Abstract
A novel approach was developed to efficiently package and deliver nucleic acids with low generation polypropylenimine (PPI) dendrimers by using Au nanoparticles as a “labile catalytic” packaging agent. The Au nanoparticles (Au NPs) helped low generation dendrimers to package nucleic acids into discrete nanoparticles but are not included in the final DNA/siRNA complexes. Therefore it becomes possible to eliminate the potential toxic problems associated with Au NPs by selectively removing the Au NPs from the resulting nucleic acid complexes before their delivery to targeted cells. This is a new concept in using inorganic engineered nanoparticles in nucleic acid packaging and delivery applications. Furthermore, compared to the siRNA nanostructures (mainly randomly aggregated nanofibers) fabricated by low generation dendrimer alone (Generation 3), the siRNA nanoparticles packaged using this novel approach (by Au NPs modified with G3 PPI) can be internalized by cancer cells and the delivered siRNAs can efficiently silence their target mRNA. The efficiency of mRNA silencing by this novel approach is even superior to higher generation dendrimers (Generation 5).
Keywords: Gene therapy, siRNA, plasmid DNA, non-viral gene delivery, Au nanoparticles, polypropylenimine (PPI) dendrimers
INTRODUCTION
The inefficient transport of nucleic acids through the cell membrane is a major limiting factor in clinical application of therapeutic nucleic acids, including antisense or anti-gene oligonucleotides, and short interference RNA (siRNA). Several approaches including packaging of the nucleic acid to nanoparticles have recently been used to facilitate transmembrane transport of nucleic acids.1-2 Viral vectors are able to efficiently accomplish such task; however, the immune response elicited by viral proteins has posed a major challenge to this approach.3 Hence, there is much interest in developing nonviral gene delivery vehicles.
Cationic polymers and lipids have been widely used for condensation of DNA/siRNA into nanoparticles for cellular delivery. Dendrimers are highly branched three-dimensional polymers with a large number of controllable peripheral functionalities, useful as gene delivery agents, drug delivery vehicles, and magnetic resonance imaging agents.4-8 The protonatable amino groups of dendrimers appear to buffer acidic endosomal compartment and release DNA to the cytoplasm.4,9-10 However, it was reported that dendrimers bearing -NH2 termini displayed generation- and concentration-dependent haemolysis, with higher generation and/or higher concentration having higher cytotoxicity11-13 and their synthesis and purification are usually tedious with low yield.14-15 In contrast, low generation dendrimers are nontoxic and easy to synthesize,16 however, the limited surface charges of low generation dendrimers lead to inefficient complexation with DNA and low cellular uptake efficacy.17-18 We recently found that only higher generations of polypropylenimine (PPI) dendrimers could enhance oligodeoxynucleotide (ODN) and siRNA uptake to cancer cells as demonstrated by confocal microscopy.19-20
There is a surge of interest in using inorganic engineered nanoparticles for medical and biological applications. They are expected to solve some drug delivery problems due to their unique properties and remarkably large surface area. Studies using inorganic engineered nanoparticles modified with cationic molecules, including dendrimers have demonstrated enhancement in DNA/siRNA condensation, delivery and transfection in mammalian cells.21-25 However, the inorganic nanoparticles were encapsulated inside the resulting DNA/siRNA complexes, and must be co-delivered together with the therapeutic DNA/siRNA. The toxicity of inorganic nanoparticles represents a major concern for practical therapeutic applications. It is reported that nanoparticles chemically modified with low generation dendrimers may act as mesomolecules inducing haemolysis comparable or even higher than that of the higher generation dendrimers.26 For Au nanoparticles, even though bulk Au was considered inert, the toxicity of Au nanparticles depends on their size, shape and surface charge.27-29 In general, smaller sized nanoparticles (< 3 nm in diameter) are more toxic than larger ones. Positively charged nanoparticles are more toxic than the neutral or negatively charged ones. Chithrani et al. studied the endocytosis and excytosis of transferrin-coated Au nanoparticles (from 14 nm to 100 nm).30 They found that smaller NPs appeared to exocytose at a faster rate and at a higher percentage than large NPs. These exciting results indicated that it is not necessary to worry the long term toxicity of Au nanoparticles anymore. However, the authors of this paper claimed that these results could not directly extend to other particles since uncoated or other surface modified Au nannoparticles may bind other proteins in serum and they may behave differently in terms of toxicity, endocytoses or exoctytosis. Indeed, Yung et al. 31 found that negatively charged, citrate-stabilized AuNPs with a diameter of 20 nm after passivation with fetal bovine serum inhibited cell proliferation by down-regulating cell cycle genes. They found that the Au NPs passivated with fetal bovine serum not only caused DNA oxidative damage but also affected genes associated with genomic stability and DNA repair, which may lead to carcinogenesis.32 Pernodet et al.33 also found that citrate-stabilized AuNPs with a diameter of 14 nm can easily cross the cell membrane of dermal fibroblast cells and accumulate into vacuoles. The presence of the particles induced abnormal actin filaments and extracellular matrix constructs in dermal fibroblasts, which in turn, caused decrease cell proliferation, adhesion, and motility. Therefore the long-term toxicity of Au nanoparticles still needs extensive study before their clinic applications.
In this work, we report our interesting finding that Au nanoparticles (NPs) can be used in combination with low generation polypropyleneimine (PPI) dendrimers to effectively assemble and package DNA/siRNA into discrete complex nanoparticles. Moreover, the Au NPs were not encapsulated inside the final DNA/siRNA complexes. Therefore it becomes possible to eliminate the potential toxic problems associated with Au NPs by selectively removing the Au NPs from the resulting nucleic acid complexes before their delivery to targeted cells. In contrast to the siRNA nanostructures (mainly random aggregated nanofibers) formed from low generation dendrimer alone (PPI Generation 3), the siRNA complex nanoparticles packaged by this novel approach (Au nanoparticles modified with G3 PPI dendrimers) can be internalized by cancer cells and the delivered siRNAs can effectively silence their targeted mRNA. The efficiency of mRNA silencing by the proposed nanoparticles is even superior to higher generation dendrimers (PPI Generation 5).
RESULTS AND DISCUSSION
A Novel Approach for Assembly and Delivery of DNA Using Labile Au/G3 PPI Dendrimer
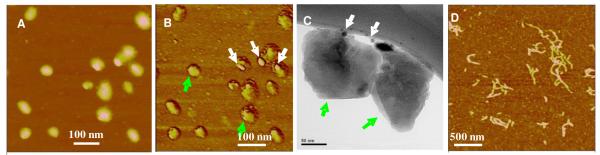
We prepared Au NPs physically attached with several generation 3 (G3) PPI dendrimers according to a protocol described by Wang and his colleagues.34-35 Briefly, 32.8 mg of G3 dendrimer was mixed with 8.8 ml of 2.45 mM HAuCl4 solution at a molar ratio of 0.9:1 and then heated at 80° C for 1 h with continuous stirring. As a result, a clear and reddish Au NP solution (Supporting information, Figure S1) was obtained with each Au NP physically anchoring several G3 dendrimers.36 The largely increased positive charges on each of the condensing agent (here Au NPs modified with G3 dendrimers) enabled the low generation dendrimers to efficiently condense DNA as higher generation dendrimers do.21-22,37-38 To remove the un-reacted HAuCl4 and unreacted dendrimers, the solution thus obtained was cooled to 22 °C and dialyzed (3.5k-Da cutoff) against 200 ml of double-distilled water twice before it was used to condense nucleic acids. UV-visible spectroscopy was used to characterize the formed nanoparticle solution (Supporting information, Figure S2). A strong absorption was observed at ~523 nm, which was due to the surface plasmon resonance of the Au NPs. The mean hydrodynamic radius of Au NPs was 8.9 ± 2.8 nm as determined by dynamic light scattering (DLS) (Supporting information, Figure S3). Using these Au NPs to condense PGL3 Plasmid DNA, we found that DNA was efficiently condensed into nanoparticles, while Au NPs were not encapsulated inside the final condensates (Figure 1a, b). Two sets of nanoparticles were observed, one set with an average height of 3.9 ± 0.7 nm and diameter of 81 ± 36 nm, which are believed to be the condensed DNA arbitrated from their daisy flower-shaped peripheral. The other set of nanoparticles have an average height of 6.4 nm, and diameter of 23 nm, very close to the size of the gold nanoparticles in DLS experiment (hydrodynamic radius of 8.9 nm and diameter of 17.8 nm, note that AFM average height is often lower than the actual diameter based on our and others observations. The possible reasons have been discussed in our previous work 19). Therefore we assign this set of nanoparticles as the gold nanoparticles, which left the DNA nanoparticles upon DNA condensation. The phase image in Figure 1b shows that the two sets of nanoparticles have different phase shifts. The phase of the small nanoparticles shifted 6.1 ± 1.1 degree (appearing brighter) and that of the larger ones only shifted 0.8-1.7 degree, further suggesting that these two sets of nanoparticles were composed of different materials. The arrow in Figure 1b indicates an Au NP leaving a condensate.
