Abstract
A plethora of epigenetic modifications have been described in the human genome and shown to play diverse roles in gene regulation, cellular differentiation, and the onset of disease. While some modifications have been linked with activity levels of different functional elements, their combinatorial patterns remain unresolved, and their potential for systematic de novo genome annotation remains untapped. In this paper, we systematically discover and characterize recurrent spatially-coherent and biologically-meaningful chromatin mark combinations, or chromatin states, in human T-cells. We describe 51 distinct chromatin states, including promoter-associated, transcription-associated, active intergenic, large-scale repressed and repeat-associated states. Each chromatin state shows specific functional, experimental, conservation, annotation, and sequence-motif enrichments, revealing their distinct candidate biological roles. Overall, our work provides a complementary functional annotation of the human genome revealing the genome-wide locations of diverse classes of epigenetic functions, including previously-unsuspected chromatin states enriched in transcription end sites, distinct repeat families, and disease-SNP-associated states.
While the primary DNA sequence of the human genome is ultimately responsible for the encoding and functioning of each cell, numerous epigenetic modifications can modulate the interpretation of this primary sequence. These lead to the diversity of function found across different human cell types, play key roles in the establishment and maintenance of cellular identity during development, and have been associated with roles in DNA repair, replication, and disease. Post-translational modifications in the tails of histone proteins that package DNA into chromatin constitute perhaps the most versatile type of such epigenetic information, with more than a dozen positions of multiple histone proteins and variants each undergoing several distinct modifications, such as acetylation and mono-, di-, or tri-methylation1, 2.
More than 100 distinct histone modifications have been described, leading to the ‘histone code hypothesis’ that specific combinations of chromatin modifications would encode distinct biological functions3. Others however have instead proposed that individual epigenetic marks act in additive ways and the multitude of modifications simply serves a role of stability and robustness4. Understanding which combinations of epigenetic modifications are biologically meaningful, and revealing their specific functional roles, are still open questions in epigenomics, with great relevance to many ongoing efforts to understand the epigenomic landscape of health and disease.
To directly address these questions, we introduce a novel approach for discovering ‘chromatin states’ (Fig. 1; Supplementary Table 1, Supplementary Fig. 1), or biologically-meaningful and spatially-coherent combinations of chromatin marks, in a systematic de novo way across a complete genome based on a multivariate Hidden Markov Model (HMM) that explicitly models mark combinations. Biologically these states may correspond to different genomic elements (e.g. transcription start sites, enhancers, active genes, repressed genes, exons, heterochromatin), even though no information about these genomic elements is given to the model as input.
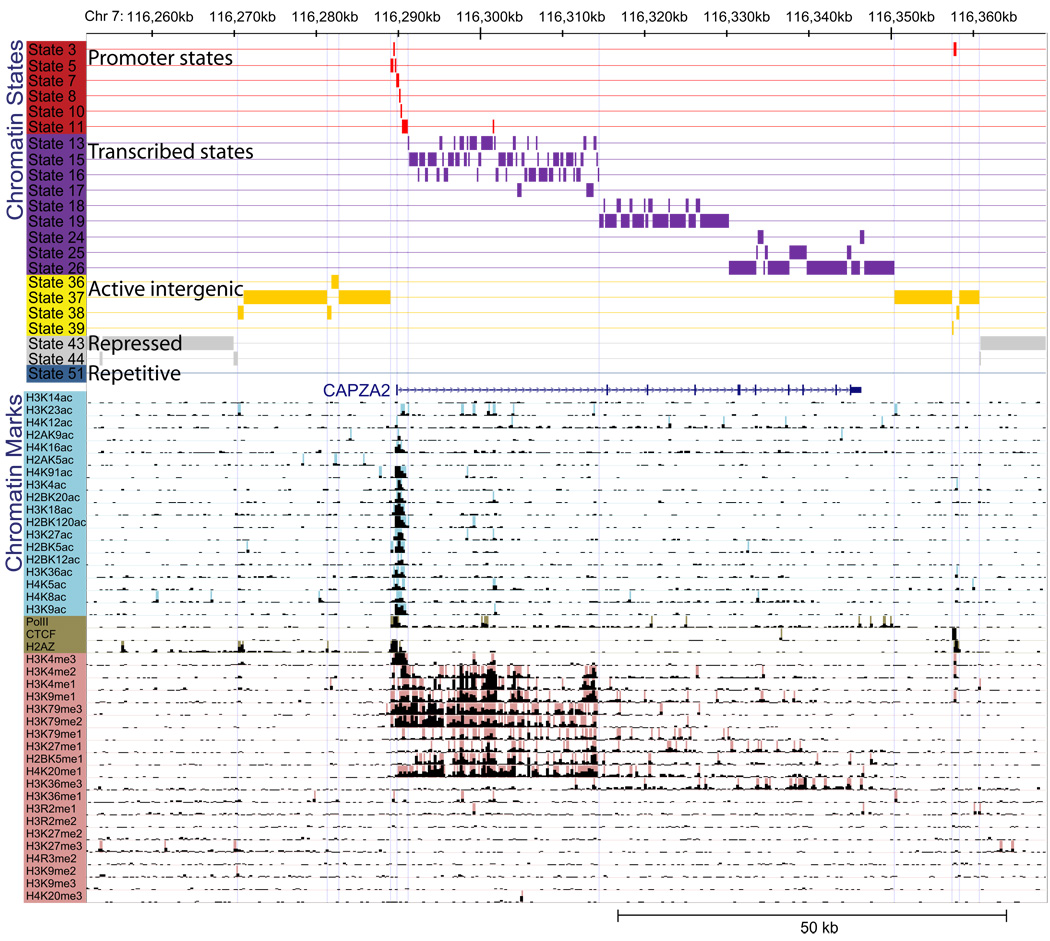
Figure 1. Example of chromatin state annotation.
Input chromatin mark information and resulting chromatin state annotation for a 120kb region of human chromosome 7 surrounding the CAPZA2 gene. For each 200-bp interval, the input ChIP-Seq sequence tag count (black bars) is processed into a binary presence/absence call for each of 18 acetylation marks (light blue), 20 methylation marks (pink), and CTCF/Pol2/H2AZ (brown). The precise combination of these marks in each interval in their spatial context is used to infer the most probable chromatin state assignment (colored boxes). Although chromatin states were learned independent of any prior genome annotation, they correlate strongly with upstream and downstream promoters (red), 5′-proximal and distal transcribed regions (purple), active intergenic regions (yellow), repressed (grey) and repetitive (blue) regions (state descriptions shown in Supplementary Table 1). This example illustrates that even when the signal coming from chromatin marks is noisy, the resulting chromatin state annotation is very robust, directly interpretable, and shows a strong correspondence with the gene annotation. Several spatially-coherent transitions are seen from large-scale repressed to active intergenic regions near active genes, from upstream to downstream promoter states surrounding the TSS, and from 5′-proximal to distal transcribed regions along the body of the gene. The frequent transitions to state 16 correlate with annotated Alu elements (57% overlap vs. 4% and 25% for states 13 and 15 respectively). Transitions to state 13 are likely due to enhancer elements in the first intron of CAPZA2, a region where regulatory elements are commonly found, and correlate with several enhancer marks. While maximum-probability state assignments are shown here, the full posterior probability for each state in this region is shown in Supplementary Figure 2.
