Abstract
RNA editing is a widespread post-transcriptional molecular phenomenon that can increase proteomic diversity, by modifying the sequence of completely or partially non-functional primary transcripts, through a variety of mechanistically and evolutionarily unrelated pathways. Editing by base substitution has been investigated in both animals and plants. However, conventional strategies based on directed Sanger sequencing are time-consuming and effectively preclude genome wide identification of RNA editing and assessment of partial and tissue-specific editing sites. In contrast, the high-throughput RNA-Seq approach allows the generation of a comprehensive landscape of RNA editing at the genome level. Short reads from Solexa/Illumina GA and ABI SOLiD platforms have been used to investigate the editing pattern in mitochondria of Vitis vinifera providing significant support for 401 C-to-U conversions in coding regions and an additional 44 modifications in non-coding RNAs. Moreover, 76% of all C-to-U conversions in coding genes represent partial RNA editing events and 28% of them were shown to be significantly tissue specific. Solexa/Illumina and SOLiD platforms showed different characteristics with respect to the specific issue of large-scale editing analysis, and the combined approach presented here reduces the false positive rate of discovery of editing events.
INTRODUCTION
Next-generation sequencing platforms (Solexa/Illumina GA, ABI SOLiD and Roche 454) are radically changing the field of genomics (1,2), allowing both re-sequencing and de novo sequencing of whole genomes (3) with notable reductions in time and cost with respect to conventional approaches. These technologies are now routinely applied to a variety of functional genomics problems, including, but not restricted to, global identification of genomic rearrangements, investigation of epigenetic modifications and single nucleotide polymorphism (SNP) discovery (4). RNA-Seq—the application of next generation sequencing to entire transcriptomes—can provide accurate gene expression profiles for coding and non-coding RNAs (5) greatly facilitating genome annotation (6).
RNA editing is a widespread post-transcriptional molecular phenomenon that can increase proteomic diversity (7) by modifying the sequence of completely or partially non-functional primary transcripts (8), through a variety of mechanistically and evolutionarily unrelated pathways. ‘Substitution’ editing by simple base modification is the most frequent type of editing and is seen both in plant organelles and in the nucleus of higher eukaryotes (8–11) as well as in sequences of viral origin (12). In land plant organelles, RNA editing consists almost exclusively of C-to-U substitutions (rarely reverse U-to-C conversions) mostly at first or second positions of codons (9)—typically leading to conservative amino-acid changes and increasing similarity to non-plant homologs. Some plant organellar RNA editing events create translation initiation or termination codons while several known editing events in tRNA or introns improve the stability of functionally relevant secondary structure motifs (13,14). The systematic identification of RNA editing events thus represents an important objective that could significantly improve our understanding of organellar and nuclear molecular genetics. Moreover, the alteration of the RNA editing pattern in plant mitochondria can lead to male sterility, also known as the CMS phenotype (15).
Classically, RNA editing events were identified experimentally by comparing cloned cDNA sequences with their corresponding genomic templates (16). This procedure allows the study of a relatively small number of sequences and does not take into account potential cloning artefacts. More recently, large-scale identification of RNA editing sites has been performed using collections of expressed sequence tags (ESTs) and full-length cDNAs mainly stored in public databases (17,18). However, the generally low quality of EST sequences, and the incomplete nature of some editing events markedly hampers such approaches. Indeed, C-to-U editing has been explored at the whole mitochondrial (mt) genome level in only four higher plants, Arabidopsis thaliana (19), Brassica napus (16), Beta vulgaris (20) and Oryza sativa (21). High-throughput transcriptome sequencing by next-generation technologies provides deep coverage per reference nucleotide and indications of base call qualities and may overcome existing limitations and improve the large-scale detection of RNA editing sites.
Recently, human RNA editing sites have been identified using massively parallel target capture and DNA sequencing employing computationally predicted A-to-I sites (22). In another approach, Life Science (Roche) 454 Amplicon Sequencing technology has been used to determine global expression of known RNA editing sites during brain development (23).
In the present work, focused on the de novo detection of C-to-U editing modifications occurring in coding and non-coding genes of the Vitis vinifera mitochondrial genome, we also present a novel strategy to investigate the landscape of RNA editing at the genome level through RNA-Seq. This strategy involves the use of millions of short reads generated by Solexa/Illumina GA and ABI SOLiD systems. Over 6 000 000 short reads (from both platforms) mapping uniquely onto the grapevine mitochondrial genome provided significant support for 401 C-to-U alterations in coding regions. Sixty percent of the identified events occurred at second codon positions. Forty-four additional editing modifications (38 C-to-U and 6 U-to-C) were identified in tRNAs and group II introns, supporting the notion of pervasive RNA editing in grape mitochondria. Interestingly, 76% out of all C-to-U conversions in coding genes represent partial RNA editing, and 28% of them were shown to be significantly tissue specific.
In this study, we prove the effectiveness of RNA-Seq data for the the global identification of RNA editing sites and the relative performances of the Solexa/Illumina GA and ABI SOLiD systems to reliably identify editing sites. The computational strategy presented here can be applied to the discovery of substitution editing events of any type in both nuclear and organellar compartments of different organisms.
MATERIALS AND METHODS
Assembly and annotation of the PN40024 mitochondrial genome
Ad-hoc perl scripts making use of the NCBI Blast URL API were used to automate similarity searches of the PN40024 genome sequencing project trace archive with overlapping 10-kb windows of the Pinot Noir ENTAV115 mitochondrial genome [GenBank:NC_012119]. Only traces showing greater than 95% identity to the ENTAV115 genome were retained. The ‘query_tracedb’ script provided by NCBI was used to recover sequences and associated quality scores (16 789 putative mitochondrial sequences of which 13 682 were identified as mate pairs). The average read length was 785 bases, implying a hypothetical redundancy of greater than 20 times. The software PCAP (24) was used, without reference to the ENTAV115 sequence, to assemble four contigs of 339 264, 132 252, 202 123 and 76 068 nt. Our assembly represented 96.37% of the reference sequence, with which it showed 99.92% identity. Similarity searches using the ENTAV115 annotation allowed the identification of all of the genes of mitochondrial origin proposed by Goremykin et al. (25). In addition, the mitochondrial origin of each coding gene was confirmed comparing grape ORFs to genomic and unedited mitochondrial genes downloaded from the specialized REDIdb database (http://biologia.unical.it/py_script/search.html) (26).
Short read sequencing and mapping
In total, 205 435 765 short reads were obtained by sequencing cDNA obtained from four tissue samples with the Solexa/Illumina technology: leaf (11 lanes), root (9 lanes), callus (9 lanes), stem (14 lanes) (6). The mRNA molecules were purified from total RNA extractions and fragmented before cDNA synthesis. The single-end reads obtained were 33-nt long, except for five lanes in the callus sample, where the reads were 35-nt long. Total RNA from PN40024 grape cultivar was sequenced with the SOLiD-2 technology, resulting in 139 467 080 short reads from leaf and 188 742 647 short reads from root. All SOLiD short reads were 35-nt long. For the construction of the SOLiD libraries we had early access to the Applied Biosystems Whole Transcriptome Shotgun procedure. Poly(A)+ RNA was enzymatically fragmented and directionally ligated to adaptors, essentially as indicated in the AMBION Small RNA Expression Kit (SREK).
