Abstract
Megasatellites are a new family of long tandem repeats, recently discovered in the yeast Candida glabrata. Compared to shorter tandem repeats, such as minisatellites, megasatellite motifs range in size from 135 to more than 300 bp, and allow calculation of evolutionary distances between individual motifs. Using divergence based on nucleotide substitutions among similar motifs, we determined the smallest distance between two motifs, allowing their subsequent clustering. Motifs belonging to the same cluster are recurrently found in different megasatellites located on different chromosomes, showing transfer of genetic information between megasatellites. In comparison, evolution of the few similar tandem repeats in Saccharomyces cerevisiae FLO genes mainly involves subtelomeric homologous recombination. We estimated selective constraints acting on megasatellite motifs and their host genes, and found that motifs are under strong purifying selection. Surprisingly, motifs inserted within pseudogenes are also under purifying selection, whereas the pseudogenes themselves evolve neutrally. We propose that megasatellite motifs propagate by a combination of three different molecular mechanisms: (i) gene duplication, (ii) ectopic homologous recombination and (iii) transfer of motifs from one megasatellite to another one. These mechanisms actively cooperate to create new megasatellites, that may play an important role in the adaptation of Candida glabrata to its human host.
INTRODUCTION
Megasatellites are a new class of large tandem repeats that were recently discovered in the Candida glabrata genome sequence (1,2). They are widespread in the genome of this hemiascomycetous yeast species, but share no significant homology with any other tandem repeat or gene sequenced so far. Among the 84 minisatellites previously reported in Saccharomyces cerevisiae (3), four harbor a tandemly repeated motif of 135 bp or larger, and may qualify as megasatellites (a motif is defined as the smallest DNA sequence that is tandemly repeated within a minisatellite or a megasatellite). These four S. cerevisiae megasatellites are found in the paralogous FLO1, FLO5, FLO9 genes (135-bp repeated motif), involved in flocculation and cellular adhesion, and in the NUM1 gene (192-bp repeated motif), encoding a protein required for nuclear migration along microtubules during cell division (3–5).
The 44 megasatellites found in C. glabrata are present in 29 different protein-coding genes and six pseudogenes. Two major families of megasatellites have been described: the ‘SFFIT family’ (due to the conservation of these five amino acids in each motif) present in 11 genes and three pseudogenes, and the ‘SHITT family’ in 12 genes and six pseudogenes. Four genes and three pseudogenes carry both types of megasatellites. The remaining 10 genes contain megasatellites that do not share significant homology with SFFIT and SHITT megasatellites. Remarkably, although minisatellites are evenly distributed in the C. glabrata genome, megasatellites show a very strong bias towards locations in subtelomeric regions (2).
The existence of tandem repeats with such long motifs, and their abundance in this yeast genome, raise the question of their very origin. Regular minisatellites in other yeasts generally contain motifs that are 9- to 81-bp long (3), the average motif size being 27 bp. In C. glabrata, the average motif size of a regular minisatellite is slightly shorter (21 bp), whereas SHITT and SFFIT megasatellites contain much longer motifs (135 and 300 bp, respectively). It was proposed, several years ago, that minisatellites are initially formed by replication slippage between two short sequences (5 bp) spaced by a few nucleotides, thus creating an initial motif duplication that could further expand by replication slippage and unequal sister-chromatid recombination between the two motifs (6). This model, however, does not explain how minisatellites (or megasatellites) propagate into new genes to form large families, as observed in C. glabrata.
In the present work, we address this question using an in silico approach to infer evolutionary relationships between megasatellite motifs both at the intra- and inter-genic levels in the C. glabrata and S. cerevisiae genomes. Minisatellite motifs are too short to measure evolutionary distances between them. By contrast, evolutionary distances can be measured between megasatellites, that contain longer DNA motifs, by classical sequence homology methods. In addition, to the expected similarity detected between motifs belonging to the same megasatellite, we also found a surprising conservation between motifs located in different megasatellites, suggesting transfer of motifs from one megasatellite to another one. We discuss possible mechanism(s) responsible for these ‘motif jumps’ among megasatellites, and their possible selection during evolution of this pathogenic yeast.
MATERIALS AND METHODS
Megasatellite sequences
The starting set of megasatellite-containing genes was extracted from the complete genomic sequence of C. glabrata (http://www.genolevures.org/). In this sequence, 16 megasatellite-containing regions were determined from BAC inserts instead of direct shotgun assembly to eliminate the risk of misassembly of repeated sequences (1). We verified here the presence of each individual motif of these megasatellites using original sequence reads of those BACs. Megasatellites present in genes CAGL0E00143g, CAGL0E01661g and CAGL0I10098g (1) were not considered in this work because they were not covered by BACs. In addition, the correctness of megasatellite sequences were verified by direct sequencing of PCR products for the genes CAGL0K13024g, CAGL0I10200g and CAGL0I10362g. Note that the sequence of CAGL0I10200g is not exactly identical to Génolevures database. In total, out of 23 megasatellite-containing genes and pseudogenes presented in Table 1, only two genes (CAGL0G10219g and CAGL0H10626g) and four pseudogenes (CAGL0B05093g, CAGL0F00110g, CAGL0H00132g and CAGL0I00110g) were not directly verified in this work. Note that the list contains five pseudogenes, annotated as such because they contain 11–69 stop codons or an extensive 3′ deletion (CAGL0A04873g). Motifs were extracted from the megasatellites as described in Thierry et al. (1). Incomplete motifs at 5′- or 3′- ends were eliminated before analysis.