HMMs are well-suited to the task of discovering unobserved ‘hidden’ states from multiple ‘observed’ inputs in their spatial genomic context (see Online Methods). In our model each state has a vector of ‘emission’ probabilities (Fig. 2 and Supplementary Figs. 2 and 3), reflecting the different frequency with which chromatin marks are observed in that state, and an associated ‘transition’ probability vector (Supplementary Fig. 4) encoding spatial relationships between neighboring positions in the genome, associated with spreading of chromatin marks, or functional transition such as between intergenic regions, promoters, and transcribed regions (see Supplementary Notes, Supplementary Figs. 5 and 6).
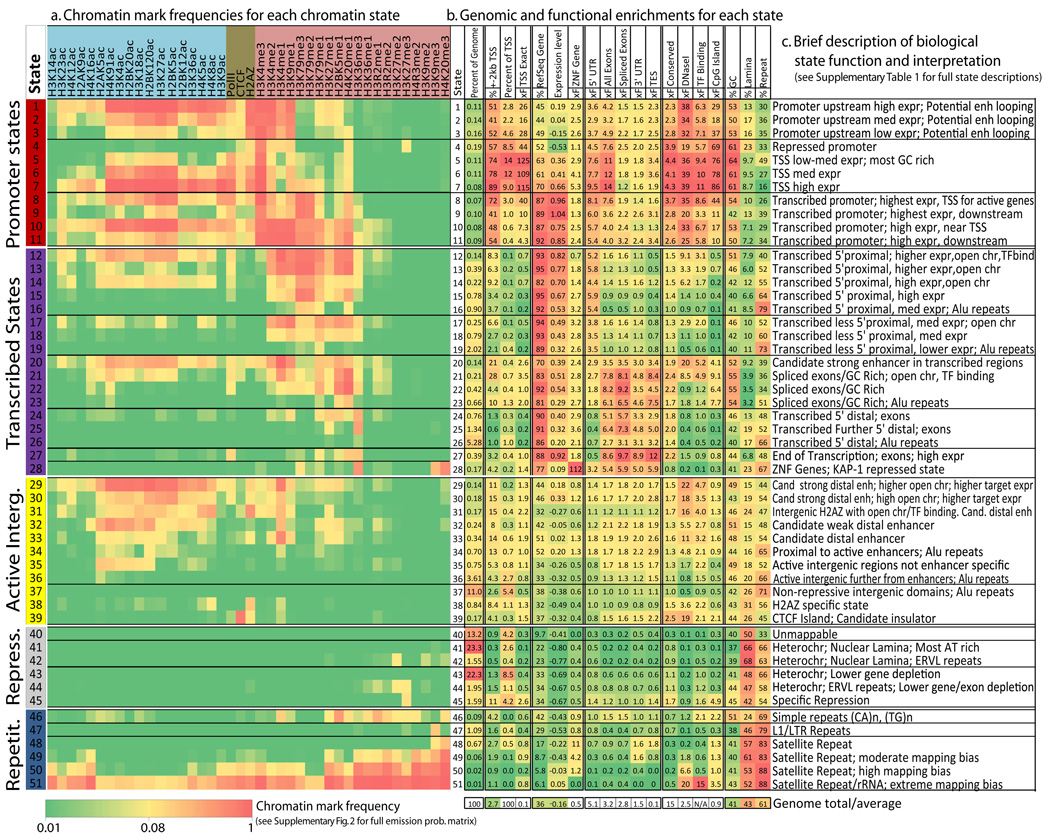
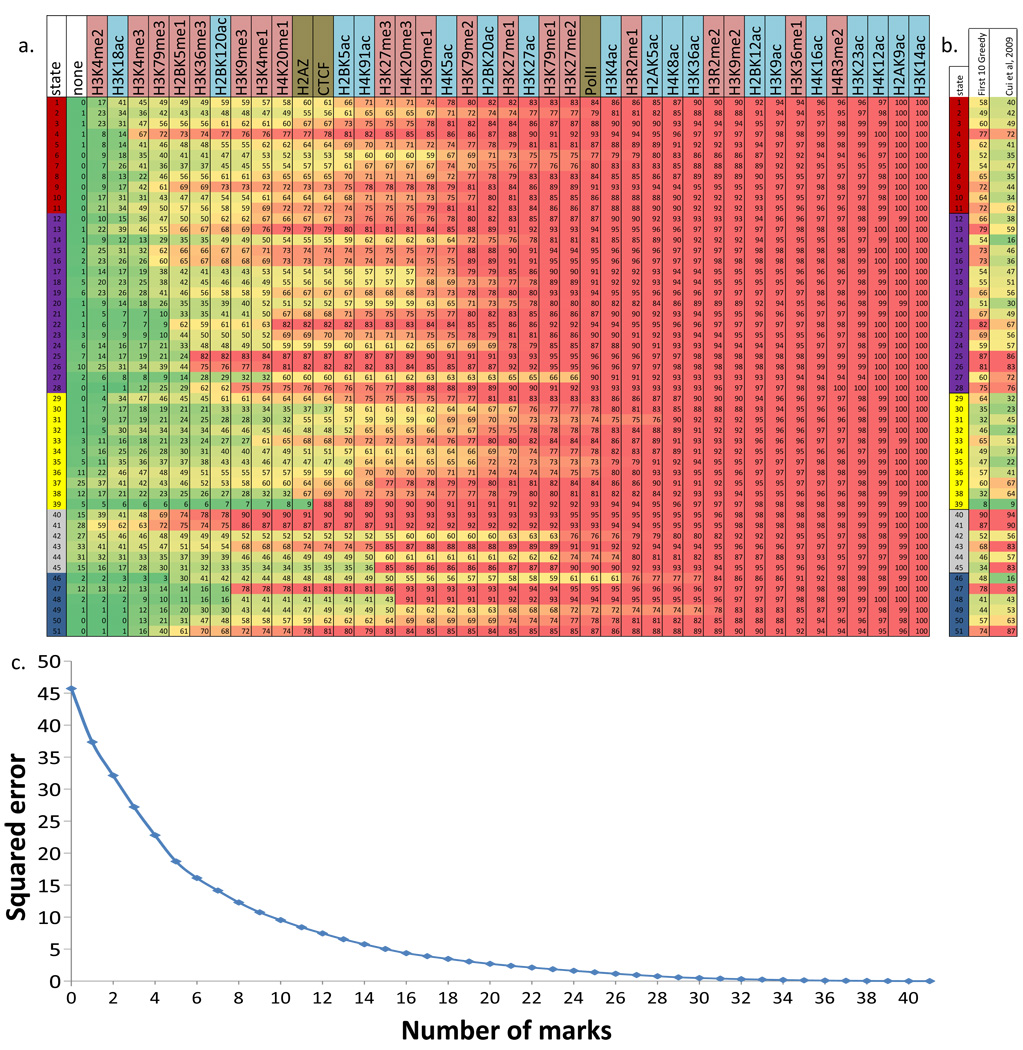
Figure 2. Chromatin state definition and functional interpretation.