Figure 1.
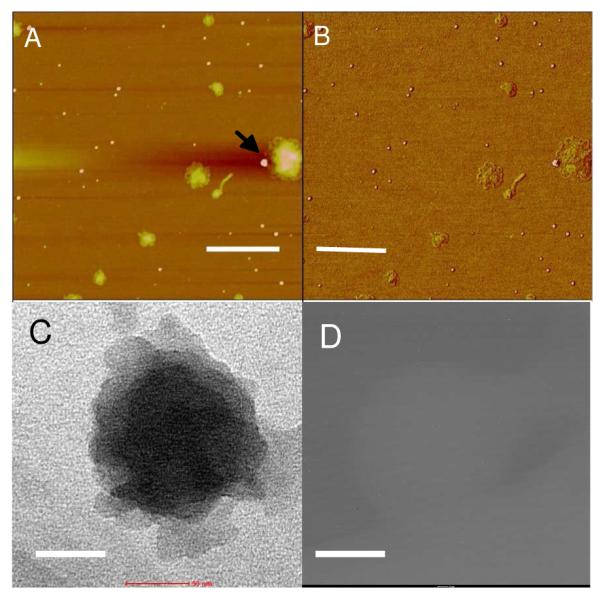
AFM images (panels A and B) and TEM images (panels C and D) of condensates formed by plasmid DNA in the presence of G3 PPI dendrimer modified Au NPs (1-h condensation) (A) Height image, z range = 18.0 nm; (B) phase image, z range=10 degree. The bar represents 200 nm in panels (A) and (B). It showed that the Au NPs were isolated from the condensed nanoparticles. (C) One DNA nanoparticle imaged by TEM at zeroloss mode. (D) The same DNA nanoparticle imaged by TEM in the energy filtering mode, the energy slit of 150 eV energy window was set around the maximum loss (2386 eV) of M5 edge of Au element. High beam current and long exposure time were employed in the energy filtering imaging mode. It showed that no Au NPs were present in the condensate of plasmid DNA. Scale bar represents 50 nm in panels (C) and (D).
To further confirm that the Au NPs were not encapsulated inside DNA condensates, the same solution sample was imaged using a TEM (Libra 120 Energy Filtering, Zeiss). If Au NPs were included in the condensates, as found in previous reports,21,39 we should be able to see them due to the native high contrast of Au in TEM. Figure 1c shows a typical TEM image of a DNA condensate. The diameter of the condensates is 100 nm ± 30 nm. The size and shape of the condensates was similar to that of the bigger flower-shaped nanoparticles in the AFM images. By imaging the same condensate in the energy filtering TEM mode,40-41 setting the energy slit to 2386 eV, which is the energy edge of Au element, we performed Au elemental analysis of the condensates. As shown in Figure 1d, the image was blank, as there was no Au signal from the DNA condensate, indicating that the Au NPs left the products upon condensation. The result found here is very different from previous reported DNA condensation by using Au NPs and other nanoparticles as condensing agents, which were encapsulated in the final DNA nanoparticles.21,24,37,39,42-43
Packaging of 21-base pair (bp) siRNAs to Nanoparticles
There is an increasing enthusiasm for developing therapies based on RNA interference (RNAi), a post-transcriptional gene silencing method, mediated by small duplex RNAs of 19-23 base pairs. The advantage of RNAi compared to other gene therapeutic strategies lies in its high specificity and potency of gene silencing, coupled with the fact that it can target every gene, and every cell, which has the necessary machinery.44-45 However, just like other gene therapeutic strategies, the main obstacle to the success of siRNA therapies is in delivering it across the cell membrane to the cytoplasm where it can enter the RNAi pathway and guide the sequence-specific mRNA degradation.45 It is reported that in general short oligonucleotides are more difficult to package into well-defined particles than the long plasmid DNA.46-47 In our recent studies,20 we found that in contrast to long plasmid DNA, siRNA required higher generation dendrimers to reach the maximum transfection efficiency (G4 PPI for siRNA vs. G2 PPI for plasmid DNA), possibly due to the less negative charge per molecule and the more rigid structure of siRNA. Recently, Huang et al reported that by adding high molecular weight calf thymus DNA into the siRNA formulation, the size of the formed nanoparticles decreased by 10-30% and the delivery efficiency increased by 20-80%.48-49
To demonstrate the wide application of the new packaging approach in gene therapy, we next tested whether the Au NPs modified with G3 PPI dendrimer could catalytically provoke siRNA nanoparticle formation. The complex formation is driven mainly by electrostatic interaction between negatively charged siRNA and positively charged dendrimers on Au NPs. Such binding leads to the formation of positively charged complexes that can be retarded in gel electrophoresis when compared with free siRNAs, which cannot. Therefore the sufficient amount of G3 PPI dendrimers needed for the complex formation can be determined from an agarose gel electrophoresis experiment. It was found that when the N/P (nitrogen/phosphate) ratios, which refer to the ratios of the positively charged primary amine groups of the G3 dendrimer on the Au-NPs to negatively charged phosphate groups from siRNAs, were equal to or higher than 1.2, the siRNA was completely retained in the sample wells with negligible electrophoretic shift corresponding to free siRNA (Supporting information, Figure S4). This indicated that all siRNAs formed stable complex with Au-G3 at N/P ratios equal to or higher than 1.2. Based on this, we prepared siRNA nanoparticles by mixing the G3 PPI dendrimer-modified Au NPs solution and siRNA solution in physiological buffer at an N/P ratio of 2.4 and then incubating for 30 min at room temperature. We then visualized nanoparticles using AFM and TEM as described in the previous section. Figure 2a shows that siRNA nanoparticles with an average height of 10 ± 3 nm and diameter of 70 ± 10 nm were formed after 30 min of condensation. siRNA nanoparticles were also observed in TEM images (Figure 2c), indicated by the green arrows. Remarkably, both AFM and TEM images clearly show that most of the Au NPs (white arrows) were not included inside the siRNA nanostructures. For comparison, we also studied the morphology of the siRNA nanostructures formed with G3 dendrimer alone under the same condensation conditions (Figure 2d). Instead of nanoparticles, most of the siRNAs formed randomly nanofibers. The height of the fiber-like structures was around 1.7 nm with the length varied from 100 nm to several μm. The relatively weak electrostatic cooperative interaction between siRNA and G3 PPI may be the reason to form this fiber shaped structures.
Figure 2.
