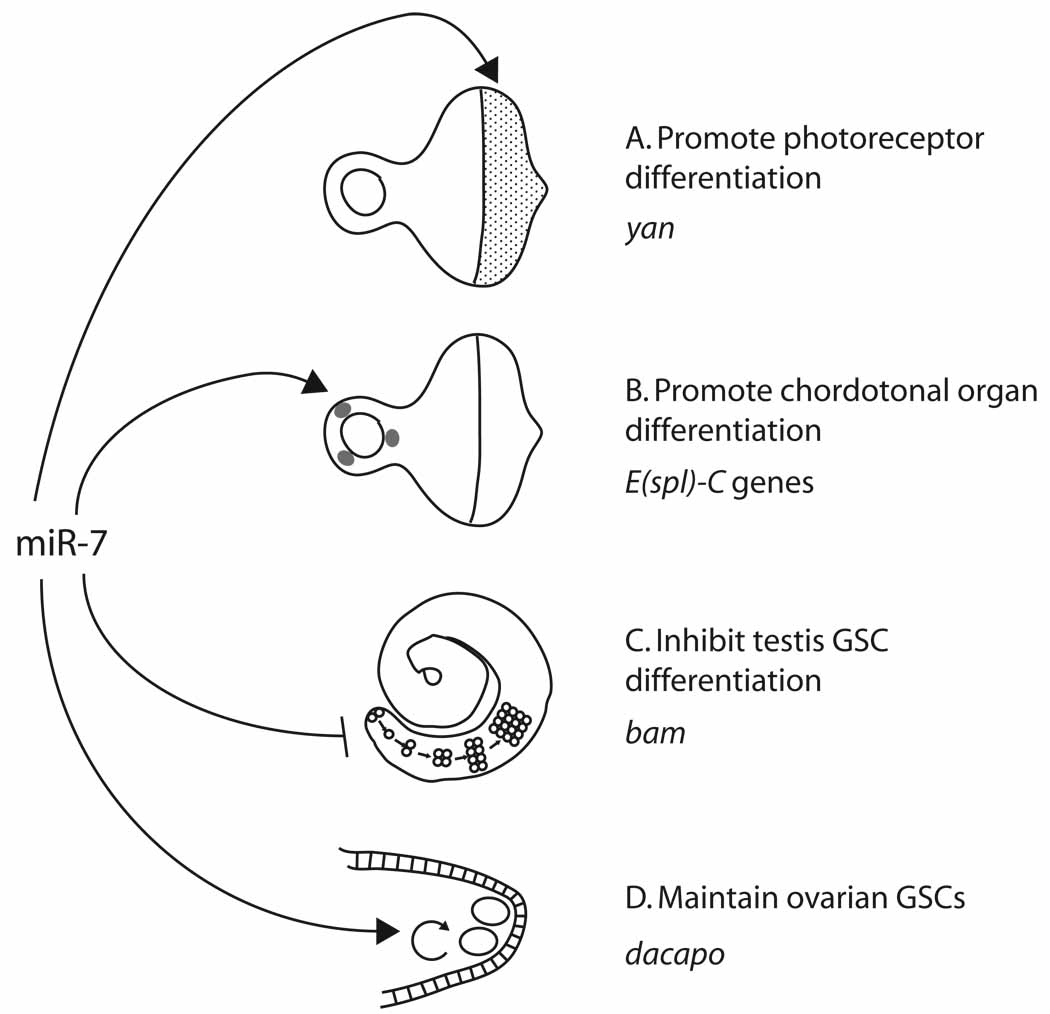
Figure 2. Processes regulated by Drosophila miR-7.
(A) In the eye imaginal disc, miR-7 is expressed in the specified photoreceptors (stippled portion) and promotes their differentiation by inhibiting yan, an Ets domain transcriptional repressor of neural differentiation. (B) miR-7 also promotes the differentiation of chordotonal organs (grey circles) by inhibiting members of the E(spl)-C family of bHLH transcriptional repressors. (C) In the testis, miR-7 represses bam to inhibit differentiation of germline stem cells (GSCs), whilst in the ovary, miR-7 represses the cell cycle regulator dacapo to promote maintenance of GSCs (D).
Following this work, the Carthew group expanded these regulatory principles by showing that Yan and miR-7 both participate in coherent feedforward loops (FFLs) (Li et al., 2009). Coherent FFLs direct their desired output by both direct and indirect means, thereby ensuring robustness of outcome. In this setting, the Pnt-P1 transcription factor directly represses yan transcription and activates mir-7 transcription, the latter of which represses yan post-transcriptionally. Reciprocally, Yan represses the transcription of both mir-7 and phyllopod, the latter of which encodes an E3 ubiquitin ligase that degrades TTK69, also a repressor of mir-7 transcription. In short, Yan represses mir-7 transcription through both direct and indirect mechanisms. The interlocking of these coherent FFLs generates the reciprocal negative feedback loop between miR-7 and Yan, which in turn helps to coordinate the orderly specification of photoreceptors.
miR-7 and chordotonal organs
miR-7 also regulates the development of proprioceptive (chordotonal) organs (Li et al., 2009) (Figure 2B), and E(spl)-C targets are relevant in this context (Lai et al., 2005; Stark et al., 2003). In contrast to retinal development, however, miR-7 participates with E(spl)-C and the proneural transcription factor Atonal in an incoherent FFL in this context (Li et al., 2009). In this schema, an upstream factor activates two targets with opposing functions. In particular, Atonal directly activates the transcription of many E(spl)-C genes as well as of mir-7, which as mentioned represses multiple E(spl)-C genes. These genes are further connected as E(spl)-bHLH repressor proteins inhibit atonal transcription. In such an incoherent FFL configuration, fluctuating Ato levels may result in back and forth repression between Atonal and E(spl)-C genes, but robust Atonal expression will result in sustained repression of E(spl)-C genes via miR-7, further strengthening Atonal expression. The objective and outcome of these complex gene regulatory networks is to permit neural specification only in response to strong Atonal activity.
