Abstract
Vector-borne microbes necessarily co-occur with their hosts and vectors, but the degree to which they share common evolutionary or biogeographic histories remains unexplored. We examine the congruity of the evolutionary and biogeographic histories of the Lyme disease system, the most prevalent vector-borne disease in North America. In the Eastern and Midwestern US, Ixodes scapularis ticks are the primary vectors of Borrelia burgdorferi, the bacterium that causes Lyme disease. Our phylogeographic and demographic analyses of the 16S mitochondrial rDNA suggest that I. scapularis populations originated from very few migrants from the Southeastern US that expanded rapidly in the Northeast and subsequently in the Midwest after the recession of the Pleistocene ice sheets. Despite this historical gene flow, current tick migration is restricted even between proximal sites within regions. In contrast, B. burgdorferi suffers no barriers to gene flow within the Northeastern and Midwestern regions but shows clear inter-regional migration barriers. Despite the intimate association of B. burgdorferi and I. scapularis, the population structure, evolutionary history, and historical biogeography of the pathogen are all contrary to its arthropod vector. In the case of Lyme disease, movements of infected vertebrate hosts may play a larger role in the contemporary expansion and homogenization of the pathogen than the movement of tick vectors whose populations continue to bear the historical signature of climate-induced range shifts.
Keywords: phylogeography, vector-borne disease, population genetics, population structure, Lyme disease, climate change, recolonization, microbial biogeography
INTRODUCTION
The geographic distribution of plants and animals is influenced as much by historical climate as by current ecological factors. The recent Pleistocene glaciations profoundly impacted northern biota and is a dominant factor affecting genetic population structure (Pielou 1992; Avise and Walker 1998; Avise 2000; Knowles 2001). Many organisms expanded their ranges northwards as the Pleistocene ice sheet retreated (Pielou 1992). Phylogeographic analyses incorporating gene frequency and sequence divergence among populations have been used to test hypotheses of vertebrate evolution, migration, and demography in the northern hemisphere (Avise and Walker 1998; Avise et al. 1998; Mila et al. 2000; Knowles 2001). The emerging correlation between contemporary climate change and the distribution of infectious diseases alludes to the impact of climate on the evolution, migration, and demography of microbial pathogens, particularly those obligately associated with animal hosts or vectors (Patz et al. 2005).
Vector-borne infectious diseases are biologically and ecologically coupled to invertebrate vector species whose own historical biogeographic distributions often bear the signature of climatic oscillations (Pielou 1992; Avise and Walker 1998; Avise 2000; Knowles 2001). Even with contemporary biological coupling of pathogen and vector, it is unclear to what extent pathogens and their vectors share common evolutionary or biogeographic histories. We reconstruct the biogeographic history, demographic history, and population genetic structure of Lyme disease bacterium Borrelia burgdorferi and its tick vector, Ixodes scapularis. We examine if both species of this biologically coupled system bear the genetic footprint of a recent and rapid expansion in the regions made available by the retreat of Pleistocene ice sheets and if their evolutionary and biogeographic history share concordant genetic footprints.
The recolonization and migration patterns of obligate parasites, such as B. burgdorferi, are contingent on the distribution of their hosts and vectors. B. burgdorferi is carried among several species of vertebrate hosts by I. scapularis in the Northeastern and Midwestern US (Burgdorfer et al. 1982; Anderson et al. 1983; Piesman et al. 1986). The continual passage of B. burgdorferi between tick vectors and vertebrate hosts is required for the existence of local B. burgdorferi populations as neither ticks nor vertebrates transmit B. burgdorferi to their offspring (Magnarelli et al. 1987; Lane et al. 1991; Mather et al. 1991; Patrican 1997). Thus, B. burgdorferi is sustainable only in a community of appropriate vertebrate host species and abundant ticks (LoGiudice et al. 2003; Brisson and Dykhuizen 2006). Although, B. burgdorferi can migrate with infected small mammals, most terrestrial mammalian hosts of B. burgdorferi have limited long-distance migration (Bowman et al., 2002). Long distance migration of B. burgdorferi in ticks carried on migrating birds has been documented and can seed a region with B. burgdorferi (Scott et al., 2001).
Ticks carrying B. burgdorferi are concentrated in multiple foci around the northern hemisphere within regions once under the Pleistocene glacier (Pielou 1992). Ticks in Northeastern US bear the genetic footprint of a recent colonization and rapid population expansion (Qiu et al. 2002). The ancestry of Northeastern tick populations can be traced to one or a few migrants originating in the Southeastern US (Qiu et al. 2002). The route of B. burgdorferi colonization is unclear but may have originated in Europe and invaded the Northeast (Margos et al. 2008; Qiu et al. 2008). The recent discovery of I. scapularis in the Midwestern US have lead to the hypotheses that ticks were introduced or reintroduced into this region in the last century (Jackson and DeFoliart 1970) or that continuous long-distance migration between the Northeastern and Midwestern regions on migrating birds (Scott et al. 2001; Ogden et al. 2008) explain the introduction and rapid increase in density of this medically important vector species. The tempo and mode of historical migration may expose the current and future migration patterns of the Lyme disease system as climate and human land-use patterns continue to change.
In North America, the majority of Lyme disease cases come from the Northeast (~75%) and the Midwest (~12%) (Orloski et al. 2000; McNabb et al. 2008). Lyme disease in the Midwest has received little attention, likely because far fewer human cases are reported in the area. This has resulted in a scientific naïveté concerning the combination of historical factors that have generated the relatively low Lyme disease incidence in the Midwest. For example, it is unknown whether Midwestern ticks, which are morphologically different in several allometric characters than their eastern conspecifics (Hutcheson et al., Keirans, 1996 #813), originated from a different founding tick population with different behaviors than Northeastern ticks. In this study we investigate the effect of current migration as well as historical distribution and demography of I. scapularis on the current distribution of B. burgdorferi.
METHODS
Sampling natural populations
Ixodes scapularis
110, 216 and 125 host-seeking adult ticks were collected using a 1m2 drag-cloth (Lane et al. 1991) from the Colfax, Stockton and Marinette, WI in 1998 (Figure 1). A 350bp fragment of the mitochondrial 16s rDNA was amplified from whole genome DNA extracted from individual ticks (QIAGEN DNeasy, Alcameda, CA) using primers 16Sa/16sB DNA (Rich et al. 1995). PCR products were subsequently sequenced and aligned with Clustal W (Thompson et al. 1994; Larkin et al. 2007).