a. Chromatin mark combinations associated with each state. Each row shows the specific combination of marks associated with each chromatin state, and the frequencies between 0 and 1 with which they occur (color scale). These correspond to the emission probability parameters of the Hidden Markov Model (HMM) learned across the genome during model training (values shown in Supplementary Fig. 2). Marks and states colored as in Figure 1. b. Genomic and functional enrichments of chromatin states. % denotes percentage, xF denotes fold enrichment. In order columns are: percentage of the genome assigned to the state; percentage of state that overlaps a 200bp-interval within 2kb of an annotated RefSeq Transcription Start Site (TSS); percentage of RefSeq TSS found in the state; fold enrichment for TSS; percentage of state overlapping a RefSeq transcribed region; average expression level of genomic intervals overlapping the state; fold enrichment for ZNF-named gene; fold enrichment for RefSeq 5′ Untranslated Region (5′-UTR) exon and introns; fold enrichment for RefSeq exons; fold enrichment for spliced exons (2nd exon or later); fold enrichment for RefSeq 3′ Untranslated Region (3′-UTR) exons and introns; fold enrichment for RefSeq transcription end sites (TES); fold enrichment for PhastCons conserved elements; fold enrichment for DNaseI hypersensitive sites; median fold enrichment for transcription factor binding sites over a set of experiments (expanded in Supplementary Fig. 23); fold-enrichment for CpG islands; percentage of GC nucleotides; percent overlapping experimental nuclear lamina data; percent overlapping a RepeatMasker element (expanded in Supplementary Fig. 31). All enrichments are based on the posterior probability assignments. Genome total indicates the total % of 200bp intersecting the feature or the genome average for expression and %GC. c. Brief biological state description and interpretation (‘chr’: chromatin, ‘enh’: enhancer, full descriptions in Supplementary Table 1).
We applied our model to the largest set of chromatin marks available to date, consisting of the genome-wide occupancy data for a set of 38 different histone methylation and acetylation marks in human CD4 T-cells, as well as histone variant H2AZ, PolII, and CTCF5, 6 obtained using chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) (Online Methods). To understand the biological importance of the resulting chromatin states, we undertook a large-scale systematic data-mining effort, bringing to bear dozens of genome-wide datasets including gene annotations, expression information, evolutionary conservation, regulatory motif instances, compositional biases, genome-wide association data, transcription-factor binding, DNaseI hypersensitivity, and nuclear lamina datasets.
This work has strong implications for genome-annotation providing an unbiased and systematic chromatin-driven annotation for every region of the genome at a 200bp resolution, which both refines previously-known classes of epigenetic states, and introduces new ones. Regardless of whether these chromatin states are causal in directing regulatory processes, or simply reinforcing independent regulatory decisions, these annotations should provide a valuable resource for interpreting biological and medical datasets, such as genome-wide association studies for diverse phenotypes, and potentially pinpointing novel classes of functional elements.
Results
Chromatin states model and comparison to previous work
Previous analyses have largely focused on identifying instances of or characterizing the marks predictive of specific classes of genomic elements defined a priori such as transcribed regions, promoters, or putative enhancers5–12, including left-to-right HMMs over locally-defined intervals12. An unsupervised local chromatin pattern discovery method13 first demonstrated that many of the patterns previously associated with promoters and enhancers could be discovered de novo, but did not discover patterns associated with broader domains and left the vast majority of the genome unannotated (Supplementary Fig. 7). Multivariate HMMs have also been used in an unsupervised fashion to model epigenomic data based on raw measured signal levels using a multivariate normal emission distribution model14–17, and a non-parametric histogram strategy18.
In contrast to previous approaches, we explicitly model the combinatorial detection of the presence of a set of marks, as opposed to modeling the range of measured experimental intensity levels for every input. This results in more directly interpretable states, is less prone to over-fit biologically-insignificant variations in signal intensity levels, makes fewer assumptions about the distribution of mark intensity levels associated with different states, and requires learning of significantly fewer parameters, thus increasing model robustness.
We also introduce a new framework for model learning and selection of the number of states that compactly and adequately describes the biological datasets (Online Methods), based on a two-stage nested initialization procedure. In the first stage, we learn a large compendium of models at varying numbers of states and from multiple random initializations, and select a best scoring model (Supplementary Fig. 8). In the second stage, we prune the selected model by removing states that are least representative of the mark combinations discovered across the compendium of models, and use the resulting pruned models as the seeds for an expectation-maximization learning procedure at each number of states. We finally selected a 51-state model that captures the biologically-interpretable states that were consistently found in larger models, while minimizing the total number of states, and further ensured that general properties of the resulting model validated our approach, including robustness to varying thresholds and different background models, and independence of marks given a chromatin state (Supplementary Notes, Supplementary Figs. 7–21, Supplementary Table 2).
We next describe the likely biological functions of the 51 discovered chromatin states, divided into five large groups.
Promoter-associated states
The first group of states, states 1–11, all had high enrichment for promoter regions: 40%–89% of each state was within 2kb of a RefSeq transcription start site (TSS), compared with 2.7% genome-wide (P<10−200, for all states). These states accounted for 59% of all RefSeq TSS while covering only 1.3% of genome. These states all had in common a high frequency of H3K4me3, but differed in terms of other associated marks, primarily H3K79me2/3, H4K20me1, H3K4me1/2, and H3K9me1, and the overall level of numerous acetylations (Fig. 2a). These correlated with varying levels of expression and varying enrichment levels for DNaseI hypersensitive sites, CpG islands, evolutionarily-conserved motifs and bound transcription factors (Fig. 2b and Supplementary Figs. 22–23).
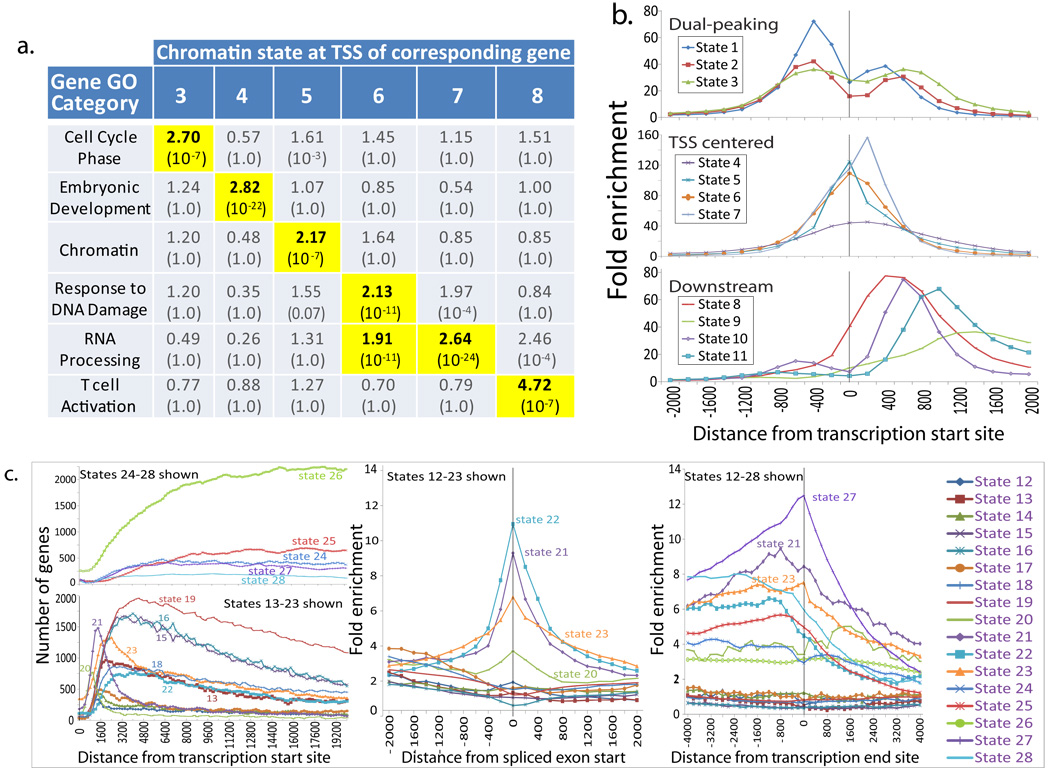
Surprisingly, promoter states differed in the Gene Ontology (GO) functional enrichments of associated genes including cell cycle, embryonic development, RNA processing, and T-cell activation (Fig. 3a). Promoter states also differed in their positional enrichments with respect to the TSS of associated genes (Fig. 3b). States 4–7 were most concentrated over the TSS (showing upwards of 100-fold enrichment), states 8–11 peaked between 400 bp and 1200 bp downstream of the TSS and corresponded to transcribed promoter regions of expressed genes, and states 1–3 peaked both upstream and downstream of the TSS.