Solexa/Illumina and SOLiD short tags, pooled from all tissues, were mapped to the assembled V. vinifera mitochondrial genome using version 0.5 of the PASS software (27) with a seed length of 12, a minimum identity of 90% and a minimum alignment length per read of 30 nt. Similar to a BLAST approach, PASS seed sequences (called long word anchors) are extended on the flanking regions using DNA words of predefined length (typically 6 or 7 bases) for which the alignment scores are pre-computed according to Needlelman–Wunch. Significant matches are then refined to improve the global alignment quality. In particular, we used a pre-computed scoring matrix (PST) based on DNA words of 7 bases long (W7M1m0G0X0.pst, downloadable from the PASS web site: http://pass.cribi.unipd.it/), filtering hits having more than 11 discrepancies. Moreover, we filtered out Solexa/Illumina and SOLiD short tags containing more than 5 bases with a quality threshold less than 15. In case of Solexa/Illumina reads, we used the -gff option to print out mapping results in the standard GFF (version 3) format (see http://www.sequenceontology.org/gff3.shtml for more details about this format).
SOLiD reads, derived from a ligation-mediated sequencing strategy, are not collected as nucleotide sequences, but instead are recorded in color space where each color provides information about two adjacent bases but their identification is not provided (a complete description of 2-base color codes can be found at the ABI web site http://www3.appliedbiosystems.com/AB_Home/). For this reason, SOLiD data need a distinct processing method, including an accurate decoding step in which color reads are converted to sequence reads. However, decoding should not be performed before mapping because sequencing errors may affect the translation to base space leading to significant inaccuracies. Therefore, we mapped SOLiD reads to the known reference within color space, again using PASS, allowing at most four color mismatches using the option -SOLiDCS. Next, resulting query-to-reference alignments in color space were parsed by custom python scripts in order to correct sequencing errors and identify isolated color changes corresponding to valid base space mismatches (main scripts are available upon request). Moreover, we performed a further modification—using SOLiD quality scores per single base to reliably call individual nucleotides. For the SOLiD technology, a quality score is assigned to each color (corresponding to a pair of adjacent nucleotides) and each nucleotide (except the first and the last) is read twice as it is included in two adjacent colors. Consequently, a per-base quality score can be reasonably assigned calculating the average quality between two adjacent colors (i.e. two overlapping dinucleotides). If two neighboring colors have high quality scores, the nucleotide in common between them has a high quality score. If two adjacent colors have very different quality scores we call the base in common between them according to a defined quality threshold. The threshold, set at 15 for both Solexa/Illumina and SOLiD reads was generated considering the distribution of detected quality scores per base and considering the fact that SOLiD quality values are also calculated using a phred-like scale.
SOLiD mapping results, in addition to potential mismatches, were finally saved in GFF format. Solexa/Illumina and SOLiD mapping data in GFF format are available upon request.
Computational identification of RNA editing sites
Solexa/Illumina and SOLiD mapping results in GFF format were used to identify C-to-U changes due to RNA editing in the grape mitochondrial genome of the cultivar PN40024 by means of ad hoc custom python scripts.
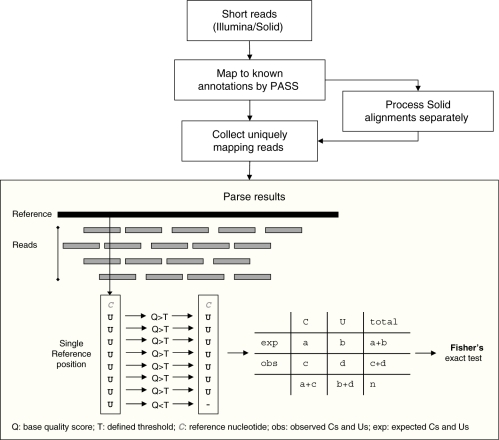
The main script, in particular, takes as input a GFF file, the reference sequence of the grape mitochondrial genome in FASTA format and a textual file containing protein-coding annotations. It collects all uniquely mapping reads (with at most two mismatches and no indels) falling in annotated genes and for each reference position calls the corresponding read nucleotide if its quality score is above the fixed threshold of 15. Finally, for each reference position, the script calculates the frequency of the modified nucleotide (if any) over the total recorded signal (sum of modified and not modified nucleotides) (Figure 1). Results obtained from Solexa/Illumina and SOLiD data are available as Supplementary Data in tab-formatted text files.
Figure 1.
Methodology overview. Graphical overview of the computational methodology used to detect RNA editing sites by short sequencing reads of next generation platforms.
RNA editing sites due to C-to-U changes were detected separately for each platform and tissue. Rates of sequencing errors were estimated for each sample as the total frequency of non-C↔U substitutions. Among the potential editing sites, corresponding to sites where a genomic C was aligned to one or more U from RNA-Seq data, statistically significant editing sites were determined by applying the Fisher’s exact test by comparing the observed and expected C and U occurrences in the aligned reads. A confidence level of 0.05 (also with FDR or Bonferroni correction) was used as cut-off.
A putative editing site is classified as ‘conserved’ if one or more homologous sites in other plants are experimentally known to be edited or if a fully conserved U is observed in all homologous sites, according to the data collected in the REDIdb database (26).
RNA editing sites in non-coding grapevine genes and group II introns were detected according to the same computational strategy. These results are also available as Supplementary Data.
Statistically significant edited sites have been classified fully or partially edited depending on if the observed fraction of RNA-Seq aligned U was above or below 90%.
All statistically significant RNA editing events have been submitted to the specialized REDIdb database (http://biologia.unical.it/py_script/search.html) (26) and can be freely consulted in their gene context under the accessions EDI_000000804–EDI_000000840. Finally, data providing additional editing information per each coding gene, tissue and platform, including short read coverage per gene and single reference position, are supplied as Supplementary Data.
Characterization of grape mitochondrial editing sites
All statistics to characterize detected RNA editing sites in grape mitochondrial protein-coding genes, including affected codon positions and amino acid changes, were calculated by custom python scripts. The effect of RNA editing alterations in tRNA genes was evaluated according to secondary structure predictions by the tRNA-Scan web server (http://lowelab.ucsc.edu/tRNAscan-SE/) (28), whereas the impact of C-to-U modifications in the domain V of the group II intron nad7i4 was manually checked.
Tissue-specific editing sites were identified by means of a chi-square statistical test comparing for each edited position the observed and expected distributions of Cs and Us in all available tissues. Three degrees of freedom were used for Solexa/Illumina data (four tissues) and one for SOLiD reads (two tissues). Significant sites were detected at 0.05 and 0.01 confidence levels, corrected for false discovery rate according to Benjamini and Hochberg (29). The Bonferroni correction, while highly conservative, was also used.
Nucleotide sequences (40-bp long) around RNA editing sites detected in protein-coding genes were examined in terms of relative entropy using windows of 1, 2 or 3 bases, according to the computational methodology described by Mulligan et al. (30). Sequence logos were generated by the WebLogo program (version 3) (31).