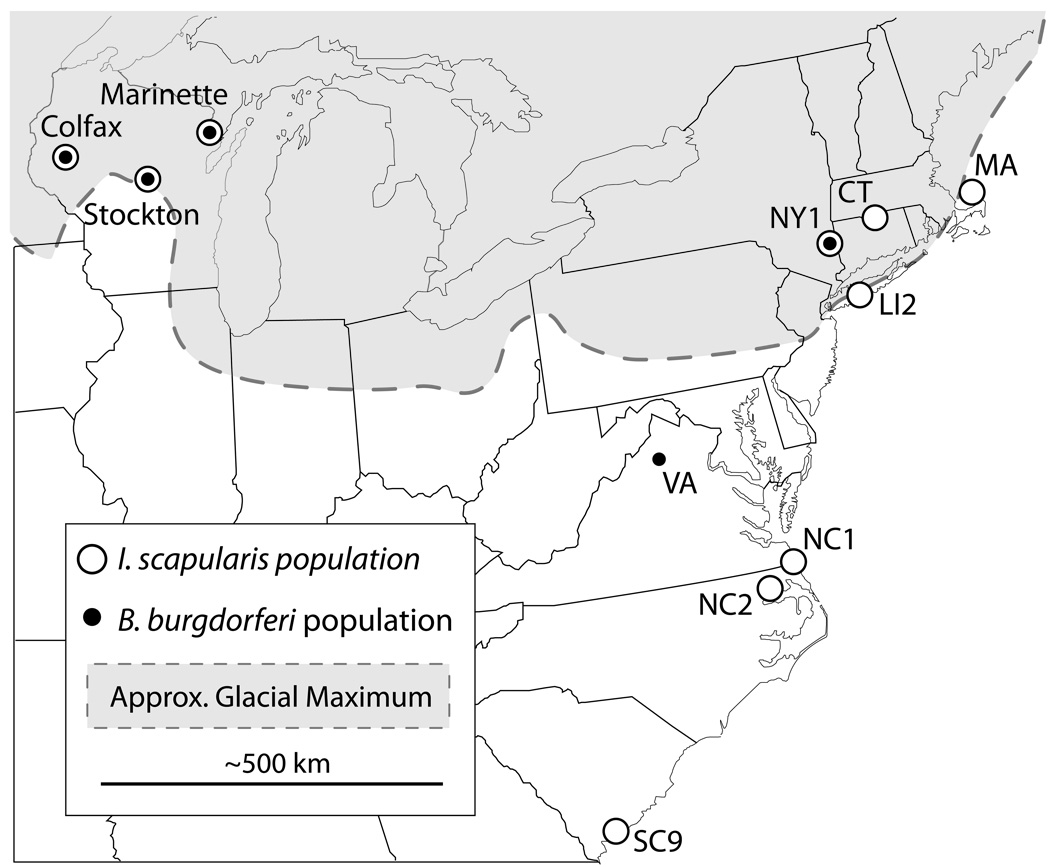
Figure 1.
Map displays collections sites for I. scapularis from the Northeastern (NY1; MA, CT, LI2), Southeastern (NC1; NC2; SC98) and Midwestern US (Stockton, Colfax, Marinette); and for B. burgdorferi populations from the Northeastern (NY1; VA) and Midwestern US (Stockton, Colfax, Marinette). Approximate glacial maximum of the Pleistocene ice sheet is drawn, after Church (2003) and Pielou (1992).
Midwestern 16s rDNA haplotypes were aligned with previously reported haplotypes (Qiu et al. 2002). Gaps were removed to safeguard against introduced SNPs resulting in an alignment of 342nt. Haplotype frequency data from three Northeastern, three mid-Atlantic, one Long Island, and three Southeastern tick populations were incorporated into this study (Qiu et al. 2002). The Northeastern sites cover a similar geographical area as the Midwestern sites and were undeniably under the last glacial ice sheet (Figure 1). Mid-Atlantic populations – those that may have been affected by the Pleistocene ice sheet – were included in analyses as either Northeastern populations and as populations from a separate “mid-Atlantic” region.
Borrelia burgdorferi
A 1kb fragment of the non-coding rrs-rrlA intergenic spacer (IGS) region was amplified as previously described (Bunikis et al. 2004). IGS sequences from 41 Midwestern B. burgdorferi were derived from a random sample of the infected Midwestern ticks originating from Stockton (16), Marinette (13), or Colfax (12), WI (Figure 1). B. burgdorferi from the sites sampled in Qiu et al. (2002) are no longer available. Eastern IGS sequences were derived from individual ticks collected from two eastern locations: Millbrook, NY (15) and Front Royal, VA (15). B. burgdorferi is too rare from regions south of Virginia to obtain appropriate frequencies of IGS haplotypes for statistical analysis (Oliver 1996; Clark et al. 2002; Qiu et al. 2002). The geographical and temporal differences between the tick and bacterial species in the sampling sites does not affect the comparisons of the among-region phylogeographical analyses. IGS PCR products from ticks were cloned into pCR2.1 cloning vector (Invitrogen) and a single transformed colony was randomly chosen for sequencing. Sequences were aligned as above and singleton polymorphisms discarded to guard against PCR and sequencing artifacts.
Population genetic analyses
Identical population genetic analyses were performed on both the I. scapularis and B. burgdorferi datasets. Unrooted Bayesian phylogenies and unrooted minimum spanning trees (MST) were constructed using MrBayes 3.1.2 and TCS1.21, respectively (Clement et al. 2000; Huelsenbeck and Ronquist 2001) to evaluate phylogenetic relationships among haplotypes within each species. These methods determine the gene network in which the total length of the branches that connect haplotypes is minimized. To discriminate among equal-length MSTs we assumed that older alleles are more common than recently derived alleles and that new mutations are more likely to be found in the same population as their ancestor (Excoffier et al. 1992; Neigel and Avise 1993).
The population genetic structure of both tick and pathogen populations was determined using analysis of molecular variance (AMOVA) and exact tests as implemented in Arlequin 3.1 (Raymond and Rousset 1995; Excoffier et al. 2005). AMOVA extends Wright's F to incorporate genetic relatedness in addition to allelic frequency to determine the degree of subdivision among populations (Excoffier et al. 1992). The effect of differences in sampling effort among the regions was investigated for all demographic parameters. One hundred datasets were created by reducing the sample size of each tick population to 24 (the smallest sample size in the dataset) by subsampling with replacement the haplotypes within each population. These datasets were investigated to estimate the robustness of the population genetic and demographic analyses.
The historical population growth rates of tick and B. burgdorferi populations were investigated using the sequence and frequency of each haplotype in the program LAMARC (Kuhner 2006). LAMARC co-estimates the effective population size in terms of θ (θ = Ne•μ) and the exponential growth rate of the population (g in units of 1•µ−1 individuals per generation). Population parameter estimates were derived using a Bayesian search strategy with uninformative priors, three replicate runs of 30,000 generations, and multiple random seeds to insure convergence.
The time to most recent common ancestor (TMCRA) was estimated from gene sequences using the coalescent-based program GENETREE (Bahlo and Griffiths 2000). Three replicate runs of 1,000,000 generations each using a random seed all converged on similar results. Estimates of TMRCA are reported in coalescent units, T, where T = real time / Ne•(generation time in years).
RESULTS
Genetic Variation and Phylogenetic Analyses
Genetic analysis of 451 I. scapularis ticks collected from three Midwestern sites yielded 29 unique 16s rDNA haplotypes (Figure 1). The most dominant Midwestern haplotypes, 1 and 2, are identical to the most dominant Northeastern haplotypes, F and D (Table 1). Six of the ten haplotypes present in the Northeast were also found in the Midwest. The majority of Midwestern haplotypes (23 of 29) occur at low frequencies (0.8 – 2.9%) and are unique to this region; 20 are unique to one Midwestern site (Table 1). Northeastern tick populations are dominated by three haplotypes (F - 46.2%, D - 23.3%, and A - 16%) and all ten of the Northeastern haplotypes found more than once are present in multiple Northeastern tick populations (Table 1). Only four haplotypes discovered in the Northeast and the Midwest are present in the Southeast (1/F, 2/D, 23/A and 26/G). Tick populations from mainland Southeastern sites have an order of magnitude more sequences diversity (πn) than Midwestern and Northeastern tick populations (Table 1). Marinette and the Eastern coastal populations (NC1 and MA) are the least diverse of all populations (Table 1).