Figure 3. Promoter and transcribed chromatin states show distinct functional and positional enrichments.
a. Distinct Gene Ontology (GO) functional enrichments (fold and corrected p-values) found for genes associated with different promoter states at their Transcription Start Site (TSS). For additional states and GO terms, see Supplementary Figure 29. b. Distinct positional biases of promoter states with respect to nearest RefSeq TSS distinguish states peaking upstream, only downstream, and centered at the TSS. c. Positional biases of Transcribed States with respect to TSS, nearest spliced exon start, and Transcription End Sites (TES). These distinguish 5′-proximal states (12–23, left panel), 5′-distal states (24–28), states strongly enriched for spliced exons (middle panel, see also Supplementary Fig. 24 for plot for States 24–28), and TES-associated states (with state 27 being particularly precisely positioned, right panel).
Transcription-associated states
The second large group of chromatin states consisted of 17 transcription-associated states. These are 70–95% contained within RefSeq-annotated transcribed regions compared to 36% for the rest of the genome (Fig. 2b, P<10−200, for all states). This group was not predominantly associated with a single mark, but instead defined by combinations of seven marks, H3K79me3, H3K79me2, H3K79me1, H3K27me1, H2BK5me1, H4K20me1, and H3K36me3 (Fig. 2a). Based on their transition frequencies the states in this group could be sub-grouped corresponding to 5′-proximal and 5′-distal states, and states associated with genes of varying expression levels (see Fig. 2c, Supplementary Notes, Supplementary Table 1, Supplementary Fig. 4).
We observed several states strongly enriched for spliced exons (States 21–25 and 27–28, 5.7–9.7 fold enrichments) (Figs. 2b and 3c, and Supplementary Fig. 24), previously reported to be enriched in several individual marks19–22. In contrast to these previous studies, the combinatorial approach we have taken here shows that individual marks in spliced exonic states are also frequently detected in several other states that only show only a modest 1.3–1.6 fold enrichment for spliced exons (e.g. states 12, 13, 14, and 17), suggesting that the chromatin signature of spliced exons is not solely the presence of previously reported H3K36me3, H2BK5me1, H4K20me1, and H3K79me1 marks but their specific combinations with the absence of H3K4me2, H3K9me1, and H3K79me2/3.
State 27 showed a 12.5-fold enrichment for transcription end sites (TES) with its enrichment peaking directly over these locations (Fig. 3c). It was characterized both by the presence of H3K36me3, PolII, and H4K20me1, and the absence of H3K4me1, H3K4me2, and H3K4me3, distinguishing it from other transcribed states with higher PolII frequencies or high H3K36me3 frequencies. This suggests a novel and distinct signature for 3′ ends of genes for which no specific chromatin signature had been described, further validated by a 3.4-fold signal enrichment specifically for the elongating form of PolII surveyed in an independent study (Supplementary Fig. 25), even though our input data did not distinguish between the elongating and non-elongating form.
State 28 showed a 112-fold enrichment in zinc finger genes (named ZNF), which comprise 58% of the state. This state was characterized by the high frequency for H3K9me3, H4K20me3, and H3K36me3 and relatively low frequency of other marks. This specific combination has been independently reported as marking regions of KAP1 binding, a ZNF-specific co-repressor, which also shows a specific 44-fold enrichment for state 2823, 24. While H3K9me3 and H4K20me3 association with ZNF genes had been previously reported5, the de novo discovery of this highly specific signature of ZNF genes illustrates the power of the methodology, and also reveals the additional likely presence of H3K36me3 and lower frequency of other marks as complementing the signature of ZNF genes.
Active intergenic states
The third broad class of chromatin states consisted of 11 active intergenic states (States 29–39), including several classes of candidate enhancer regions, insulator regions, and other regions proximal to expressed genes (Supplementary Notes). These states were associated with higher frequencies for H3K4me1, H2AZ, numerous acetylation marks, and/or CTCF, and lower frequencies for other methylation marks (Fig. 2a; Supplementary Fig. 2 and 3). They occurred primarily away from promoter regions (85–97% outside 2kb of a TSS), and outside of transcribed genes (48–64% outside of RefSeq annotations, Fig. 2b). When they overlapped gene annotations, it was on average for regions that were repressed or lowly expressed (see expression column in Fig. 2b).
States 29–33 were notable since they corresponded to smaller fractions of the genome specifically associated with greater DNaseI hypersensitivity, transcription factor binding, and regulatory motif instances and are likely to represent enhancer regions (Fig 2 and Supplementary Fig. 23). Surprisingly, even though these candidate enhancer states shared higher H3K4me1 frequencies, they showed differences in the expression levels of downstream genes associated with subtle differences in the specific mark combinations (Supplementary Fig. 22). For instance, genes downstream of State 30 had a consistently higher average expression level than genes downstream of state 31 (P<0.001 at 10kb, 2-sided t-test). The two states differed in the frequency of several acetylation marks (State 30 relative to 31 showed higher frequency for H2BK120ac, H3K27ac, and H2BK5ac and lower frequency for H4K5ac, H4K8ac), and also in the level of H2AZ (higher in state 31 than 30), suggesting that these marks may be playing a more complex role than previously thought in enhancer regions.
