Abstract
Higher expression of human telomerase reverse transcriptase (hTERT) and subsequent activation of telomerase occur during cellular immortalization and are maintained in cancer cells. To understand the mode of hTERT expression in cancer cells, we identified cancer-specific trans-regulatory proteins that interact with the hTERT promoter, using the promoter magnetic precipitation assay coupled to mass spectrometry (PMS-MS). The identified proteins include MutS homologue 2 (MSH2), heterogeneous nuclear ribonucleoprotein (hnRNP) D, hnRNP K, and Grainyhead-like 2 (GRHL2). We noticed higher expression of these proteins in human oral squamous cell carcinoma (OSCC) cells than in normal cells, which do not exhibit telomerase activity. Knockdown of MSH2, hnRNP D and GRHL2 resulted in notable reduction of the hTERT promoter activity in tested cancer cells. Silencing of the above genes resulted in the significant reduction of telomerase activity in OSCC cells. Interestingly, among the four identified genes, silencing of GRHL2 was essential in reducing telomerase activity and viability of tested cancer cells. These results suggest a possible role of GRHL2 in telomerase activation during cellular immortalization.
Keywords: hTERT, telomerase, GRHL2, hnRNPs, MSH2, oral cancer
Introduction
Telomerase is a ribonucleoprotein complex composed of the catalytic protein subunit hTERT (human telomerase reverse transcriptase) and RNA subunit hTR (human telomerase RNA), which serves as the template for the synthesis of telomeric DNA (Cairney and Keith, 2008). Most human tumor cells demonstrate high level of telomerase activity, while normal somatic cells display undetectable level of activity (Kim et al., 1994). This negligible telomerase activity is sharply enhanced when the cells convert to malignant phenotype (Kim et al., 2007). The enzyme is also required for cancer cells to maintain their malignant phenotype and thus the enzyme would be an ideal target for the development of anticancer drugs (Shay and Keith, 2008). Therefore, detailed understanding of telomerase regulation is necessary to determine the mechanisms of carcinogenesis and to develop novel armaments against cancer.
Telomerase activity is closely associated with hTERT expression, while hTR is constitutively expressed in cells (Kyo et al., 2000; Yan et al., 2001). Thus, many studies have focused on the regulation of hTERT expression to investigate the mechanisms of carcinogenesis. These efforts have revealed that hTERT promoter is enriched with transcription factor binding sites (Poole et al., 2001). Several trans-regulatory factors of the proximal hTERT promoter, including Myc/Max heterodimer, HIF-2α, Cbfa1, AP-2β, TAK1 and Sp1/Sp3, have been identified (Fujiki et al., 2007; Lou et al., 2007; Deng et al., 2007; Isenmann et al., 2007; Lebel et al., 2007). These factors might impose their regulatory roles through direct physical interactions with the cis-elements of the hTERT promoter.
In the present study, we identified new cancer-specific trans-regulatory proteins of the hTERT promoter using the promoter magnetic precipitation coupled with mass spectrometry (PMP-MS). This approach allowed us to identify numerous proteins that co-precipitate with the hTERT promoter. Among them, MutS homologue 2 (MSH2), heterogeneous nuclear ribonucleoproteins K and D (hnRNPs K and D), and Grainyhead-like 2 (GRHL2) were confirmed to be associated with the hTERT promoter in vitro and in vivo. These proteins bind the hTERT promoter preferentially in human oral squamous cell carcinoma (OSCC) cells, which express high level of telomerase activity, compared with senescent, telomerase-deficient normal human oral keratinocytes (NHOK) or fibroblasts (NHOF).
MSH2 is a component of highly conserved post-replicative DNA mismatch repair system. Its deficiency leads to genetic instability, telomere capping defect, and telomerase-independent cell proliferation, indicating its possible role in regulation of telomerase function (Rizki and Lundblad, 2001; Campbell et al., 2006). hnRNP D contains two RNA recognition motifs that regulate the stability of specific mRNAs containing AUUUA repeats (Wagner et al., 1998). hnRNP K is known to function as a transcriptional regulator by direct binding to the cis-regulatory elements within the promoter sequences (Ostrowski et al., 2004; Lynch et al., 2005). Notably, its expression is upregulated in OSCC (Roychoudhury and Chaudhuri, 2007), indicating its involvement in oral carcinogenesis. GRHL2 is a mammalian homologue of Grainyhead (GRH), a Drosophila protein implicated in development (Wilanowski et al., 2002). The GRH and GRHL possess the DNA binding domains and are involved in the transcriptional regulation of protective layer formation, such as the outer cuticle of Drosophila and stratified epithelium in mammals (Stramer and Martin, 2005). In the current study, we show the involvement of MSH2, hnRNP K, hnRNP D, and GRHL2 in the transcriptional regulation of the hTERT promoter.
Materials and methods
Cells and cell culture
Primary NHOK and NHOF were prepared from keratinized oral epithelial tissues according to the methods described elsewhere (Park et al., 1991; Kang et al., 1998). Normal human epidermal keratinocytes (NHEK) were prepared from foreskin epithelium. Primary NHOK and NHEK were maintained in Keratinocyte Basal Medium (KBM) supplemented with the growth factor bullet kit (Lonza Biologicals, Inc., Portsmouth, NH) or EpiLife supplemented with HKGS (Invitrogen, Carlsbad, CA). These cells were serially subcultured until they reached the senescent stage as previously reported (Kang et al., 1998). The pre- and post-crisis human oral keratinocytes (HOK) were obtained from NHOK infected sequentially with the retroviral vectors expressing Bmi-1 and the E6 oncoprotein of human papillomavirus (HPV) type 16 (Kim et al., 2007). Human OSCC cell lines (SCC4, SCC9, SCC15, HEp-2, FaDu, BaP-T, and 1483) were cultured according to the methods previously described (Kang et al., 1998).
Promoter magnetic precipitation (PMP) assay
PMP was performed with the late passage (senescing) NHOK culture and SCC4 cells, which represented telomerase negative and positive cells, respectively (Kang et al., 1998). Biotinylated hTERT promoter (549 bp) was PCR-amplified using the forward primer 5’-CTC CGT CCT CCC CTT CAC-3’ and the reverse primer 5’-ATC GAT CAG CGC TGC CTG AAA CTC-3’ with biotin conjugation on the 5’-end of the forward primer. The hTERT promoter fragment was conjugated with Dynabeads® (Invitrogen). The PMP assay was performed by incubating the immobilized hTERT promoter fragment with the nuclear extracts (20 or 200 µg) precleared with unconjugated Dynabeads® in the presence of poly(dI:dC). After 1hr incubation at room temperature (RT), the beads were collected by the magnetic apparatus and washed with the binding buffer containing 0.5% nonidet P-40. Proteins that were coprecipitated were eluted in the SDS sample buffer or 2D sample buffer and fractionated in 8–16% gradient SDS-PAGE or by 2 dimensional gel electrophoresis.
For the regional analysis of the hTERT promoter, the 549 bp hTERT promoter was amplified into six overlapping regions using specific primer sets. The sequence of the primers and PCR conditions can be available upon request. The PMP analysis was performed using the nuclear extracts of SCC4 cells. Subsequently, Western blotting was performed to identify specific proteins in the magnetic precipitates.