Domain searches in edited and unedited grapevine mitochondrial genes were performed through the Pfam webserver (http://pfam.sanger.ac.uk/search) using 1.0e–05 as E-value cut-off (32).
RNA editing in A. thaliana mitochondria
To detect C-to-U changes in mitochondria of A. thaliana, we used 63 850 661 Solexa/Illumina short reads (distributed over five runs) from floral tissue of Col-0 ecotype downloaded from NCBI Short Read Archive under the accession SRX002554. All these reads, each of 50 nt in length, were mapped onto the reference Arabidopsis mitochondrial genome [GenBank:NC_001284] using PASS with settings as described above. Potential RNA editing sites were identified according to the computational strategy previously explained. Known Arabidopsis C-to-U substitutions were downloaded from REDIdb database and used to identify new editing sites.
RESULTS
Grapevine mitochondrial genome assembly and annotation
The complete mitochondrial genome sequence of the Pinot Noir, clone ENTAV115 was recently presented by Goremykin et al. (25). The genome is, at over 773 kb in length, the largest sequenced higher plant mitochondrial genome. Notably, Goremykin et al. (25) estimate that >42% of the Vitis plastid genome has been incorporated into the mitochondrial sequence, and the high similarity of such sequences to their plastidic forbearers (25) indicates that such transfers have occurred recently. While plant mitochondrial-coding regions tend to show extremely high levels of conservation (33), for the purposes of the current study, we wished to compare transcriptome reads to genomic templates derived from identical cultivars (PN40024). Accordingly, we used overlapping windows along the Goremykin et al. sequence (25) to perform similarity searches against the PN40024 genome sequencing project trace archive (Sanger sequencing reads) (34). Assembly of 16 789 putatively mitochondrial reads yielded four contigs covering 96.37% of the Vitis mitochondrial template. Interestingly, the positions where assembly of contigs was not possible consistently corresponded to regions containing large plastid-like insertions in the Goremykin et al. assembly (25), suggesting either that some such insertions occurred after the divergence of the two cultivars in question or that some such regions have undergone elimination or rearrangement after the divergence of the two clones. Unsurprisingly, similarity searches allowed us to confidently identify all 37 mitochondrial genes (24 components of the respiratory chain and 13 ribosomal proteins) previously annotated (25), in addition, we were able to identify 13 tRNA genes of mitochondrial origin and a number of potentially functional tRNAs of plastidic origin. Protein-coding regions were almost identical to those previously identified by Goremykin et al. (25). Indeed within the 37 protein-coding genes of mitochondrial origin studied in the current work, only a single potential synonymous polymorphism was identified between the two clones. A detailed description of patterns of variability between non-coding portions of grapevine mitochondrial genomes will be presented elsewhere. The PN40024 mitochondrial genome contigs are available through Genbank under accessions GQ220323, GQ220324, GQ220325 and GQ220326.
Computational strategy to detect RNA editing sites by short sequencing reads
The strategy proposed here is conceptually simple, computationally tractable, and suitable for Solexa/Illumina and SOLiD short sequencing RNA reads. In the first part of our approach, depicted in Figure 1, we mapped and aligned short reads to the reference genome using the PASS software (27) (see ‘Materials and Methods’ section for more details). To reduce inconsistent results, we retained only alignments of at least 30 nt in length with a minimum identity of 90% and no indels. In addition, problematic reads were discarded a priori by setting PASS (27) quality parameters as described in ‘Materials and Methods’ section. We recovered only reads mapping once to the reference sequence with at most two mismatches. For each reference position we collected all corresponding reads, scoring hits only if their corresponding quality scores were above a defined threshold (Figure 1). In this way, potential sequencing errors are minimized obtaining a high confidence set of bases per reference position.
RNA editing sites are finally detected by interrogating the reference position by position. A site is considered potentially edited if a C is observed in the reference genome and one or more U in the aligned reads at the same position. The Fisher’s exact test has been carried out, as described in ‘Material and Methods’ section, to assess the statistical significance of each potentially edited site. This statistical assessment was performed separately for every tissue and platform to account for tissue specificity and the different features of Solexa/Illumina and SOLiD systems. Indeed, Solexa/Illumina and SOLiD platforms show different behaviours in terms of base substitution pattern (see below for details) and coverage per base that may affect the identification of genuine editing sites increasing the false discovery rate.
Editing of grapevine mitochondrial RNAs is revealed by Solexa/Illumina and SOLiD RNA-seq
RNA editing in higher plant mitochondria (predominantly C-to-U conversions) represents one of the most investigated types of editing (9), although its molecular mechanism is yet largely unknown (11). Data stored in primary and specialized databases indicate that the mitochondrial genomes of A. thaliana, B. napus, B.vulgaris and O. sativa contain 441, 427, 357 and 491 C-to-U edited sites, respectively. We analyzed 205 million reads obtained by Solexa/Illumina technology from four different tissues (stem, root, callus and leaf) as well as 328 million reads produced by SOLiD technology from leaf and root tissues of the highly homozygous PN40024 clone.
We aligned Solexa/Illumina and SOLiD reads to grape PN40024 mitochondrial contigs, recovering 939 554 unique Solexa/Illumina alignments and 5 207 827 unique SOLiD alignments. The different fraction of uniquely aligned reads (0.45 and 1.59% for Solexa/Illumina and SOLiD, respectively) also reflect quite different coverage patterns, which seem much more biased for SOLiD (Supplementary Table S1). We noted that despite the much higher overall fold coverage of SOLiD (158×) than SOLEXA (35×) both platforms provided a similar percentage of covered nucleotides in the coding regions, 96.9 and 96.6%, respectively (see Supplementary Table S1). Furthermore, 16 out of 37 annotated mitochondrial coding genes were fully covered by Solexa/Illumina reads while only 11 were fully supported by SOLiD data (Supplementary Figures S1–S3). Looking at reads distribution along the reference sequence, we also noted local maxima in SOLiD reads in which several mitochondrial regions appeared deeply covered.
While the patterns of coverage seem to indicate a notable bias in the per-site distribution of the coverage depth across coding genes for the SOLiD data, a moderate, but highly significant (r = 0.25, P < 0.0001) correlation was observed between per base coverage by SOLiD and Solexa/Illumina sequencing for individual positions in the coding sequences of the 37 genes of mitochondrial ancestry—possibly due to a known dependence of recovery of fragmented cDNA (by gel elution) on GC content (35). However, distinct coverage patterns by these different sequencing strategies contribute to a substantially higher coverage when both technologies were combined—complete coverage of 25 genes out of the 37 and an overall coverage of 98.3% of all coding nucleotides (see an example in Figure 2 or extended images in Supplementary Figures S1–S3).