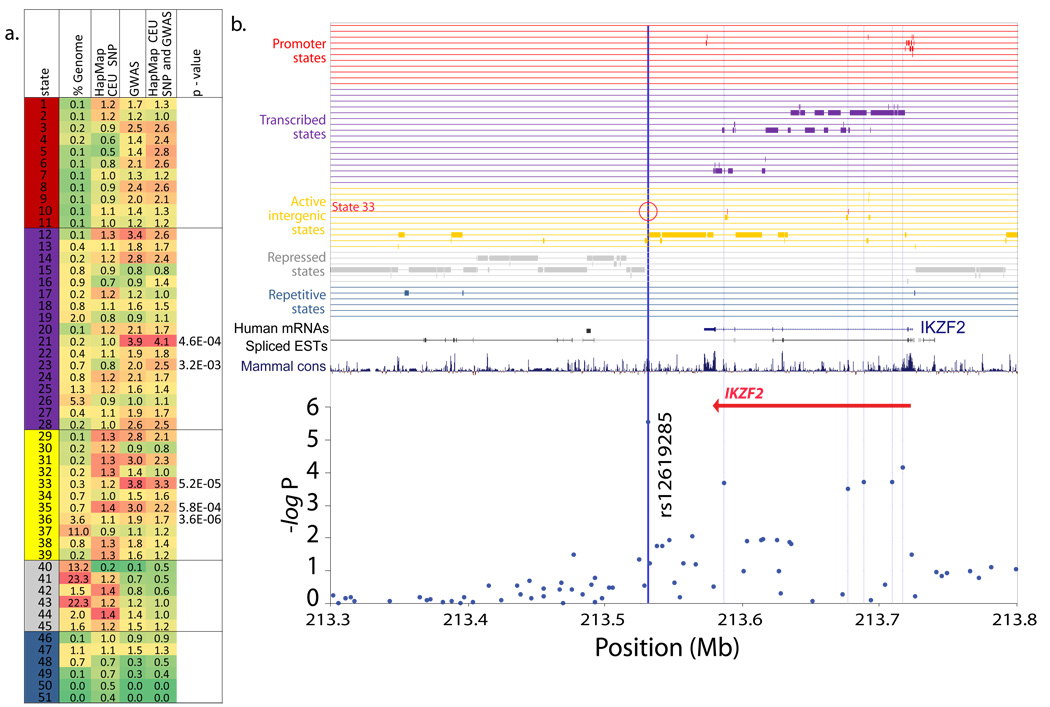
Several active intergenic states showed significant enrichments for genome-wide association studies (GWAS) hits (e.g. 3.3-fold for candidate enhancer state 33, Fig. 4a), based on a curated database25 of top-scoring Single-Nucleotide Polymorphisms (SNPs) in a range of diseases and traits. These states thus provide a likely common functional role and means of refining many intergenic SNPs even in absence of other annotations. As an example (Fig. 4b), we show one of the SNPs reported to be most associated with plasma eosinophil count levels found associated with inflammatory diseases (rs12619285)26. This SNP is located 40kb downstream of IKZF2, in an intergenic region devoid of annotations, but the chromatin state annotation overlapping this SNP reveals its association with the GWAS-enriched state 33. In contrast, the surrounding region of the genome is assigned to other active or repressed intergenic states with no significant GWAS association.
Figure 4. SNP and GWAS enrichments for chromatin states.
a. Several chromatin states show enrichments for disease association datasets. For each state is shown: genome percentage; fold enrichment for SNPs from the HapMap CEU population; fold enrichment from a collection of 1640 genome-wide association study (GWAS) Single Nucleotide Polymorphisms (SNPs) associated with a variety of diseases and traits from numerous studies (Hindorff et al, 2009)25; fold enrichment of GWAS SNPs relative to the HapMap CEU SNP enrichment; significance of GWAS SNPs relative to the underlying SNP frequency (when the corrected p-value <0.01). b. Example of intergenic SNP in GWAS-enriched state 33, found 40kb downstream of the IKZF2 gene and associated with plasma eosinophil count levels26. SNP significance as reported in the supplement of (Gudbjartsson, et al, 2009)26 is shown for each SNP in the region (blue circles) and associated chromatin state annotation (similar to Fig. 1). Red circle denotes top SNP and its overlap with state 33. In addition to top SNPs, secondary SNPs were also frequently found at or near GWAS-enriched states in several cases.
Large-scale repressed states
The next group of states (40–45) marked large-scale repressed and heterochromatic regions, representing 64% of the genome. The two most frequently detected modifications in total for all the states in this group were H3K27me3 and H3K9me3. State 40, covering 13% of the genome, was essentially devoid of any detected modifications, States 41–42 (25% of the genome) had a higher frequency for H3K9me3 than H3K27me3, while States 43–45 (26% of the genome) had a higher frequency for H3K27me3. States 41–42 as compared to States 43–45 showed a stronger depletion for genes, promoters, and conserved elements, and stronger association with nuclear lamina regions27, A/T nucleotides, and the darkest-staining chromosomal bands28 (Fig. 2b; Supplementary Fig. 26–28).
State 45 likely corresponds to targeted gene repression. It showed the highest frequency for H3K27me3 and was unique amongst repressed states to show enrichment for TSS. The corresponding genes were enriched for development-related GO categories (Supplementary Fig. 29), similar to the repressed promoter state 4 marked by H3K4me3. However, in contrast to States 4, State 45 showed almost no change in acetylation levels in response to histone deacetylase inhibitor (HDACi) treatment (Supplementary Fig. 30) suggesting that state 4 is poised for activation while State 45 is stably repressed29.
Repetitive states
The final group of six states (46–51) showed strong and distinct enrichments for specific repetitive elements (Supplementary Fig. 31). State 46 had a strong enrichment of simple repeats, specifically (CA)n, (TG)n, or (CATG)n (44, 45, and 302 fold respectively) possibly due to sequence biases in ChIP-based experiments30. State 47 was characterized specifically by H3K9me3 and enriched for L1 and LTR repeats. State 48–51 all had a higher frequency of H4K20me3 and H3K9me3, and heavily enriched for satellite repeat elements. States 49–51 showed seemingly high frequencies for numerous modifications, but also IgG control31 (Supplementary Fig. 20), suggesting these enrichments are likely due to a lack of coverage for the additional copies of these repeat elements in the reference genome assembly32 illustrating the power of our model to capture such potential artifacts by considering all marks jointly.
Predictive power for genome annotation
We next set out to study the predictive power of chromatin states for the discovery of novel elements. We focused on two classes of elements that benefit from ample experimental information independent of chromatin marks, transcription start sites (TSS) and transcribed regions. We found that chromatin states consistently outperformed predictions based on individual marks (Fig. 5a,b) emphasizing the importance of using mark combinations and spatial genomic information (see also Supplementary Notes and Supplementary Fig. 32 for a comparison to k-means clustering and a supervised classifier). The prediction performance based on just CD4 T cells was surprisingly similar to that of cap analysis of gene expression (CAGE) tags and expressed sequence tags (ESTs) data, even though these were obtained across many diverse cell types, enabled by active and inactive states together capturing information spanning cell type boundaries (Fig. 5, Supplementary Figs. 33–35). Moreover, the TSS and transcribed region predictive power held when our 51-state model was applied to a subset of 10 chromatin marks in CD36 erythrocyte precursors and CD133 hematopoietic stem cells (Supplementary Fig. 36).