Western blotting
Whole cell extracts (WCE) from cultured cells were fractionated by SDS-PAGE and transferred to Immobilon protein membrane (Millipore, Billerica, MA). Immobilized membrane was incubated with the primary antibodies, i.e., hnRNP K (Zymed Laboratories, South San Francisco, CA), hnRNP D (Aviva Systems Biology, San Diego, CA), MSH2 (Calbiochem, San Diego, CA), GRHL2 (Abnova, Taipei, Taiwan), Sp1 (Santa Cruz Biotech., Santa Cruz, CA), and β-actin (Santa Cruz Biotech.) and probed with the respective secondary antibodies.
Two dimensional gel electrophoresis (2DGE)
The proteins coprecipitated with the hTERT promoter were subjected to 2DGE, which involved first dimension separation by isoelectric focusing (IEF) using immobilized pH gradient strips (IPG strip, 7-cm long, linear pH 3–10/pH 5–8) from Bio-Rad (Hercules, CA) and second dimension separation by gradient SDS-PAGE. IEF followed a stepwise incremental voltage program: 250 V for 15 min, 250V~4000V for 2 hrs, 4000 V for 5 hrs, and hold at 500V using the IEF cell (Bio-Rad). After IEF, the strips were subjected to two-step equilibration in equilibration buffers containing 6 M urea, 30% glycerol, 2% SDS, and 50 mM Tris-HCl, pH 8.8 with 1% DTT w/v for the first step, and 2.5% (w/v) iodoacetamide for the second step. The strips were then transferred onto 8–16% gradient SDS-PAGE gel (Invitrogen) and electrophoresed. The protein spots were stained with Sypro Ruby (Bio-Rad).
Protein identification by mass spectrometry
The protein spot patterns from 2DGE were compared between NHOK and SCC4 cells. The protein spots were excised by the ProteomWorks spot cutter system and analyzed by Applied Biomics, Inc. (Hayward, CA). Proteins of interest were digested in-gel with modified porcine trypsin protease (Trypsin Gold, Promega, Madison, WI). MALDI-TOF MS and TOF/TOF tandem MS/MS were performed on an ABI 4700 mass spectrometer (Applied Biosystems, Framingham, MA). The peptide mass and associated fragmentation spectra were submitted to GPS Explorer workstation equipped with MASCOT search engine (Matrix science) to search National Center for Biotechnology Information non-redundant (NCBInr). Candidates with either protein score C.I.% or Ion C.I.% greater than 95 were considered significant.
Reverse Transcription (RT)-PCR
Total RNA was isolated from the cultured cells using RNeasy Mini Kit (Qiagen, Chatsworth, CA). The RT reaction was performed with 5 µg total RNA, and the PCR reactions were performed for hTERT, hnRNP D, GRHL2, and GAPDH in semi-quantitative manner. The PCR amplification was performed with primer sequences were 5’-GCC TGA GCT GTA CTT TGT CAA-3’(forward) and 5’-CGC AAA CAG CTT GTT CTC CAT GTC-3’(reverse) for hTERT with the annealing temperature at 48°C; 5’-TGG ATC CAT GTC GGA GGA GCA GTT CGG-3’ (forward) and 5’-ATC CTG CAG CTA CAC GCT CAG CTG CCA AC-3’ (reverse) for hnRNP D at 63°C; 5’-GTG GTA CCA TGT CAC AAG AGT CGG ACA A-3’ (forward) and 5’- TTC TAG ACT AGA TTT CCA TGA GCG TGA-3’ (reverse) for GRHL2 at 55°C; 5’-GAC CCC TTC ATT GAC CTC AAC-3’(forward) and 5’-CTT CTC CAT GGT GGT GAA GA- 3’(reverse) for GAPDH at 55°C. The PCR products were fractionated by agarose gel electrophoresis and stained with ethidium bromide.
Chromatin immunoprecipitation assay
We performed ChIP assay using the commercial kit (Upstate, Temecula, CA) according to the manufacturer’s guidelines. Rapidly proliferating SCC4 cells were exposed to 1% formaldehyde, and chromatin was fragmented by sonication. Immunoprecipitation was performed using IgG (negative control) or with the primary antibodies for hnRNP K, hnRNP D, MSH2, or GRHL2 as described above. DNA was purified from the immunoprecipitates using the PCR purification kit (Qiagen). Subsequently, PCR was performed to amplify the hTERT promoter region using the primers 5'-TTT GGG CTA GTC TGG GGC GGG G-3' (forward) and 5'-GGC GCC CCC CAG CAG CTT AGG C-3' (reverse). The PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide.
Protein function analysis by RNAi
Endogenous GRHL2 expression was knocked down by RNAi using short hairpin RNA (shRNA). GRHL2 shRNA target sequences were identified by Ambion – siRNA Database and Design Tools (http://www.ambion.com/techlib/misc/siRNA_tools.html). We constructed a lentiviral vector expressing GRHL2 shRNA (LV-GRHL2i) for the internal GRHL2 sequence of 5’-GAG ATT GCA TAT AAT GCT G-3’ using pLentilox 3.7 plasmid (AddGene Inc., Cambridge, MA). LV-GRHL2i and the LV-EGFP (empty vector) were produced and concentrated as previously described (Kang et al., 2007). These viral vectors were used to infect SCC4 or SCC15 cells. Virus infection was allowed for 4 hrs in the presence of 6 µg/ml polybrene (Sigma, St. Louis, MO). Infected SCC4 cells were cloned by limiting dilution in 96-well plates. Knockdown of endogenous hnRNP K, hnRNP D, and MSH2 was performed using the validated double-stranded small interfering RNAs (siRNAs) (Santa Cruz Biotech.). The siRNAs were transfected into the cultured cells according to the manufacturer’s guidelines. Successful knockdown of gene expression was confirmed after 72 hrs post-transfection.
Transfection and reporter gene assay
A pGL3B-TRTP containing the 1670-bp fragment (− 1665 to + 5) of the hTERT promoter region linked to firefly luciferase cDNA (gift of Dr. J.C. Barrett, NIEHS/NIH, Bethesda, MD) was used for the hTERT promoter reporter assay. Prior to transfection, a six-well plate with approximately 5 × 104 cells per well was inoculated and cultured for 24 hrs. pGL3B-TRTP (1 µg per each well) was introduced into cells using Lipofectin Reagent (Invitrogen). To control for the different transfection efficiencies, pRL-SV40 plasmid (1 ng per each well) containing the Renilla luciferase cDNA under the control of SV40 enhancer/promoter was also transfected into each cell group. Cells were collected after 48 hrs post-transfection, and the cell lysates were prepared using the Dual Luciferase Reporter Assay System (Promega, Madison, WI). Luciferase activity was measured using a luminometer (Turner Designs, Sunnyvale, CA).
We also constructed a mutant hTERT promoter with deletion from −49 to +5 (pGL3-hTERTpΔ(−42 to +5), which contains the GRHL2 binding region. Due to existing Xho I site immediately downstream of the hTERT promoter sequence, the deletion was made by first creating a new Xho I site at −50 using QuikChange multi site-directed mutagenesis kit (Stratagene, La Jolla, CA). The plasmid was then digested with Xho I and allowed to self-ligate. The wild-type hTERT promoter construct (pGLB-TRPT) and the mutant promoter construct were transfected into rapidly proliferating SCC4 cells, and the promoter activities were compared by luciferase activity. Transfection efficiencies were normalized by the Renilla luciferase activity.
