Abstract
Chronic infection by Hepatitis C virus (HCV) causes liver fibrosis, which is accelerated by unknown mechanisms in patients with HIV-1 coinfection. The evolution of HCV quasispecies in this setting of coinfection is not fully understood. To compare HCV quasispecies between HIV-HCV coinfection and HCV monoinfection, we sequenced 340 HCV clones from the HVR-1 and NS3 regions at two different time points in two groups of treatment-naïve patients with HCV-1a infection: (1) HIV-HCV positive (n=6); and (2) HIV negative-HCV positive (n=3). In HCV/HIV coinfection, we found a trend for reduced HCV genetic complexity and diversity, and a trend towards reduced dN/dS ratios in the HVR-1 region, especially in those patients with CD4<200 cells/mm3, who lost positive selective immune pressure in the HVR-1 region. Differences in immune regulation of HCV quasispecies in HIV coinfected individuals deserve further exploration to clarify the different outcomes of chronic hepatitis C noted between the immunocompromised and the immunocompetent host.
Keywords: Immune pressure, coinfection, AIDS, envelope, NS3, CD4 counts
Hepatitis C virus (HCV, Flaviviridae family, genus Hepacivirus) coinfection in Human Immunodeficiency virus-1 (HIV, Retroviridae family, genus Lentivirus) infected individuals is a clinical problem worldwide, and results from studies on the evolution of HCV in HIV-HCV coinfection are controversial (Thomas, 2002; Devereux et al., 1997; Sherman et al., 1996). We analysed two groups of treatment-naïve patients infected with HCV-1a: (i) HIV-HCV coinfection and (ii) HCV monoinfection. We studied HCV clones from the hypervariable region-1 (HVR-1) target for neutralising antibodies, and from the non-structural protein 3 (NS3), target for the cellular immune response. Our endpoint was to determine the effects of HIV-coinfection on HCV genetic variation.
Two groups of male individuals infected with HCV subtype 1a (Inno-LIPA HCV II, Innogenetics, Zwijndrecht, Belgium) were selected from a natural history cohort attending the San Francisco Veterans Administration Medical Center (SFVAMC) and San Francisco General Hospital (SFGH). Group 1 included six patients with HIV-HCV coinfection selected on basis of the following criteria: anti-HIV, anti-HCV, and HCV-RNA positive; CD4 counts <200 cells/mm3 (n=3) or >200 cells/mm3 (n=3). Group 2 included anti-HIV negative individuals (n=3) with chronic hepatitis C (anti-HIV negative, anti-HCV and HCV-RNA positive) as controls. All the patients had clinically-proven chronic hepatitis C (abnormal ALT and detectable HCV-RNA by nested-PCR over time), negative hepatitis B surface antigen (HBsAg), and negative anti-hepatitis B core IgM antibodies (anti-HBc IgM). Serum samples were obtained at two different time points (mean interval between samples 13.8 months). Patients did not receive antiviral therapy prior to or during the observation period. The study was approved by the University of California San Francisco (UCSF) Board, and informed consent was obtained in writing in every case.
HCV-RNA extraction, reversed transcription and polymerase chain reaction (RT-PCR) were performed using primers from the HCV HVR-1 or NS3 regions (table 1), as described (Chazouilleres et al., 1994 ; Lopez-Labrador et al., 2004; Weiner et al., 1991). Purified PCR products (Wizard PCR, Promega, Madison, WI) were cloned by using the TA Cloning Kit (Invitrogen, Carlsbad, CA), plasmids extracted with the Wizard Plus Minipreps (Promega) and sequenced at the UCSF Medical Center Sequencing facility using BigDye chemistry on an ABI 3700 DNA sequencer (Applied Biosystems, Foster City, CA). HCV-RNA was quantitated with the Amplicor HCV Monitor test v2.0 (Roche diagnostics, Branchburg, NJ), after 1:10 dilution (Martinot-Peignoux et al., 2000).
Table 1. Oligonucleotide primers and conditions used for PCR amplification of HCV E2 and NS3 regions.