AFM images (panels a and b) and TEM image (panel C) of siRNA nanoparticles formed from 0.4 μM 21 bp siRNA in the presence of Au NPs modified with G3 PPI dendrimer (2.5 μM). (A) AFM image, z range= 20.0 nm; (B) phase image, z range=40 degree. The bar represents 100 nm in panels (A) and (B). (C) TEM image of NPs. The bar represents 50 nm in panel C. AFM phase image and TEM image clearly show the Au NPs (indicated by white arrows) were not included in the siRNA nanoparticles (indicated by green arrows). (D) AFM images of siRNA nanostructures formed from G3 alone with the same concentration of siRNA and G3 PPI as in the case of Au NPs modified with G3 PPI dendrimer. The bar represents 500 nm in panels (D)
Au nanoparticles can be selectively removed from the siRNA complexes solution
We found that although the Au nanoparticles were not included inside of the DNA/siRNA complexes (Figure 1 and 2), the Au nanoparticles still existed in the DNA/siRNA solution, which may cause long-term toxicity. To selectively remove the Au nanoparticles, we studied the effect of ionic strength and pH on the aggregation/precipitation characteristics of Au nanoparticles in the siRNA complex solution prepared from Au-G3 NPs. Au-G3 NPs were first complexed with siRNA at an N/P ratio of 1.2 to prepare the Au-G3/siRNA complex. Then the obtained complex solution was diluted 12 times into H2O, 100 mM pH=7.4, 6.0 and 4.5 phosphate buffers respectively. After storing at room temperature for 1 h and then at 4 °C for 21 h, supernatants from each sample were collected and analyzed by UV-Vis directly.
The UV-Vis spectra of all four supernatants are shown in Figure 3a. It showed that under all three 100 mM phosphate buffers with different pHs, significant amount of Au NPs precipitated, as indicated by the decreased absorbance of supernatant at ~523 nm, which is the characteristic absorption of Au NPs in solution. It further demonstrated that the amount of Au precipitation is inversely proportional to the pH, with more precipitation at lower pH. We wondered whether the siRNA complexes also experienced aggregation and then precipitated out of the solution with the Au nanoparticles. We overlaid the UV-Vis spectra of supernatants collected from suspensions of Au-G3/siRNA in H2O and from suspensions of same concentration of Au NPs in H2O after stored at RT for 1 h and at 4 °C for 21 h (Figure 3a inset). It showed that the difference of absorbance spectra of Au-G3/siRNA complex as compared to that of Au-G3 is mainly in the region of 200-310 nm, where the siRNA has absorbance. To relatively compare the amount of siRNA remaining in each supernatant, the absorbance at 260 nm was subtracted by the absorbance at 320 nm (where the siRNA has no absorbance). After correction, the absorbance at 260 nm of each supernatant was normalized to that of the supernatant collected from suspensions of Au-G3/siRNA in H2O and then plotted in Figure 3b (green bars). Similarly, the absorbance at 523 nm of each supernatant is also normalized to that of the supernatant collected from suspensions of Au-G3/siRNA in H2O at 523 nm. It showed that the absorbance at 260 nm of supernatant from suspension of complex in three different pH buffers was only slightly reduced when compared to that in H2O (Figure 3b green bars). This was in sharp contrast to the absorbance at 523 nm (Figure 3b pink bars), which was significantly decreased for supernatants from suspensions of complex in three different pH buffers when compared to that in H2O. This study demonstrated that the Au NPs can be selectively removed from the siRNA nanoparticle solutions, which further demonstrated that the Au nanoparticles were not encapsulated in the final siRNA complexes.
Figure 3.
(a). UV-Vis spectra of supernatants collected from suspensions of complex of Au NPs with siRNA in H2O, 100 mM pH=7.4, 6.0, and 4.5 buffer respectively after stored at RT for 1 h and at 4 °C for 21 h. Inset. UV-Vis spectra of supernatants collected from suspensions of complex of Au NPs with siRNA in H2O vs. from suspensions of same concentration of Au NPs in H2O after stored at RT for 1 h and at 4 °C for 21 h. (b). Absorbance at 260 nm and at 523 nm of supernatants collected from suspensions of Au-G3/siRNA complex in H2O, 100 mM pH=7.4, 6.0, and 4.5 buffer respectively after stored at RT for 1 h and at 4 °C for 21 h. The absorbance at 260 nm was reported after subtracting the absorbance at 320 nm. After correction, the reported absorbance at 260 nm can be approximately correlated to the amount of siRNA in each supernatant.
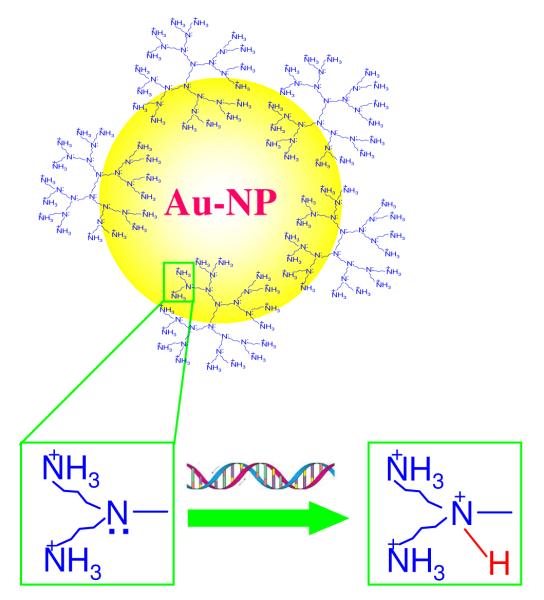
The mechanism of this unique DNA/siRNA packaging is still under study. We hypothesize that it relies on the competing affinities of Au and nucleic acids for the amine sites of low generation polypropyleneimine (PPI) dendrimers. In neutral pH solutions, the primary amines of the dendrimers are protonated (pKa~9.8), while most of their tertiary amines (pKa~5.9-7.0) are not,50-53 which provide strong Au/amine interactions via coordination interactions between the empty d orbital of Au and the free pair of electrons on the N atom of the non-protonated tertiary amines. As a result, each Au NP anchors several low generation dendrimers through multiple Au/amine collective coordination interactions.54 The largely increased positive charges on each of the delivery vehicles (here Au NPs modified with low-generation dendrimers) resemble higher generation dendrimers (Scheme 1) and enable effective packaging of nucleic acids (both long plasmid DNA and siRNA) into nanoparticles. However, when multiple nucleic acids interact with the delivery vehicle, the local pH effects caused by the presence of the multiple nucleic acids increase the local acidity of the dendrimers. As a result, the number of non-protonated tertiary amine sites on dendrimer is decreased, which in turn decreases the affinity of dendrimer to Au while increasing the affinity to nucleic acids.54-58 The large local pH effect has been used to extend the conductivity of polyaniline to neutral or slightly basic solutions, including our previous work.59
Scheme 1.
An Au NP anchored with several low generation dendrimers through Au-amine coordination bonds. Addition of DNA/siRNA leads to an increase of local acidity, which protonates the tertiary amines, weakening the Au-amine interactions. As a result, the Au NPs are released from the dendrimers and are not included in the final DNA/siRNA nanoparticles.
Another possible mechanism involved pH dependent hydrogen bonding between tertiary amines and prorogated primary amines. Instigated by the recent observation of dendrimer aggregation, we assume that the PPI G3 dendrimer may be arranged in multiple layers on the Au nanoparticle surface due to the hydrogen bonding between tertiary amine groups from one dendrimer and the prorogated primary amines from another dendrimer in the outer layer ( or another way around). When multiple nucleic acids come close to this delivery vehicle, the strong interaction between the primary amines groups and nucleic acids would weaken and break the hydrogen binding, leaving Au nanoparticles outside of the formed DNA/siRNA complexs. If this was the case, the Au nanopartiles would be still covered with a few dendrimer molecules. The fact that the Au nanoparticles were still stable in water solution after complexation with DNA/siRNA, but precipitated out in phosphate buffer solution, supports this assumption. However, more Au nanoparticles were precipitated out in lower pH solutions than those in higher pH solutions, indicating the first mechanism we hypothesized. More detailed and systematic study is needed to further uncover the exact mechanism for this interesting packaging approach.