Telomerase assay
Telomerase activity was detected using the TRAP-eze Telomerase Detection Kit (Chemicon, Temecula, CA) according to manufacturer's guidelines. This kit allows for semi-quantitative detection of telomerase activity and amplification of the internal controls. Whole cell extract isolated in 1 × CHAPS buffer was used with the TRAP reaction mixture in the two-step telomerase assay involving the PCR amplification (Kim et al., 1994). The PCR products were separated by 12.5% polyacrylamide gel electrophoresis. The radioactive signal was detected by Phosphor Imager (Molecular Dynamix Inc., Sunnyvale, CA).
Results
Identification of the putative trans-regulatory proteins of the hTERT promoter
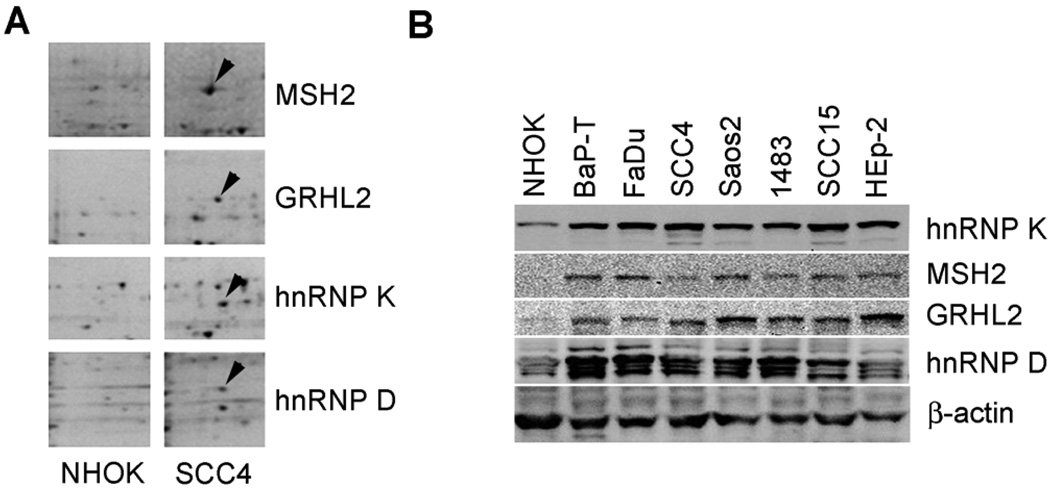
We identified the proteins that associate with the hTERT promoter preferentially in telomerase-positive cells by performing the PMP assay. The nuclear extracts from SCC4 and senescent NHOK were incubated with the hTERT promoter fragment immobilized onto paramagnetic Dynabeads® Separation of the DNA-bound proteins revealed numerous nuclear proteins differentially precipitated between NHOK and SCC4 cells (Figure 1A). We selectively identified the proteins that were present more abundantly in the precipitates of SCC4 compared to those from NHOK. These proteins were excised from the 2D gel and identified by mass spectrometry to include MSH2, hnRNPs K and D, and GRHL2.
Figure 1.
Identification of hTERT promoter binding proteins. (A) Nuclear extracts prepared from senescent NHOK and actively proliferating SCC4 were incubated with the hTERT promoter DNA linked to Dynabeads®. Protein-DNA complexes were allowed to form under native conditions and precipitated magnetically. The bound proteins were washed, eluted, fractionated by 2DGE, and stained with Sypro Ruby. Proteins that were more abundantly precipitated in SCC4 compared with NHOK were identified and excised for mass spectrometric analysis. These proteins were identified as MSH2, hnRNP K, hnRNP D, and GRHL2 (arrowheads). (B) Western blotting was performed with NHOK (pre-senescent and senescent cultures) and OSCC cell lines (BaP-T, FaDu, SCC4, 1483, SCC15, and HEp-2) to detect hnRNP K, MSH2, GRHL2, and hnRNP D. β-actin was used as loading control.
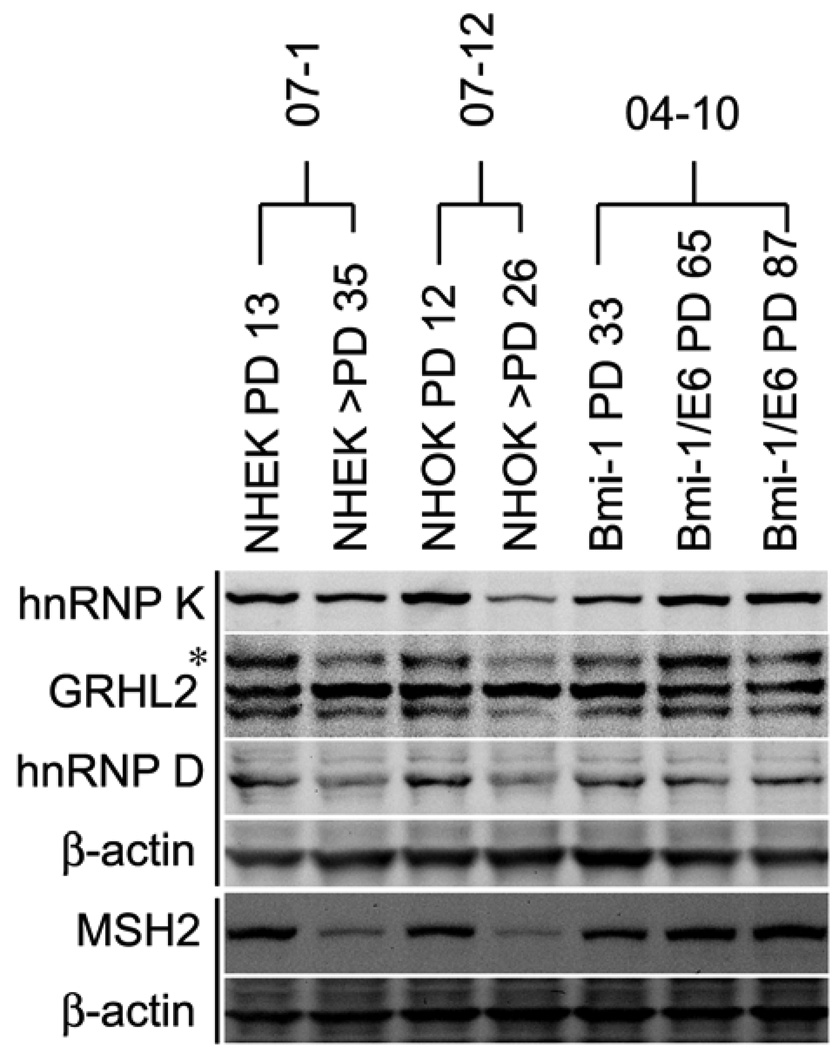
All four identified proteins were expressed at higher levels in the OSCC cells compared to senescent NHOK (Figure 1B). Among the normal cells (NHOK and NHEK), actively proliferating cells (population doubling [PD] 12–13) showed higher protein expression levels compared to the senescent cultures (PD 26–36) (Figure 2). During cellular immortalization by Bmi-1 and HPV E6 (Kim et al., 2007), MSH2, hnRNP K, and GRHL2 expression levels were increased in post-crisis cells (at PDs 65 and 87) compared to the pre-crisis cells (PD 33). The enhanced expression of these proteins coincided with the increased telomerase activity in the post-crisis cells (Kim et al., 2007). These results indicate that the identified proteins may contribute to telomerase activation during cellular immortalization by physical interaction with the hTERT promoter regions.