Table 1.
Frequency distribution of I. scapularis 16s rDNA haplotypes
| Region: | Midwest | Northeast | Southeast | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population: | Co | Sto | Mar | Sum | NY1 | LI2 | MA | CT | Sum | NC1 | NC2 | SC98 | Sum | Totals | |
| Hap | Qiu 2002 | ||||||||||||||
| 1 | F | 70 | 115 | 109 | 294 | 9 | 30 | 18 | 18 | 75 | 14 | 19 | 6 | 39 | 408 |
| 2 | D | 20 | 38 | 58 | 4 | 6 | 3 | 29 | 42 | 8 | 2 | 10 | 110 | ||
| 8 | 1 | 1 | 2 | 2 | |||||||||||
| 9 | 4 | 9 | 13 | 13 | |||||||||||
| 17 | 1 | 2 | 3 | 3 | |||||||||||
| 21 | 2 | 2 | 2 | ||||||||||||
| 22 | 1 | 1 | 1 | ||||||||||||
| 23 | A | 2 | 2 | 13 | 4 | 6 | 10 | 33 | 2 | 1 | 3 | 38 | |||
| 25 | 3 | 3 | 3 | ||||||||||||
| 26 | G | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 4 | ||||||
| 29 | 2 | 2 | 2 | ||||||||||||
| 31 | 1 | 1 | 1 | ||||||||||||
| 32 | 1 | 1 | 1 | ||||||||||||
| 33 | 1 | 1 | 1 | ||||||||||||
| 4 | 1 | 1 | 1 | ||||||||||||
| 5 | 4 | 4 | 4 | ||||||||||||
| 6 | 12 | 12 | 12 | ||||||||||||
| 7 | 2 | 2 | 2 | ||||||||||||
| 10 | 1 | 1 | 1 | ||||||||||||
| 11 | 7 | 7 | 7 | ||||||||||||
| 13 | 6 | 6 | 6 | ||||||||||||
| 15 | 13 | 13 | 13 | ||||||||||||
| 16 | 2 | 2 | 2 | ||||||||||||
| 18 | 1 | 1 | 1 | ||||||||||||
| 19 | 2 | 2 | 2 | ||||||||||||
| 12 | 3 | 3 | 3 | ||||||||||||
| 34 | H | 4 | 4 | 1 | 1 | 2 | 6 | ||||||||
| 35 | B | 8 | 8 | 3 | 3 | 11 | |||||||||
| 36 | 1 | 1 | 1 | ||||||||||||
| E | 4 | 4 | 4 | ||||||||||||
| C | 5 | 1 | 6 | 6 | |||||||||||
| I | 3 | 1 | 4 | 4 | |||||||||||
| MA1 | 1 | 1 | 1 | ||||||||||||
| K | 1 | 3 | 4 | 4 | |||||||||||
| NC6 | 1 | 1 | 1 | ||||||||||||
| L | 1 | 1 | 1 | ||||||||||||
| O | 6 | 6 | 6 | ||||||||||||
| NC29 | 1 | 1 | 1 | ||||||||||||
| M | 2 | 2 | 2 | ||||||||||||
| N | 18 | 18 | 18 | ||||||||||||
| TOTALS | 110 | 216 | 125 | 451 | 30 | 53 | 28 | 60 | 171 | 24 | 35 | 28 | 87 | 709 | |
| N haps | 14 | 16 | 5 | 29 | 4 | 8 | 4 | 6 | 10 | 4 | 8 | 5 | 11 | 40 | |
| πn*102 | 0.24 | 0.28 | 0.09 | - | 0.35 | 0.33 | 0.21 | 0.29 | - | 0.22 | 1.42 | 1.95 | - | - | |
Co=Colfax, Sto=Stockton, Mar=Marinette, Wisconsin, US. πn = average pairwise nucleotide divergence
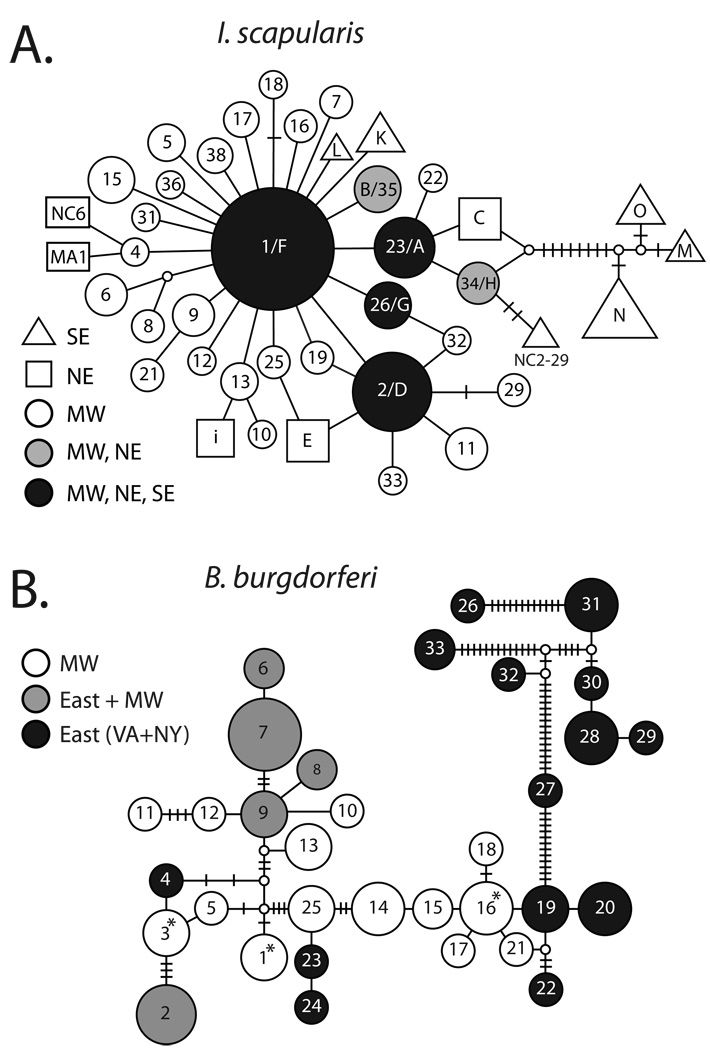
The minimum-spanning tree (MST) reconstructed from tick 16s rDNA haplotypes depicts the phylogenetic similarity between Midwestern and Northeastern tick populations (Figure 3a). The majority of haplotypes differ by exactly one nucleotide from other extant haplotypes, a common phylogenetic pattern of rapidly expanding populations. Haplotype pairs that differ by one mutational step are found within the same sampling location (e.g. 10 and 13 both found in Stockton, WI; Figure 3a, Table 1). The closest related haplotype to several Northeastern haplotypes is often found in the Midwest (Figure 3a). For example, haplotype I from the Northeast is most closely related to Midwestern haplotype 13. The Midwestern and Northeastern tick populations are highly divergent from Southern populations. Southern tick haplotypes O, M and N form a distinct clade (Clade B of Qiu et al, 2002) that is separated from Northeast and Midwest haplotypes by several mutational steps (Figure 3a).