It is relevant to bear in mind that miR-7, in stark contrast to Yan, Ato, and the E(spl)-C, is mostly dispensable for eye and chordotonal organ development under standard laboratory conditions (Li and Carthew, 2005). Instead, it was proposed that many regulatory connections of miR-7 in feed-forward and feedback loops provide robustness to the precise nature of nervous system specification. To test this theory, mir-7 mutants were subjected to environmental perturbation using temperature cycling. Indeed, the combination of mir-7 deletion and fluctuating temperature resulted in reduction of Ato, increased and irregular accumulation of Yan, and defects in eye and sensory organ development (Li et al., 2009).
miR-7 and germline stem cells
A distinct role for miR-7 in the differentiation of germline stem cells (GSCs) was uncovered through analysis of mutants in maelstrom (mael), which encodes an HMG-box transcription factor (Findley et al., 2003; Pek et al., 2009). In wild-type testes, GSCs maintain contact with the somatic hub and divide asymmetrically to produce two cells; a differentiating gonialblast (GB), which loses contact with the hub, and the stem cell, which retains contact with the hub. In male flies lacking mael, differentiation fails in the cells that should have become the GBs, leading to strong accumulation of GSC-like cells in the testes and a decline in fertility with age.
This phenotype is reminiscent of loss of function mutations in bag of marbles (bam), a novel gene that regulates GSC progression, and Bam is in fact reduced at the post-transcriptional level in mael testes (Pek et al., 2009). The bam 3’ UTR contains binding sites for several miRNAs, including miR-7, and a bam 3' UTR sensor was sensitive to miR-7 dosage in vivo. A regulatory cascade was extended by the observation that miR-7 levels are increased in mael testes, concomitant with reduction of a repressive histone mark at the mir-7 locus. Since Mael binds to consensus HMG binding sites upstream of the mir-7 promoter, Mael may directly repress mir-7 during GB differentiation. The significance of miR-7 upregulation in mael mutants was shown by the finding that heterozygosity for mir-7 partially suppressed the differentiation defect in mael testes (Figure 2C). Recently, miR-7 was also implicated in the development of ovarian GSCs through the regulation of the cyclin-dependent kinase inhibitor dacapo (Figure 2D), providing evidence for roles of miR-7 in both the male and female germline (Yu et al., 2009).
Altogether, these studies of miR-7 introduce the concept that a single miRNA can have different key targets in different developmental contexts. However, while miR-7 is identical between flies and humans, these data are not necessarily informative of key miR-7 targets in vertebrates. Indeed, while many miRNA targets can be defined on the basis of their evolutionary conservation amongst flies, amongst nematodes, or amongst vertebrates, there turn out to be very few targets shared by miRNAs that are conserved in all three of these species clades (Chen and Rajewsky, 2006). This has led to the notion that miRNA targets are acquired and lost quite frequently across large evolutionary distances. Indeed, vertebrate orthologues of yan and E(spl)bHLH genes do not contain miR-7 sites that are conserved amongst vertebrates, and miR-7 is not apparently expressed in vertebrate chordotonal organs (Li et al., 2009). In the case of the GSC requirement for miR-7, despite the evidence of a functional role for miR-7 in repressing Bam expression in GBs, the binding site analyzed is not even present in the closest sister species to D. melanogaster, namely D. simulans and D. sechellia (Pek et al., 2009). These results suggest that mRNAs can readily evolve new regulatory relationships with conserved miRNAs.
miR-8: deeply conserved roles of a conserved miRNA
Another miRNA well conserved through animal evolution is miR-8. There is a single copy of mir-8 in flies, but mammalian genomes have 5 paralogues in two distinct genomic clusters that are known as the miR-200 family. Interestingly, in contrast to the studies to date with miR-7, several important miR-8 targets and biological activities appear to be conserved between invertebrates and vertebrates.
miR-8 and neurodegeneration
The first study of miR-8 loss of function reported flies with reduced survival, malformed legs and progressive neurodegeneration (Karres et al., 2007) (Figure 3A). As with most other miRNAs, there are many (in this case perhaps >300) targets of miR-8 that are predicted to be broadly conserved amongst the Drosophilids (e.g., http://www.targetscan.org/). If the derepression of all of these contributed equivalently to the mir-8 mutant phenotype, then it would be unlikely that any particular gene would emerge as a key target responsible for phenotype. Amongst genes upregulated in mir-8 mutants, the transcriptional corepressor encoded by atrophin has several miR-8 binding sites in its 3' UTR and was responsive to miR-8 in sensor tests. Remarkably, ectopic expression of atrophin in miR-8-expressing cells phenocopied mir-8 mutants, while heterozygosity for atrophin could partially rescue the survival and leg defects of mir-8 mutants. These data therefore support a particularly substantial role for miR-8-mediated repression of atrophin, out of the hundreds of genes that bear conserved miR-8 binding sites. Notably, the human ortholog of atrophin, RERE, has three potential binding sites for mammalian miR-8 members, and is responsive to their activity in cultured cell assays (Karres et al., 2007). Therefore, this regulatory linkage may be deeply conserved across animals.