Figure 2.
Expression of the identified hTERT promoter binding proteins was enhanced during cellular immortalization. Western blotting was performed with replicating and senescing cultures of NHEK (07-1) and NHOK (07–12) at the indicated PD levels. We also included Bmi-1-expressing NHOK at PD 33 (pre-crisis) and those immortalized cells expressing exogenous Bmi-1 and HPV 16 E6 at PDs 65 and 87 (post-crisis). Western blotting was performed for intracellular hnRNP K, GRHL2, hnRNP D, and MSH2. β-actin was probed for loading control.
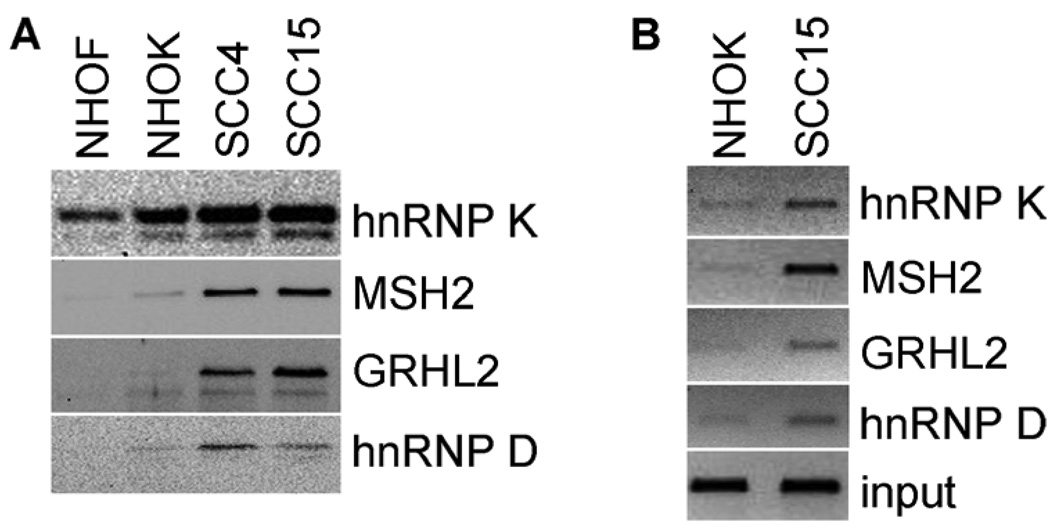
We confirmed the binding of MSH2, hnRNPs K and D, and GRHL2 with the hTERT promoter in vitro by performing the PMP assay followed by Western blotting. PMP was performed with the nuclear extracts from NHOF, NHOK, SCC4, and SCC15. As shown in Figure 3A, all four tested proteins were detected in the precipitates, and the level of binding was higher in SCC4 and SCC15 compared with NHOK. It was further reduced or undetectable in NHOF, which completely lack telomerase activity (Kang et al., 1998), compared to NHOK. These proteins were expressed at comparable levels in NHOK and NHOF and yet yielded differential binding capacity to the hTERT promoter. Thus, the protein expression level in cells may not be the sole factor determining the promoter binding capacity.
Figure 3.
The identified proteins bind the hTERT promoter in vitro and in vivo preferentially in SCC4 cells compared with NHOK. (A) PMP assay was performed with NHOF, NHOK (senescent), SCC4, and SCC15 cells and the amount of precipitated hnRNP K, MSH2, GRHL2, and hnRNP D was determined by Western blotting. (B) In vivo DNA-protein interaction was confirmed in senescent NHOK and SCC15 cells by ChIP assay. These cells were treated with formaldehyde to crosslink DNA-protein complexes and immunoprecipitated using specific primary antibodies for hnRNP K, MSH2, GRHL2, and hnRNP D. The presence of hTERT promoter sequence in the immunoprecipitates was determined by PCR amplification. Input control was constituted from the cell lysates before immunoprecipitation.
We also confirmed in vivo DNA-protein interactions by ChIP assay for MSH2, hnRNPs K and D, and GRHL2 in NHOK and SCC15 (Figure 3B). The hTERT promoter region was detected in the immunoprecipitates of all four proteins by PCR amplification. The level of DNA-protein association was notably higher in SCC15 compared to those in NHOK. This result is consistent with the in vitro DNA-protein binding revealed by the PMP assay (Figure 3A). These experiments confirm the binding of MSH2, hnRNPs K and D, and GRHL2 to the hTERT promoter and indicate the enhanced binding in SCC4 compared to NHOK or NHOF.
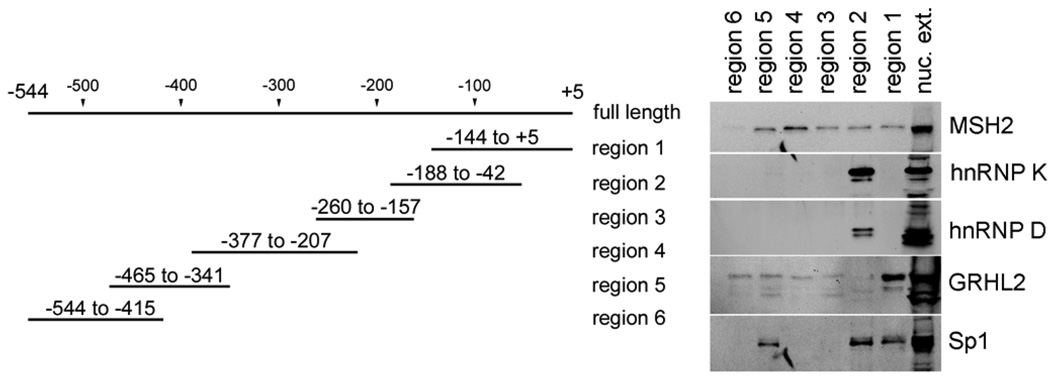
To further characterize the association between the identified proteins and the hTERT promoter, we dissected the full-length hTERT fragment (−544 to +5) into six overlapping regions and determined the level of protein binding to each region by PMP assay (Figure 4). GRHL2 bound strongly in the region 1 (−144 to +5) containing the transcription start site. hnRNPs K and D both were localized exclusively in the region 2 (−188 to −42). MSH2 bound most strongly with the region 4 (−377 to −207) and weakly with the regions 1–3, and 5, while the region 6 remained unbound. We also explored the binding pattern for Sp1, a protein known to interact with the hTERT promoter (Won et al., 2002). Sp1 exhibited binding in the region 5 (−465 to −341) and the regions 1 and 2 (together covering from −188 to +5). These results indicate that MSH2, hnRNPs K and D, and GRHL2 bind the hTERT promoter regions in sequence-specific manner.
Figure 4.