Figure 3.
Minimum Spanning Tree (MST) of I. scapularis and B. burgdorferi haplotypes. Each line connecting haplotypes represents one mutational change. Haplotype size reflects relative sampling frequency. A. I. scapularis haplotype MST. SE=Southeast, NE=Northeast, MW=Midwest. Haplotypes 10 and 22 from the Midwest, haplotypes K, L, LI-4; LI-9; and NY-11 from the Northeast, and haplotypes NC2-22 and NC2-29 from the Southeast are each one mutation away from 1/F and are not shown. B. B. burgdorferi IGS haplotype MST. Haplotype named according to Figure 2c where possible. VA=Virginia, NY=New York, MW=Midwest. Haplotypes 16 (2d), 3 (6a), 1 (7a) are not present in the Northeastern sample although initially described in a previous Northeastern sample are marked with an asterisks (Bunikis et al. 2004).
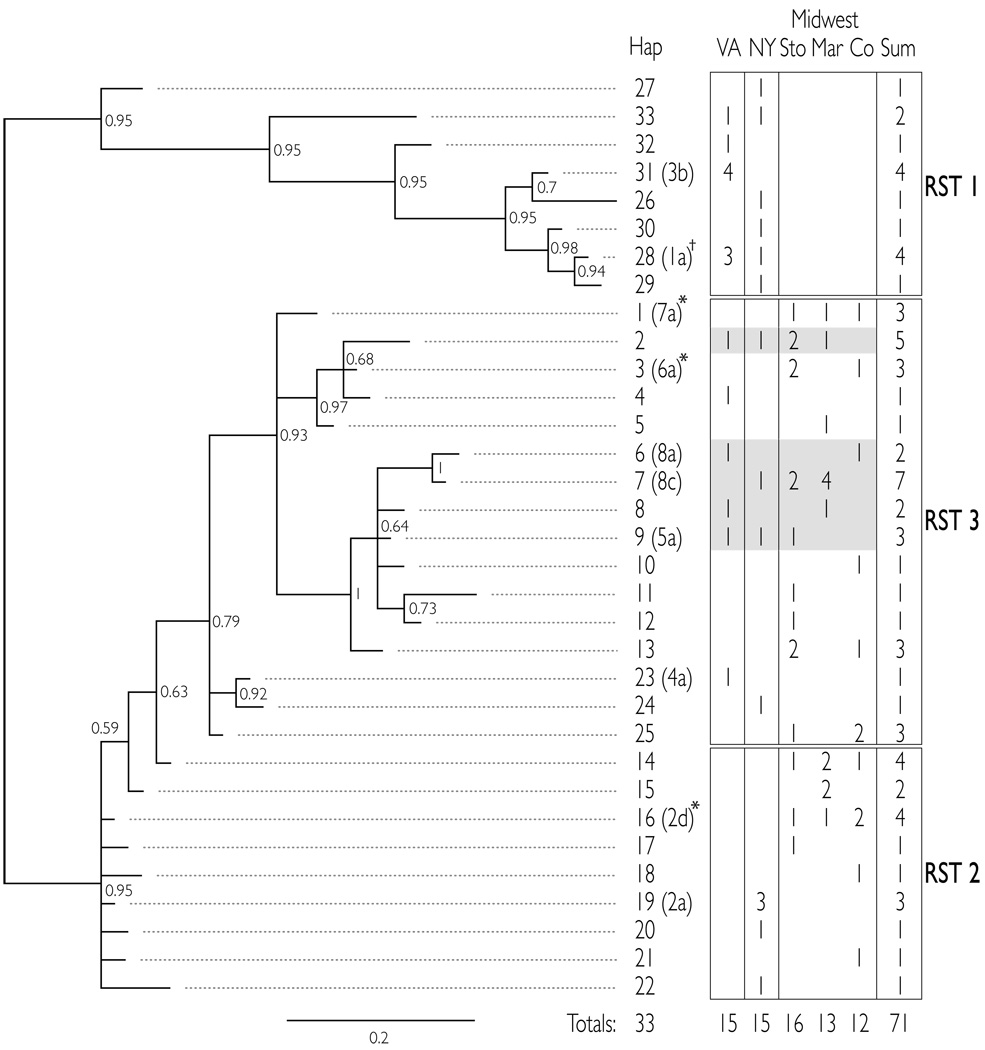
In a sample of 71 B. burgdorferi isolates from ticks collected in the Midwestern (41) and Northeastern sites (30), 33 distinct B. burgdorferi (IGS) haplotypes were recovered (Figure 1), ten of which are identical to previously described IGS sequences from the Northeast (Figure 2c). B. burgdorferi infection prevalence in ticks was similar at all sites (25–30%). Sequence diversity in the Northeastern B. burgdorferi populations (NY=0.028; VA=0.027) is more than double that in Midwestern populations (Co=0.01; Sto=0.011; Mar=0.012), despite the roughly equal numbers of unique sequences found in each population. The differences in nucleotide diversity among the regions is driven by the absence of RST-1 sequences in the Midwestern samples.
Figure 2.
Cladogram and frequency distribution of B. burgdorferi IGS haplotypes collected from Virginia (VA), New York (NY), and three Midwestern sites (Sto, Mar, Co). A. Bayesian cladogram of IGS haplotypes with posterior probability values reported for each node. B. Haplotype frequency distribution with gray shading indicating types shared between Northeastern and Midwestern sites. C. Previously described haplotypes in Bunikis et al. 2004. Haplotypes found only in our Midwestern samples but previously found in Northeastern samples (Bunikis et al. 2004) are marked with an asterisks. The haplotype of the B31 type strain is marked with a cross.
The Northeastern B. burgdorferi samples are dominated by a strongly supported clade of haplotypes corresponding to rDNA intergenic spacer type I (RST-I) (Liveris et al. 1996; Bunikis et al. 2004) that comprise 60% and 40% of all samples from Virginia and New York, respectively. No RST-I sequence was detected in the 41 B. burgdorferi samples taken from the three Midwestern sites (Figure 2a–c). The additional internal nodes are weakly supported but substantiate the previous RST designations (Figure 2a).
Northeastern and Midwestern RST-II and III haplotypes share a recent common ancestor. Eight identical haplotypes from these clades are found in both regions and the closest related haplotype to several Northeastern haplotypes was often sampled only in the Midwest (Figure 2b, Figure 3b). The absence of RST-I haplotypes in the Midwest distinguishes populations in the two regions. RST-I haplotypes are divergent - often separated by tens of mutational steps - while all other haplotypes are separated by one to five mutations (Figure 3).