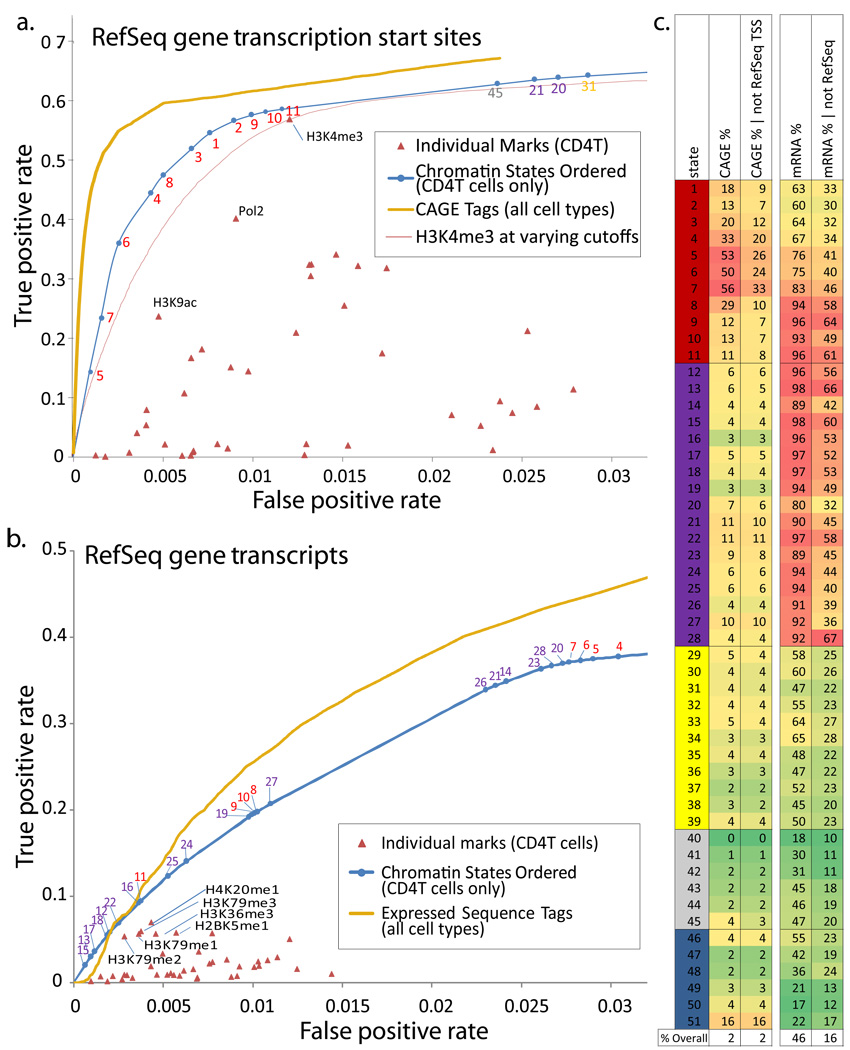
Figure 5. Discovery power of chromatin states for genome annotation.
a. Comparison of the power to discover transcription start sites (TSS) for individual chromatin marks (red), chromatin states (blue) ordered by their TSS enrichment, and a directed experimental approach based on CAGE sequence tag data read counts from all available cell types48 (gold), while the chromatin-states and marks only use data from CD4 T-cells. Both chromatin states and CAGE tags are compared using a Receiver Operating Characteristic (ROC) curve that shows the false positive (x-axis) and true positive (y-axis) rates at varying prediction thresholds in the task of predicting if a 200bp interval intersects a RefSeq TSS. Thin red curve compares performance of H3K4me3 mark at varying intensity thresholds. b. Comparison of the power to detect RefSeq transcribed regions for chromatin states and marks as in a, and directed experimental information coming from Expressed Sequence Tag (EST) data (gold) based on sequence counts from all available cell types38, 49. c. Independent experimental and comparative information provides support that a significant fraction of ‘false positives’ in panels a and b are genuine novel unannotated TSS and transcribed regions currently missing from RefSeq. Percentage of each state supported by a CAGE tag (column 1), and the same percentage for locations at least 2kb away from a RefSeq TSS (column 2), suggests that many promoter-associated states outside RefSeq promoters are supported by CAGE tag evidence. Similarly, percentage of each state overlapping a GenBank mRNA (column 3), and the same percentage specifically outside RefSeq genes (column 4), suggest that transcription-associated states outside RefSeq genes are supported by mRNA evidence. Similar support is found by GenBank Expressed Sequence Tags (ESTs) and evolutionarily-conserved predicted new exons (Supplementary Fig. 33).
We also found that chromatin states revealed candidate novel promoter and transcribed regions. Candidate promoters overlapped CAGE tags (Fig. 5c) and intergenic Pol2 (Supplementary Fig. 37), and candidate transcribed regions overlapped GenBank mRNAs (Fig. 5c) and EST data (Supplementary Fig. 33). A number of promoter and transcribed states outside known genes were also strongly enriched for novel protein-coding exons predicted using evolutionary comparisons of 29 mammals (Lin and Kellis, in preparation) (Supplementary Fig. 33). We note that some candidate promoters may represent distal enhancers, sharing promoter associated marks potentially due to looping of enhancer to promoter regions7.
Recovery of chromatin states using different combinations of marks and in additional cell types
As the large majority of chromatin states were defined by multiple marks, we next sought to specifically study the contribution of each mark in defining chromatin states. First, we found several notable examples of both additive, such as acetylation marks in promoter regions, and combinatorial relationships, such as methylation marks associated with repressive and repetitive elements (Supplementary Notes, Supplementary Fig. 38). We also evaluated varying subsets of chromatin marks in their ability to distinguish between chromatin states (Supplementary Notes and Supplementary Figs. 39–41). More generally, we sought to provide guidelines for selecting subsets of chromatin marks to survey in new cell types that would be maximally informative.
As a proof of principle, we evaluated the recovery power for increasing numbers of marks in a greedy way, weighing each state equally and penalizing mismatches uniformly (see Online Methods, Fig. 6), providing an initial unbiased recommendation of marks to survey for a new cell type. We find that increasing subsets of marks rapidly converge to a fairly accurate annotation of chromatin states (Fig. 6c), providing cost-efficient recommendations for new cell types. In addition to an overall error score, this analysis provides information on the proportion of each state accurately recovered, and specific pairwise state mis-assignments, which could be incorporated to provide targeted chromatin mark recommendations based on the subset of chromatin states that are of particular biological interest, or the particular state distinctions that are most important to each study.
Figure 6. Recovery of chromatin states with subsets of marks.
a. The figure shows the ordering of marks based on a greedy forward selection algorithm to optimize a squared error penalty on state mis-assignments (Online Methods). Conditioned on all the marks to the left having already been profiled, the mark listed is the optimal selection for one additional mark to be profiled based on the target optimization function. Below each mark is the percentage of a state with identical assignments using the subset of marks. b. Comparison of the percentage of each state recovered between the first 10 marks based on the greedy method and the 10 marks used in (Cui et al, 2009)50 (Supplementary Fig. 39). The two columns after the state IDs are the proportion of the states recovered using the greedy algorithm and the set used in (Cui et al, 2009)50. c. The figure shows a progressive decrease in squared error for state mis–assignment as a function of the number of marks selected based on the greedy algorithm.
Discussion
The discovery and systematic characterization of chromatin states presented here has given a systematic view of the epigenomic landscape in human cells, revealing a surprising diversity of function in 51 chromatin states with distinct biological interpretations. While the exact number of chromatin states can vary based on the number of chromatin marks surveyed and the desired resolution at which state differences are studied, our results suggest that the genome annotation resulting from these states can dramatically extend the landscape of interpretable regions in the human genome especially outside protein-coding genes. The definition of the states themselves revealed numerous new insights into the combinatorial and additive roles of chromatin marks, while the genome-wide annotation of these states revealed many novel candidate elements of distinct functions and candidate new chromatin functions.