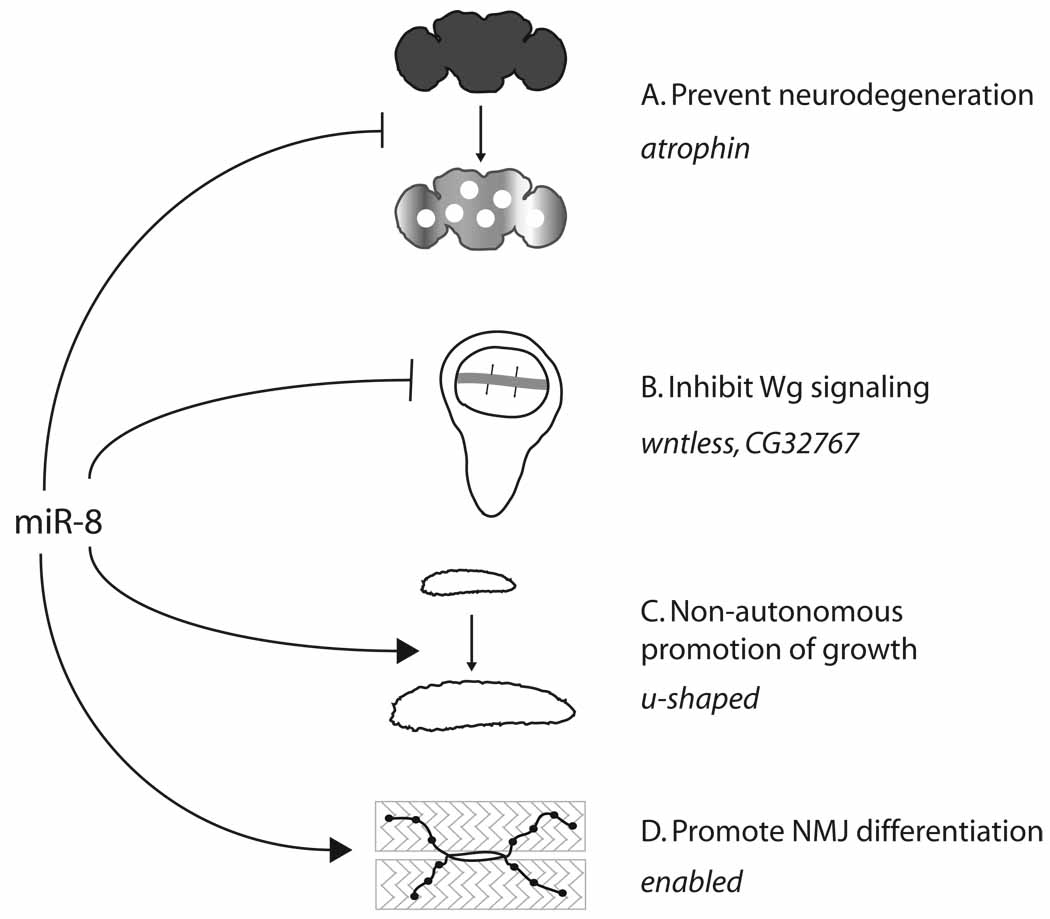
Figure 3. Processes regulated by Drosophila miR-8.
(A) miR-8 prevents neurodegeneration in the brain by limiting levels of the transcriptional corepressor atrophin. (B) Though not demonstrated under endogenous circumstances, miR-8 is able to repress Wg/Wnt signalling in the developing wing and eye by inhibiting of wntless, which encodes a transmembrane protein required for secretion of Wg; it can also repress CG32767, a positive regulator of Wg/Wnt signalling. (C) During larval growth, miR-8 represses the zinc finger protein encoded by u-shaped in the fat body to promote organismal growth. (D) Finally, miR-8 controls the morphology of neuromuscular junctions by acting in muscles to repress enabled, an EVH1/PH domain regulator of synapse development.
miR-8 and Wingless signaling
An overexpression screen for negative regulators of the Wingless (Wg)/Wnt pathway in Drosophila also identified miR-8 (Kennell et al., 2008). In this role, miR-8 directly represses wntless (wls), a gene required for export of the Wg morphogen (Figure 3B). Overexpression of miR-8 in vivo decreased levels of Wls cell-autonomously, with a concomitant decrease of extracellular Wg. A direct regulatory relationship with miR-8 was also reported for the positive regulator of the Wg pathway CG32767 (DasGupta et al., 2005; Kennell et al., 2008), suggesting that miR-8 negatively regulates the Wg pathway at multiple levels. Using a mouse cell line in which inhibition of Wnt signalling leads to adipogenesis, ectopic expression of mammalian miR-200 family members also induced adipogenesis marker genes and increased lipid accumulation. Although the endogenous influence of these miRNAs on signaling remains to be demonstrated, these studies suggest that a role for miR-8 in repressing Wg/Wnt signaling may be conserved in mammals.
miR-8 and growth control
Identification of miR-200 family miRNAs as growth promoting factors when ectopically expressed in human cell culture led to re-analysis of the original mir-8 mutant flies (Hyun et al., 2009; Park et al., 2009). This uncovered a significant growth defect at larval stages, slightly delayed eclosion and reduced size in adults for flies lacking miR-8 (Figure 3C). This appeared to be due to reduced cell number, with no obvious difference in cell size. In addition to previously reported expression patterns (Karres et al., 2007), mature miR-8 was strongly expressed in the fat body, and fat body-specific expression of miR-8 was sufficient to rescue the size defect of mir-8 mutants (Hyun et al., 2009). Notably, neuronal expression of miR-8 did not rescue the growth defect, demonstrating that the growth and neuronal phenotypes of mir-8 mutants are distinct.
To identify the relevant target(s) of miR-8 in the fat body, predicted targets of Drosophila miR-8 and the human miR-200 family were pooled from multiple computational schema, and mined for shared targeting of orthologous transcripts. Out of 15 orthologous pairs, seven were found responsive to their cognate miRNAs in their respective cell assays. Fat body-specific knockdown of these candidates using inverted repeat transgenes was used to assess whether reduction of any of them could rescue the small body phenotype of mir-8 mutants. Interestingly, depletion of the zinc finger protein encoded by u-shaped (ush) rescued mir-8 flies to wild-type weight. Notably, ush knockdown did not increase the weight of wild-type flies, indicating that the rescue is not simply an additive effect but is specific to the mir-8 mutant condition. Further analysis showed that Ush is elevated in mir-8 larvae, and that Ush and its human orthologue FOG2 regulate the PI3 kinase pathway by directly interacting with the regulatory subunit of PI3 kinase (Hyun et al., 2009).
miR-8 and NMJ development
Amongst the varied phenotypes associated with manipulation of miR-8, the most recent finding is its control over neuromuscular junction (NMJ) formation (Loya et al., 2009). In flies null for mir-8, NMJs are smaller, less branched, and have fewer boutons than in wild-type (Figure 3D). This phenotype was recapitulated by ubiquitous expression of a miR-8 transgenic sponge, a decoy target that acts as a sequence-specific competitive inhibitor of miRNA function (Ebert et al., 2007). Transgenic knockdown of miRNA activity permitted a test of whether miR-8 was required pre- or post-synaptically. In fact, the mir-8 mutant phenotype was obtained when the sponge was expressed post-synaptically in the target muscles, but not when expressed in the neurons. These phenotypes resembled manipulation of Enabled, an EVH1/PH domain regulator of synapse development. Indeed, enabled is a direct miR-8 target whose protein output is substantially increased in mir-8 mutants. More to the point, reduction of enabled dosage, either systemically or specifically in the muscle, detectably alleviated mir-8 mutant and miR-8-sponge phenotypes. These data provide strong support that deregulation of Enabled in muscles is causative for the NMJ phenotype of mir-8 mutants.