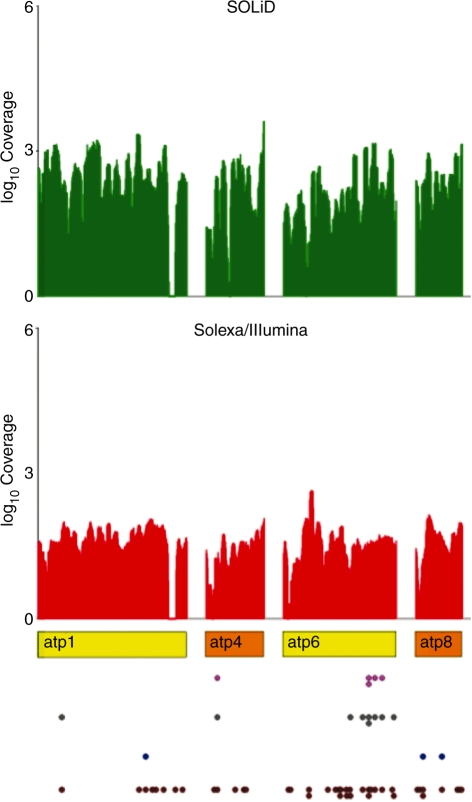
Figure 2.
Solexa/Illumina and SOLiD coverage for four mitochondrial atp genes. Plot showing the coverage depth for Solexa/Illumina and SOLiD reads in four mitochondrial genes coding for subunits of the atp synthase. Rectangles in colour indicate protein-coding genes (orange if the strand is forward and yellow if the strand is reverse). RNA editing sites are drawn as brown colored dots below each gene. Blue and grey dots designate the subset of tissue specific sites at 0.05 and 0.01 confidence levels, respectively. Sites passing the Bonferroni test at 1% confidence level are indicated in magenta. Coverage is reported in log10 scale.
Both Vitis mitochondrial assemblies harbor two identical copies of rps19, one upstream of the rps3 and rpl16 genes and another downstream of a pseudo atp1 gene. Experimental data suggest that the evolutionarily conserved cluster rps19, rps3 and rpl16 is transcribed as a polycistronic RNA in land plants (36). When only reads that map uniquely to the genome were considered, the rps19 gene was, unsurprisingly, not covered. When we allowed the use of reads mapping on at most two genome locations, we found eight C-to-U modifications in the rps19 coding region, three occurring at the third codon position and the remaining five in non-synonymous positions that were also conserved across different land plants, except for an event at position 260 that seems to be grapevine specific. We cannot, with confidence, establish if one or both the copies of rps19 are expressed, although the confirmed expression of rps3 and rpl16 genes suggest that at least the rps19 copy completing the canonical gene cluster should be transcribed.
In total we identified 401 significantly supported editing sites in grapevine mitochondrial coding regions with a 5% confidence level in the Fisher’s exact test. To evaluate the effectiveness of the statistical assessment we determined the percentage of conserved edited sites (see ‘Materials and Methods’ section) of putative editing sites (Supplementary Figure S4). Interestingly, >90% of significantly detected edited sites were conserved, supporting the reliability of the statistical test. Indeed, only a slight increase was observed with more stringent cut-offs (5% confidence level with FDR or Bonferroni correction). To be noted that a remarkable level of conservation was also observed for putative editing sites filtered out by the statistical test. It is highly likely that the read coverage at these positions is not deep enough to provide statistical support. Including all 314 additional putative edited sites with conserved homologous counterparts in other plants, more than 700 sites may be edited in the grapevine mitochondrion (P-values for all C residues falling in annotated coding genes are available in Supplementary Data).
All 401 significantly detected editing events were collected in the REDIdb database (26) under accessions EDI_000000804–EDI_000000840. Of these editing events 24.6% were supported by Solexa/Illumina reads and 75.4% were supported by SOLiD data.
A survey of mismatches identified by short reads
In addition to the C-to-U changes, marking editing events in the mitochondrial coding regions, we also analyzed other mismatch types (Table 1). The mismatch distribution, also used for carrying out the statistical tests (see ‘Materials and Methods’ section), resulted strikingly different between Solexa/Illumina and SOLiD data. In particular, G-to-U, C-to-A substitutions appeared overrepresented by Solexa/Illumina reads with respect to SOLiD data, likely reflecting typical miscalls of Solexa/Illumina reads (37). For the vast majority of G-to-U and C-to-A mismatches at positions covered by both technologies, SOLiD provided no evidence of variation between genomic and transcribed sequences. The established base call quality threshold (>15) likely reduced SOLiD and Solexa/Illumina false mismatches as we observed a slight overrepresentation of mismatches in reads where the corresponding base showed a relatively low quality score (Supplementary Figures S5 and S6). The lower frequencies of non-canonical mismatches recovered by SOLiD data (Table 1) suggest that this sequencing technology shows a higher overall accuracy. However, the combination of SOLiD and Solexa/Illumina data seems particularly suitable for the reliable detection of editing sites.
Table 1.
Base substitution frequencies detected by Solexa/Illumina, SOLiD and both technologies
| From | Into |
||||
|---|---|---|---|---|---|
| A | C | G | U | Any | |
| Solexa/Illumina | |||||
| A | – | 0.0078 | 0.0129 | 0.0037 | 0.0244 |
| C | 0.0177 | – | 0.0025 | 0.8768 | 0.8970 |
| G | 0.0187 | 0.0039 | – | 0.0273 | 0.0499 |
| U | 0.0057 | 0.0127 | 0.0102 | – | 0.0286 |
| Any | 0.0421 | 0.0244 | 0.0256 | 0.9078 | |
| SOLiD | |||||
| A | – | 0.0022 | 0.0112 | 0.0042 | 0.0176 |
| C | 0.0015 | – | 0.0017 | 0.9215 | 0.9247 |
| G | 0.0255 | 0.0029 | – | 0.0096 | 0.0380 |
| U | 0.0019 | 0.0151 | 0.0028 | – | 0.0198 |
| Any | 0.0289 | 0.0202 | 0.0157 | 0.9353 | |
| Both | |||||
| A | – | 0.0041 | 0.0118 | 0.0040 | 0.0199 |
| C | 0.0069 | – | 0.0020 | 0.9064 | 0.9064 |
| G | 0.0232 | 0.0032 | – | 0.0156 | 0.0420 |
| U | 0.0032 | 0.0143 | 0.0053 | – | 0.0228 |
| Any | 0.0333 | 0.0216 | 0.0191 | 0.9260 | |
A survey of nuclear sequences showing more than 95% identity with mitochondrial coding regions revealed, in almost all cases, a cytosine in the detected edited positions. Indeed, rather than resulting from retro-transcription of potentially edited mitochondrial transcripts, mitochondria-like sequences in the nuclear genome derive from mitochondrial genomic fragments. Interestingly, apart from editing sites, differences between mitochondrial genes and their corresponding nuclear pseudogenes were predominantly transitions to A and T in the nuclear compartment [consistent with the high AT content of non-coding regions of the Vitis nuclear genome (34)]. Thus, cross matching reads derived from background transcription of nuclear mitochondrial pseudogenes might also account for a proportion of observed G-to-A and A-to-G mismatches (results not shown).
Overall, we find no compelling evidence for editing events other than the canonical C-to-U.