We only expect the discovery power of these methods to increase as additional genome-wide epigenetic datasets become available, and as additional cell types are surveyed systematically. Chromatin states can be inferred with virtually any type of epigenetic and related information, including histone variants, DNA methylation, DNaseI hypersensitivity, and binding of chromatin-associated and sequence-specific transcription factors. While we focused on a single cell type in human, the methods are generally applicable to any species and any number of cell types, and even whole embryos albeit in mixed cell populations mutually-exclusive marks found in different subsets of cells could potentially be interpreted as co-occurring.
Specifically for understanding epigenomic dynamics, chromatin states can play a central role going forward, as they provide a uniform language for interpreting and comparing diverse epigenetic datasets, for selecting and prioritizing chromatin marks for additional cell types, and for summarizing complex relationships of dozens of marks in directly-interpretable chromatin states. As several large-scale data production efforts are currently underway to map the epigenomes of many more cell types, exemplified by the ENCODE33, modENCODE34, and Epigenome Roadmap projects (roadmapepigenomics.org), chromatin states will likely play a key role towards a systematic understanding of the human epigenome and its role in development, health, and disease.
Methods
Input Data for Modeling
The initial unprocessed data were bed files containing the genomic coordinates and strand orientation of mapped sequence reads from ChIP-seq experiments5, 6. There was a separate bed file for each of the 18 acetylations, 20 methylations, H2AZ, CTCF, and PolII in CD4 T cells. We used the updated version of the H3K79me1/2/3 data reported in (Wang et al, 2008)6, which differs from the version first reported in (Barski et al, 2007)5.
To apply the model we first divided the genome into 200-base-pair non-overlapping intervals within which we independently made a call as to whether each of the 41 marks was detected as being present or not based on the count of tags mapping to the interval. Each tag was uniquely assigned to one interval based on the location of the 5′ end of the tag after applying a shift of 100 bases in the 5′ to 3′ direction of the tag. The threshold, t, for each mark was based on the total number of mapped reads for the mark (Supplementary Table 2), and was set to be the smallest integer t such that P(X>t)<10−4 where X is a random variable with a Poisson distribution with mean parameter set to the empirical mean of the number of tags per interval.
The Probabilistic Model
The probabilistic model is based on a multivariate instance of a Hidden Markov Model (HMM)35. The model assumes a fixed number of hidden states K. In each hidden state, the emission distribution, that is the probability distribution over each combination of marks, is modeled with a product of independent Bernoulli random variables. Formally, for each of the K states, and M=41 input marks, there is an emission parameter pk,m denoting the probability in state k (k=1,…,K) that input mark m (m=1,…,M) has a present call. Let c ∈ C denote a chromosome where C is the set of all chromosomes. Let ct denote an interval on chromosome c where t=1,…,Tc corresponds sequentially to the 200bp intervals on chromosome c. c1 is the interval corresponding to base pairs 1–200 on chromosome c and Tc is the number of non-overlapping 200bp intervals on chromosome c. Let vct,m be ‘1’ if there is a present call for input mark m and ‘0’ otherwise at location ct. Denote the specific combination of marks at interval ct as vct = (vct,1,…, vct,m). Let bij denote the probability of transitioning from state i to j where i=1,…,K and j=1,…,K. We also have parameters ai (i=1,…,K) which denote the probability that the state of the first interval on the chromosome is i. Let sc ∈ SC be an unobserved state sequence through chromosome c and SC be the set of all possible state sequences. Let sct denote the unobserved state on chromosome c at location t for state sequence sc. The full likelihood of all of the observed data v for the parameters a, b, and p can then be expressed as:
Model Learning
We first used an iterative learning expectation-maximization approach to infer state emission and transition parameters that best summarize the observed genome-wide chromatin mark information using a fixed number of randomly-initialized hidden states, varying from 2 up to 80 states. To minimize the number of states and facilitate recovery of a robust and comparable set of states across models of varying complexity, we then used a nested initialization procedure that seeded parameters of lower-complexity models with states of higher-complexity models.
From an initial set of parameters we found a local optimum of the parameter values using a variant of the standard expectation-maximization based Baum-Welch algorithm for training HMMs35. Our variant after the first full iteration over all the chromosomes, used an incremental expectation-maximization procedure36, which would update the parameters through a maximization step after performing an expectation over any chromosome. This allowed improved parameter estimates from the maximization step to be more quickly incorporated in the more time consuming expectation step. Also for computational considerations, if a transition parameter fell below 10−10 during training we set the parameter value to 0, which allowed faster training with virtually no impact on the final model learned. The transitions were initialized to be fully connected, and except for the 10−10 criterion there was no regularization forcing them closer to 0. We would terminate the training after 300 passes over all the chromosomes, which was sufficient for the likelihood to demonstrate convergence (Supplementary Fig. 8).
The procedure for determining the initial parameters used to learn the final set of HMMs was to first learn in parallel for every number of states from 2 to 80 three HMM models based on three different random initializations of the parameters. Each model was scored based on the log likelihood of the model minus a penalization on the model complexity determined by the Bayesian Information Criterion (BIC) of one-half the number of parameters times the natural log of the number of intervals. We then selected the model with the best BIC score among these 237 models, which had 79 states (Supplementary Figs. 8 and 12). We then iteratively removed states from this 79 state model. When removing a state the emission probabilities would be removed entirely, and any state that transitioned to it would have that transition probability uniformly re-distributed to all the remaining states. This resulting set of models was then used as the initial parameters of the HMMs in the final model learning. During this final model learning, one HMM was learned for every number of states between 2 and 79 in parallel (Supplementary Fig. 13).
The criterion for selecting a state to remove from a model was based on first forming a set E containing all the emission vectors from all the 237 models learned from the random initializations. The procedure would then remove a state such that the elements in E had in total the least distance from their closest emission vector among the remaining states. Formally for a set of emission vectors Cn corresponding to states in a model the method would form a set Cn−1 and corresponding model by removing r defined by
where here we used (1−ρ) where ρ is the standard correlation coefficient as the distance d, though the method is general and could be used with other distance measures.
The entire procedure discovered models with comparable or superior likelihood scores to randomly initialized models, while also having sets of parameters that would be more directly comparable (Supplementary Figs. 8 and 13). The number of states for a model to analyze can then be selected by choosing the model trained from a nested initialization with the smallest number of states that sufficiently captures all states of interest in larger models (Supplementary Notes).
Associating Genomic Locations with States
After a model is learned, a posterior probability distribution over the state of each interval is computed using a forward-backward algorithm35. Unless otherwise noted, the analysis was based on the ‘soft’ state assignments of the posterior distribution. We also formed hard assignments of states to locations by using the maximum-posterior state assignment at a location. Both the full posterior and hard assignments are available on the supplementary website http://compbio.mit.edu/ChromatinStates/.
External Fold Enrichments and Percentage Overlaps
For a state the sum of posterior probability over all 200bp intervals was computed, denoted by a. For an external data source the total number of 200bp intervals that it intersects at least one base was computed, denoted by b. For the state and the external data source the total sum of the posterior for the state in intervals intersecting the external data source were computed, denoted by c. Also the total number of 200bp intervals is denoted by d. The percentage of a state’s overlap with an external data source is defined as (c/a*100) while the fold enrichment is (c/a)/(b/d). p-values of the overlap were computed based on the hypergeometric distribution.