Cellular Uptake of siRNA by Fluorescence Microscopy and Flow Cytometry
We next studied the ability of the siRNA nanoparticles to undergo facile cellular uptake in A549 human lung cancer cell line. Au NPs modified with G3 PPI dendrimers (Au-G3) and G3 PPI dendrimer were respectively complexed with siGLO green siRNA transfection indicator (fluorescein amidite (FAM)-labeled) and then added to A549 lung cancer cells and incubated for 24 h. The cells were then washed with PBS buffer and then fresh medium was added for fluorescence imaging.
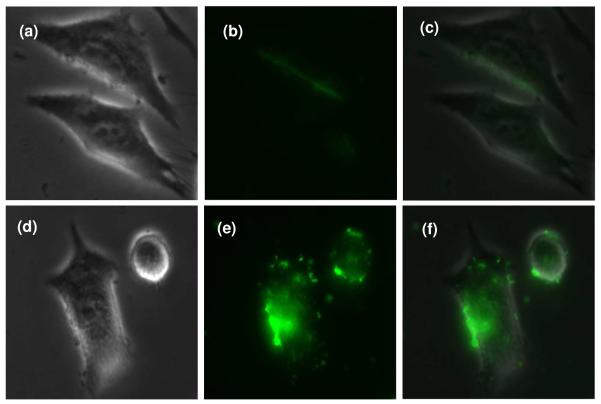
As shown in Figure 4, cells dosed with siRNA nanoparticles fabricated by G3 PPI alone showed very mild green fluorescence on the surface and inside of the cells (Figure 4b, c), indicating a low level of siRNA uptake. Similar result was obtained when G3 PPI dendrimer was used to deliver antisense oligodeoxynucleic acids in our previous study.19 In contrast, the siRNA nanoparticles fabricated by Au-G3 was highly effective in transporting siRNA to the cell, as shown by the green fluorescence in the cells (Figure 4e, f). This result clearly demonstrated that the Au NPs can help the low generation dendrimers effectively deliver siRNA into cancer cells.
Figure 4.
Representative fluorescence microscopic images of cellular uptake of FAM-labeled siRNA complexed with G3 PPI dendrimers (a, b, c) or Au NPs modified with G3 PPI dendrimers (d, e, f). An N/P ratio of 2.4 was used and the final siRNA concentration was 0.25 μM. Incubation was 24 h at 37 °C. Light images of cells are shown in panels a and d. Detection of FAM-labeled siRNA (b and e) and overlay of images (c and f) are shown.
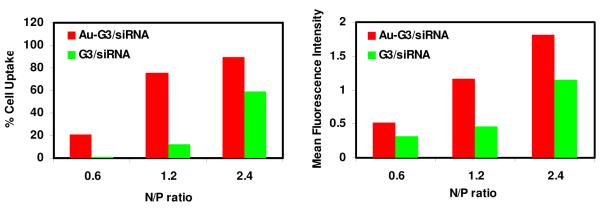
To gain more quantitative information on the cellular uptake of siRNA delivered by Au-G3 versus G3 PPI dendrimers, the cell uptake of siRNA was further studied by flow cytometry. A549 cells were placed in 6-well plates at a density of 500K cells/well and cultured for 24 h. The cells were then treated with siRNA (FAM-labeled siGLO green siRNA transfection indicator) nanoparticles fabricated by Au-G3 and G3 PPI dendrimers respectively for 24 h. Cells were then trypsinized and collected for flow cytometry analysis. As shown in Figure 5, when an N/P ratio of 0.6 was used, only 20% of cells treated with Au-G3/siRNA were transfected and none of the cells treated with G3/siRNA were transfected. When N/P ratio was increased to 1.2, siRNA internalized into ~80% of cells in the case of treatment with Au-G3/siRNA, while only ~10% of cells treated with G3/siRNA was transfected with siRNA. As N/P ratio further increased to 2.4, the cell uptake reached maximum in the case of treatment with Au-G3/siRNA, with 90% cells transfected and the mean green fluorescence intensity of each cell reaching 1.82. The transfection of cells treated with G3/siRNA also significantly increased when N/P ratio increased to 2.4, with ~60% cells transfected and the mean fluorescence intensity of each cell reaching 1.15. This data clearly indicated that the siRNA nanoparticles fabricated by Au-G3 are more easily to internalize into cells. To achieve same percentage of cell transfection, a much lower N/P ratio was required in the case of Au-G3/siRNA due to the higher efficiency of Au-G3 to complex with siRNA and to compact siRNA into nanoparticles. Particularly at N/P ratio of 2.4, not only were more cells transfected in the case of Au-G3/siRNA than the case of G3/siRNA (90% vs. 60%), the mean green fluorescence intensity of each cell is ~ 60% higher in the case of Au-G3/siRNA than the case of G3/siRNA. This result quantitatively confirms that the Au-G3 is significantly more efficient in delivering siRNA into cancer cells.
Figure 5.
Cell uptake of siRNA nanoparticles fabricated by Au-G3 vs. G3 PPI dendrimers at different N/P ratios by flow cytometry. (a). Percentage of cell uptake as a function of N/P ratio. (b). Mean green fluorescence intensity of each individual cell as a function of N/P ratio. A549 cells were incubated by Au-G3/siRNA and G3/siRNA respectively at 37 °C for 24 h at N/P ratios of 0.6, 1.2, and 2.4. The percentage of cell uptake and the mean green fluorescence intensity of each individual cell were calculated based on data collected from a population of 10000 gated cells.
Gene Knockdown
The ability of the siRNA nanoparticles fabricated by Au-G3 to silence the target mRNA expression were studied with quantitative reverse transcriptase-polymerase chain reaction (RT-PCR). 21-bp siRNA designed to silence BCL2 mRNA to suppress non-pump drug resistance was used. For comparison, we also performed the same experiment on the siRNA nanoparticles fabricated by G3 PPI dendrimers as well as higher generation G5 PPI dendrimers. G5 PPI dendrimers have been demonstrated much more efficient in packaging and delivering antisense ODNs into cancer cells compared to G3 PPI dendrimers.19 The same trend was also demonstrated in our recent work in siRNA delivery.20 A549 cancer cells were incubated for 24 h with siRNA nanoparticles fabricated by G3 PPI, G5 PPI and Au-G3 respectively. Gene expression was calculated as a ratio of band intensity of BCL2 mRNA to that of internal standard, β2-m and then the ratio of each sample was normalized to that of sample without treatment. As shown in Figure 6, Au-G3, G3 PPI, G5 PPI alone without siRNA showed similar level of BCL2 mRNA (lanes 2, 3, 4) as that of control cells without treatment (lane 1). This indicated minimal influence of Au-G3, G3 PPI or G5 PPI on the expression of BCL2 gene. In contrast, after incubation with G3/BCL2 siRNA and G5/BCL2 siRNA, the BCL2 mRNA level was suppressed to ~ 80% (Figure 6, lane 6) and 52% (Figure 6, lane 7) respectively. However, the BCL2 siRNA nanoparticles fabricated by Au-G3 are even more efficient than those fabricated by G5 PPI in inhibiting the BCL2 mRNA expression. The BCL2 mRNA level was significantly suppressed to ~28% after incubated with Au-G3/BCL2 siRNA nanoparticles. These results are striking, which soundly demonstrated that the Au NPs can help the low generation dendrimers effectively deliver siRNAs into cancer cells and efficiently inhibit their target mRNA expression. To further study the gene knockdown specificity of the BCL2 siRNA delivered by Au-G3, Au-G3 was complexed with a scrambled siRNA and the BCL2 mRNA level of the cell was analyzed after incubated with Au-G3/scramble siRNA for 24 h. As shown in Figure 6 (lane 8), there was no inhibition on BCL2 gene expression after the treatment of A549 cells with control scrambled siRNA complexed with Au-G3, confirming the specificity of BCL2 siRNA delivered by Au-G3 to knockdown its target mRNA.