Characterization of editing sites affecting coding genes in mitochondria of V. vinifera
The 401 C-to-U editing modifications detected in coding regions in Vitis mitochondria are unevenly distributed across different genes, ranging from 0.8% (rpl2) to 18.2% (rps19) of total cytosines (Supplementary Table S2) although no significant correlation was observed between sequencing fold-coverage and percentage of edited cytosines (data not shown). Our data also confirm a degree of species specificity of RNA editing. For example, the Vitis rps3 transcript is edited at 10 sites, whereas the homologs from B. vulgaris and Cycas revoluta are edited at 8 and 28 positions, respectively (16,36). In grapevine mitochondria, genes coding for subunits of complex I seem to be more edited than genes coding for other subunits. However, the editing extent for each gene of a given mitochondrial complex is quite variable (Supplementary Table S2 and Supplementary Figure S7). The cob gene, encoding the cytochrome b of complex III, is the most edited gene, whereas the sdh3, a member of the complex II, is the least edited gene (see Supplementary Table S2). Some variability in the extent of editing can be also observed among gene groups belonging to the same complex, with genes of Complex I showing the highest level of edited sites (6.5% of total C) and genes of Complex II showing the lowest level (4.2% of total C) (Supplementary Table S2 and Supplementary Figure S7).
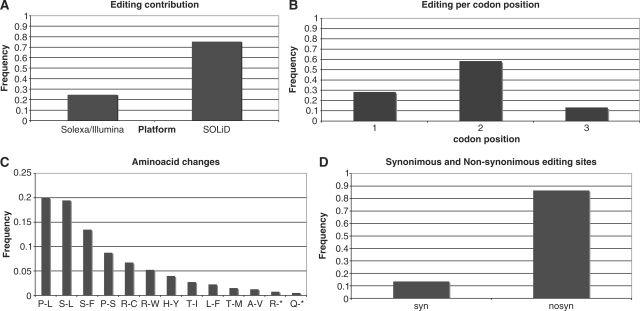
In total, 87% of the 401 editing modifications occurred at the first and second positions of codons, almost invariably resulting in replacement of the encoded amino acid (Figure 3). Indeed, only 1 out of 114 events affecting the first codon position resulted in synonymous changes. All non-synonymous editing conversions could modify the biochemical nature of the affected proteins. As observed in mitochondria of A. thaliana (19), the most frequent amino acid changes induced by RNA editing in grapevine were P-to-L (20.0%), S-to-L (19.4%) and S-to-F (13.5%) (Figure 3) increasing the proportion of hydrophobic amino acids and suggesting a real functional role for RNA editing through protein modifications in predominantly membrane-localized proteins. Additionally, S-to-L or S-to-F substitutions potentially increase the hydrophobicity of interface residues while P-to-L conversions occurring in secondary structures can contribute to protein functionality by avoiding defects in 3D structures (38).
Figure 3.
Principal statistics of detected RNA editing sites in V. vinifera mitochondria. (A) The contribution of each sequencing platform to editing detection; (B) distribution of C-to-U editing conversions across codon positions; (C) distribution of amino acids changes induced by detected RNA editing; (D) frequencies of synonymous and non-synonymous editing changes.
Besides the non-random distribution of editing with respect to codon positions, we observed a preference of RNA editing towards specific codons. In particular, the three most frequently edited codons were UCA, CCA and UCC, accounting for 32.7% of all edited codons. The only C-containing codons never affected by editing were GGC, AGC and UGC in which editing could only lead to synonymous substitutions. C-to-U variations at specific codons were uncorrelated with codon usage, according to the correlation factor proposed by Giegé and Brennicke (19) (the ratio between the frequency of edited codons and the analogous proportion in the total population of C-containing codons of all investigated grapevine mitochondrial mRNAs). RNA editing in grape mitochondria creates three start codons (for cox1, nad4L and rps10 genes), and generates the site of termination of translation in atp6, ccmFC and rps10 transcripts. In the rpl16 mRNA an additional editing event introducing a stop codon in frame with an upstream AUG was found. This suggests that the RPL16 protein is likely translated using a GTG codon just downstream of the edit-generated upstream ORF as initiator. Strikingly, this editing pattern, affecting the protein annotation, is highly conserved across mitochondria of land plants (39).
Although a strict consensus motif for sequences surrounding RNA editing sites has not been identified, bias towards pyrimidines at positions –2 and –1, and a bias towards purines at position +1 have been demonstrated (30). This behavior is also observed in the grapevine mitochondrial genome when the relative entropy in the 40 nt flanking edited and unedited cytidines was calculated. In particular, our data indicate that the relative entropy is extremely high in the immediate vicinity of the editing site (nt from –4 to +1), exceeding the 1% confidence interval calculated by 1000 iterations of random assignment of RNA editing sites. Interestingly, high relative entropy at the 5′-end of edited sites was also evident when it was calculated for 2- and 3-nt windows. Therefore, this region could be directly involved in editing site recognition, especially at position from –5 to –1 and from –18 to –14 as found in computational analyses conducted on four complete plant mitochondrial genomes by Mulligan et al. (30). The relative entropy for the 40-nt flanking grapevine editing sites is shown in the Supplementary Figure S8.
RNA editing in coding regions tends to increase cross-species conservation at the protein level and a correlation between amino acids modified by RNA editing and functional residues at protein structure has been shown (38). We performed domain searches of Pfam using either the protein conceptually translated from genomic or edited sequences (32). Interestingly, amino acid changes induced by RNA editing increased the scores of matches to individual Pfam domains from an average of 133.92 to 144.73.
Partial editing and tissue specificity of grape RNA editing sites
Twenty four percent of the 401 C-to-U conversions were classified as fully edited sites while 76% were considered partially edited sites—supporting the hypothesis that partial RNA editing is common in higher plant mitochondria (16,40). A proportion of partial editing might be due to transcripts where editing was not yet complete, while other partial events might derive from tissue-specific edits derived from mixed tissue samples (41).
Our Solexa/Illumina short sequencing reads were generated from total cellular RNA extracted from four different grapevine tissues: stem, leaf, root and callus; while SOLiD short reads were produced from leaf and root RNA (see ‘Materials and Methods’ section). Therefore, these data offered a unique opportunity to investigate the issue of RNA editing tissue specificity on a large scale. We compared the observed and expected distributions of Cs and Us in all available tissues by means of the chi-square test. 112 editing events were identified as significantly tissue specific at the 5% confidence interval corrected for false discovery rate, whereas 77 of them were selected as significant at 1% corrected confidence level. The Bonferroni correction were also applied at 1% confidence level resulting in a highly conservative estimate of 35 significant tissue specific editing sites (a list of tissue specific editing events is available in the Supplementary Table S4; see also Supplementary Figure S1).
Our findings indicate that tissue specificity accounts for a fraction of the observed partial RNA editing. Tissue specific editing might be required to modulate protein functionality in response to cell-type specific requirements. The high depth of coverage afforded by the SOLiD data resulted in the recovery of the majority of the significantly tissue specific edits by this technology. In summary, using the information from both sequencing technologies we discovered that 71% of all tissue-specific C-to-U changes occurred in leaf, whereas only a small fraction (0.4%) occurred in stem. Tissue specific editing events occurring in root and callus, instead, constituted 21 and 7.6%, of the total, respectively (Supplementary Figure S9).