Population Genetic Structure
Gene flow among tick populations within and between regions is significantly restricted (Table 2). Strong genetic structure is detectable among tick populations both within (ΦSC = 0.1579; p<0.001) and between the Midwestern, Northeastern, and Southern regions (ΦCT = 0.9688; p<0.001). The among region genetic structure is driven by differences between the Northeastern and Southeastern regions and between the Midwestern and Southeastern regions (p<0.004). No significant structure was detected between the Northeastern and Midwestern region (p=0.059). Exact test of population differentiation showed strong genetic structure among the tick populations in the Midwest, indicating that barriers to gene flow exist even over short distances (100km) within this region (p<0.001). A similar degree of genetic structure is found for tick populations within the Northeast (p<0.001) and the Southeast (p<0.001). The mid-Atlantic region including sites in Pennsylvania, New Jersey, and Maryland is not significantly different from the Northeastern region (p=0.76). Including these sites in the analysis as either their own region or including the sites in the Northeastern region does not affect the outcome of the analyses reported.
Table 2.
Population genetic structure for I. scapularis and B. burgdorferi within and between regions
| Treatment | ΦSC | p | ΦCT | p |
|---|---|---|---|---|
| I. scapularis | ||||
| NEvMWvSE | 0.1579 | <0.001 | 0.9688 | <0.001 |
| NEvMW | 0.0788 | <0.001 | 0.0443 | 0.059 |
| NEvSE | 0.2412 | <0.001 | 0.2024 | <0.001 |
| MWvSE | 0.2055 | <0.001 | 0.1899 | 0.004 |
| B. burgdorferi | ||||
| NYvVAvMW | 0.0045 | 0.4236 | 0.223 | <0.001 |
| EastvMW | 0.0190 | 0.1069 | 0.276 | <0.001 |
sc - Among populations within regions
ct - Between regions
The large differences in sampling effort between the Eastern tick populations and the Midwestern populations does not affect the significance of the analyses. Significant genetic structure was found for all 100 subsampled datasets within and between the Northeastern, Midwestern, and Southeastern regions as well as between the Northeastern and Southeastern regions and the Midwestern and Southeastern regions (p<0.05). Significant structure was found between the Northeastern and Midwestern regions in 13 of the 100 subsampled datasets. These analyses, along with the nearly significant structure found using the entire dataset, suggest that given greater sampling in the Northeast, significant genetic structure would be found. Nevertheless, the identity and frequency of tick haplotypes in the Northeastern and the Midwestern regions are much more similar to each other than either is to the Southeastern region.
Population genetic structure is not detected among B. burgdorferi populations within either the Midwestern or Northeastern US (ΦSC=0.034; p=0.11; Table 2). The majority of haplotypes are present in every sampled population indicating that high genetic diversity is maintained within sampling areas (Table 2). Significant differences in the identity and frequency of IGS haplotypes between Midwestern and Northeastern regions (ΦCT=0.276; p<0.0001) indicate inter-regional barriers to gene flow. Regional differentiation is driven by the undetectably low frequency of RST-I haplotypes in the Midwest. No population differentiation was detected when RST-I samples were removed Northeastern populations (p=0.34), suggesting that the barrier to gene flow between these two regions is restricted to RST-I haplotypes.
Time to the most recent common ancestor (MRCA)
Midwestern tick populations coalesce three times more quickly than those in the Northeast and more than ten times faster than Southeastern tick populations (Table 3). Two Eastern sites, MA in the Northeast and NC1 in the Southeast, coalesce more rapidly than populations in the Midwest. The time to coalescence was not estimated for B. burgdorferi populations because the pattern of nucleotide substitutions among IGS haplotypes violates the assumption of infinite sites inherent to the coalescent estimation procedures.
Table 3.
Time to most recent common ancestor (TMRCA) estimates for I. scapularis populations
| Region | Population | TMRCA | SD |
|---|---|---|---|
| Midwest | Stockton | 0.898 | 0.388*10−3 |
| Marinette | 0.616 | 0.318*10−3 | |
| Colfax | 0.898 | 0.996*10−3 | |
| Northeast | NY1 | 2.20 | 0.087 |
| LI2 | 2.18 | 0.102 | |
| CT | 2.24 | 0.108 | |
| MA | 0.525 | 0.0495 | |
| Southeast | SC98 | 7.48 | 0.027 |
| NC2 | 11.9 | 0.0318 | |
| NC1 | 0.592 | 1.5*10−3 |
TMRCA estimates in units of T where T = real time / (Ne* generation time in years).
SD=standard deviation
Demographic History
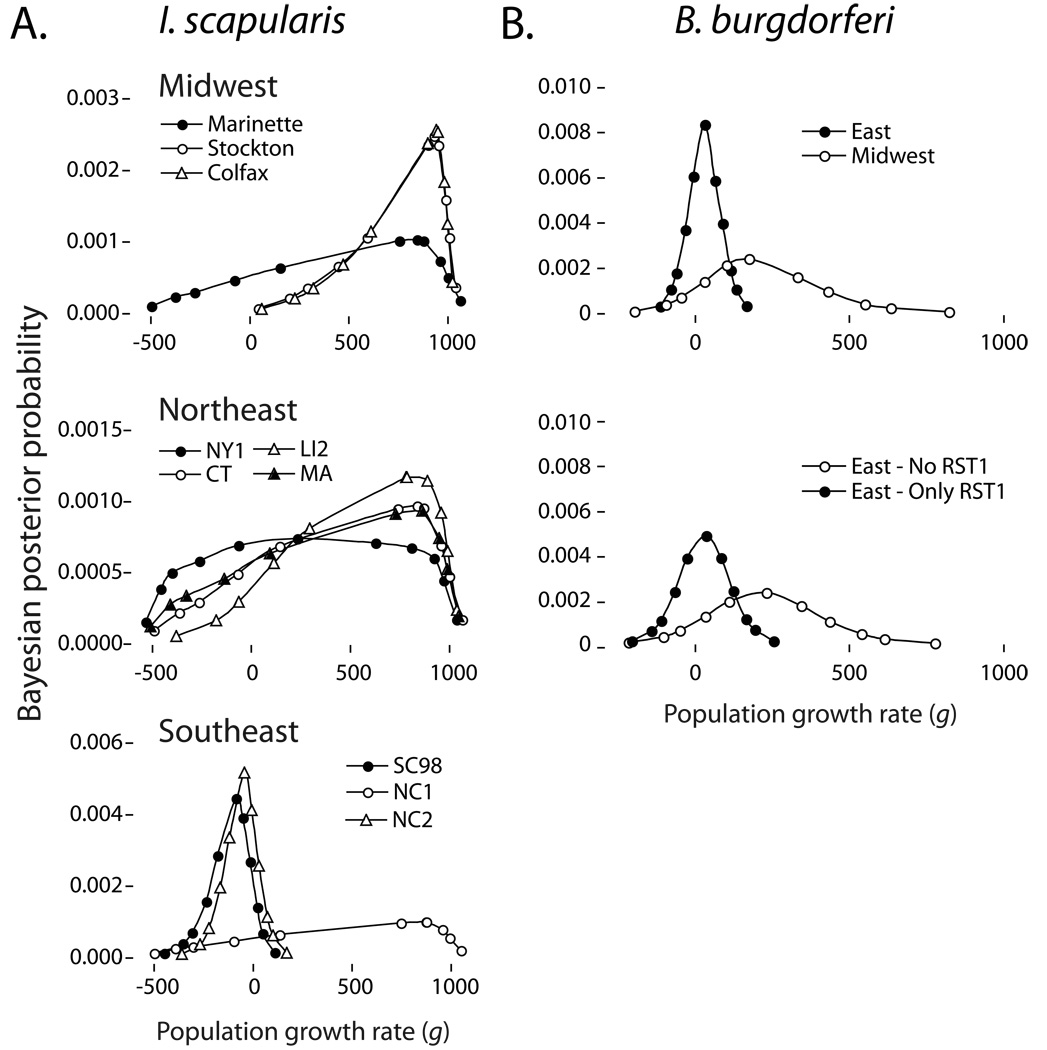
Tick populations in the Midwest and Northeast experienced recent and rapid increases in population size (Figure 4a). By contrast, Southeastern tick populations SC98 and NC2 show no evidence of changes in historical population size. NC1 remains an exception to this pattern, showing a high estimated growth rate. Midwestern B. burgdorferi also show strong evidence of historical population growth (g=185), while Eastern B. burgdorferi samples show support for little historical population growth (g=43; Figure 4b upper panel). The finding of limited population growth in the Northeastern sites is driven by the RST-I haplotypes that dominate these samples (g=39 for RST-I alone). Estimates using only RST-II and III result in strong support for population growth very similar to that found for the Midwest (g=234; Figure 4b lower panel). Estimates for ticks and B. burgdorferi remained unchanged when singleton haplotypes are removed from the analysis.