Figure 6.
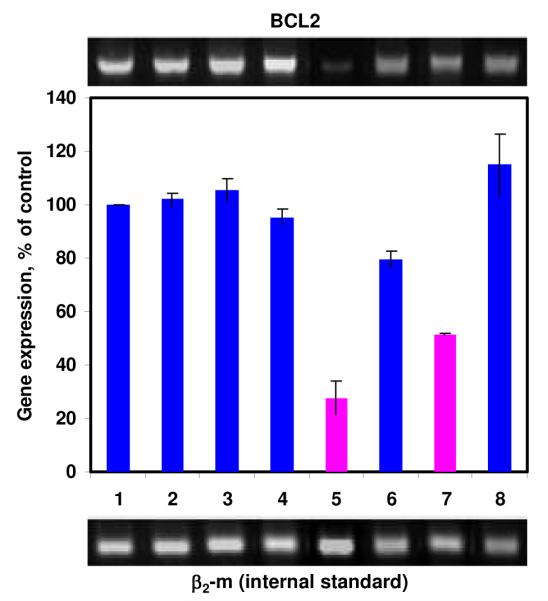
Effect of different formulations on the expression of BCL2 mRNA in A549 lung cancer cells. (1) No treatment; (2) Au-G3; (3) G3; (4). G5; (5) Au-G3/BCL2 siRNA; (6) G3/BCL2-siRNA; (7) G5/BCL2-siRNA; (8) Au/G3-scrambled siRNA. Gene expression was calculated as a ratio of band intensity of BCL2 gene to that of internal standard, β2-m and then the ratio of each sample was normalized to that of sample without treatment. In samples (5-8), an N/P ratio of 2.4 was used and final concentration of siRNA in cell medium was 0.25 μM. The concentration of primary amine in all samples (2-8) was 25 μM.
In vitro Cytotoxicity of G3 PPI vs. Au-G3
In order to examine whether Au-G3 influenced the cell viability of A549 cancer cells, the cytotoxicity of Au-G3 was determined by MTT assay and compared with that of G3 PPI dendrimers alone. As shown in Figure 7, at a concentration of G3 PPI dendrimers up to 4 μM, both G3 PPI dendrimers and Au NPs modified by G3 PPI dendrimers are non toxic. In all our experiments, a concentration of G3 PPI dendrimer up to 1.57 μM has been used.
Figure 7.
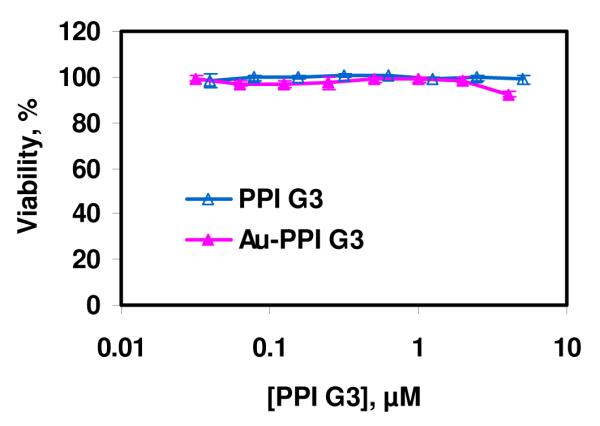
Viability of A549 cells after incubation with G3 PPI dendrimers or Au NPs modified with G3 PPI dendrimers for 24 h at 37 °C. It showed that at a concentration of G3 PPI dendrimer up to 4 μM (which is relevant to our cell uptake study), both G3 PPI dendrimer and Au NPs modified by G3 PPI dendrimers are non-toxic.
In conclusion, we report a novel approach to efficiently package and deliver nucleic acids with low generation dendrimers by using Au NPs as a “labile catalytic” packaging agent. The Au NPs helped low generation dendrimers to package nucleic acids into discrete nanoparticles but are not included in the final DNA/siRNA complexes. Compared to the siRNA nanostructures (mainly nanofibers) fabricated by low generation dendrimer alone (G3 PPI), the siRNA nanoparticles packaged using this novel approach (by Au NPs modified with G3 PPI) can be internalized by cancer cells and the delivered siRNAs can efficiently silence their target mRNA. The efficiency of mRNA silencing by the proposed approach is even superior to higher generation dendrimers (G5 PPI). More importantly, the Au NPs can be selectively separated from the siRNA complex solution without influencing the integrity of the siRNA complexes, which provides a possibility to remove the Au NPs before the nucleic acid NPs are delivered, preventing cytotoxicity of the Au NPs. This is a new concept in using inorganic engineered NPs in nucleic acid packaging and delivery applications. Currently, we are carrying out detailed studies on the mechanism of this novel packaging approach and the cellular uptake, gene knockdown efficiency, and the toxicity of the nucleic acid complex after removal of Au NPs.
METHODS
Chemicals
The PGL-3 luciferase control vector (5256 bp), which contains the SV40 promoter and enhancer sequences, was purchased from Promega (Madison, WI). Polypropyleneimine hexadecaamine dendrimer (PPI dendrimer) generation-3 (G3) and generation-5 (G5) and other chemicals used in this study were purchased from Aldrich (Milwaukee, WI), and used without further purification. Fluorescent RNA duplex, siGLO green siRNA transfection indicator, was obtained from Applied Biosystems (Ambion, Inc., Foster City, CA). siRNA that are sequence specific for human BCL2 mRNA was custom synthesized by Ambion (Austin, TX). The sequence of the siRNA used as follows: sense strand, 5′-GUGAAGUCAACAUGCCUGC-dTdT-3′; antisense strand, 5′-GCAGGCAUGUUGACUUCAC-dTdT-3′.
Fabrication of Au NPs modified with G3 PPI dendrimers
32.8 mg of G3 PPI dendrimer was mixed with 8.8 ml of 2.45 mM HAuCl4 solution with a molar ratio of 0.90:1 and then heated at 80° C for 1 h with continuous stirring. The clear solution thus obtained was cooled to RT and dialyzed (3.5k Da cutoff regenerated cellulose tubular membrane) against 200 ml of double-distilled water twice for a total of 4 h. The dialyzate thus obtained was used as stock solution.
Determination of PPI G-3 dendrimer concentration in the Au NPs modified with PPI G-3 dendrimers
To determine the PPI G-3 dendrimer concentration in the Au NP modified with PPI G-3 dendrimer stock solution, an improved 2, 4, 6-Trinitrobenzenesulfonic (TNBS) acid method was used. Briefly, 75 μl of aqueous 0.03 M TNBS is added to 4.0 ml of 0.042 M sodium tetraborate buffer containing different amount of samples. Thus-obtained solution was shaken in a shaking block for 1 h and then ~ 3 ml of the solution was transferred into a quartz cuvette for UV-Vis analysis. 75 μl of aqueous 0.03 M TNBS in 4 ml of 0.042 M sodium tetraborate buffer was used as blank solution. The absorbance at 420 nm was used to obtain the calibration curve and determine the dendrimer concentration in the Au stock solution.
Plasmid DNA Condensation by the Au NPs modified with G3 PPI dendrimers
Condensation experiments were conducted in DI water by adding a suitable amount of stock solution of Au NPs to a diluted plasmid DNA solution in water. The resultant mixture was mixed by a vertex machine for several seconds. The final DNA phosphate concentration is 2.0 μM and the ratio of peripheral nitrogen of PPI G-3 dendrimer to the phosphorus of plasmid DNA (N/P value) is 6:1.
Packaging siRNA with G5 PPI dendrimers and with Au NPs modified with G3 PPI dendrimers for cell uptake experiment
The condensed siRNA complexes were prepared at N/P ratio of 2.4 either in DI water or 10 mM Hepes buffer (pH 7.2) by adding a stock solution of PPI G5 dendrimer or a stock solution of Au-G3 into prepared siRNA solution. The samples were vortexed briefly, and the solutions were then incubated at room temperature for 30 min before being dosed.