RNA editing in non-coding regions of grapevine mitochondrial genome
While RNA editing by C-to-U modification occurs mainly in coding regions of land plant mitochondrial transcripts, several alterations to non-coding RNAs have also been described (14). In Oenothera berteriana mitochondria, a C-to-U transition at position 4 of the trnF gene corrects a mispairing in its acceptor stem improving the corresponding folding (42). Applying our computational strategy to 13 tRNA genes known to be of mitochondrial origin, we identified two C-to-U editing events, one in the anticodon stem of the trnC gene altering a C–U mismatch to U–U supported by Solexa/Illumina reads and the other occurring at position 4 of the trnF acceptor stem, replacing a C–A mismatch with a conventional U–A Watson–Crick base pair, supported by SOLiD reads from root tissue (Supplementary Figure S10). Although the former editing event does not significantly change the stability of the trnC secondary structure, it occurs in the first 3 nt of the acceptor stem, a region that normally provides major identity elements and specific contact points for the cognate aminoacyl–tRNA synthetase. Moreover, this modification has also been described in trnC of Oenothera mitochondria (42). Notably, Solexa/Illumina reads also identified a reverse U-to-C editing event affecting the trnP at position 73. As a consequence, a native G-U match was replaced by a more stable G–C base pair. SOLiD data from leaf and root tissues, instead, supported another U-to-C change located at the first nucleotide 5′ of the anticodon, most likely contributing to codon–anticodon recognition.
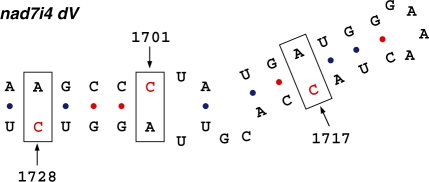
RNA editing can also modify C residues in intronic sequences of plant mitochondrial genomes (13). Several C-to-U transitions have been described for group II introns, where they generally stabilize folding (13). Many such modifications occur in intron domains I, V and VI that are important for the excision reaction. We also investigated the extent of RNA editing in grape mitochondrial group II introns (excluding trans-splicing introns). Surprisingly, we observed 36 C-to-U modifications and four potential U-to-C reverse events. Moreover, 10 out of 36 conversions affected the nad1 intron containing the matR gene. Several such editing modifications, as expected, occurred in domains V and VI improving the relative folding and, thus, the intron functionality in terms of self-splicing efficiency. We have analyzed three editing sites occurring in the domain V of the nad7 intron 4 in detail. These modifications correct three C–A mispairings affecting the folding of this functionally indispensable domain (Figure 4). Several C-to-U events were also conserved across known group II introns of diverse land plants (13). Taken together, our findings indicate that the extent of RNA editing in mitochondrial introns of land plants could be higher than anticipated by previous genome wide studies.
Figure 4.
Secondary structure of the domain V of nad7i4. In the grapevine nad7i4 domain V, cytosines subjected to RNA editing are indicated by arrows and included in a rectangle.
RNA editing in A. thaliana
To further confirm the reliability of our computational strategy, we also investigated RNA editing by C-to-U conversions in mitochondria of A. thaliana, for which the complete editing landscape has been estimated according to standard experimental procedures based on the Sanger methodology (19). In particular, we used ≈64 million short reads (each of 50 nt in length) generated through the Solexa/Illumina technology from total RNA of floral tissue belonging to Columbia Arabidopsis ecotype (43). After the first mapping step, however, we obtained only 241 359 reads uniquely located across Arabidopsis mitochondrial protein-coding genes. While the number of mapped reads was limited, we identified 76 C-to-U fully edited sites. Ten of these are new editing sites not previously described by Giegé and Brennicke (19). Three occur at third codon positions and the remaining seven at the first two positions. Several of these changes increase the conservation of the affected protein across land plants. Surprisingly, we found an edited site in position 1277 of the cox1 transcript, for which no editing sites have been yet observed in Arabidopsis. This modification causes the amino acid transition T-to-I for which a hydrophilic residue is replaced by a hydrophobic one. However, the effect of this change on the protein functionality is unknown. A specific protein modulation through RNA editing could also be required in floral tissue. However, this editing position, in addition to another C-to-U change at position 787 of the rps3 mRNA, are supported by a very limited number of independent reads (<4) and, thus, more investigations are needed to verify the existence of such modifications.
In addition, we also checked for editing sites in non-coding RNAs of Arabidopsis mitochondria. According to Giegé and Brennicke, no C-to-U sites were found in tRNAs, whereas new editing conversions where discovered in group II introns. In particular, we detected two new C-to-U changes occurring at the first and third intron of the nad4 gene other than one additional event in the unique rpl2 intron. Such editing modifications, however, were again supported by a limited number of short reads (<4).
DISCUSSION
Detecting editing sites by RNA-Seq technology
RNA editing sites are usually identified by direct comparison of transcribed sequences with their related templates (44). Target cDNAs have typically been amplified by gene specific primers or isolated from cDNA libraries and sequenced using the standard Sanger methodology. cDNA sequences are aligned onto their corresponding genomic loci and all detected variations are scored as RNA editing sites (16,44). However, the restricted number of cDNAs per locus, in addition to potential sequencing artefacts, can lead to false positives and prevent the detection of genuine C-to-U editing events. Moreover, poor cDNA sampling can preclude the assessment of tissue specificity of editing modifications and the evaluation of their statistical support. In contrast, deep sequencing can overcome these limitations allowing the characterization of the RNA editing landscape of a given reference annotation. To date, however, no computational approaches have been developed to this end. To fill this gap and to benefit from RNA-Seq technology for the investigation of editing, we propose a simple strategy that can efficiently handle short reads obtained by massive sequencing of RNAs by using either the Solexa/Illumina GA or ABI SOLiD platforms. Initially, short reads are mapped to a reference sequence using stringent quality criteria and allowing at most two mismatches and no indels. Subsequently we filter mapping results, considering only reads mapping to unique reference locations. This set of alignments is employed to generate a distribution of high quality nucleotides supporting each base of the reference. Unlike previous methodologies based on Sanger sequencing, short reads offer a high coverage depth per reference position and improve the detection of RNA editing sites. We have tested our approach, identifying C-to-U editing modifications occurring in the mitochondrial genome of V. vinifera. Plant mitochondrial RNA editing has been extensively studied and many C-to-U substitutions have been characterized in different organisms (9,26). The precise molecular mechanism is unknown but likely depends on nuclear factors belonging to PPR protein family (45). Moreover, the availability of well-annotated mitochondrial editing sites through specialized databases provides a valid benchmark with which to compare grape C-to-U modifications (26).
The availability of genome and RNA-Seq data from the same source, in our study the highly homozygous PN40024 grapevine genotype, is a fundamental requisite for a reliable editing detection. Indeed, nucleotide changes detected by comparing genome and transcript data may be genuine editing events or sequencing errors. In this respect, in addition to the expected C-to-U alterations, the Solexa/Illumina technology identified several potential non-canonical edits that were not supported by SOLiD reads—leading us to believe that for our data at least, the Solexa/Illumina reads are more prone to errors than those generated by the SOLiD technology. The frequencies of base substitutions shown in Table 1 support this hypothesis. The peculiar features of the color-space based SOLiD technology are particularly suitable for a reliable discrimination of real mismatches (two-color changes) from sequencing errors (single-color changes). Coverage depth could also influence the pattern of observed substitutions and contribute to the correction of potential mismatches occurring at low frequency. In our case, SOLiD data provided a mean per-base coverage depth that was three times higher than the Illlumina data (Supplementary Table S1). Indeed, despite the average 3-fold higher coverage, SOLiD data covered a similar number of bases to Solexa/Illumina (Supplementary Table S1).