MSH2, the hnRNPs, and GRHL2 bind the hTERT promoter in sequence specific manner. The hTERT promoter (−544 to +5) was amplified into six overlapping regions with the forward primer conjugated with biotin at 5’ end. The hTERT promoter fragments representing the six regions were used to determine the regions at which the identified proteins interact with the hTERT promoter. PMP assay was performed with nuclear extracts isolated from actively dividing SCC4 cells. The precipitated proteins were eluted, fractionated by SDS-PAGE, and subjected to Western blotting for MSH2, hnRNP K, hnRNP D, and GRHL2. For comparison, we also included Sp1, which is known to interact with the proximal hTERT promoter region (Won et al., 2002).
The hTERT promoter binding proteins regulate the hTERT promoter activity
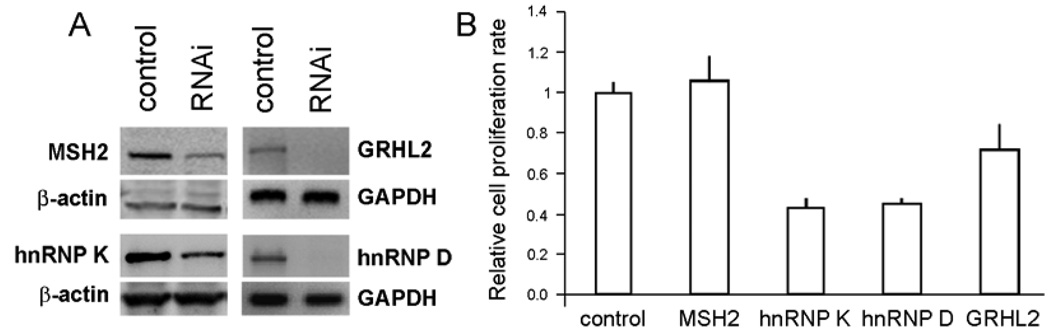
The biological functions of MSH2, hnRNP K, hnRNP D and GRHL2 were investigated for their role in the regulation of hTERT expression. We knocked down their expression in actively proliferating SCC4 cells, which express these genes at high levels (Figure 1B). We utilized the validated siRNAs targeting MSH2, hnRNP K, or hnRNP D. To knockdown the expression of GRHL2, we constructed a lentiviral vector expressing shRNA against GRHL2 (LV-GRHL2i). SCC4 cells were either transfected with the siRNAs or infected with LV-GRHL2i. After three days of RNAi, the cells were harvested, and successful knockdown was confirmed by either Western blotting or semi-quantitative RT-PCR (Figure 5A). We also determined the short-term effects of RNAi for the individual genes on cell proliferation. With an exception of MSH2, knockdown of all other gene expression individually led to varying degrees of cell proliferation arrest (Figure 5B).
Figure 5.
Validation of RNAi against MSH2, the hnRNPs, and GRHL2. (A) SCC4 cells were transiently transfected with the siRNAs targeting MSH2, hnRNP K, or hnRNP D, or infected with lentiviral vector expressing shRNA against GRHL2 (LV-GRHL2i) or with LV-EGFP as control. The cells were harvested at 72 hrs after siRNA transfection. Semi-quantitative RT-PCR was performed to determine the level of their respective mRNA expression after RNAi. (B) Cell numbers were determined before and after 72 hrs post-transfection of the siRNAs to compare the effects of gene knockdown on short-term proliferation of cells. Bar indicates standard deviation.
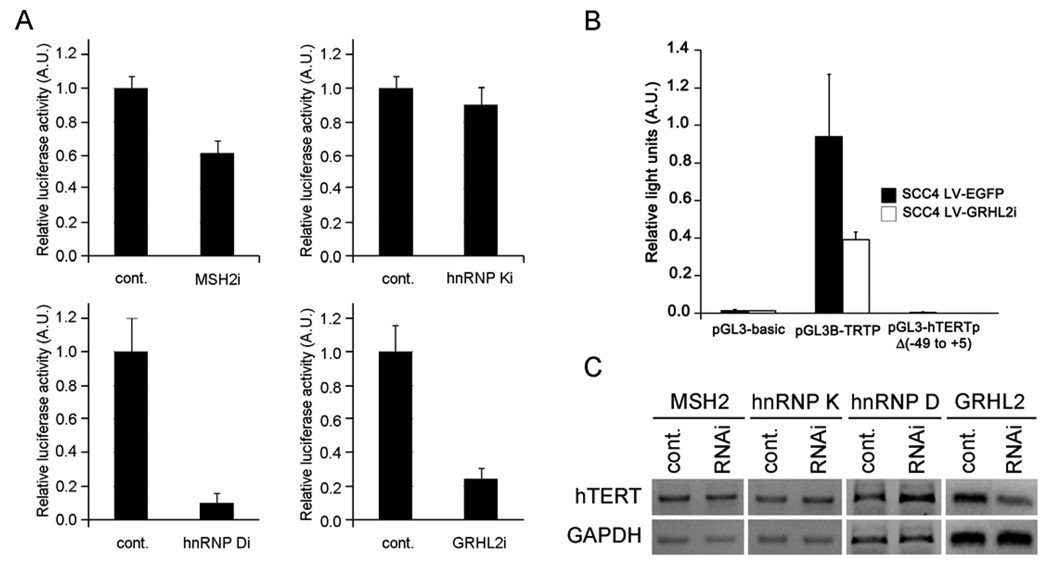
To determine the role of these genes in hTERT promoter regulation, we utilized the hTERT promoter-luciferase reporter construct (pGL3B-TRTP) transfected into SCC4 cells after the transfection of the siRNAs targeting MSH2, hnRNP K, or hnRNP D. The reporter construct was also transfected into the cells infected with LV-EGFP or LV-GRHL2i. After two days post-transfection, firefly luciferase reporter activity was determined. Knockdown of MSH2, the hnRNPs, or GRHL2 all led to reduction of the luciferase activity, albeit to varying degrees ranging from approximately 10 – 90% (Figure 6A). We also constructed mutant hTERT promoter lacking the sequences from −49 to +5 (pGL3-hTERTpΔ[−49 to +5]), which contains the region that GRHL2 binds (Figure 4). To determine the effects of deleting these sequences on hTERT promoter activity, we transfected the wild-type (pGL3B-TRTP) or the mutant hTERT promoter-luciferase constructs into SCC4 cells with or without GRHL2 knockdown. The deletion mutant completely abolished the hTERT promoter activity (Figure 6B). Furthermore, GRHL2 knockdown led to significant diminution of the hTERT mRNA expression although knockdown of MSH2 or the hnRNPs did not (Figure 6C).
Figure 6.
Effects of gene knockdown on hTERT expression level and the promoter activity. (A) SCC4 cells after RNAi (either through transfection of siRNAs or infection with LV-GRHL2i) were used to determine the hTERT promoter activity. The cells were transfected with pGL3B-TRTP containing the hTERT promoter driving the expression of firefly luciferase reporter gene, whose activity was determined at 48 hrs transfection of the reporter plasmid. Transfection efficiency was normalized against the Renilla luciferase activity driven by SV40 promoter. Relative luciferase activity from individual gene knockdown is shown as mean of triplicates. Bar indicates standard deviation. (B) SCC4 cells stably infected with LV-EGFP or LV-GRHL2i were transfected with either pGL3-basic (promoterless luciferase vector), pGL3B-TRTP (wild-type hTERT promoter luciferase), or pGL3-hTERTpΔ(−49 to +5) containing the mutant hTERT promoter sequences. Firefly and Renilla luciferase activities were determined at 48 hrs post-transfection and plotted as normalized values. Error bars are standard errors. (C) SCC4 cells were transfected with the siRNAs (MSH2, hnRNP K, or hnRNP D) or infected with LV-GRHL2i. After 72 hrs post-transfection, level of hTERT mRNA expression was determined by RT-PCR. GAPDH is used to control the starting amount of cDNA.