Primer positions refer to the HCV-1 prototype (Genbank accession M62321).
| Primer | Sequence | Nucleotide position |
|---|---|---|
| HVR-1 | ||
| First Round RT-PCR: | ||
| {43°C for 30 min; (94°C for 10 sec; 55°C for 30 sec; and 72° for 30 sec) × 35 cycles} | ||
| 244 bp PCR product | ||
| Outer Sense | 5′-GGTGCTCACTGGGGAGTCCT-3′ | 1389–1408 |
| Outer Antisense | 5′-CATTGCAGTTCAGGGCAGTCCTG-3′ | 1632–1610 |
| Second Round PCR: | ||
| {(94°C for 10 sec; 50°C for 20 sec; 72°C for 30 sec) × 35 cycles} | ||
| 176 bp PCR product | ||
| Inner Sense | 5′-TCCATGGTGGGGAACTGGGC-3′ | 1428–1447 |
| Inner Antisense | 5′-TGCCAACTGCCATTGGTGTT-3′ | 1603–1584 |
| NS3 | ||
| First Round RT-PCR: | ||
| {43°C for 30 min; (94°C for 10 sec; 50°C for 20 sec; 72°C for 30 sec;) × 35 cycles} | ||
| 537 bp PCR product | ||
| Outer Sense | 5′-ACGTACTCCACCTACGGCAA-3′ | 4215–4234 |
| Outer Antisense | 5′-AAGGTAGGGTCAAGGCTGAA-3′ | 4750–4731 |
| Second Round PCR: | ||
| {(94°C for 10 sec; 50°C for 20 sec; 72°C for 30 sec) × 35 cycles} | ||
| 289 bp PCR product | ||
| Inner Sense | 5′-CATCCCAACATCGAGGAGGT-3′ | 4417–4435 |
| Inner Antisense | 5′-TTGCAGTCTATCACCGAGTC-3′ | 4705–4686 |
Both positive controls (HCV-RNA positive sera with known amounts of HCV-RNA) and negative controls (HCV-RNA negative sera) were included in every extraction-RT-PCR run, to ensure that sufficient viral RNA was extracted and subjected to RT-PCR amplifications prior to cloning. Only runs with positive HCV-RNA results for positive controls and negative results for negative controls were accepted, both for the HVR-1 and NS3 PCRs, and all runs were checked for sensitivity by a nested PCR of the 5′NC region performed in parallel. In addition, all samples were HCV-RNA positive in separate runs from different aliquots, used for clinical routine determinations (Amplicor HCV Monitor v2.0, and viral genotype by LiPA). Since all patients included in this study were infected with HCV subtype 1a, we designed subtype-1a specific primers after inspection of HCV genotype-1 aligned sequences from Genebank, in order to avoid selective amplification bottlenecks of certain HCV variants in a given sample. To avoid underestimation of HCV variants present in samples from patients with low viremia levels, approximately the same amount of PCR products (amplicons) from each sample was used for cloning experiments, reducing the possibility for underestimation of the viral complexity from the original sample. Amplified PCR products were quantitated by comparison with DNA standards of known concentration.
A total of 340 HCV clones were analysed: 156 for the HVR-1 and 184 for NS3 (Genebank accession numbers EF208216-EF208558). Shannon entropy (S) and normalised entropy (Sn) were calculated to measure complexity (Wolinsky et al., 1996). Genetic distances (diversity) were calculated with a modified Kimura-2 parameter method with gamma correction for multiple hits (Kimura, 1983). Average non-synonymous (dN) and synonymous (dS) substitution frequencies were calculated using a modified Nei and Gojobori algorithm with the Jukes and Cantor correction (Nei and Kumar, 2000). Genetic distances and substitution frequencies were all determined with the MEGA program, version 2.1, both within-sample and between samples (Kumar et al., 2001). Statistical comparisons were performed using non-parametric tests (Mann Whitney U or Kruskal-Wallis tests, when appropriate), and the SPSS for Windows package, version 8.0 (SPSS, Chicago, IL). P values lower than 0.05 were considered significant.
The main characteristics of the patients are summarised in table 2. Mean HCV-RNA levels were slightly higher in HIV-HCV coinfected compared to HCV monoinfected patients (5.67 ± 0.34 vs. 5.08 ± 0.09 log10 IU/mL; P= 0.025). Mean HCV viral load was 5.72 ± 0.41 and 5.62 ± 0.34 in patients from group 1 with CD4 counts <200 cells/mm3 or >200 cells/mm3, respectively (difference NS; P=0.094).
Table 2.