Cell Lines
Human lung carcinoma cell line A549 was obtained from the ATTC (Manassas, VA, USA). Cells were cultured in RPMI 1640 medium (Sigma Chemical Co., Louis, MO) supplemented with 10% fetal bovine serum (Fisher Chemicals, Fairlawn, NJ). Cells were grown at 37°C in a humidified atmosphere of 5% CO2 (v/v) in air. All of the experiments were performed on the cells in exponential growth phase.
Cytotoxicity
The cellular cytotoxicity of G3 PPI dendrimers and Au-G3 was assessed using a modified MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Cells were seeded into 96-well microtiter plates at the density of 10,000 cells per well. After incubation for 24 h, medium were aspirated and various concentrations of G3 or Au-G3 in fresh cell growth medium (200 μl/well) were added. Control cells were added with equivalent volume of fresh media. Cells were cultured for 24 h before the cell viability assay was performed. The old medium was removed and 100 μl of fresh medium and 25 μl of a 5 mg/ml MTT (Fluka) solution in DPBS was added to each well. Plates were then incubated under cell culture conditions for 3 h. Every well was then added with 100 μl of 50% (v/v) dimethylformamide in water containing 20% (w/v) sodium doecyl sulfate (with pH adjusted to 4.7 by acetic acid) and incubated overnight to dissolve the formazan crystals. The absorbance of each sample was measured at 570 nm with a background correction at 630 nm. Based on these measurements, IC50 doses of G3 PPI dendrimers and Au-G3 (the concentrations of G3 necessary to inhibit the cell growth by 50%) were calculated.
Cellular Internalization
Cellular internalization of FAM-labeled siRNA nanoparticles packaged with Au NPs modified with G3 PPI and G3 PPI alone were analyzed by fluorescence microscope (Olympus America Inc., Melville, NY). A549 cells were plated (20,000 cells/well) in 6-well tissue culture plate and cultured for 24 h. The cells were then treated with siRNA nanoparticles packaged with Au NPs modified with G3 PPI or siRNA nanoparticles packaged with G3 PPI alone respectively for 24 h. The concentration of G3 PPI dendrimer for both cases was 1.57 μM and the concentration of siRNA was 0.25 μM. After 24 h of treatment, cells were washed three times with phosphate buffered saline (PBS) and 1 mL of media was added to each well.
Gene Knockdown
The ability of the siRNA delivered by Au NPs modified with G3 PPI, G3 PPI dendrimers and G5 PPI dendrimers to silence the target mRNA expression were studied with quantitative reverse transcriptase-polymerase chain reaction (RT-PCR). A549 cells were placed in small flasks at a density of 750,000 cells/flask and cultured for 24 h. Then the cells were treated with different siRNA nanoparticles. Cells were also treated with Au-G3 complex with scrambled siRNA, Au-G3, G3 PPI, and G5 PPI respectively as control. After 24hrs of incubation of A-549 lung cancer cells with different formulations, the total cellular RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA). First strand cDNA was synthesized by Ready-To-GO You-Prime First-Strand Beads (Amersham Biosciences, Piscataway, NJ) with 4mg of total cellular RNA and 100 ng of random hexadeoxynucleotide primer (Amersham Bioscience). After synthesis, the reaction mixture was immediately subjected to polymerase chain reaction, which was carried out using GenAmp PCR System 2400. The pairs of BCL2 and 2-m primers used to amplify each type of cDNA. PCR products were seperated in 4% NuSieve 3:1 Reliant agarose gels in 1×TBE buffer (0.089 M Tris/Borate, 0.002 M EDTA, pH 8.3; Research Organic Inc., Cleveland OH) by submarine electrophoresis. The gels were stained with ethidium bromide, digitally photographed and scanned using Gel Documentation System 920 (NucleoTech, San Mateo, CA). Gene expression was calculated as the ratio of mean band density of analyzed RT-PCR product to that of the internal standard (β2-m). In preparing all siRNA complex, an N/P ratio of 2.4 was used and final concentration of siRNA in cell medium was 0.25 μM. The concentration of primary amine of PPI dendrimers in all samples was 25 μM.
Atomic Force Microscopy
AFM images were obtained using Nanoscope IIIA equipment (Digital Instruments, Santa Barbara, CA) in tapping mode, operating in ambient air. A 125 μm long rectangular silicon cantilever/tip assembly was used with a spring constant of 40 N/m, resonance frequency of 315-352 kHz, and tip radius of 5-10 nm. The applied frequency was set on the lower side of the resonance frequency. The image was generated by the change in amplitude of the free oscillation of the cantilever as it interacted with the sample. The height differences on the surface are indicated by the color code, lighter regions indicating increase in height of the NPs. In order to image DNA or siRNA nanoparticles, 5 μL of the prepared complex solutions were deposited on a freshly cleaved mica surface. After 3-6 minutes of incubation, the surface was rinsed with 2-3 drops of nanopure water (Barnstead), and dried under a flow of dry nitrogen. Height and outer diameter of nanoparticles were measured using the Nanoscope software. Data are given as mean ± standard error of the mean.
UV-Vis Absorbance
An appropriately diluted solution of Au NPs was transferred to a UV-Vis cuvette for analysis by Cary 300 UV-Vis Spectrophotometer (Varian Inc., CA). The sample was scanned from 800 nm to 200 nm.
Transmission Electron Microscopy (TEM)
TEM analyses of siRNA/DNA condensates were performed using Libra 120 Energy Filtering TEM (Carl Zeiss). 5 μl of the siRNA/DNA condensate solution was deposited on a carbon coated 200 mesh copper grid that was glow discharged for 1 minute. After 3 minutes, the sample was drained off with a filter paper and further dried with a flow of N2 gas. Then the sample was viewed on the electron microscope and photographs were taken using a SSCCD camera attached to the microscope. Some samples were also imaged in the energy filtering TEM mode60 and analyzed by Au elemental analysis by setting the energy slit to 2386 eV, which is the M5 edge of Au element.
Flow Cytometry
Flow cytometry analyses were performed on a Cytomics FC500 cytometer (Beckman-Coulter). An air-cooled argon laser was used to excite the siGLO green siRNA (FAM-labeled) at 488 nm and the emitted light was collected at 525 nm after filtered by a 525 nm band-pass filter. Under this excitation condition, the emission from Au/PPI nanoparticles alone was barely detected. The cells were gated by forward and side-scatter parameters, and data were collected from a population of 10000 gated cells. The data were analyzed using the Cytomics FC500 RXP software.
Supplementary Material
ACKNOWLEDGEMENT
The research was supported in part by the National Science Foundation under Grant CBET-0933966, and NIH grants CA080163, CA100098, CA111766 from the National Cancer Institute, and grants from the Foundation of the University of Medicine and Dentistry of New Jersey (64-09 and 30-09). A. M. Chen acknowledges Merck & Co., Inc. for support. H. He also acknowledges the Trustees Research Fellowship Program at Rutgers, The State University of New Jersey.