Effects of MSH2, the hnRNPs, and GRHL2 RNAi on telomerase activity in SCC4 cells
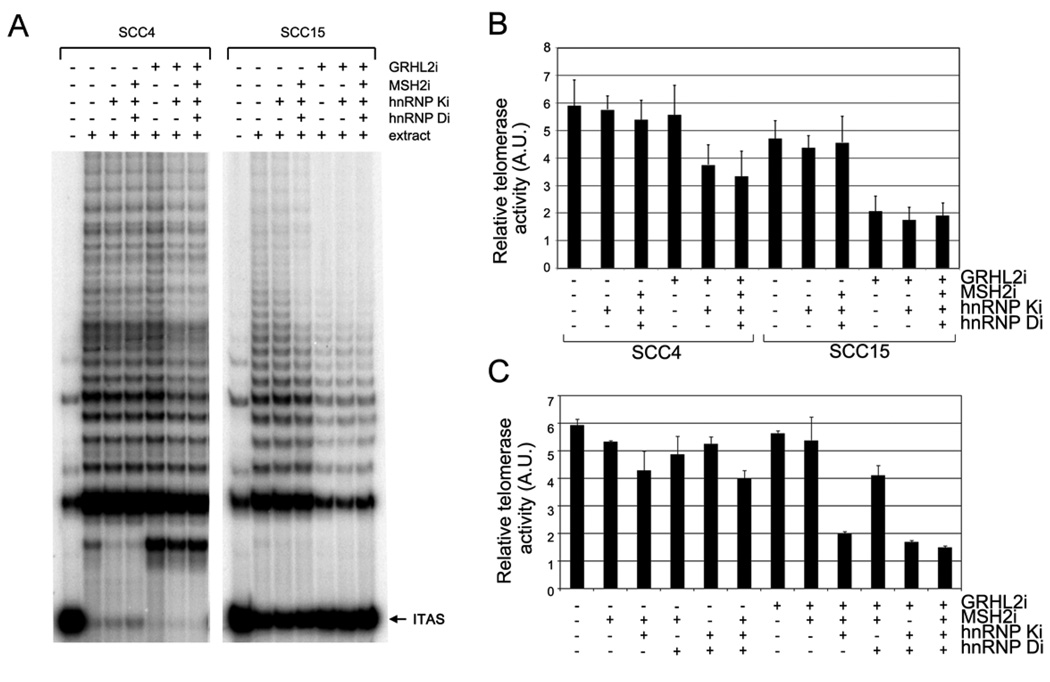
We determined whether MSH2, the hnRNPs, and GRHL2 play a role in the regulation of telomerase activity by knocking down their expression in SCC4 cells. After three days of individual gene knockdown of MSH2, hnRNP K, or hnRNP D, there was little or no significant change in telomerase activity in SCC4 cells (data not shown). Subsequently, we administered combined RNAi by utilizing the SCC4 cells stably infected with LV-EGFP or LV-GRHL2i, which were then transfected with varying combinations of siRNAs. Marked reduction of telomerase activity was noted in SCC4 cells infected with LV-GRHL2i after knockdown of hnRNP K, while the other siRNA combinations showed little effect (Figures 7A – C). This experiment was repeated using a different OSCC line, SCC15. In these cells, however, GRHL2 knockdown alone led to notable reduction of telomerase activity. Thus, RNAi of GRHL2 alone appears to be sufficient for telomerase inhibition in some OSCC cells.
Figure 7.
Telomerase activity in SCC4 cells was inhibited by multiple gene knockdown involving at least GRHL2 and hnRNP K. (A) SCC4 and SCC15 cells stably infected with LV-EGFP or LV-GRHL2i were transfected with various combinations of siRNAs targeting MSH2, hnRNP K, or hnRNP D. The cells were harvested after 48 hrs post-transfection, and TRAP assay was performed to determine telomerase activity. ITAS, internal telomere amplification standard. (B) Three independent TRAP assay results were quantitated by phosphometric analyses using ImageQuant software (Molecular Dynamix) and normalized against each control group (without the extract). Bar represents the mean and standard errors. (C) Independent experiment was performed with SCC4 stably infected with LV-EGFP or LV-GRHL2i and subsequently transfected with varying combinations of siRNAs targeting MSH2 and/or the hnRNPs. After 72 hrs post-transfection, the cells were harvested and TRAP assay performed. Telomerase activity was quantitated and normalized against the control group (without the extract). Reduction of telomerase activity was evidenced in the groups in which both GRHL2 and hnRNP K were knocked down.
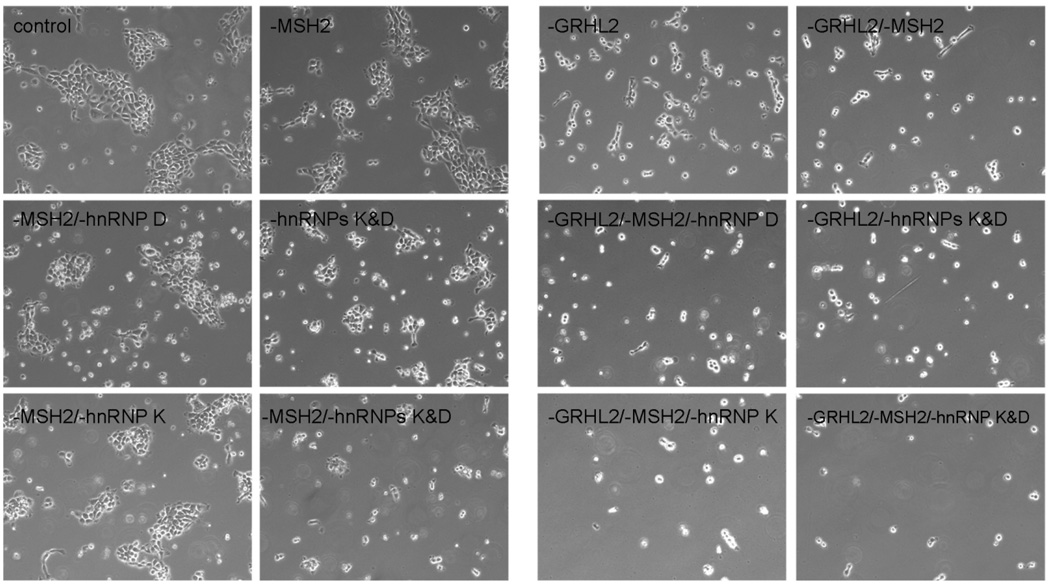
The inhibitory effect of GRHL2 and hnRNP K on telomerase activity was in part correlated with the proliferation and viability status of SCC4 cells (Figure 8). The cells demonstrated significant reduction in cell proliferation after infection with LV-GRHL2i compared to those infected with LV-EGFP. Cell proliferation and viability were further inhibited after additional knockdown of MSH2, hnRNP K, and/or hnRNP D. Taken together, the above findings confirm the biological functions of the identified hTERT promoter binding proteins in regulation of the hTERT promoter activity, telomerase activity, cell proliferation, and viability of SCC4 cells.