Main clinical and demographical characteristics of the study subjects.
| Patient | HIV status | CD4+ T-cell Status (cells/mm3) | Age (Yrs.) | Gender (M/F) | HCV type | CD4+ T-cell count (cells/mm3) | HCV-RNA (log10) IU/mL | Months between samplesb | ||
|---|---|---|---|---|---|---|---|---|---|---|
| T0a | T1a | T0a | T1a | |||||||
| Group 1 | ||||||||||
| 1-1 | Positive | <200 | 43 | M | 1a | 153 | 133 | 6.15 | 5.64 | 15 |
| 1-2 | Positive | <200 | 56 | M | 1a | 165 | 177 | 5.35 | 5.61 | 17 |
| 1-3 | Positive | <200 | 44 | M | 1a | 111 | N.D.d | 5.66 | 5.79 | 15 |
| 1-4 | Positive | >200 | 33 | M | 1a | 663 | 585 | 5.85 | 5.26 | 14 |
| 1-5 | Positive | >200 | 49 | M | 1a | 489 | 731 | 5.79 | 5.57 | 11 |
| 1-6 | Positive | >200 | 41 | M | 1a | 722 | 1024 | 5.23 | 5.24 | 10 |
| Group 2 | ||||||||||
| 2-1 | Negative | N.A.c | 54 | M | 1a | N.D.d | N.D.d | 4.97 | 4.89 | 12 |
| 2-2 | Negative | N.A.c | 43 | M | 1a | N.D.d | N.D.d | 5.14 | 4.82 | 14 |
| 2-3 | Negative | N.A.c | 37 | M | 1a | N.D.d | N.D.d | 5.14 | 5.54 | 9 |
T0 = sample at baseline (time point 0); T1 = sample at follow-up (time point 1).
Mean interval between samples was 13.9 ± 0.8 months (median 14 months, range 9–17).
NA = non applicable.
ND = not done.
Our analysis of HCV clones is summarised in Table 3. We noted several differential trends for HCV quasispecies evolution in HIV-HCV coinfection that deserve to be examined at the clonal level in larger cohorts. Coinfecion seemed to be associated with a trend towards decreased complexity (Sn) for the HVR-1 region, in agreement with data from Mao et al. (Mao et al., 2001) and with recent studies using heteroduplex-tracking assays (Shuhart et al., 2006). There was a also a trend for higher within-sample diversity in the HVR-1 region at baseline (nucleotide and amino acid distances), and for lower net genetic distances between samples in patients with HIV-HCV coinfection, especially in those with CD4 <200 cells/mm3. Mean dN/dS ratios at baseline were < 1 in coinfected patients with CD4 counts <200, and regardless of the CD4 counts after 1 year follow-up, which suggests that immune pressure to the HVR-1 might be diminished with HIV coinfection. In contrast, dN/dS for the HVR-1 was always >1 in HCV monoinfection. When analysing changes with time in the HVR-1 region, coinfection was coincident with a trend towards decreasing