The gene annotations used were the RefSeq annotations37 as of December 14th, 2008 obtained from the UCSC genome browser browser38 and are based on hg18. The sequence data for computed nucleotide frequencies, CpG islands, repeats39, and conservation data were also obtained from the UCSC genome browser. The conservation data was based on PhastCon conserved elements using the 44-way vertebrate alignment40, 41 (Lindblad-Toh et al, in preparation). Transcription factor binding enrichments were computed for 18 experiments from numerous publications (Supplementary Fig. 23), the median enrichment over all these experiments is reported in Figure 2b. The DNaseI hypersensitivity data was from (Boyle et al, 2008)42 obtained from the UCSC genome browser. The nuclear lamina data of human fibroblasts was obtained from the supplementary materials of (Guelen et al., 2008)27. The ZNF genes were defined as those that had ‘ZNF’ at the beginning of the gene symbol in the RefSeq gene table. For published coordinates that were in hg17 we converted them to hg18 using the liftover tool from the UCSC genome browser43.
Expression, Motif, and Gene Ontology Analyses
We obtained the processed CD4 T expression data from (Su et al, 2004)44 for both replicates. We then averaged the two replicates. After averaging the two replicates we performed a natural log transform of the average values. We then standardized all values by subtracting the mean log transformed value, and then dividing by the standard deviation of the log transform values. The genome coordinates of each probe set were obtained from the UCSC genome browser. Each 200bp interval that overlapped a probe set obtained the transformed expression score. If multiple probe sets overlapped the same 200bp then the average of the expression values associated with these were taken.
We generated transcription factor motif enrichments as described in (Kheradpour et al., 2007)45, extended for Position-Weight Matrices (PWMs) (Kheradpour and Kellis, in preparation) based on the hard state assignments.
Gene ontology enrichments were based on the hard state assignment of the interval containing the RefSeq annotated TSS of the gene. Enrichments were computed using the STEM software (v.1.3.4) and the Bonferroni corrected p-values are reported46.
SNP and GWAS Analysis
The HapMap CEU47 data was downloaded from the UCSC genome browser. Significant GWAS hits were taken from (Hindorff et al, 2009)25. SNPs listed as occurring multiple times were only counted once, and for the SNP set listed as a 17-marker haplotype only the first SNP was used giving 1640 SNPs. In computing enrichment for HapMap and GWAS SNPs if two SNPs mapped to the same interval they were counted multiple times. To determine if the number of GWAS SNPs in a chromatin state was more significant than would be expected based on the general SNP frequency in the state we used a binomial distribution where n=1640 and p is the proportion of HapMap CEU SNPs assigned to the state. We applied a Bonferonni correction for testing multiple states and only reported those p-values significantly enriched with p<0.01.
RefSeq TSS and Gene Transcripts Discovery
The ROC curve for the CAGE data was based on the number of CAGE tags mapping to a 200bp interval retrieved from the Fantom database and converted from hg17 to hg18 using the UCSC genome browser lift over tool48. The overlap with EST was based on those EST listed in the UCSC genome browser all_est table as of Nov 29th, 200938, 49. The overlap with Genbank mRNA is based on the overlap with the UCSC genome browser mRNA listed in the table as of October 31st, 200938, 49. The novel exon predictions are from (Lin and Kellis, in preparation).
Mark subset evaluation and selection
When evaluating the coverage of a specified subset of marks, first a posterior distribution over the states at each interval is computed using the model learned on the full set of marks, except that the marks not in the subset are omitted when computing emission probabilities. For an interval t we define here st,k and ft,k to be the posterior assignment to state k at interval t based on the subset and full set of marks respectively. The proportion of state k recovered with a subset of marks is defined as:
where the sum is over all intervals t in the genome. The ordering of marks presented without any prior biological knowledge was based on a greedy forward selection algorithm designed to select marks that would minimize this function:
where the sum is over all states. At each step the algorithm would then choose the one additional mark, conditioned on all the other previously selected marks that would cause this function to be minimized. We note that this target function considers all non-identical state assignments to have equal loss. An extension of this approach would be to apply target functions that weigh different mis-assignments differently. The proportion of state k with the full set of marks that is mis-assigned to state i using a subset of marks, mk,i, as is presented in Supplementary Figures 39 and 40, is defined as:
The first term in the sum in the numerator represents for an interval t the amount of posterior probability assigned to state k using the full set of marks not assigned using the subset of marks. The second term represents the portion of this posterior probability that will be credited to state i. The portion credited to state i is the proportion of the surplus posterior state i received with the subset of marks in the interval relative to the total surplus posterior all states received in the interval.
Supplementary Material
Acknowledgments
We thank Bradley Bernstein, Noam Shoresh, Charles Epstein, Tarjei Mikkelsen for helpful discussions, Adam Siepel, Kerstin Lindblad-Toh, and Eric Lander for use of comparative information on 29 mammals, Mike Lin for predicted new exons, Pouya Kheradpour for regulatory motif instances, Loyal Goff, Chris Bristow and Rachel Sealfon for comments and feedback, and all members of the MIT CompBio Group for feedback and support. This material is based upon work supported by the National Science Foundation under Award No. 0905968 and funding from the US National Human Genome Research Institute (NHGRI).
Footnotes
Author Contributions. JE and MK developed the method, analyzed results, and wrote the paper.
Competing financial interests. The authors declare no competing financial interests.
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions
References
- 1.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 5.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 8.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput. Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Xuan Z, Zhao X, Li Y, Zhang MQ. High-resolution human core-promoter prediction with CoreBoost_HM. Genome Res. 2009;19:266–275. doi: 10.1101/gr.081638.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Won KJ, Chepelev I, Ren B, Wang W. Prediction of regulatory elements in mammalian genomes using chromatin signatures. BMC Bioinformatics. 2008;9:547. doi: 10.1186/1471-2105-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput. Biol. 2008;4:e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day N, Hemmaplardh A, Thurman RE, Stamatoyannopoulos JA, Noble WS. Unsupervised segmentation of continuous genomic data. Bioinformatics. 2007;23:1424–1426. doi: 10.1093/bioinformatics/btm096. [DOI] [PubMed] [Google Scholar]
- 15.Jia L, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurman RE, Day N, Noble WS, Stamatoyannopoulos JA. Identification of higher-order functional domains in the human ENCODE regions. Genome Res. 2007;17:917. doi: 10.1101/gr.6081407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuettengruber B, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaschek R, Tanay A. Spatial clustering of multivariate genomic and epigenomic information. Proceedings of the 13th Annual International Conference on Research in Computational Molecular Biology, Springer.2009. [Google Scholar]
- 19.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 21.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Geen H, et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. A catalog of published genome-wide association studies. [Accessed July 22, 2009]; Available at: www.genome.gov/gwastudies.
- 26.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 27.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 28.Furey TS, Haussler D. Integration of the cytogenetic map with the draft human genome sequence. Hum. Mol. Genet. 2003;12:1037–1044. doi: 10.1093/hmg/ddg113. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DS, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis. Cambridge Univ. Press; 1998. [Google Scholar]
- 36.Neal RM, Hinton GE. A view of the EM algorithm that justifies incremental, sparse, and other variants. Learning in graphical models. 1998;89:355–368. [Google Scholar]
- 37.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996 [Google Scholar]
- 40.Miller W, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.International HapMap Consortium et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 49.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic Acids Res. 2004;32:D23–D26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell. Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.