The collected analyses of miR-8 provide a curious contrast to those with miR-7. Both miRNAs are conserved between invertebrates and vertebrates, but the major targets of Drosophila miR-7 to date (yan, multiple E(spl)-C and Brd-C genes, bam) are not similarly conserved. In contrast, the orthologs of two different major confirmed targets of Drosophila miR-8 (atrophin and ush) were reported as targets of the orthologous miR-200 family in vertebrates, and there is some evidence suggesting that miR-8 represses Wg signalling in both flies and mammals. The meta-analysis of combined outputs from many prediction algorithms yielded only a handful of targets for miR-7 (Li et al., 2009) and miR-8/miR-200 (Hyun et al., 2009) that are even conceivably shared between mammals and flies (9 for miR-7, 15 for miR-8/miR-200), in agreement with previous conclusions on a paucity of deeply conserved miRNA:target reglationships (Chen and Rajewsky, 2006). The analysis of such deeply conserved targets of miR-7 may conceivably reveal important biological interactions, as should investigation of additional clade-specific targets of miR-8 (such as enabled).
miR-9a: multiple roles in developmental patterning
Drosophila miR-9a was originally described as a gene that exhibited strong target avoidance in its expression domain (Stark et al., 2005). In the early embryo, mir-9a is specifically expressed in the presumptive ectoderm and neuroectoderm, but is later maintained only in the ectoderm and is excluded from neural tissue. mRNAs with the ontological classification "neurogenesis" are enriched for the shared target sites of the related miR-9a/b/c miRNAs, suggesting that expression of miR-9 family miRNAs is mutually exclusive to many of its targets. It was inferred that a role for miR-9a is to suppress the neural potential of ectodermal derivatives (Stark et al., 2005).
miR-9a and sensory bristles
Flies lacking miR-9a exhibit a mild ectopic sensory organ phenotype in the larva, on the anterior wing margin, and on the thorax, consistent with a failure to correctly repress neural identity (Li et al., 2006) (Figure 4A). These flies also exhibited a completely penetrant loss of posterior wing margin. Here, the zinc finger proneural gene senseless (sens) was reported as a critical target of miR-9a. In particular, heterozygosity for sens restored the correct number of sensory organs on the anterior wing margin and could partially rescue the loss of posterior wing margin in mir-9a mutants (Li et al., 2006). Based on the ectopic but variable increase of sensory organs in mir-9a mutant, it was proposed that miR-9a helps prevent in appropriate SOP specification by keeping sens levels below a threshold needed to lock in the SOP fate robustly (Cohen et al., 2006; Li et al., 2006). Therefore, sens appears to be one of the key neurogenesis targets of miR-9a, amongst a cohort of neural targets of this miRNA.
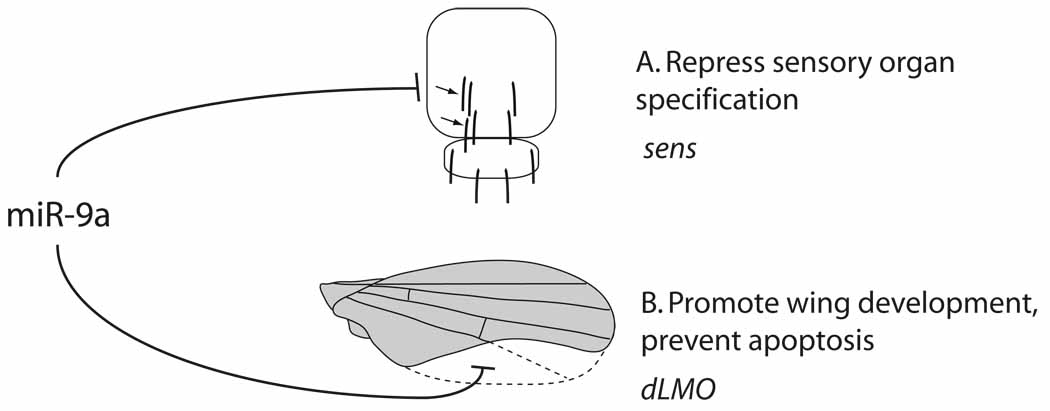
Figure 4. Processes regulated by Drosophila miR-9a.
(A) During development of the peripheral nervous system, miR-9a prevents ectopic specification of sensory organ bristles (arrows) through repression of the zinc finger transcription factor encoded by senseless (and to a lesser extent via the LIM protein encoded by dLMO). (B) miR-9a also controls margin specification and limits apoptosis in the developing wing through regulation of dLMO (and to a lesser extent, senseless); the dashed portion of the wing represents tissue that fails to develop in the mir-9a mutant.
miR-9a and wing development
Although the strong wing defect of the mir-9a mutant was reported earlier (Li et al., 2006), its developmental basis was not examined. Recent studies showed that mir-9a mutant wing pouches, assayed during larval development, showed defects in wing margin gene expression, broad ectopic apoptosis, and upregulation of the transcriptional regulator Drosophila LIM-only (dLMO) (Bejarano et al., 2009; Biryukova et al., 2009) (Figure 4B). Indeed, dLMO proved to the critical target of miR-9a in this setting. In contrast to heterozygosity for sens, removal of one copy of dLMO strongly reduced ectopic apoptosis during wing development and completely rescued the loss of posterior margin seen in mir-9a mutant flies (Bejarano et al., 2009; Biryukova et al., 2009). A further twist is that dLMO has an additional pre-pattern function to specify certain proneural territories (Asmar et al., 2008); in this context, it acts developmentally upstream of Sens. Heterozygosity for dLMO can partially suppress the ectopic sensory bristles present on the notum of mir-9a mutants, although not to the same extent as the reduction of sens (Bejarano et al., 2009). Therefore targeting of both Sens and dLMO by miR-9a are relevant to both PNS and wing development, but Sens may be more critical in the PNS and dLMO more critical in the wing.