complexity, and with HCV dN/dS ratios between samples lower than one. When the coinfection group was subdivided, dN/dS at baseline and dN/dS between samples were >1 for the HVR-1 in coinfected patients with CD4>200 cells/mm3 and in HCV monoinfected patients, but <1 in those coinfected with CD4<200 cells/mm3. Finally, no specific trend for coinfection was noted in NS3.
Table 3.
Mean genetic estimates of HCV quasispecies in the HVR-1 and NS3 regions. The number clones sequenced for each HCV region is indicated before the estimates. dN/dS ratios suggestive of positive immune pressure are bolded. (1) All HIV(+) vs. HIV(−); Mann-Whitney U test. (2) Comparison between HIV(+) CD4<200, HIV(+)>200, and HIV(−); Kruskal-Wallis test.
| Patient Group |
|||||||
|---|---|---|---|---|---|---|---|
| HIV(+) |
|||||||
| Sample | CD4<200 (cells/mm3) | CD4>200 (cells/mm3) | All HIV(+) | HIV(−) | P value | ||
| HVR-1 | (1) | (2) | |||||
| No. clones | T0 | 5.67 0.88 | 5.67 1.45 | 5.67 0.76 | 9.00 5.00 | 1.000 | 0.989 |
| T1 | 5.33 0.67 | 8.00 3.51 | 6.67 1.71 | 9.67 4.18 | 0.381 | 0.431 | |
| Sn | T0 | 0.5830 | 0.6220 | 0.6026 | 0.8330 | 0.548 | 0.304 |
| T1 | 0.1680 | 0.4461 | 0.3067 | 0.8230 | 0.95 | 0.126 | |
| ΔSn (btw.) | −0.4160 | −0.1760 | −0.2959 | −0.0090 | 0.714 | 0.733 | |
| d (nt) | T0 | 0.1225 | 0.0263 | 0.0744 | 0.0067 | 0.167 | 0.288 |
| T1 | 0.1332 | 0.0493 | 0.0912 | 0.0812 | 0.548 | 0.739 | |
| Between | 0.0180 | 0.0130 | 0.0152 | 0.1482 | 0.381 | 0.491 | |
| d (aa) | T0 | 0.2019 | 0.0676 | 0.1347 | 0.0068 | 0.381 | 0.651 |
| T1 | 0.2646 | 0.0881 | 0.1764 | 0.1972 | 0.262 | 0.413 | |
| Between | 0.2461 | 0.0365 | 0.1407 | 0.3024 | 0.714 | 0.587 | |
| dN/dS | T0 | 0.5702 | 1.4373 | 0.9170 | 1.2584 | 0.857 | 0.915 |
| T1 | 0.8493 | 0.7904 | 0.8297 | 1.9837 | 0.200 | 0.304 | |
| Between | −5.6136 | 3.1432 | −1.2350 | 2.3371 | 1.00 | 0.213 | |
| NS3 | |||||||
| No. clones | T0 | 10.67 0.33 | 11.00 0.58 | 10.83 0.31 | 10.00 0.58 | 0.216 | 0.99 |
| T1 | 11.67 0.88 | 10.67 0.33 | 11.17 0.48 | 16.00 3.46 | 0.291 | 0.476 | |
| Sn | T0 | 0.5200 | 0.8420 | 0.6812 | 0.6000 | 0.905 | 0.179 |
| T1 | 0.5760 | 0.5590 | 0.5675 | 0.6400 | 0.548 | 0.714 | |
| ΔSn (btw.) | 0.0560 | −0.2830 | −0.1138 | 0.0410 | 0.348 | 0.061 | |
| d-nt | T0 | 0.0044 | 0.0147 | 0.0096 | 0.0133 | 0.381 | 0.193 |
| T1 | 0.0051 | 0.0147 | 0.0096 | 0.0091 | 0.262 | 0.066 | |