Furthermore, >99% of SOLiD reads map on the sense strand, while Solexa/Illumina reads are equally distributed between the two strands (Supplementary Table S3 and Supplementary Figures S2 and S3). This is mainly due to the experimental protocol used to generate Solexa/Illumina reads (at the time of this work, the protocol to get strand specific Solexa/Illumina reads was not yet available). Considering the SOLiD data in isolation, we were able to exclude the possibility that the observed partial editing of some sites was a result of noise derived from non-edited antisense transcripts.
However, combining the information from both sequencing technologies we observed a significant increase in coverage and reduction of potential erroneous substitutions (Table 1).
The relatively high frequency for A-to-G and G-to-A mismatches can also be explained by cross mapping of short reads. Sequence similarity searches of the PN40024 nuclear genome revealed a number of regions showing high similarity to genes of mitochondrial origin. Interestingly, these sequences consistently showed higher identity to mitochondrial genome sequences than to edited mitochondrial transcripts. For high scoring segment pairs longer than 100 bases and showing >95% identity with mitochondrial coding regions (∼32 000 bases of nuclear DNA), over 400 positions indicated that nuclear insertions were comprised of unedited rather than edited sequences, while only three mismatches with mitochondrial genome sequences suggested the presence of edited sequences. Interestingly, among other mismatches of nuclear to mitochondrial sequences, transitions to A and T were predominant (245/341 of the remaining substitutions). This observation is consistent with the known strong AT bias of intergenic regions of the Vitis genome (34) and corroborates our suspicion that some G-to-A changes are due to cross mapping of reads derived from background transcription of nuclear sequences.
Mitochondrial RNA editing in grapevine
The complete RNA editing pattern has been experimentally detected for four higher plant mitochondrial genomes. In total, 441 C-to-U modifications have been found in Arabidopsis mitochondria (19) and 427 in B. napus (20). Coding genes of O. sativa are modified at 491 positions (21), while only 357 editing sites have been found in mitochondria of B. vulgaris (16). While we found 401 significantly supported C-to-U editing modifications in 37 mitochondrial protein-coding genes of V. vinifera, an additional 314 sites showing non-significant levels of editing corresponded to editing sites in other species. Thus, it is likely that >700 sites are edited in grape mtDNA, and that our test is rather conservative—potentially due to overestimation of sequencing error rates. This implies that editing in Vitis is slightly more pervasive than in other plants or that many sites remain undiscovered in other species.
The extremely high level of identity of the PN40024 and ENTAV 115 mitochondrial consensus sequences—particularly those corresponding to coding regions, coupled with the fact that our RNA-Seq data derive from one of these clones (PN40024) lead us to discount the possibility that Single Nucleotide Polymorphisms between the individuals used for genome sequencing and transcriptome analysis should account for a substantial number of inferred editing events.
For the 401 statistically significant events, we found a remarkably conserved pattern of editing: 91% of the grape mtDNA edited sites (366/401) were either edited in the same position in at least one other species (327/401) or the editing event increased conservation at the genomic level by introducing a uridine/thymine (39/401). For the remaining cases editing was prevalently observed at the third codon position (17/35, 48.6%), a much higher value than the 13.2% observed overall (Figure 3B).
An interesting finding concerns the extent of partially edited sites (in Vitis 76% of all detected modifications), and the observation that >85% of edited sites falling at silent (third codon) positions are partially edited. The predominance of partial editing at silent sites could be due to non-specific binding of editing specificity factors rather than an inefficiency of a putative ‘editosome’ machinery (16).
Partially edited sites may derive from immature transcripts or from differential (and possibly tissue-specific—see below) efficiency of the editing process in different positions. The impact of immature transcripts has been demonstrated by Verbitskiy and colleges (41) who showed that partially edited RNAs are intermediates of RNA editing in plant mitochondria. Moreover, we detected 36 editing sites in grape mitochondrial intervening sequences and all group II introns appeared well supported by short reads, indicating that incompletely processed messages are present in our samples. However, the observed range of variability—from 10 to 90%—of the percentage of unedited reads observed for the subset of deeply covered partially edited sites (>100 reads per site), is suggestive of differential editing efficiency at different sites.
A limited fraction of partially edited sites were shown to be significantly tissue specific. It should be noted that a high per base coverage depth is indispensable for statistical validation of the tissue specificity. Notably, the average per base coverage increases with the level of stringency of the statistical validation (i.e. FDR < 0.05, 165.23 reads per site; FDR < 0.01, 186.02 reads per site; Bonferroni correction, 248.60 reads per site). Therefore, we expect that additional tissue-specific sites would be identified by increasing the sequencing depth.
Considering all detected editing positions, our results are consistent with editing data from other land plants. Ninety percent of all grape RNA editing sites are non-synonymous, occurring with the highest frequency at the second codon position. Moreover, a large proportion of resulting amino acid changes fall in three categories P-to-L, S-to-L and S-to-F. Our results, therefore, validate the proposed computational approach based on next generation of sequencing reads.
Moreover, the detection of RNA editing sites has also been extended to mitochondria of A. thaliana. In spite of the restricted number of available short reads (the search for new editing events in Arabidopsis mitochondria was limited to fully supported sites in order to avoid potential noise due to false substitutions), ten new C-to-U changes were found in protein-coding genes, in addition to three modifications occurring in group II introns. Such new editing sites could be specific to the floral tissue since previous investigations have been conducted on cell-suspension culture only (19). However, the Arabidopsis mitochondrial genome and the Solexa/Illumina data of the accession SRX002554 belong to the same ecotype but not to the same individual and there is evidence that raises the possibility that the ecotype of the accession NC_001284 used by Giegé and Brennicke (19) is not Columbia (46), we can not therefore exclude the possibility that some of the novel Arabidopsis editing sites result from genomic polymorphisms.
Finally, we investigated the nucleotide context of edited sites in Vitis mitochondria and confirmed previously reported biases towards pyrimidines in nucleotides immediately upstream of edited cytidines and the frequent presence of a purine (generally a G) immediately following edited sites (Supplementary Figures S8 and S9). Thus, our data support the contention that groups of nucleotides in specific locations are important in the recognition of editing sites (30).
CONCLUSIONS
New high-throughput sequencing strategies offer unprecedented opportunities to investigate key molecular mechanisms at the genome level. In particular, RNA-Seq is a powerful tool for high-throughput transcriptome analysis including the investigation of basic post-transcriptional events such as alternative splicing and RNA editing. Editing by base conversion has been extensively studied in animal nuclei and land plant organelles where it seems to be essential for regular gene expression and genome variability maintenance. Indeed, organellar RNA editing may compensate for Muller’s ratchet in genomes where nucleotide substitution rates are very low. However, the identification of edited sites is often time-consuming and costly, precluding genome wide investigations.