In retrospect, the realization of a profound requirement for regulation of dLMO by miR-9a was the culmination of nearly 100 years of Drosophila genetics. dLMO was hit genetically in the 1920s by dominant Beadex mutations that induced wing loss (Morgan, 1925) similar to mir-9a mutants, and these proved to be transposon insertions that disrupted the 3' UTR of dLMO (Milán et al., 1998; Shoresh et al., 1998; Zeng et al., 1998). In fact, dLMO levels must be so carefully controlled that even 2x dosage resulting from duplication of the dLMO locus can induce the Beadex mutant phenotype, i.e. wing margin loss (Green, 1952; Lifschytz and Green, 1979). This history serves to remind us that while the general appreciation of the importance of miRNA regulation has come quite recently, the genetic evidence for important miRNA targets has been with us for much, much longer.
Interestingly, miR-9a appears to have quite different relationships with its two best-characterised targets. With sens, the thresholding model suggests that before cell fate decisions have been made, miR-9a and sens are co-expressed in the same cells, where miR-9a acts to keep sens levels below a threshold that would trigger SOP formation without other appropriate signalling events. Once SOPs are defined, the expression patterns become mutually exclusive. The relationship with dLMO appears quite different. miR-9a is detected by in-situ hybridisation throughout the entire wing imaginal disc (Biryukova et al., 2009; Li et al., 2006), and dLMO protein is similarly quite broadly expressed (Milán et al., 1998). In addition, clonal loss of mir-9a leads to upregulation of both dLMO and a GFP sensor for miR-9a activity throughout the whole wing disc (Bejarano et al., 2009), indicating that miR-9a is expressed and acts in the same tissues as one of its primary targets. This is more indicative of a tuning relationship than a mutually exclusive relationship. Although the thresholding model also requires co-expression of miRNA and target, once the threshold is exceeded, the expression patterns are expected to become mutually exclusive. Therefore, the same miRNA may act in different manners with different targets in different tissues.
Bantam: many essential roles of an invertebrate-restricted miRNA
The three previous examples concerned miRNAs that are deeply conserved between invertebrates and vertebrates. Even so, some of the phenotypes that have been characterized might arguably be considered subtle, and dependent on the expertise of particular laboratories for their elucidation. For example, the notum peripheral nervous system defect of mir-9a mutants comprises an extra bristle or two in only a subset of animals, whereas the eye phenotype of mir-7 mutants is only revealed by sensitized genetic backgrounds or environmental perturbations. Since miRNAs that are more deeply conserved appear to generally have more targets than those that are more recently evolved, one might suspect that miRNAs with lesser evolutionary range might correspondingly be of more subtle biological importance. However, this is clearly not the case. For instance, miR-279 is insect-restricted but is mostly lethal prior to adult hatching, and causes a strong defect in olfactory nervous system specification (Cayirlioglu et al., 2008). miR-iab-8 is similarly insect-restricted, but its loss results in complete sterility in both sexes (Bender, 2008).
bantam and growth control
Perhaps the most compelling example of a Drosophila miRNA with dramatic biological functions, but lacking vertebrate homologs, is that of bantam. Amongst the first miRNAs to be characterized genetically, bantam was initially defined as an enigmatic locus that regulated growth (Hipfner et al., 2002; Raisin et al., 2003) and only later appreciated to encode a miRNA (Brennecke et al., 2003). Ectopic bantam promotes overgrowth of various tissues such as eyes and wings, while flies carrying hypomorphic alleles of bantam are small. The complete loss of bantam is lethal at larval and pupal stages, and clonal analysis of tissues homozygous for a null allele revealed their inability to proliferate substantially (Figure 5A). The pro-apoptotic gene head involution defective (hid) was a particularly compelling target of bantam, with 5 conserved target sites in its 3' UTR (Brennecke et al., 2003). Interestingly, suppression of apoptosis does not induce overt tissue overgrowth, as seen with bantam overexpression, suggesting that additional growth targets of bantam exist. These inferred targets remain elusive to date.
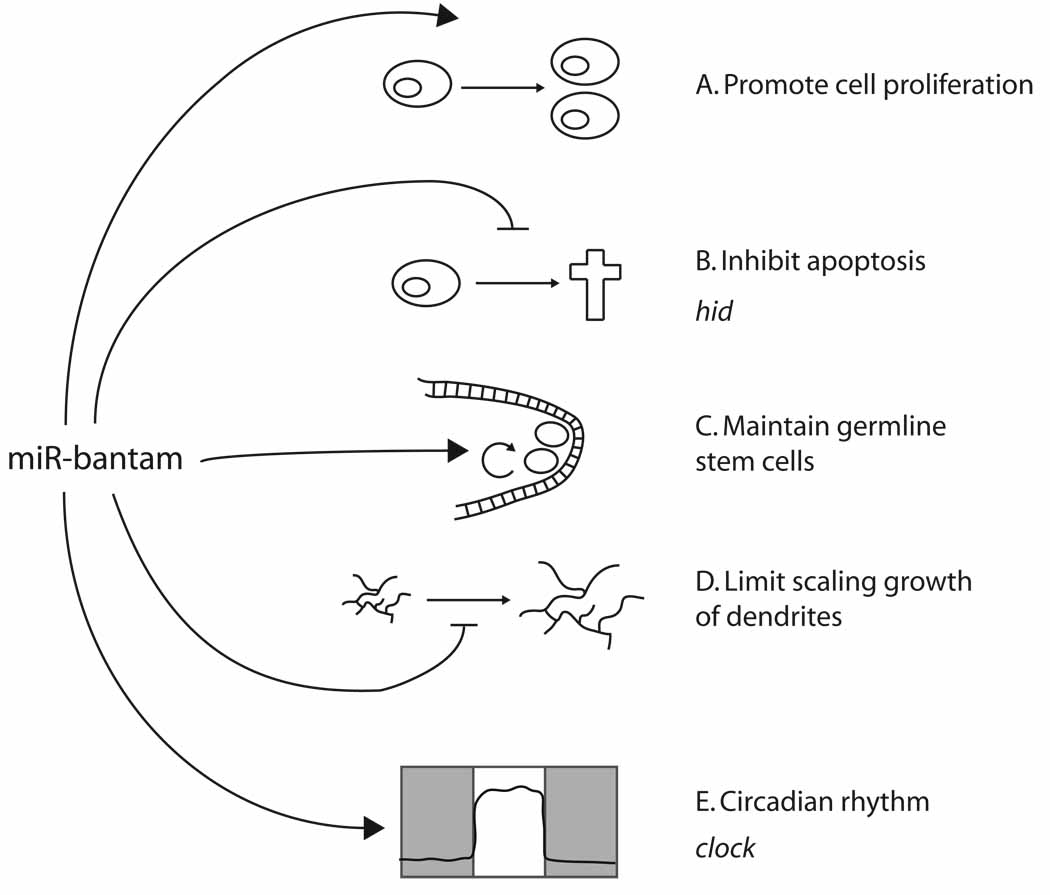
Figure 5. Processes regulated by Drosophila miR-bantam.