| Between | 0.0034 | 0.0063 | 0.0048 | 0.0040 | 0.714 | 0.488 | |
| d-aa | T0 | 0.0041 | 0.0107 | 0.0074 | 0.0097 | 0.381 | 0.193 |
| T1 | 0.0057 | 0.0114 | 0.0086 | 0.0089 | 0.905 | 0.252 | |
| Between | 0 | 0.00065 | 0.0032 | 0.0021 | 0.714 | 0.039 | |
| dN/dS | T0 | 0.2949 | 0.1972 | 0.2461 | 0.1623 | 0.905 | 0.957 |
| T1 | 0.2832 | 0.4144 | 0.3488 | 0.2254 | 0.381 | 0.561 | |
| Between | 0.0911 | 0.3401 | 0.2156 | 0.3855 | 0.548 | 0.298 | |
NOTE: Change in HCV sequence complexity (ΔSn) was calculated as ΔSn = Sn(T1)−Sn(T0). Genetic distances and substitution frequencies were determined both in every given sample (within-sample) and between samples of the same patient at the two different time points (net distance between samples, defined as the net change in intrasubject distances between baseline and follow-up groups of HCV clones dA=(dXY−(dX−dY)/2), were dXY is the average distance between groups of clones from baseline (X) and follow-up samples (Y), and dX and dY are the mean within-group distances).
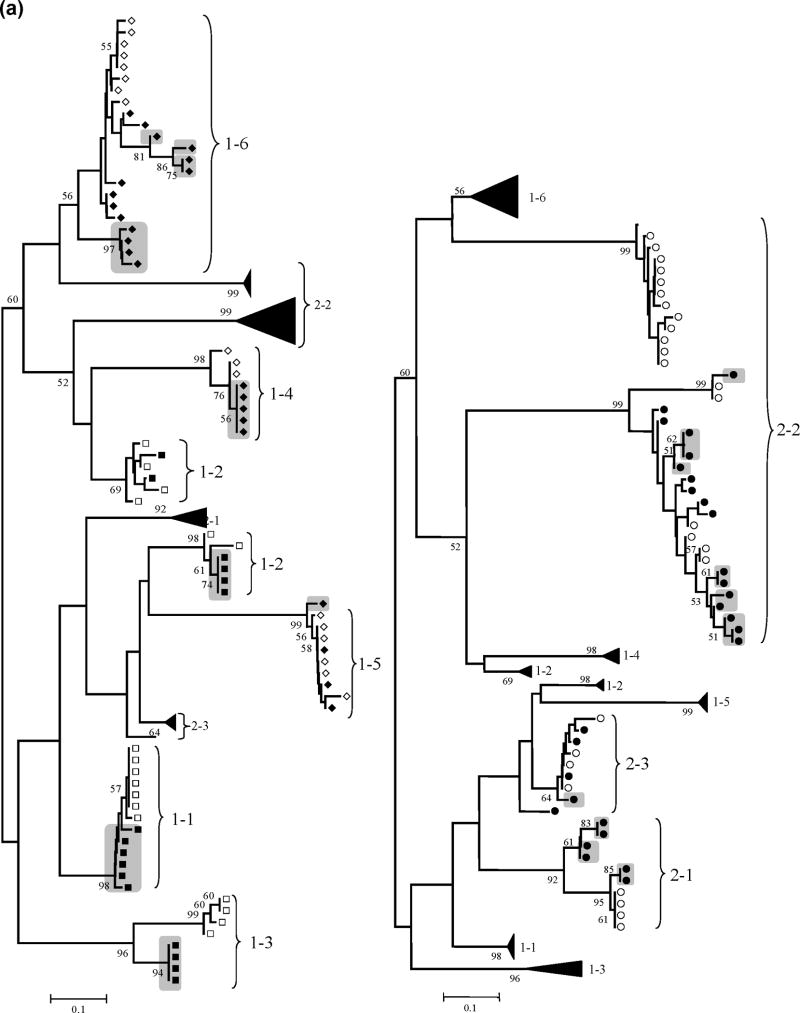
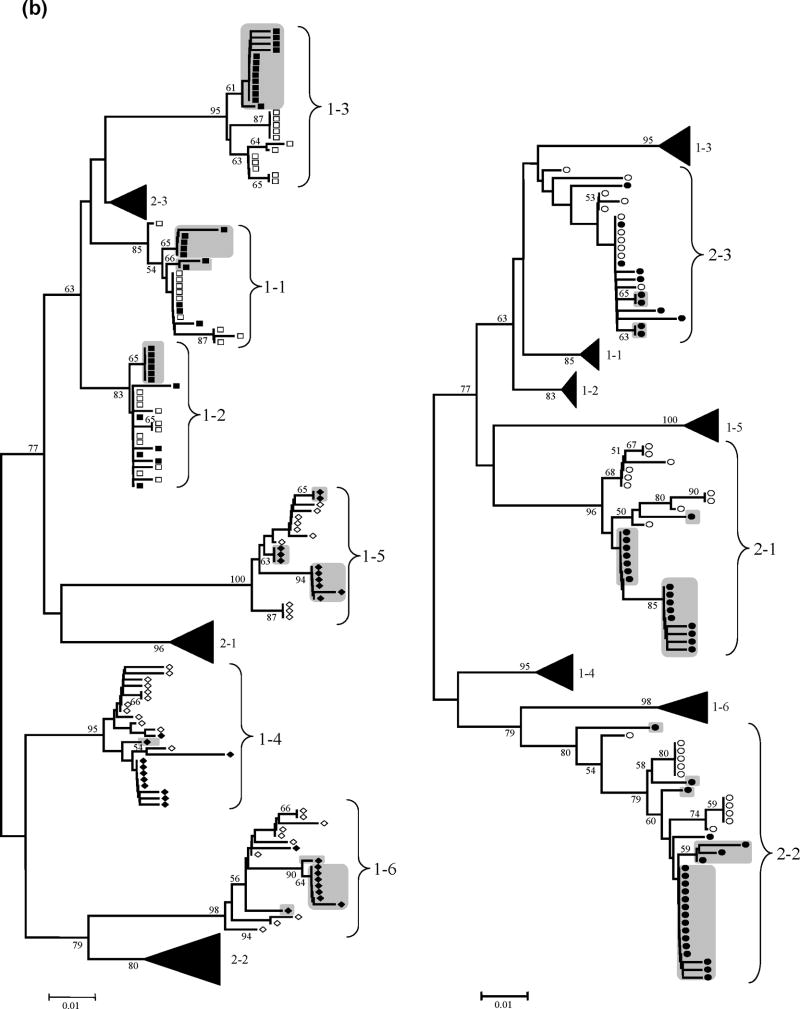
Figure 1 shows the phylogenetic trees for HVR-1 and NS3, reconstructed with the nucleotide sequences from all HCV clones by means of the neighbour-joining method with the MEGA program with bootstrap resampling (1,000 replicates). Sequences from the same individuals clustered together, indicating the absence of PCR cross-contamination. There were two exceptions: HVR-1 sequences from patients 1–2 and 2-2 were divided in two separate independent clusters, but they did not overlap with HCV sequences isolated from other individuals, suggesting mixed infection with different HCV envelope variants rather than PCR contamination. Overall, there was a trend for a more homogeneous population of HCV variants in HIV-HCV coinfected patients: less new variants appeared with time and dominated the viral population (means 1,5 vs. 3,3 in the HVR-1 and 1,8 vs. 3,3 in NS3, coinfected vs. HCV monoinfection; P=NS), especially in those coinfected patients with CD4 cell counts <200 cells/mm3 (means 1,0 vs. 2,0 in the HVR-1 and 1,3 vs. 2,3 in NS3, CD4<200 cells/mm3 vs. CD>200 cells/mm3; P=NS). Remarkably, HVR-1 variants presents at baseline were completely replaced after one year follow-up in the three coinfected patients with CD4 cell counts <200 cells/mm3 (Figure 1).