Recently, high throughput approaches have been used to identify A-to-I sites in human (22) and detect the efficiency of editing for 28 different sites during the development of the mouse brain (23). Such approaches, however, are not based on RNA-Seq and potential editing sites are known from the literature or computational analyses. In this work, we have presented a novel computational strategy that greatly facilitates the discovery of RNA editing sites at the genome level using short sequencing reads. We show that a combined approach including short reads from both Solexa/Illumina and SOLiD technologies may greatly improve the detection of reliable C-to-U editing sites in grapevine mitochondria, significantly reducing the discovery of false substitutions, particularly for editing sites supported by both platforms. However, it should be pointed out that our approach depends on the quality of short reads and should be performed on the same organism and individual. When the last request cannot be satisfied, results must be filtered for known SNPs and the conservation should be taken into account to identify candidate sites.
Although our procedure has been assessed in mitochondria of V. vinifera and A. thaliana, it can be applied to discover RNA editing events occurring on chloroplast or nuclear genomes, and to investigate the alterations of RNA editing patterns in diverse mammalian diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Ministero dell'Istruzione, dell'Università e della Ricerca (Fondo Italiano Ricerca di Base: ‘Laboratorio Internazionale di Bioinformatica’ (LIBI); Laboratorio di Bioinformatica per la Biodiversità Molecolare (MBLAB); VIGNA Consortium (Ministero delle Politiche Agricole, Alimentari e Forestali).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Carmela Gissi for helpful suggestions on the data analysis, Scott Kuersten and Alain Rico (Applied Biosystems) for help and advice with the SOLiD libraries preparations, and Davide Campagna, CRIBI, Padua University for advices in using PASS software.
REFERENCES
- 1.Mardis ER. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 2.Schuster SC. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 3.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denoeud F, Aury JM, Da Silva C, Noel B, Rogier O, Delledonne M, Morgante M, Valle G, Wincker P, Scarpelli C, et al. Annotating genomes with massive-scale RNA sequencing. Genome Biol. 2008;9:R175. doi: 10.1186/gb-2008-9-12-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gott JM. Expanding genome capacity via RNA editing. C. R. Biol. 2003;326:901–908. doi: 10.1016/j.crvi.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 9.Gray MW. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life. 2003;55:227–233. doi: 10.1080/1521654031000119425. [DOI] [PubMed] [Google Scholar]
- 10.Steinhauser S, Beckert S, Capesius I, Malek O, Knoop V. Plant mitochondrial RNA editing. J. Mol. Evol. 1999;48:303–312. doi: 10.1007/pl00006473. [DOI] [PubMed] [Google Scholar]
- 11.Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8:35–46. doi: 10.1016/j.mito.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Casey JL. RNA editing in hepatitis delta virus. Curr. Top. Microbiol. Immunol. 2006;307:67–89. doi: 10.1007/3-540-29802-9_4. [DOI] [PubMed] [Google Scholar]
- 13.Carrillo C, Chapdelaine Y, Bonen L. Variation in sequence and RNA editing within core domains of mitochondrial group II introns among plants. Mol. Gen. Genet. 2001;264:595–603. doi: 10.1007/s004380000345. [DOI] [PubMed] [Google Scholar]
- 14.Brennicke A, Marchfelder A, Binder S. RNA editing. FEMS Microbiol. Rev. 1999;23:297–316. doi: 10.1111/j.1574-6976.1999.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 15.Kempken F, Howard W, Pring DR. Mutations at specific atp6 codons which cause human mitochondrial diseases also lead to male sterility in a plant. FEBS Lett. 1998;441:159–160. doi: 10.1016/s0014-5793(98)01538-5. [DOI] [PubMed] [Google Scholar]
- 16.Mower JP, Palmer JD. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol. Genet. Genomics. 2006;276:285–293. doi: 10.1007/s00438-006-0139-3. [DOI] [PubMed] [Google Scholar]
- 17.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 19.Giege P, Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 22.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 23.Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Yang SP. Curr. Protoc. Bioinformatics. 2005. Generating a genome assembly with PCAP. Chapter 11, Units 11 13. [DOI] [PubMed] [Google Scholar]
- 25.Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 2009;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 26.Picardi E, Regina TM, Brennicke A, Quagliariello C. REDIdb: the RNA editing database. Nucleic Acids Res. 2007;35:D173–D177. doi: 10.1093/nar/gkl793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campagna D, Albiero A, Bilardi A, Caniato E, Forcato C, Manavski S, Vitulo N, Valle G. PASS: a program to align short sequences. Bioinformatics. 2009;25:967–968. doi: 10.1093/bioinformatics/btp087. [DOI] [PubMed] [Google Scholar]
- 28.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 30.Mulligan RM, Chang KL, Chou CC. Computational analysis of RNA editing sites in plant mitochondrial genomes reveals similar information content and a sporadic distribution of editing sites. Mol. Biol. Evol. 2007;24:1971–1981. doi: 10.1093/molbev/msm125. [DOI] [PubMed] [Google Scholar]
- 31.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coggill P, Finn RD, Bateman A. Curr. Protoc. Bioinformatics. 2008. Identifying protein domains with the Pfam database. Chapter 2, Unit 25. [DOI] [PubMed] [Google Scholar]
- 33.Palmer JD, Herbon LA. Plant mitochondrial DNA evolves rapidly in structure, but slowly in sequence. J. Mol. Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 34.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 35.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat. Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Regina TM, Picardi E, Lopez L, Pesole G, Quagliariello C. A novel additional group II intron distinguishes the mitochondrial rps3 gene in gymnosperms. J. Mol. Evol. 2005;60:196–206. doi: 10.1007/s00239-004-0098-4. [DOI] [PubMed] [Google Scholar]
- 37.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yura K, Go M. Correlation between amino acid residues converted by RNA editing and functional residues in protein three-dimensional structures in plant organelles. BMC Plant Biol. 2008;8:79. doi: 10.1186/1471-2229-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bock H, Brennicke A, Schuster W. Rps3 and rpl16 genes do not overlap in Oenothera mitochondria: GTG as a potential translation initiation codon in plant mitochondria? Plant Mol. Biol. 1994;24:811–818. doi: 10.1007/BF00029863. [DOI] [PubMed] [Google Scholar]
- 40.Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M. Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion. 2008;8:319–327. doi: 10.1016/j.mito.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Verbitskiy D, Takenaka M, Neuwirt J, van der Merwe JA, Brennicke A. Partially edited RNAs are intermediates of RNA editing in plant mitochondria. Plant J. 2006;47:408–416. doi: 10.1111/j.1365-313X.2006.02794.x. [DOI] [PubMed] [Google Scholar]
- 42.Binder S, Marchfelder A, Brennicke A. RNA editing of tRNA(Phe) and tRNA(Cys) in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet. 1994;244:67–74. doi: 10.1007/BF00280188. [DOI] [PubMed] [Google Scholar]
- 43.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takenaka M, Brennicke A. RNA editing in plant mitochondria: assays and biochemical approaches. Methods Enzymol. 2007;424:439–458. doi: 10.1016/S0076-6879(07)24020-0. [DOI] [PubMed] [Google Scholar]
- 45.Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21:558–567. doi: 10.1105/tpc.108.064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bentolila S, Elliott LE, Hanson MR. Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics. 2008;178:1693–1708. doi: 10.1534/genetics.107.073585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.