Figure 4.
Bayesian posterior probability surface of the estimate of the population growth rate (g) for I. scapularis and B. burgdorferi populations. The Most-Probable Estimates (MPE) of g lie at apex of each curve. A. Estimates of g for individual I. scapularis populations from each region. Growth estimates for Northeast and Southeast populations coincide with previous reports (Qiu et al. 2002). B. Estimates of g for B. burgdorferi populations grouped by region. The lower chart displays estimates of g for Northeastern B. burgdorferi with RST-I haplotypes removed (n=15) and RST-I haplotypes alone (n=15).
DISCUSSION
The evolutionary and biogeographic history of vector-borne parasites like B. burgdorferi are necessarily influenced by the evolutionary and biogeographic history of their vectors; B. burgdorferi occurs almost exclusively where I. scapularis is present (Dennis et al. 1998). However, the historical demographic, migratory, and population genetic patterns of B. burgdorferi do not reflect that its obligate tick vector. The I. scapularis populations show considerable population genetic structure within the Midwestern and Northeastern regions but no significant structure was detected between the regions (Table 2). B. burgdorferi populations, on the other hand, show no population genetic structure within regions, but significant structure between regions (Table 2). Human infectious RST-I strains are absent from the Midwestern samples but constitute roughly half of all samples from the Northeastern US (Figures 2; 3). The Northeastern I. scapularis populations are derived from a small number of migrants from the Southeastern region after the recession of the Pleistocene ice sheet. Phylogenetic similarity among northern tick populations suggests that ticks already established in the post-glacial Northeastern US colonized the Midwest. The B. burgdorferi populations in both the Midwest and Northeast are derived from the same pool of highly variable strains with the exception of RST-I strains that likely face an ecological or geographic barrier to migration.
I. scapularis Phylogeography
The mitochondria in both the Midwestern and Northeastern I. scapularis populations are derived from a single colonizing tick that originated from refugia located to the south of the Pleistocene ice sheet. The recent common ancestry of the Midwestern and Northeastern tick populations is evinced by their phylogenetic similarity and the lack of significant genetic structure detected among populations from these regions (Table 2). Neither Northeastern nor Midwestern ticks are monophyletic suggesting a recent shared history and appreciable migration (Fig 3). The Midwestern populations are much younger than Northeastern populations and both are an order of magnitude younger than Southeastern populations (Table 3), suggesting that Midwestern populations were the most recently colonized by I. scapularis. Northeastern migrants colonized the Midwest, either by long-distance migration on avian hosts or by range expansion through regions currently supporting very low tick densities (Diuk-Wasser et al. 2006; McNabb et al. 2008).
After the initial migration across the northern region of the United States, populations have been largely isolated as evidenced by the extensive genetic differentiation among populations within each of the three regions (Table 2). Although the most frequent haplotype in all three Midwestern populations is 1/F, all but four of the remaining haplotypes are found in only one population (Table 1). Haplotypes that differ by one nucleotide are always observed in the same tick population in the Midwest and are likely recently derived mutations (Figure 3). Four haplotypes found in only one Midwestern population are also found in the Northeastern region, indicating either recurrent mutation, different founders for each Midwestern population, or haplotype loss by genetic drift. Northeastern and Southeastern populations also show evidence of limited migration among populations within regions (Qiu et al. 2002). However, the population structure of Northeastern I. scapularis populations differs from the Midwestern populations in that each population has a different most-frequent haplotype and the majority of haplotypes are found in every population, indicating less subdivision in the Northeast than the Midwest. The Southeastern region has the most genetically divergent tick populations, differing in both frequency and composition of haplotypes, a common pattern for populations with long histories of isolation (Avise 2000).
The Southeastern NC1 population sampled from Pea Island National Wildlife Refuge in North Carolina appears much younger than any Northeastern population except MA and shows evidence of rapid population growth. Pea Island National Wildlife Refuge is located on a coastal barrier island (max elevation 8ft) that may have been colonized or re-colonized recently by I. scapularis. Coastal populations like NC1 and MA, located at the tip of Cape Cod, may have been subjected to repeated population bottlenecks due to changes in ocean levels and severe weather. To the contrary, inland and Long Island tick populations are at elevations at least five-fold higher than the highest point of either the NC1 or MA (max elevation 32ft) populations.
The recent discovery of I. scapularis in Midwestern US have led to the hypothesis that ticks were introduced or reintroduced in this region in the last century (Jackson and DeFoliart 1970). Although the density and recognition of this medically important vector species has likely expanded rapidly over the past five decades, Midwestern tick populations have accrued dozens of novel haplotypes necessitating many hundreds of generations. Further, sites as close as 100km are significantly genetically differentiated (Figure 3a, Table 2). Rapid population growth can increase the rate of introduction of new mutations into a population thereby accelerating genetic differentiation among populations (Maruyama and Fuerst 1984; Kimmel et al. 1998). Untenable rates of mutation would have to be invoked to produce the population genetic patterns seen for ticks over 50 years.
B. burgdorferi Population Structure
The evolutionary and biogeographic history of B. burgdorferi does not reflect that of its tick vector. B. burgdorferi populations show no population genetic structure within each region but strong barriers to gene flow between the Northeastern and Midwestern region. The between-region structure is driven by the lack of RST-I genotypes in the Midwestern dataset (RST-1 has recently been detected at very low frequencies in the Midwest (Gatewood et al. 2009)). RST-II and III strains from the Northeast and Midwest display no genetic structure and appear to share a recent common ancestor. The undetectably low frequency of RST-I isolates at Midwestern sites likely has arisen through either selective processes that prevent RST-I lineages from establishing in the Midwest or due to lower migratory rates than RST-II and III.