Figure 1. HCV HVR-1 and NS3 phylogenetic analyses.
Panel (a): Neighbour-Joining phylogenetic tree obtained with all HCV HVR-1 sequences from the study subjects at the two time points. (Left) Viral isolates from patient group 1 (HIV-HCV coinfected). Branches corresponding to isolates from patient group 2 are shown collapsed for clarity. (Right) Viral isolates from patient group 2 (HIV negative). Branches corresponding to isolates from patient group 1 are shown collapsed for clarity. Panel (b): Neighbour-Joining phylogenetic tree obtained with all HCV NS3 sequences from the study subjects at the two time points. (Left) Viral isolates from patient group 1 (HIV-HCV coinfected). Branches corresponding to isolates from patient group 2 are shown collapsed for clarity. (Right) Viral isolates from patient group 2 (HIV negative). Branches corresponding to isolates from patient group 1 are shown collapsed for clarity.
NOTE: Symbols: empty squares: HCV-HIV CD4<200 T0; filled squares: HCV-HIV CD4<200 T1; empty diamonds: HCV-HIV CD4>200 T0; filled diamonds: HCV-HIV CD4>200 T1; empty circles: HCV monoinfection T0; filled circles: HCV monoinfection T1; grey boxes: newly emerged variants after one-year follow-up, with bootstrap values >50 (indicated in the corresponding branches). HCV variants with bootstrap support >50% were considered divergent because the phylogenetic signal in the HCV genomic regions sequenced was relatively low.
The main limitation of the current study (small sample size) hampers our ability to demonstrate differences that reach statistical significance. However, for most research groups clonal analysis of large patient cohorts can be cumbersome. In a recent report, Suhuart et al. faced a similar problem, and analysed only 57 clones in only 6 out of 69 coinfected patients from their study based in heteroduplex assays (Shuhart et al., 2006). In concordance with our data, they found immune selective pressure to HCV (HVR-1 dN/dS > 1) in only one out of the three coinfected patients naïve for HAART. However, CD4 counts that study cohort were higher than 300 cells/mm3, and data on HCV quasispecies in patients with low CD4 counts, such as those included in our study, is still scarce. Our results should be interpreted with caution because of statistical limitations: the data are not really consistent across time points in all cases, which may be a function of the small sample size and sampling variability. Nevertheless, our observations complement other data suggesting that HIV-HCV coinfection is associated with a differential evolution of HCV (Babik and Holodniy, 2003; Blackard et al., 2004; Mao et al., 2001; Qin et al., 2005; Sherman et al., 1996). We analysed 340 HCV clones sequenced bidirectionally (total of 680 sequences), but sequencing a more extensive number of clones would also reduce the chance for sampling variability. Besides, it is possible that more than two time points will be needed to achieve a more accurate estimate of quasispecies dynamics over time. Unfortunately, extension of our study to a larger number of patients, HCV clones, or time points was not feasible. Several explanations can be given to the trend for a reduced HCV complexity and evolution with time in HIV coinfected patients. The low CD4 counts per se could drive HCV evolution in this setting, maybe by an alteration of CD4 help to HCV-specific CD8 cells, or to B cells. Alternatively, a differential selection of HCV quasispecies populations present in other compartments, such as PBMC or the liver, (Ducoulombier et al., 2004; Roque-Afonso et al., 2005) may give raise to a differential viral evolution in HIV-HCV coinfected patients. Whether this differential evolution of HCV may contribute to, or be responsible for, the accelerated liver fibrosis progression seen in coinfected individuals still remains to be determined.
Acknowledgments
This work was supported in part by the National Institute of Drug Abuse (DA RO1 DA13737) (T.W.), the Robert Wood Johnson Foundation (L.D.) and the Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Spanish Ministry of Health (proj. 05-0981 to M.B.; and CP04/0020 to F.X.L.). F.X.L. holds a P.I. position supported by the Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Spanish Ministry of Health.