The bantam miRNA controls a wide variety of processes. (A) The most overt defect of bantam mutants are their small size due to failure of cell proliferation, although the relevant target(s) are not known. (B) bantam also prevents apoptosis (cross) by repressing hid, which encodes a pro-apoptotic factor. bantam maintains ovarian germline stem cells (C), and limits the scaling growth of dendrites during larval development (D); the targets mediating these functions are not yet known. (E) Finally, bantam maintains the appropriate length of the circadian clock (represented by an activity trace of animal movements across the day) by regulating clock, which encodes a bHLH-PAS transcription factor.
The first evidence for regulation of bantam by pathways that promote cell growth was the ability of bantam to modify the small, blistered eye phenotype of ectopic expression of the tumour suppressor expanded (Hipfner et al., 2002). Co-expression of bantam suppressed the phenotype resulting in almost wild-type morphology, whereas loss of one copy of bantam in this background resulted in a smaller eye with more pronounced blistering (Hipfner et al., 2002). Subsequently, Expanded was shown to be a key apical component of the Salvador/Warts/Hippo (SWH) growth control pathway (Cho et al., 2006; Hamaratoglu et al., 2006; Silva et al., 2006), for which loss-of-function of various cytoplasmic and transmembrane components results in massive tissue overgrowth and resistance to apoptosis.
The phenotypic similarity of bantam gain-of-function and loss-of-function of various SWH pathway components raised the question of whether bantam was a transcriptional target of the SWH pathway. Indeed, bantam proved to be a target of the Yorkie transcriptional co-activator (Nolo et al., 2006; Thompson and Cohen, 2006), whose nuclear function is suppressed by activity of the SWH pathway (Huang et al., 2005). Accordingly, overexpression of Yorkie in clones mutant for bantam failed to yield overgrowth, suggesting that bantam is required for Yorkie-induced overgrowth. Conversely, forced activation of bantam in cells lacking yorkie resulted in overgrowth, suggesting that ban is a key growth-promoting transcriptional target of Yorkie (Nolo et al., 2006; Thompson and Cohen, 2006). More recently, bantam was shown to be a crucial downstream target of the SWH pathway in the proliferating cells of the anterior eye imaginal disc, where Yorkie complexes with different transcription factors compared to the wing (Peng et al., 2009). Therefore, bantam is a crucial target of growth regulatory pathways in a variety of cellular and developmental contexts.
bantam and apoptosis
The most well characterised bantam:target interaction is with the pro-apoptotic gene hid (Brennecke et al., 2003; Nahvi et al., 2009), but only recently has this been analysed in the context of endogenous levels of the two genes (Figure 5B). In addition to their growth defects (Brennecke et al., 2003), bantam mutants exhibit ectopic apoptosis, and animals homozygous for hypomorphic alleles of bantam also show significantly increased levels of apoptosis in response to ionizing radiation (IR) (Jaklevic et al., 2008). Bantam also prevents rbf mutant interommatidial cells from undergoing cell death in the developing eye disc (Tanaka-Matakatsu et al., 2009), demonstrating that cell autonomous anti-apoptotic activity mediated by bantam is also present in the eye disc.
Consistent with a role in limiting excess apoptosis in response to IR, bantam activity is strongly increased soon after IR, as measured by decreased levels of a bantam-responsive sensor, however, this does not appear to correspond to significant changes in levels of mature bantam (Jaklevic et al., 2008). Suppression of apoptosis in irradiated cells by expression of the baculovirus inhibitor of apoptosis protein p35 prevents the downregulation of the bantam sensor, suggesting that the upregulation of bantam activity is due to a signal from apoptotic cells and is not a direct response to DNA damage. p53, which is a crucial gene in DNA damage response, is required for the increase in bantam activity induced by IR (Jaklevic et al., 2008). On the other hand, IR exposure is also expected to result in p53-mediated activation of the SWH pathway activation, which is expected to decrease bantam transcription leading to apoptosis (Colombani et al., 2006). These apparently contradictory data may be reconciled by the non-autonomous role of apoptosis in inducing bantam activity. Perhaps p53 is inducing SWH pathway activation in cells that have sustained DNA damage beyond a certain threshold, leading to apoptosis and a growth-promoting signal to neighbouring cells, which then increase their bantam activity. This may explain the absence of an obvious increase in bantam levels in these irradiated discs.