Ecological barriers to gene flow may be present in the form of unsuitable host species as the relative fitness of B. burgdorferi genotypes differs among vertebrate species in the Northeast (Brisson and Dykhuizen 2004; Hanincova et al. 2006; Brisson et al. 2008). However, host species commonly infected by RST-I strains in the Northeast such as Peromyscus leucopus are present in the Midwest, suggesting RST-I strains can colonize the Midwest if introduced. The recent shared history of Northeastern and Midwestern ticks makes it unlikely that evolution in the Midwestern vector impinges on the invasion success of RST-I B. burgdorferi. Prevailing abiotic conditions in the Midwest differ from those in the Northeast such that the activity period of the immature stages of I. scapularis are synchronous in the Midwest and asynchronous in the Northeast. The asynchronous activity periods in the Northeast may enrich for the purportedly longer-lived RST-I infections (Diuk-Wasser et al. 2006; Ogden et al. 2007; Gatewood et al. 2009).
RST-I lineages of B. burgdorferi are highly invasive in humans in the Northeastern US (Seinost et al. 1999; Dykhuizen et al. 2008; Wormser et al. 2008) and are typically the most common strains found in Northeastern ticks (Figure 2). The relative paucity of RST-I B. burgdorferi genotypes in the Midwest explains why the reported Lyme disease incidence in this region is relatively reduced compared to the Northeast, despite similar tick densities and infection rates among host-seeking ticks (McNabb et al. 2008; Gatewood et al. 2009).
Demographic Diversity within B. burgdorferi
The genetic diversity and stable population size of RST-I haplotypes (Fig 4b) suggests that the RST-I clade is much older than other B. burgdorferi groups (Slatkin and Hudson 1991). Thus, RST-I appears to be the ancestral clade from which RST-II and III were derived. Two recent reports suggest that B. burgdorferi arose in Europe and recently migrated to North America (Margos et al. 2008; Qiu et al. 2008). However, all RST types are present in European populations suggesting that either RST-II and III are older than suggested in this paper or that RST-II and III strains migrated from North America to Europe (Margos et al. 2008; Qiu et al. 2008).
Range expansion of I. scapularis and B. burgdorferi – and thus range expansion of human Lyme disease – may occur as human land use patterns and global climate continue to change. Tick populations are capable of enormous rates of sustained population growth after colonizing an area (Figures 3a, 4). Both long- and short-range migration may allow tick populations to expand their range and to colonize distant regions (Ogden et al. 2007; Ogden et al. 2008; Gray et al. 2009). Migration of B. burgdorferi among localites with established and newly settled tick populations may be limited only by ecological factors inhibiting colonization.
ACKNOWLEDGEMENTS
We thank DE Dykhuizen and MJ Voordouw for helpful comments on both the project and the manuscript. This work was supported by the National Institute of Health grants AI076342 (DB), and the Center for Diseases Control and Prevention grant U01CK000170 (DB).
REFERENCES
- Anderson JF, Magnarelli LA, Burgdorfer W, Barbour AG. Spirochetes in Ixodes-Dammini and Mammals from Connecticut. American Journal of Tropical Medicine and Hygiene. 1983;32:818–824. doi: 10.4269/ajtmh.1983.32.818. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard University Press; 2000. [Google Scholar]
- Avise JC, Walker D. Pleistocene phylogeographic effects on avian populations and the speciation process. Proc Biol Sci. 1998;265:457–463. doi: 10.1098/rspb.1998.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Walker D, Johns GC. Speciation durations and Pleistocene effects on vertebrate phylogeography. Proc Biol Sci. 1998;265:1707–1712. doi: 10.1098/rspb.1998.0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahlo M, Griffiths RC. Inference from gene trees in a subdivided population. Theor Popul Biol. 2000;57:79–95. doi: 10.1006/tpbi.1999.1447. [DOI] [PubMed] [Google Scholar]
- Brisson D, Dykhuizen DE. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics. 2004;168:713–722. doi: 10.1534/genetics.104.028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Dykhuizen DE. A modest model explains the distribution and abundance of Borrelia burgdorferi strains. American Journal of Tropical Medicine and Hygiene. 2006;74:615–622. [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Dykhuizen DE, Ostfeld RS. Conspicuous impacts of inconspicuous hosts on the Lyme disease epidemic. Proceedings of the Royal Society, Biological Sciences. 2008;275:227–235. doi: 10.1098/rspb.2007.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis J, Garpmo U, Tsao J, Berglund J, Fish D, Barbour AG. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology. 2004;150:1741–1755. doi: 10.1099/mic.0.26944-0. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease - a tick-borne spirochetosis. Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Church SA, Kraus JM, Mitchell JC, Church DR, Taylor DR. Evidence for multiple Pleistocene refugia in the postglacial expansion of the eastern tiger salamander, Ambystoma tigrinum tigrinum. Evolution Int J Org Evolution. 2003;57:372–383. doi: 10.1554/0014-3820(2003)057[0372:EFMPRI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Clark KL, Oliver JH, Jr, James AM, Durden LA, Banks CW. Prevalence of borrelia burgdorferi sensu lato infection among rodents and host-seeking ticks in South Carolina. J Med Entomol. 2002;39:198–206. doi: 10.1603/0022-2585-39.1.198. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Dennis DT, Nekomoto TS, Victor JC, Paul WS, Piesman J. Reported distribution of Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae) in the United States. Journal of Medical Entomology. 1998;35:629–638. doi: 10.1093/jmedent/35.5.629. [DOI] [PubMed] [Google Scholar]
- Diuk-Wasser MA, Gatewood AG, Cortinas MR, Yaremych-Hamer S, Tsao J, Kitron U, Hickling G, Brownstein JS, Walker E, Piesman J, Fish D. Spatiotemporal patterns of host-seeking Ixodes scapularis nymphs (Acari: Ixodidae) in the United States. Journal of Medical Entomology. 2006;43:166–176. doi: 10.1603/0022-2585(2006)043[0166:spohis]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Dykhuizen DE, Brisson D, Sandigursky S, Wormser GP, Nowakowski J, Nadelman R, Schwartz I. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. American Journal of Tropical Medicine and Hygiene. 2008;78:806–810. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood AG, Liebman KA, Vourc'h G, Bunikis J, Hamer SA, Cortinas R, Melton F, Cislo P, Kitron U, Tsao J, Barbour AG, Fish D, Diuk-Wasser MA. Climate and tick seasonality are predictors of Borrelia burgdorferi genotype distribution. Appl Environ Microbiol. 2009;75:2476–2483. doi: 10.1128/AEM.02633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JS, Dautel H, Estrada-Pena A, Kahl O, Lindgren E. Effects of climate change on ticks and tick-borne diseases in europe. Interdiscip Perspect Infect Dis. 2009 doi: 10.1155/2009/593232. 2009: 593232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanincova K, Kurtenbach K, Diuk-Wasser M, Brei B, Fish D. Epidemic spread of Lyme borreliosis, northeastern United States. Emerging Infectious Diseases. 2006;12:604–611. doi: 10.3201/eid1204.051016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hutcheson HJ, Oliver JH, Jr, Houck MA, Strauss RE. Multivariate morphometric discrimination of nymphal and adult forms of the blacklegged tick (Acari: Ixodidae), a principal vector of the agent of Lyme disease in eastern North America. Journal of Medical Entomology. 1995;32:827–842. doi: 10.1093/jmedent/32.6.827. [DOI] [PubMed] [Google Scholar]