The authors are indebted to Dr. Fernando González-Candelas, Vicente Sentandreu (Evolutionary Genetics Unit, Institut Cavanilles de Biodiversitat, University of Valencia), and Satish K. Pillai, Ph.D (UCSF), for their critical review of the manuscript and helpful discussions and suggestions. F.X.L. is very thankful to Dr. Andrés González-Molina and Dr. José M. Ribera (Hospital Universitari La Fe and Research Centre) for their continuing help and support.
List of Abbreviations
- HCV
Hepatitis C Virus
- HIV
human immunodeficiency virus-1
- HVR-1
hypervariable region-1
- NS3
non-structural protein-3
- RT-PCR
reverse-transcription polymerase chain reaction
- IVDU
intravenous drug users
- dS
genetic distance (synonymous substitutions)
- dN
genetic distances (non-synonymous substitutions)
- S
Shannon entropy
- Sn
normalised Shannon entropy
Footnotes
Previous abstract disclosure: Some preliminary data of this work was presented in abstract form in the 50th Annual Meeting of the American Association for the Study of Liver Diseases. Dallas, TX. November 1999.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babik JM, Holodniy M. Impact of highly active antiretroviral therapy and immunologic status on hepatitis C virus quasispecies diversity in human immunodeficiency virus/hepatitis C virus-coinfected patients. J Virol. 2003;77(3):1940–50. doi: 10.1128/JVI.77.3.1940-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackard JT, Yang Y, Bordoni P, Sherman KE, Chung RT. Hepatitis C virus (HCV) diversity in HIV-HCV-coinfected subjects initiating highly active antiretroviral therapy. J Infect Dis. 2004;189(8):1472–81. doi: 10.1086/382959. Epub 2004 Mar 29. [DOI] [PubMed] [Google Scholar]
- Chazouilleres O, Kim M, Combs C, Ferrell L, Bacchetti P, Roberts J, Ascher NL, Neuwald P, Wilber J, Urdea M, et al. Quantitation of hepatitis C virus RNA in liver transplant recipients. Gastroenterology. 1994;106(4):994–9. doi: 10.1016/0016-5085(94)90759-5. [DOI] [PubMed] [Google Scholar]
- Devereux H, Brown D, Dusheiko G, Emery V, Lee C. Long-term evolution of the 5′UTR and a region of NS4 containing a CTL epitope of hepatitis C virus in two haemophilic patients. J Gen Virol. 1997;78(3):583–590. doi: 10.1099/0022-1317-78-3-583. [DOI] [PubMed] [Google Scholar]
- Ducoulombier D, Roque-Afonso AM, Di Liberto G, Penin F, Kara R, Richard Y, Dussaix E, Feray C. Frequent compartmentalization of hepatitis C virus variants in circulating B cells and monocytes. Hepatology. 2004;39(3):817–25. doi: 10.1002/hep.20087. [DOI] [PubMed] [Google Scholar]
- Garcia-Samaniego J, Soriano V, Castilla J, Bravo R, Moreno A, Carbo J, Iniguez A, Gonzalez J, Munoz F. Influence of hepatitis C virus genotypes and HIV infection on histological severity of chronic hepatitis C. The Hepatitis/HIV Spanish Study Group. Am J Gastroenterol. 1997;92(7):1130–4. [PubMed] [Google Scholar]
- Goedert JJ, Hatzakis A, Sherman KE, Eyster ME. Lack of association of hepatitis C virus load and genotype with risk of end-stage liver disease in patients with human immunodeficiency virus coinfection. J Infect Dis. 2001;184(9):1202–5. doi: 10.1086/323665. Epub 2001 Sep 13. [DOI] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17(12):1244–5. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lauer GM, Nguyen TN, Day CL, Robbins GK, Flynn T, McGowan K, Rosenberg ES, Lucas M, Klenerman P, Chung RT, Walker BD. Human immunodeficiency virus type 1-hepatitis C virus coinfection: intraindividual comparison of cellular immune responses against two persistent viruses. J Virol. 2002;76(6):2817–26. doi: 10.1128/JVI.76.6.2817-2826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Labrador FX, He XS, Berenguer M, Cheung RC, Gonzalez-Candelas F, Wright TL, Greenberg HB. Genetic variability of hepatitis C virus non-structural protein 3 and virus-specific CD8+ response in patients with chronic hepatitis C. J Med Virol. 2004;72(4):575–85. doi: 10.1002/jmv.20036. [DOI] [PubMed] [Google Scholar]