Bantam and germline stem cells
Animals that are hypomorphic for bantam exhibited defects in GSC maintenance, with most animals having no remaining GSCs after 14 days (Yang et al., 2009) (Figure 5C). Furthermore, clones of larval primordial germ cells (PGCs) that are null for bantam showed branched fusome structures, which are markers of PGC differentiation, suggesting that bantam limits PGC differentiation. Interestingly, in this study, GSC clones of bantam mutants did not exhibit any significant defects in maintenance, suggesting that the requirement for bantam in GSC maintenance is non-autonomous and may potentially reside in the GSC niche (Yang et al., 2009). Curiously, a separate study described a clear phenotype of defective maintenance for bantam mutant GSC clones (Shcherbata et al., 2006). Both studies utilized similar techniques involving null deletion alleles for bantam, so the reason for this discrepancy is unclear. A separate study demonstrated that the bantam sensor is strongly downregulated in GSCs compared to 16 cell cysts, suggesting that bantam is active in GSCs and inactive in the differentiating cysts (Neumuller et al., 2008).
Bantam and dendrite tiling
Very recent studies of bantam revealed its unexpected influence on the differentiation of post-mitotic neurons. In the early Drosophila larva, sensory neurons tile across the body wall to create a sensory field. As the larva grows, a fixed number of sensory neurons must expand their fields of coverage by extending dendrites in proportion to the growth of the organism as a whole, a process referred to as scaling growth. Amongst a panel of mutants that exhibit defective larval growth, animals null for bantam established normal coverage of the receptive field in early larvae, but failed to attenuate scaling growth after cessation of larval growth (Parrish et al., 2009) (Figure 5D). Strikingly, this phenotype was rescued by expression of bantam in the overlying epithelial cells, and not in the sensory neurons, demonstrating that bantam is required in these epithelial cells to provide a signal that attenuates scaling growth of sensory neurons (Parrish et al, 2009). Therefore, as hinted at by studies in GSCs, bantam also has important non-autonomous roles during dendrite tiling.
Bantam and circadian rhythm
A distinct role for bantam in neural function was revealed during studies of the circadian clock. The eponymous clock gene encodes a bHLH-PAS transcription factor that is a central generator of circadian rhythms, and is also a substantial target of bantam (Kadener et al., 2009). Overexpression of bantam in timeless-expressing cells, which comprise the central pacemaker, resulted in lengthening of the circadian period by three hours (Figure 5E). This phenotype is similar to the periodicity phenotype caused by loss of clock function, and the annotated clock 3’ UTR is indeed predicted to have a single bantam binding site. However, further studies revealed that the predominantly expressed clock 3' UTR is substantially longer and bears three functional bantam binding sites, which are responsive to bantam repression in cultured cell assays. This provides a motivation to be cautious in assessing computational target predictions, which do not necessarily account for all in vivo transcript isoforms.
Unlike other characterized settings of bantam function, it is technically challenging to remove bantam specifically from pacemaker cells to check for a circadian phenotype. Still, an informative test can be done in cis to assess the functional impact of bantam sites in the clock transcript. Compared to a wild-type clock transgene, a clock transgene bearing specific mutations in its bantam binding sites was unable to provide full genetic rescue of the clock null mutant background (Kadener et al., 2009). This constituted compelling evidence for the endogenous regulation of clock by bantam to generate normal circadian rhythm.
The wide range of severe phenotypes resulting from loss of bantam are notable in light of the widely-held view that miRNAs are primarily fine tuning factors. Through regulation of its so far described targets hid and clock, bantam regulates processes as diverse as apoptosis and circadian rhythm. Since bantam has additional significant roles in promoting growth, maintaining GSCs and non-autonomously attenuating dendrite scaling, it will be interesting to see what other key targets it has, and whether bantam functions via "a few" or "many" targets in these various settings.
Perspectives
Through the analysis of miRNA mutants and the regulatory relationships they reveal with target genes, we are beginning to understand more about the complex and varied roles of miRNAs in animal development. The collected studies in the Drosophila model affirm the conclusion that there is no single rule or reason for miRNA regulation of a given process.
miRNAs undoubtedly have many, perhaps hundreds of direct targets, and bioinformatic studies reveal underlying logic to these networks. One conclusion is that miRNAs may suppress broad transcript cohorts that are leftover from progenitor states, or that may arise spuriously because of alternative developmental potentials of a given celltype. In this view, miRNA regulation is secondary to transcriptional regulation and serves as a general backup to reinforce a particular cell state. If the individual regulation of many targets is not critical for gross phenotypes, this might permit a great deal of evolutionary fluidity in the target networks for otherwise highly conserved miRNAs. On the other hand, there is also abundant evidence that miRNAs can have key target genes that explain a great deal about the genetic function of the miRNA gene. And at least some of these cases require the function of the miRNA in target-expressing cells, so that miRNAs play instructive roles to deliver just the right amount of target activity.
Although some of key miRNA targets were revealed as "top" predictions that contain multiple binding sites for the miRNA (e.g. hid with 5 highly conserved binding sites for bantam), other key target genes have only single target sites (e.g. dLMO with only one clear site for miR-9a) or have target sites that did not exist in annotated 3' UTRs (e.g. clock with seemingly downstream bantam sites). For many functional analyses of miRNAs, then, genetic phenotypes were critical for understanding the role of the miRNA and providing a rationale to pursue individual targets in depth.
Early evidence for the breadth of the miRNA target network came from focusing on conserved targets, and certainly conservation implies a likelihood of beneficial function. However, highly conserved features do not necessarily imply that they mediate processes that yield obvious phenotypes in the mutant situation. Certainly there are many highly conserved putative transcriptional regulatory regions for which no activity could be defined in vivo, and there are many highly conserved protein-coding genes whose deletion similarly causes no obvious defect. Nevertheless, small quantitative deviations in gene activity can result in dramatic costs to fitness, especially when measured not in a few generations in optimal laboratory conditions, but instead across evolutionary scales of time and on the population level. Therefore, the consequences of deleting some conserved miRNA genes, and potentially of removing many conserved miRNA binding sites, may need to be studied under challenging environmental conditions, under sensitized genetic backgrounds, or using highly quantitative strategies.
On the other hand, the simple lack of deep conservation may not be taken as strict evidence for lack of useful functions. We have discussed that the knockouts of several miRNAs that are conserved between invertebrates and vertebrates have relatively modest effects in Drosophila, whereas the invertebrate-specific miRNA bantam plays crucial roles in a dazzling variety of developmental and physiological settings. bantam is essential in both proliferating and post-mitotic tissues, and regulates both cell-autonomous and non-cell autonomous processes. Other insect-specific miRNAs seem to have crucial functions even as measured by such crude measures as viability and fertility. Going to the more "micro" level, there are now cases of newly-evolved miRNA:target relationships that appear to play important roles in specific Drosophilid species. Perhaps changing miRNA:target relationships have played active roles in creating species-specific characteristics.