- Jackson JO, DeFoliart GR. Ixodes scapularis Say in northern Wisconsin. J Med Entomol. 1970;7:124–125. doi: 10.1093/jmedent/7.1.124. [DOI] [PubMed] [Google Scholar]
- Kimmel M, Chakraborty R, King JP, Bamshad M, Watkins WS, Jorde LB. Signatures of population expansion in microsatellite repeat data. Genetics. 1998;148:1921–1930. doi: 10.1093/genetics/148.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles LL. Did the pleistocene glaciations promote divergence? Tests of explicit refugial models in montane grasshopprers. Mol Ecol. 2001;10:691–701. doi: 10.1046/j.1365-294x.2001.01206.x. [DOI] [PubMed] [Google Scholar]
- Kuhner MK. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- Lane RS, Piesman J, Burgdorfer W. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annual Review of Entomology. 1991;36:587–609. doi: 10.1146/annurev.en.36.010191.003103. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Liveris D, Wormser GP, Nowakowski J, Nadelman R, Bittker S, Cooper D, Varde S, Moy FH, Forseter G, Pavia CS, Schwartz I. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. Journal of Clinical Microbiology. 1996;34:1306–1309. doi: 10.1128/jcm.34.5.1306-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:567–571. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnarelli LA, Anderson JF, Fish D. Transovarial transmission of Borrelia burgdorferi in Ixodes dammini (Acari:Ixodidae) Journal of Infectious Diseases. 1987;156:234–236. doi: 10.1093/infdis/156.1.234. [DOI] [PubMed] [Google Scholar]
- Margos G, Gatewood AG, Aanensen DM, Hanincova K, Terekhova D, Vollmer SA, Cornet M, Piesman J, Donaghy M, Bormane A, Hurn MA, Feil EJ, Fish D, Casjens S, Wormser GP, Schwartz I, Kurtenbach K. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2008;105:8730–8735. doi: 10.1073/pnas.0800323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Fuerst PA. Population bottlenecks and nonequilibrium models in population genetics. I. Allele numbers when populations evolve from zero variability. Genetics. 1984;108:745–763. doi: 10.1093/genetics/108.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather TN, Telford SR, 3rd, Adler GH. Absence of transplacental transmission of Lyme disease spirochetes from reservoir mice (Peromyscus leucopus) to their offspring. Journal of Infectious Disease. 1991;164:564–567. doi: 10.1093/infdis/164.3.564. [DOI] [PubMed] [Google Scholar]
- McNabb SJ, Jajosky RA, Hall-Baker PA, Adams DA, Sharp P, Worshams C, Anderson WJ, Javier AJ, Jones GJ, Nitschke DA, Rey A, Wodajo MS. Summary of notifiable diseases--United States, 2006. MMWR Morb Mortal Wkly Rep. 2008;55:1–92. [PubMed] [Google Scholar]
- Mila B, Girman DJ, Kimura M, Smith TB. Genetic evidence for the effect of a postglacial population expansion on the phylogeography of a North American songbird. Proc Biol Sci. 2000;267:1033–1040. doi: 10.1098/rspb.2000.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigel JE, Avise JC. Application of a random walk model to geographic distributions of animal mitochondrial DNA variation. Genetics. 1993;135:1209–1220. doi: 10.1093/genetics/135.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden NH, Bigras-Poulin M, O'Callaghan C J, Barker IK, Kurtenbach K, Lindsay LR, Charron DF. Vector seasonality, host infection dynamics and fitness of pathogens transmitted by the tick Ixodes scapularis. Parasitology. 2007;134:209–227. doi: 10.1017/S0031182006001417. [DOI] [PubMed] [Google Scholar]
- Ogden NH, Lindsay LR, Hanincova K, Barker IK, Bigras-Poulin M, Charron DF, Heagy A, Francis CM, O'Callaghan CJ, Schwartz I, Thompson RA. Role of migratory birds in introduction and range expansion of Ixodes scapularis ticks and of Borrelia burgdorferi and Anaplasma phagocytophilum in Canada. Appl Environ Microbiol. 2008;74:1780–1790. doi: 10.1128/AEM.01982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JH., Jr Lyme borreliosis in the southern United States: a review. J Parasitol. 1996;82:926–935. [PubMed] [Google Scholar]
- Orloski K, Hayes E, Campbell GL, Dennis DT. Surveillance for Lyme disease -- United States, 1992–1998. MMWR CDC Surveill Summ. 2000;49:1–11. [PubMed] [Google Scholar]
- Patrican LA. Absence of Lyme disease spirochetes in larval progeny of naturally infected Ixodes scapularis (Acari:Ixodidae) fed on dogs. J Med Entomol. 1997;34:52–55. doi: 10.1093/jmedent/34.1.52. [DOI] [PubMed] [Google Scholar]
- Patz JA, Campbell-Lendrum D, Holloway T, Foley JA. Impact of regional climate change on human health. Nature. 2005;438:310–317. doi: 10.1038/nature04188. [DOI] [PubMed] [Google Scholar]
- Pielou EC. After the Ice Age: The Return of Life to Glaciated North America. Chicago: University Of Chicago Press; 1992. [Google Scholar]
- Piesman J, Donahue JG, Mather TN, Spielman A. Transovarially acquired Lyme disease spirochetes (Borrelia burgdorferi) in field-collected larval Ixodes dammini (Acari, Ixodidae) Journal of Medical Entomology. 1986;23 doi: 10.1093/jmedent/23.2.219. 219-219. [DOI] [PubMed] [Google Scholar]
- Qiu WG, Bruno JF, McCaig WD, Xu Y, Livey I, Schriefer ME, Luft BJ. Wide distribution of a high-virulence Borrelia burgdorferi clone in Europe and North America. Emerg Infect Dis. 2008;14:1097–1104. doi: 10.3201/eid1407.070880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu WG, Dykhuizen DE, Acosta MS, Luft BJ. Geographic uniformity of the Lyme disease spirochete (Borrelia burgdorferi) and its shared history with tick vector (Ixodes scapularis) in the northeastern United States. Genetics. 2002;160:833–849. doi: 10.1093/genetics/160.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Rich SM, Caporale DA, Telford SR, 3rd, Kocher TD, Hartl DL, Spielman A. Distribution of the Ixodes ricinus-like ticks of eastern North America. Proceedings of the National Academy of Sciences of the United States of America U S A. 1995;92:6284–6288. doi: 10.1073/pnas.92.14.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JD, Fernando K, Banerjee SN, Durden LA, Byrne SK, Banerjee M, Mann RB, Morshed MG. Birds disperse ixodid (Acari: Ixodidae) and Borrelia burgdorferi- infected ticks in Canada. Journal of Medical Entomology. 2001;38:493–500. doi: 10.1603/0022-2585-38.4.493. [DOI] [PubMed] [Google Scholar]
- Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang IN, Wormser GP, Schriefer ME, Luft BJ. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infection and Immunity. 1999;67:3518–3524. doi: 10.1128/iai.67.7.3518-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR. Pairwise Comparisons of Mitochondrial-DNA Sequences in Stable and Exponentially Growing Populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormser GP, Brisson D, Liveris D, Hanincova K, Sandigursky S, Nowakowski J, Nadelman RB, Ludin S, Schwartz I. Borrelia burgdorferi Genotype Predicts the Capacity for Hematogenous Dissemination during Early Lyme Disease. J Infect Dis. 2008;198:1358–1364. doi: 10.1086/592279. [DOI] [PMC free article] [PubMed] [Google Scholar]