Footnotes
SUPPORTING INFORMATION AVAILABLE. A digital picture of an aqueous solution of Au nanoparticles, UV-Vis Spectra of the Au NPs before and after dialysis, DLS of the Au NPs, and Electrophoretic mobility shift assays of Au-G3/siRNA complex at different N/P ratios. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Santhakumaran LM, Chen A, Pillai CKS, Thomas T, He HX, Thomas TJ. Nanotechnology in Non-Viral Gene Delivery. In: Hormes J, Kumar C, Leuschner C, editors. Nanofabrication Towards Biomedical Applications. Wiley-VCH; Germany: 2005. pp. 253–287. [Google Scholar]
- 2.Vijayanathan V, Thomas T, Thomas TJ. DNA Nanoparticles and Development of DNA Delivery Vehicles for Gene Therapy. Biochemistry. 2002;41:14085. doi: 10.1021/bi0203987. [DOI] [PubMed] [Google Scholar]
- 3.Bessis N, GarciaCozar FJ, Boissier MC. Immune Responses to Gene Therapy Vectors: Influence on Vector Function and Effector mechanisms. Gene Ther. 2004;11:S10. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 4.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR., Jr. Efficient Transfer of Genetic Material into Mammalian Cells Using Starburst Polyamidoamine Dendrimers. Proc. Natl. Acad. Sci, USA. 1996;93:4897. doi: 10.1073/pnas.93.10.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dufès C, Uchegbu IF, Schätzlein AG. Dendrimers in Gene Delivery. Adv. Drug Deliv. Rev. 2005;57:2177. doi: 10.1016/j.addr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Boas U, Heegaard PMH. Dendrimers in Drug Research. Chem. Soc. Rev. 2004;33:43. doi: 10.1039/b309043b. [DOI] [PubMed] [Google Scholar]
- 7.Samad A, Alam MI, Saxena K. Dendrimers: A Class of Polymers in the Nanotechnology for the Delivery of Active Pharmaceuticals. Current pharmaceutical design. 2009;15:2958. doi: 10.2174/138161209789058200. [DOI] [PubMed] [Google Scholar]
- 8.Ali MM, Yoo B, Pagel MD. Tracking the Relative In vivo Pharmacokinetics of Nanoparticles with PARACEST MRI. Molecular Pharmaceutics. 2009;6:1409. doi: 10.1021/mp900040u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang MX, Redemann CT, Szoka FC., Jr In vitro Gene Delivery by Degraded Polyamidoamine Dendrimers. Bioconjugate Chem. 1996;7:703. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- 10.Dennig J. Gene Transfer in Eukaryotic Cells Using Activated Dendrimers. Top. Curr. Chem. 2003;2003:227. doi: 10.1007/b11013. [DOI] [PubMed] [Google Scholar]
- 11.Malik N, Wiwattanapatapee R, Klopsch R, Lorenz K, Frey H, Weener JW, Meijer EW, Paulus W, Duncan R. Relationship between Structure and Biocompatibility in vitro, and Preliminary Studies on the Biodistribution of 125I-labelled Polyamidoamine Dendrimers in vivo. Journal of Controlled Release. 2000;65:133. doi: 10.1016/s0168-3659(99)00246-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhang ZY, Smith BD. High-Generation Polycationic Dendrimers Are Unusually Effective at Disruptig Anioinic Vesicles: Membrane Bending Model. Bioconjugate Chem. 2000;11:805. doi: 10.1021/bc000018z. [DOI] [PubMed] [Google Scholar]
- 13.Hong S, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi X, Balogh L, Orr BG, Baker JRJ, Hall M. M. Banaszak. Interaction of Poly(amidoamine) Dendrimers with Supported Lipid Bilayers and Cells: Hole Formation and the Relation to Transport. Bioconjugate Chem. 2004;15:774. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 14.De Brabander-van den Berg EMM, Meijer EM. Poly(propylene imine) Dendrimers: Large-scale Synthesis via Heterogeneously Catalyzed Hydrogenation. Angew. Chem. Int. Ed. 1993;32:1308. [Google Scholar]
- 15.Tomalia DA, Baker H, Dewald J, Hall M, Kallos G, Martin S, Roeck J, Ryder J, Smith P. Dendritic Macromolecules: Synthesis of Starburst Dendrimers. Macromolecules. Macromolecules. 1986;19:2466. [Google Scholar]
- 16.Newkome GR, Moorefield CN, Vogtle F, editors. Dendrimers and Dendrons: Concepts, Syntheses, Applications. Wiley-VCH; New York, NY: 2001. [Google Scholar]
- 17.Dennig J, Duncan E. Gene Transfer into Eukaryotic Cells using Activated Polyamidoamine Dendrimers. Rev. Mol. Biotechnol. 2002;90:339. doi: 10.1016/s1389-0352(01)00066-6. [DOI] [PubMed] [Google Scholar]
- 18.Braun CS, Vetro JA, Tomalia DA, Koe GS, Koe JG. Structure/Function Relationships of Polyamidoamine/DNA Dendrimers as Gene Vehicles. J. Pharm. Sci. 2005;94:423436. doi: 10.1002/jps.20251. [DOI] [PubMed] [Google Scholar]
- 19.Chen A, Santhakumaran LM, Nair SK, Amenta PS, Thomas T, He HX, Thomas TJ. Oligodeoxynucleotide Nanostructure Formation in the Presence of Polypropyleneimine Dendrimers and Their Uptake in Breast Cancer Cells. Nanotechnology. 2006;17:5449. [Google Scholar]
- 20.Taratula O, Savla R, Pandya I, Kirkpatrick P, He HX, Minko T. PPI Dendrimers as Potential siRNA Delivery Vehicles for Efficient Cancer Therapy: Structure - Function Relationship. Internat. J. Nanotech. 2010 In Press. [Google Scholar]
- 21.Thomas M, Klibanov AM. Conjugation to Gold Nanoparticles Enhances Polyethylenimine’s Transfer of Plasmid DNA into Mammalian Cells. Proc. Natl. Acad. Sci. USA. 2003;100:9138. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radu DR, Lai C-Y, Jeftinija K, Rowe EW, Jeftinija S, Lin CS-Y. A Polyamidoamine Dendrimer-Capped Mesoporous Silica Nanosphere-Based Gene Transfection Reagent. J. Am. Chem. Soc. 2004;126:13216. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Tabakman S, Welsher K, Dai HJ. Carbon Nanotubes in Biology and Medicine: In vitro and in vivo Detection, Imaging and Drug Delivery. Nano Res. 2009;2:85. doi: 10.1007/s12274-009-9009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yezhelyev MV, Qi L, O’Regan RM, Nie SM, Gao X. Proton-Sponge Coated Quantum Dots for siRNA Delivery and Intracellular Imaging. J. Am. Chem. Soc. 2008;130:9006. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung W, Pontoriero F, Taratula O, Chen AM, He HX. DNA and Carbon Nanotubes as Medicine. Adv. Drug Deliv. Rev. 2010 doi: 10.1016/j.addr.2010.03.007. In Press. [DOI] [PubMed] [Google Scholar]
- 26.Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Toxicity of Gold Nanoparticles Functionalized with Cationic and Anionic Side Chains. Bioconjugate Chem. 2004;15:897. doi: 10.1021/bc049951i. Bioconjugate Chem. 2004, 15, 897-900. [DOI] [PubMed] [Google Scholar]
- 27.Lewinski L, Colvin V, Drezek R. Cytotoxicity of Nanoparticles. Small. 2008;4:26. doi: 10.1002/smll.200700595. [DOI] [PubMed] [Google Scholar]
- 28.Pan Y, Neuss S, Leifert A, Fischler M, Wen F, Simon U, Schmid G, Brandau W, Jahnen-Dechent W. Size-Dependent Cytotoxicity of Gold Nanoparticles. Small. 2007;3:1941. doi: 10.1002/smll.200700378. [DOI] [PubMed] [Google Scholar]
- 29.Bar-Ilan O, Albrecht RM, Fako VE, Furgeson DY. Toxicity Assessments of Multisized Gold and Silver Nanoparticles in Zebrafish Embryos. Small. 2009;5:1897. doi: 10.1002/smll.200801716. [DOI] [PubMed] [Google Scholar]