- Mao Q, Ray SC, Laeyendecker O, Ticehurst JR, Strathdee SA, Vlahov D, Thomas DL. Human immunodeficiency virus seroconversion and evolution of the hepatitis C virus quasispecies. J Virol. 2001;75(7):3259–67. doi: 10.1128/JVI.75.7.3259-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinot-Peignoux M, Boyer N, Le Breton V, Le Guludec G, Castelnau C, Akremi R, Marcellin P. A new step toward standardization of serum hepatitis C virus-RNA quantification in patients with chronic hepatitis C. Hepatology. 2000;31(3):726–9. doi: 10.1002/hep.510310324. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Puoti M, Bonacini M, Spinetti A, Putzolu V, Govindarajan S, Zaltron S, Favret M, Callea F, Gargiulo F, Donato F, Carosi G. Liver fibrosis progression is related to CD4 cell depletion in patients coinfected with hepatitis C virus and human immunodeficiency virus. J Infect Dis. 2001;183(1):134–7. doi: 10.1086/317644. Epub 2000 Nov 16. [DOI] [PubMed] [Google Scholar]
- Qin H, Shire NJ, Keenan ED, Rouster SD, Eyster ME, Goedert JJ, Koziel MJ, Sherman KE the Multicenter Hemophilia Cohort Study Group. HCV quasispecies evolution: association with progression to end-stage liver disease in hemophiliacs infected with HCV or HCV/HIV. Blood. 2005;105(2):533–541. doi: 10.1182/blood-2004-04-1452. [DOI] [PubMed] [Google Scholar]
- Roque-Afonso AM, Ducoulombier D, Di Liberto G, Kara R, Gigou M, Dussaix E, Samuel D, Feray C. Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells. J Virol. 2005;79(10):6349–57. doi: 10.1128/JVI.79.10.6349-6357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque-Afonso AM, Robain M, Simoneau D, Rodriguez-Mathieu P, Gigou M, Meyer L, Dussaix E. Influence of CD4 cell counts on the genetic heterogeneity of hepatitis C virus in patients coinfected with human immunodeficiency virus. J Infect Dis. 2002;185(6):728–33. doi: 10.1086/339297. [DOI] [PubMed] [Google Scholar]
- Seibert SA, Howell CY, Hughes MK, Hughes AL. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1) Mol Biol Evol. 1995;12(5):803–13. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- Sherman KE, Andreatta C, O’Brien J, Gutierrez A, Harris R. Hepatitis C in human immunodeficiency virus-coinfected patients: increased variability in the hypervariable envelope coding domain. Hepatology. 1996;23(4):688–94. doi: 10.1002/hep.510230405. [DOI] [PubMed] [Google Scholar]
- Shuhart MC, Sullivan DG, Bekele K, Harrington RD, Kitahata MM, Mathisen TL, Thomassen LV, Emerson SS, Gretch DR. HIV Infection and Antiretroviral Therapy: Effect on Hepatitis C Virus Quasispecies Variability. J Infect Dis. 2006;193(9):1211–1218. doi: 10.1086/502974. [DOI] [PubMed] [Google Scholar]
- Soto B, Sanchez-Quijano A, Rodrigo L, del Olmo JA, Garcia-Bengoechea M, Hernandez-Quero J, Rey C, Abad MA, Rodriguez M, Sales Gilabert M, Gonzalez F, Miron P, Caruz A, Relimpio F, Torronteras R, Leal M, Lissen E. Human immunodeficiency virus infection modifies the natural history of chronic parenterally-acquired hepatitis C with an unusually rapid progression to cirrhosis. J Hepatol. 1997;26(1):1–5. doi: 10.1016/s0168-8278(97)80001-3. [DOI] [PubMed] [Google Scholar]
- Thomas DL. Hepatitis C and human immunodeficiency virus infection. Hepatology. 2002;36(5 Suppl 1):S201–9. doi: 10.1053/jhep.2002.36380. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Fukuda Y, Koyama Y, Takamatsu J, Saito H, Hayakawa T. Effect of immunosuppression on composition of quasispecies population of hepatitis C virus in patients with chronic hepatitis C coinfected with human immunodeficiency virus. J Hepatol. 1997;26(5):975–82. doi: 10.1016/s0168-8278(97)80105-5. [DOI] [PubMed] [Google Scholar]
- Weiner AJ, Brauer MJ, Rosenblatt J, Richman KH, Tung J, Crawford K, Bonino F, Saracco G, Choo QL, Houghton M, et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180(2):842–8. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- Wolinsky SM, Korber BT, Neumann AU, Daniels M, Kunstman KJ, Whetsell AJ, Furtado MR, Cao Y, Ho DD, Safrit JT. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272(5261):537–42. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]