Finally, we are fascinated by the fact that the continued study of individual Drosophila miRNA mutants continues to reveal additional phenotypes caused by the deregulation of newly defined target transcripts. Pleiotropic functions of miRNAs may provide a contributing rationale for the existence of extensive target networks for individual miRNAs. As is the case for many genes encoding transcription factors and other regulatory factors, we presume that there will continue to be more to miRNA activities than is revealed at first glance.
Acknowledgments
We thank Zhigang Jin for critical reading. Work in E.C.L.'s group was supported by the Bressler Scholars Fund, the Burroughs Wellcome Fund, and the NIH (R01-GM083300 and U01-HG004261).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Asmar J, Biryukova I, Heitzler P. Drosophila dLMO-PA isoform acts as an early activator of achaete/scute proneural expression. Dev Biol. 2008;316:487–497. doi: 10.1016/j.ydbio.2008.01.040. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bejarano F, Smibert P, Lai EC. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 2008;22:14–19. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryukova I, Asmar J, Abdesselem H, Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev Biol. 2009;327:487–496. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam Encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA-Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P, Kadow IG, Zhan X, Okamura K, Suh GS, Gunning D, Lai EC, Zipursky SL. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science. 2008;319:1256–1260. doi: 10.1126/science.1149483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3'UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71:149–156. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding--a new role for microRNAs. Genes Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- Colombani J, Polesello C, Josue F, Tapon N. Dmp53 activates the Hippo pathway to promote cell death in response to DNA damage. Curr Biol. 2006;16:1453–1458. doi: 10.1016/j.cub.2006.05.059. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MM. The Beadex Locus in Drosophila Melanogaster: The Genotypic Constitution of Bx. Proc Natl Acad Sci U S A. 1952;38:949–953. doi: 10.1073/pnas.38.11.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161:1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Jaklevic B, Uyetake L, Wichmann A, Bilak A, English CN, Su TT. Modulation of ionizing radiation-induced apoptosis by bantam microRNA in Drosophila. Dev Biol. 2008;320:122–130. doi: 10.1016/j.ydbio.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci U S A. 2008;105:15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, Macmenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai EC. microRNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3' UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. Regulation of Drosophila neurogenesis by RNA:RNA duplexes? Cell. 1998;93:1103–1104. doi: 10.1016/s0092-8674(00)81454-3. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Leviten MW, Lai EC, Posakony JW. The Drosophila gene Bearded encodes a novel small protein and shares 3' UTR sequence motifs with multiple Enhancer of split Complex genes. Development. 1997;124:4039–4051. doi: 10.1242/dev.124.20.4039. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA Mediates EGF Receptor Signaling and Promotes Photoreceptor Differentiation in the Drosophila Eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & Development. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E, Green MM. Genetic identification of dominant overproducing mutations: the Beadex gene. Mol Gen Genet. 1979;171:153–159. doi: 10.1007/BF00270001. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Sandler L, Baker BS, Carpenter ATC, Denell RF, Hall JC, Jacobs PA, Miklos GL, Davis BK, Gethman RC, et al. Segmental aneuploidy and the genetic structure of the Drosophila genome. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M, Diaz-Benjumea FJ, Cohen SM. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev. 1998;12:2912–2920. doi: 10.1101/gad.12.18.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. The genetics of Drosophila melanogaster. Biblphia Genet. 1925;2 [Google Scholar]
- Nahvi A, Shoemaker CJ, Green R. An expanded seed sequence definition accounts for full regulation of the hid 3' UTR by bantam miRNA. Rna. 2009;15:814–822. doi: 10.1261/rna.1565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008 doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron. 2009;63:788–802. doi: 10.1016/j.neuron.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Lim AK, Kai T. Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Dev Cell. 2009;17:417–424. doi: 10.1016/j.devcel.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, Rokhsar DS, Aboobaker AA. Evidence for a microRNA expansion in the bilaterian ancestor. Dev Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- Raisin S, Pantalacci S, Breittmayer JP, Leopold P. A new genetic locus controlling growth and proliferation in Drosophila melanogaster. Genetics. 2003;164:1015–1025. doi: 10.1093/genetics/164.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007b;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shcherbata HR, Hatfield S, Ward EJ, Reynolds S, Fischer KA, Ruohola-Baker H. The MicroRNA Pathway Plays a Regulatory Role in Stem Cell Division. Cell Cycle. 2006;5:172–175. doi: 10.4161/cc.5.2.2343. [DOI] [PubMed] [Google Scholar]
- Shoresh M, Orgad S, Shmueli O, Werczberger R, Gelbaum D, Abiri S, Segal D. Overexpression Beadex mutations and loss-of-function heldup-a mutations in Drosophila affect the 3' regulatory and coding components, respectively, of the Dlmo gene. Genetics. 1998;150:283–299. doi: 10.1093/genetics/150.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Smibert P, Lai EC. Lessons from microRNA mutants in worms, flies and mice. Cell Cycle. 2008;7 doi: 10.4161/cc.7.16.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA Targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Xu J, Cheng L, Du W. Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev Biol. 2009;326:347–356. doi: 10.1016/j.ydbio.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xu S, Xia L, Wang J, Wen S, Jin P, Chen D. The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009;5:e1000444. doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Reynolds SH, Hatfield SD, Shcherbata HR, Fischer KA, Ward EJ, Long D, Ding Y, Ruohola-Baker H. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development. 2009;136:1497–1507. doi: 10.1242/dev.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Justice NJ, Abdelilah S, Chan Y-M, Jan LY, Jan YN. The Drosophila LIM-only gene, dLMO, is mutated in Beadex alleles and might represent an evolutionarily conserved function in appendage development. Proc Natl Acad Sci USA. 1998;95:10637–10642. doi: 10.1073/pnas.95.18.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]