- 30.Chithrani BD, Chan WCW. Elucidating the Mechanism of Cellular Uptake and Removal of Protein-Coated Gold Nanoparticles of Different Sizes and Shapes. Nano Lett. 2007;7:1542. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 31.Li JJ, Zou L, Hartono D, Ong C-N, Bay H-H, Yung LYL. Gold Nanoparticles Induce Oxidative Damage in Lung Fibroblasts In vitro. Adv. Mater. 2008;20:138. [Google Scholar]
- 32.Zhang Y, Zhou J, C. X, Zhang Q, Lim CU, Ullrich RL, M. S, Bailey HLL. Cancer Lett. 2007;250:63. doi: 10.1016/j.canlet.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 33.Pernodet N, Fang X, Sun Y, Bakhtina A, Ramakrishnan A, Sokolov J, Ulman A, Rafailovich M. Adverse Effects of Citrate/Gold Nanoparticles on Human Dermal Fibroblasts. Small. 2006;2:766. doi: 10.1002/smll.200500492. [DOI] [PubMed] [Google Scholar]
- 34.Sun X, Dong S, Wang EK. One-Step Preparation and Characterization of Poly(propyleneimine) Dendrimer-Protected Silver Nanoclusters. Macromolecules. 2004;37:7105. [Google Scholar]
- 35.Sun X, Jiang X, Dong S, Wang EK. One-Step Synthesis and Size Control of Dendrimer-Protected Gold Nanoparticles: a Heat-Treatment-Based Strategy. Macromol. Rapid Commun. 2003;24:1024. [Google Scholar]
- 36.Garcia ME, Baker LA, Crooks RM. Preparation and Characterization of Dendrimer-Gold Colloid Nanocomposites. Anal. Chem. 1999;71:256. doi: 10.1021/ac980588g. [DOI] [PubMed] [Google Scholar]
- 37.Bharali DJ, Keljbor I, Stachowiak EK, Dutta P, Roy I, Kaur N, Bergey EJ, Prasad PN, Stachowiak MK. Oganically Modified Silica Nanoparticles: A Nonviral Vector for in vivo Gene Delivery and Expression in the Brain. Proc. Natl. Acad. Sci, USA. 2005;102:11539. doi: 10.1073/pnas.0504926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandhu KK, McIntosh CM, Simard JM, Rotello VM. Gold Nanoparticle-Mediated Transfection of Mammalian Cells. Bioconjugate Chem. 2002;13:3. doi: 10.1021/bc015545c. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya R, Mukherjee P, Xiong Z, Atala A, Soker S, Mukhopadhyay D. Gold Nanoparticles Inhibit VEGF165-Induced Proliferation of HUVEC Cells. Nano Lett. 2004;4:2479. [Google Scholar]
- 40.Park C, Crooks RE, Siochi E, Harrison JS, Evans N, Kenik E. Adhesion Study of Polyimide to Single Wall Carbon Nanotube Bundles by Energy-Filtered Transmission Electron Microscopy. Nanotechnology. 2003;14:L11. [Google Scholar]
- 41.Bleher R, Meyer DA, Albrecht RM, editors. High Resolution Multiple Labeling for Immuno-EM applying Metal Colloids and Energy Filtering Transmission Electron Microscopy (EFTEM) Cambridge University Press; 2005. [Google Scholar]
- 42.Pantarotto D, Singh R, McCarthy D, Erhardt M, Briand J-P, Prato M, Kostarelos K, Bianco A. Functionalized Carbon Nanotubes for Plasmid DNA Gene Delivery. Angew. Chem. 2004;116:5354. doi: 10.1002/anie.200460437. [DOI] [PubMed] [Google Scholar]
- 43.Tan WB, Jiang S, Zhang Y. Quantum-dot Based Nanoparticles for Targeted Silencing of HER 2/neu Gene Via RNA Interference. Biomaterials. 2007;28:1565. doi: 10.1016/j.biomaterials.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 44.Uprichard SL. The Therapeutic Potential of RNA Interference. FEBS Lett. 2005;579:5996. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dykxhoorn DM, Lieberman J. The Silent Revolution: RNA Interference as Basic Biology, Research Tool, and Therapeutic. Annu. Rev. Med. 2005;56:401. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- 46.Sarkar T, Conwell CC, Harvey LC, Santai CT, Hud NV. Condensation of Oligonucleotides Assembled into Nicked and Gapped Duplexes: Potential Structures for Oligonucleotide Delivery. Nucleic Acids Res. 2005;33:143. doi: 10.1093/nar/gki156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gary DJ, Puri N, Won Y-Y. Polymer-based siRNA Delivery: Perspectives on the Fundamental and Phenomenological Distinctions from Polymer-based DNA Delivery. J. Controlled Release. 2007;121:64. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 48.Li S-D, Huang L. Targeted Delivery of Antisense Oligodeoxynucleotide and Small Interference RNA into Lung Cancer Cells. Mol. Pharm. 2006;3:579. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 49.Li S-D, Chen Y-C, Hackett MJ, Huang L. Tumor-targeted Delivery of siRNA by Self-assembled Nanoparticles. Molecular Ther. 2008;16:163. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koper GJM, van Genderen MHP, Elissen-Roman C, Baars MWPL, Meijer EW, Bordovec M. Protonation Mechanism of Poly(propylene imine) Dendrimers and Some Associated Oligo Amines. J. Am. Chem. Soc. 1997;119:6512. [Google Scholar]
- 51.van Duijvenbode RC, Borkovec M, Koper GJM. Acid Base Properties of Poly(propylene imine) Dendrimers. Polymer. 1998;39:2657. [Google Scholar]
- 52.Cakara D, Kleimann J, Borkovec M. Microscopic Protonation Equilibria of Poly(amidoamine) Dendrimers from Macroscopic Titrations. Macromolecules. 2003;36:4201. [Google Scholar]
- 53.Sun J, Paik H-Y, Hwang BY. Ionization of Poly(ethylenimine) and Poly(allylamine) at Various pH’s. Bioorg. Chem. 1994;22:318. [Google Scholar]
- 54.Tokuhisa H, Zhao M, Baker LA, Phan VT, Dermody DL, Garcia ME, Peez RF, Crooks RM, Mayer TM. Preparation and Characterizatoin of Dendrimer Monolayers and Dendrimer-Alkanethiol Mixed Monolayers Adsorbed to Gold. J. Am. Chem. Soc. 1998;120:4492. [Google Scholar]
- 55.Sideratou Z, Tsiourvas D, Paleos CM. Quaternized Poly(propylene imine) Dendrimers as Novel pH-Sensitive Controlled-Release Systems. Langmuir. 2000;16:1766. [Google Scholar]
- 56.Liu W, Cholli AL, Nagarajan R, Kumar J, Tripathy S, Bruno FF, Samuelson L. The Role of Template in Enzymatic Synthesis of Conducting Polyaniline. J. Am. Chem. Soc. 1999;121:11345. [Google Scholar]
- 57.Nagarajan R, Liu W, Kumar J, Tripathy SK, Bruno FF, Samuelson LA. Manipulating DNA Conformation Using Intertwined Conducting Polymer Chains. Macromolecules. 2001;34:3921. [Google Scholar]
- 58.Nagarajan R, Roy S, Kumar J, Tripathy SK, Dolukhanyan T, Sung C, Bruno F, Samuelson LA. Enzymatic Synthesis of Molecular Complexes of Polyaniline with DNA and Synthetic Oligonucleotides: Thermal and Morphological Characterization. J. Mocromo. Sci---Pure. Appl. Chem. 2001;A38:1519. [Google Scholar]
- 59.Ma YF, Zhang JM, Zhang GJ, He HX. Polyaniline Nanowires on Si surfaces Fabricated with DNA Templates. J. Am. Chem. Soc. 2004;126:7097. doi: 10.1021/ja039621t. [DOI] [PubMed] [Google Scholar]
- 60.Park C, Crooks RE, Siochi EJ, Harrison JS, Evans N, Kenik E. Adhesion study of polyimide to single-wall carbon nanotube bundles by energy-filtered transmission electron microscopy. Nanotechnology. 2003;14:L11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.