Figure 8.
Morphology and growth of human oral cancer cells transfected with different combination of siRNA. The SCC4 cells infected with LV-EGFP or LV-GRHL2i were transfected with various combinations of siRNAs targeting MSH2, hnRNP K, or hnRNP D. The cells were examined under phase-contrast photomicrographs at 48 hrs after transfection. Original magnification, 100×.
Discussion
In the present study, we show that MSH2, the hnRNPs K and D, and GRHL2 are the novel proteins that physically associate with the hTERT promoter and regulate the promoter activity. This study confirmed the validity of PMP-MS in identification of DNA binding proteins. This unique combination of high throughput techniques is a powerful approach for the identification of new hTERT trans-regulatory proteins through the specific protein-DNA interactions. PMP served as the primary screening to enrich the candidate DNA binding proteins from the nuclear extract. This resulted in dramatic reduction of the sample complexity, as evinced in Figure 1A, when the magnetically precipitated proteins were fractionated by 2DGE. The data also showed the presence of numerous low abundance proteins in both NHOK and SCC4 samples that could not reliably be identified. These proteins would require additional concentration or fractionation of the PMP samples for further characterization.
We found that the wild-type hTERT promoter activity was significantly reduced by knockdown of GRHL2. Also, the mutant hTERT promoter activity was completely abolished in the presence or absence of GRHL2 expression in cells. This data indicate that the region from − 49 to +5 is critical for the hTERT promoter activity. It remains possible that this region interacts with other trans-regulatory proteins than GRHL2 which are necessary for hTERT expression. Since the exact hTERT promoter binding site of GRHL2 is unknown, further studies are needed to elucidate the effects of GRHL2 binding alone on the hTERT promoter activity.
Our study revealed the inhibitory effects of the gene knockdown on cell proliferation and the hTERT promoter luciferase activity for all four genes tested, albeit to varying degrees (Figures 5B and 6A). Knockdown of hnRNP K, hnRNP D, or GRHL2 alone resulted in significant reduction of cell proliferation in SCC4 cells. GRHL2 knockdown led to congruent loss of telomerase activity (Figure 7). We also found that overexpression of GRHL2 in primary NHOK prevented the senescence-associated loss of telomerase activity (Kang et al., 1998; Kang et al., 2004), and the cells expressing exogenous GRHL2 demonstrated enhanced cellular proliferation (data not shown). However, knockdown of the hnRNPs revealed no effect on telomerase activity. Thus, reduced cell proliferation by silencing of the hnRNPs may represent telomerase-and/ or telomere-independent event. Furthermore, with an exception of GRHL2, knockdown of MSH2 or the hnRNPs did not result in congruent reduction of the mRNA expression level. These genes possess diverse biological functions that may lead to altered RNA stability and post-transcriptional processing. This is especially true for the hnRNPs which are known as RNA binding proteins. hnRNP D is an RNA turnover factor that modulates the decay of AU-rich mRNAs (Kiledjian et al., 1997; He and Schneider, 2006). hnRNP K regulates the multitude of gene expression, including that of p53 (Moumen et al., 2005), as a transcription factor or by alteration of mRNA stability and translation (Ostareck-Lederer and Ostareck, 2004). Thus, we cannot rule out the possibility that RNAi against the hnRNPs indirectly led to stabilization of hTERT mRNA, resulting in the apparent discrepancy between the promoter activity and the mRNA level.
We found that MSH2, the hnRNPs, and GRHL2 bind to the hTERT promoter in sequence-specific manner (Figure 4). With an exception of MSH2, binding of the hnRNPs and GRHL2 to the hTERT promoter occurred within the regions 1 and 2 covering the sequences from −188 to +5 from the transcription start site. Knockdown of these genes led to notable reduction of the promoter activity, consistent with an earlier report showing the minimum sequence requirement for the full promoter activity within these regions (Cong et al., 1999). The PMP assay also revealed Sp1 binding in these regions in agreement with other studies that showed the proximal location of the Sp1 binding sites (Cong et al., 1999; Kyo et al., 2000). We also identified a novel binding site for Sp1 distally located in the region 5 (−465 to −341) although its functional significance for the promoter regulation remains to be determined. The Sp1 binding sites within the proximal regions are necessary for the full activity of the hTERT promoter, which apparently lack TATA or CAAT box (Cong et al., 1999; Kyo et al., 2000). Sp1 interacts with the basal transcription machineries, such as TATA box binding protein (TBP) and TBP-associated factors (TAFs), to help initiate the transcription of TATA-less promoters (Emili et al., 1994; Suzuki et al., 2003). However, Sp1 may also impose inhibitory effects on the promoter by recruiting histone deacetylases (HDAC) (Won et al., 2002). HDAC recruitment and abundance on the hTERT promoter are associated with transcriptional repression, presumably by maintaining the condensed chromatin state (Zhu et al., 2008). These results point out the complexity in the modes by which Sp1 regulates the hTERT promoter activity.
While individual gene knockdown had no detectable effect on telomerase activity, multiple gene silencing involving at least GRHL2 and hnRNP K led to notable reduction of telomerase activity in SCC4 cells. GRHL2 knockdown alone led to significant loss of telomerase activity in SCC15 cells. The reason for the different requirement for telomerase activity in SCC4 and SCC15 cells is unknown. In both cells, however, GRHL2 appears to be a critical factor for hTERT expression and telomerase activity. This notion is also reinforced by the result showing that GRHL2 binds the hTERT promoter DNA in close proximity to the transcription start site (Figure 4). GRHL2 knockdown in NHOK alone led to reduced hTERT mRNA expression and telomerase activity (data not shown). Further studies will elucidate the role of GRHL2 in regulation of hTERT expression and telomerase activity.
RNAi-mediated loss of telomerase activity in SCC4 cells occurred with reduced cell viability and proliferative status. This does not appear to be an artifact of nonspecific toxicity as it can be evinced in the cells infected with LV-GHRL2i and treated with MSH2 siRNA. These cells showed similar extent of proliferation defect as those treated with hnRNP K RNAi (Figure 8) but expressed intact telomerase activity (Figure 7). Similarly, those infected with LV-EGFP and exposed to the siRNAs showed varying degrees of cell proliferation arrest and loss of viability, yet retained intact telomerase activity in the adherent cells. Thus, these groups of cells illustrate a disconnect between telomerase activity and the cell viability status, and support the notion that the loss of telomerase activity reported herein is not an artifact of nonspecific toxicity from multiple gene knockdown.
In summary, we report MSH2, the hnRNPs, and GRHL2 as novel hTERT promoter binding proteins identified by the PMP assay. This approach coupled with proteomic techniques is a powerful combination of HTS for DNA binding proteins, ideally suited for exploring the trans-regulatory factors. These proteins were highly overexpressed in the OSCC cells compared with NHOK, indicating their aberrant expression associated with oral carcinogenesis. Elevation of their expression was observed during the transition from pre- to post-crisis, at which telomerase activation occurred (Kim et al., 2007). Since these proteins were necessary for the intact expression of the hTERT promoter activity in OSCC cells, we speculate that they are in part responsible for the elevated hTERT expression and telomerase activation during oral carcinogenesis.
Acknowledgements
The authors thank Dr. J.C. Barrett (NIEHS/NIH) for providing the pGL3B-TRTP vector. This study was supported in part by the grants (K22DE15316, R01DE18295, and K02DE18959 to M.K.K. and R01DE14147 to N.-H.P.) from NIDCR/NIH.
References
- Cairney CJ, Keith WN. Integrated regulation of hTR and hTERT for telomere maintenance and telomerase activity. Biochimie. 2008;90:13–23. doi: 10.1016/j.biochi.2007.07.025. [DOI] [PubMed] [Google Scholar]
- Campbell MR, Wang Y, Andrew SE, Liu Y. MSH2 deficiency leads to chromosomal abnormalities, centrosome amplification, and telomere capping defect. Oncogene. 2006;25:2531–2536. doi: 10.1038/sj.onc.1209277. [DOI] [PubMed] [Google Scholar]
- Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8:137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- Deng WG, Jayachandran G, Wu G, Xu K, Roth JA, Ji L. Tumor-specific activation of human telomerase reverses transcriptase promoter activity by activating enhancer-binding protein-2β in human lung cancer cells. J Biol Chem. 2007;282:26460–26470. doi: 10.1074/jbc.M610579200. [DOI] [PubMed] [Google Scholar]
- Emili A, Greenblatt J, Ingles CJ. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol Cell Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki T, Miura T, Maura M, Shiraishi H, Nishimura S, Imada Y, et al. TAK1 represses transcription of the human telomerase reverse transcriptase gene. Oncogene. 2007;26:5258–5266. doi: 10.1038/sj.onc.1210331. [DOI] [PubMed] [Google Scholar]
- He C, Schneider R. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenmann S, Cakouros D, Zannettino A, Shi S, Gronthos S. hTERT transcription is repressed by Cbfa1 in human mesenchymal stem cell populations. J Bone Miner Res. 2007;22:897–906. doi: 10.1359/jbmr.070308. [DOI] [PubMed] [Google Scholar]
- Kang MK, Guo W, Park NH. Replicative senescence of normal human oral keratinocytes is associated with the loss of telomerase activity without shortening of telomeres. Cell Growth Differ. 1998;9:85–95. [PubMed] [Google Scholar]
- Kang MK, Kameta A, Shin KH, Baluda MA, Park NH. Senescence occurs with hTERT repression and limited telomere shortening in human oral keratinocytes cultured with feeder cells. J Cell Physiol. 2004;199:364–370. doi: 10.1002/jcp.10410. [DOI] [PubMed] [Google Scholar]
- Kang MK, Kim RH, Kim SJ, Yip FK, Shin KH, Dimri GP, et al. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br J Cancer. 2007;96:126–133. doi: 10.1038/sj.bjc.6603529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, DeMaria CT, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonuceoprotein D) as a component of the α-globin mRNA stability complex. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RH, Kang MK, Shin KH, Oo ZM, Han T, Baluda MA, et al. Bmi-1 cooperates with human papillomavirus type 16 E6 to immortalize normal human oral keratinocytes. Exp Cell Res. 2007;313:462–472. doi: 10.1016/j.yexcr.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kyo S, Takakura M, Taira T, Kanaya T, Itoh H, Yutsudo M, et al. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nuc Acids Res. 2000;28:669–677. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel R, McDuff FO, Lavigne P, Grandbois M. Direct visualization of the binding of c-Myc/Max heterodimeric b-HLH-LZ to E-Box sequences on the hTERT promoter. Biochemistry. 2007;46:10279–10286. doi: 10.1021/bi700076m. [DOI] [PubMed] [Google Scholar]
- Lou F, Chen X, Jalink M, Zhu Q, Ge N, Zhao S, et al. The opposing effect of hypoxia-inducible factor-2α on expression of telomerase reverse transcriptase. Mol Cancer Res. 2007;5:793–800. doi: 10.1158/1541-7786.MCR-07-0065. [DOI] [PubMed] [Google Scholar]
- Lynch M, Chen L, Ravitz MJ, Mehtani S, Korenblat K, Pazin MJ, et al. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol Cell Biol. 2005;25:6436–6453. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumen A, Masterson P, O’Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH. Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol Cell. 2004;96:407–411. doi: 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Ostrwoski J, Klimek-Tomczak K, Wyrwicz LS, Mikula M, Schullery DS, Bomsztyk K. Heterogeneous nuclear ribonucleoprotein K enhances insulin-induced expression of mitochondrial UCP2 protein. J Biol Chem. 2004;279:54599–545609. doi: 10.1074/jbc.M406753200. [DOI] [PubMed] [Google Scholar]
- Park NH, Min BM, Li SL, Huang MZ, Cherick HM, Doniger J. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis. 1991;12:1627–1631. doi: 10.1093/carcin/12.9.1627. [DOI] [PubMed] [Google Scholar]
- Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 2001;269:1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- Rizki A, Lundblad V. Defects in mismatch repair promote telomerase-independent proliferation. Nature. 2001;411:713–716. doi: 10.1038/35079641. [DOI] [PubMed] [Google Scholar]
- Roychoudhury P, Chaudhuri K. Evidence for heterogeneous nuclear ribonucleoprotein K overexpression in oral squamous cell carcinoma. Br J Cancer. 2007;97:574–575. doi: 10.1038/sj.bjc.6603911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Keith WN. Targeting telomerase for cancer therapeutics. Br J Cancer. 2008;98:677–683. doi: 10.1038/sj.bjc.6604209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B, Martin P. Cell biology: master regulators of sealing and healing. Curr Biol. 2005;15:R425–R427. doi: 10.1016/j.cub.2005.05.034. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Muto S, Miyamoto S, Aizawa K, Horikoshi M, Nagai R. functional interaction of the DNA-binding transcription factor Sp1 through its DNA-binding domain with the histone chaperone TAF-I. J Biol Chem. 2003;278:28758–28764. doi: 10.1074/jbc.M302228200. [DOI] [PubMed] [Google Scholar]
- Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- Wilanowski T, Tuckfield A, Cerruti L, O’Connell S, Saint R, Parekh V, et al. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech Dev. 2002;114:37–50. doi: 10.1016/s0925-4773(02)00046-1. [DOI] [PubMed] [Google Scholar]
- Won J, Yim J, Kim TK. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J Biol Chem. 2002;277:38230–38238. doi: 10.1074/jbc.M206064200. [DOI] [PubMed] [Google Scholar]
- Yan P, Saraga EP, Bouzourene H, Bosman FT, Benhattar J. Expression of telomerase genes correlates with telomerase activity in human colorectal carcinogenesis. J Pathol. 2001;193:21–26. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH728>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Liu C, Ge Z, Fang X, Zhang X, Straat K, et al. Lysine-specific demethylase 1 (LSD1) is required for the transcriptional repression of the telomerase reverse transcriptase (hTERT) gene. PLos ONE. 2008;3:e1446. doi: 10.1371/journal.pone.0001446. [DOI] [PMC free article] [PubMed] [Google Scholar]