Table 1.
Megasatellites studied in this work and their corresponding motifs
| Organism | Gene Name | MS number | Motif Family | IS |
|---|---|---|---|---|
| C. glabrata | CAGL0A04873g | 114/230 | 31xSHITT/3xSFFIT | +/– |
| CAGL0B05093g | 231 | 11xSHITT | – | |
| CAGL0E00231g | 206/207 | 6xSHITT/3xSFFIT | –/– | |
| CAGL0F00110g | 115 | 4xSHITT | + | |
| CAGL0G10219g | 209 | 4xSHITT | – | |
| CAGL0H00132g | 234 | 4xSHITT | – | |
| CAGL0H010626g | 211 | 2xSHITT | – | |
| CAGL0I00110g | 235/236 | 2xSHITT/8xSFFIT | –/– | |
| CAGL0I00157g | 222/223 | 10xSHITT/3xSFFIT | –/– | |
| CAGL0L00227g | 112 | 4xSHITT | + | |
| CAGL0I10246g | 215 | 5xSFFIT | – | |
| CAGL0I10340g | 216 | 2xSFFIT | – | |
| CAGL0I10200g | 237 | 9xSFFIT | – | |
| CAGL0J01774g | 108 | 6xSHITT | + | |
| CAGL0K13024g | 221 | 5xSHITT | – | |
| CAGL0L13310g (EPA11) | 225/226 | 10xSHITT/6xSFFIT | –/– | |
| CAGL0L13332g (EPA13) | 227 | 5xSHITT | + | |
| CAGL0C00253g | 205 | 5xSFFIT | – | |
| CAGL0I10147g | 214 | 32xSFFIT | – | |
| CAGL0I10362g | 217/218 | 3xSHITT/5xSHITT | –/– | |
| CAGL0J05170g | 202 | 10xSHITT | + | |
| CAGL0L09911g | 224 | 5xSFFIT | – | |
| CAGL0L10092g | 229/228 | 5xSHITT/2xSFFIT | +/– | |
| S. cerevisiae | YAR050W (FLO1) | – | 7xTFTST | – |
| YHR211W (FLO5) | – | 17xTFTST | – | |
| YAL063C (FLO9) | – | 12xTFTST | – |
Genes in which megasatellites are found are indicated by their names. Pseudogenes are underlined. Paralogous gene families are indicated with vertical lines on the left (2). Megasatellite are numbered according to Thierry et al. (1) for C. glabrata (two numbers are used to indicate the presence of two distinct families in the same gene). S. cerevisiae FLO megasatellites are those from (3). Corresponding motifs and number of repeats are indicated in column 4. Motifs are designated by the five conserved amino-acid sequence found at their beginning. IS: Intervening Sequences within the megasatellite (see text).
Saccharomyces cerevisiae FLO megasatellites are described in (3). FLO1, FLO5 and FLO9 sequences were retrieved from SGD (http://www.yeastgenome.org/).
Calculation of distances using a transition/transversion-based model (TN93)
DNA sequences of megasatellite motifs were aligned using ClustalW program (7). N and C terminal ends were manually trimmed so that all motifs have exactly the same length [alignments are shown using the ClustalX colour scheme (8) in Supplementary Figures S1–S3]. The PAML yn00 program (9) was then executed to calculate nucleotide substitutions, using the Tamura and Nei substitution model (10) with default parameters. This model provides independent rate parameters for A <-> G and C <-> T transitions (in addition to the transversion rate parameters) and is more tractable than other one- or two-parameter models (11–14). All pairwise comparisons were computed, resulting in sequence-based distances between all motifs (Supplementary Table S1). In order to compare the distances, we used the non-parametric Wilcoxon rank test (15), as implemented in the R software (16).
Determination of shortest paths and cluster visualization
From the distances between megasatellite motif pairs, we constructed a directed weighted complete graph, with nodes representing motifs and edges representing weighted links between couples of motifs, as determined by distance calculation. In this complete graph, we identified the shortest path between any pair of motifs using the Dijkstra algorithm (17), as implemented in Networkx python package (http://networkx.lanl.gov/). In this complete matrix of shortest paths, one or more edges carrying the smallest value were kept for each motif. This led to the formation of 13–20 clusters, depending on the motif family. We re-used this same algorithm to identify the second smallest shortest path, in order to define super clusters. In order to measure the robustness of the clusters obtained, we applied the same strategy to 1000 replicates, in which the original sequences were randomly mutated using Seqboot program from PHYLIP package (18). In these 1000 replicates, we calculated the number of times each original edge of the graph appeared, and used it as a bootstrap value. Shortest paths, super clusters and bootstrap information are provided in Supplementary Table S2. Graph visualizations were obtained using the Cytoscape program (19), providing a circular graphical layout, helping cluster visualization.
Single linkage hierarchical clustering of megasatellite motifs
In addition to the graph approach, a single linkage analysis was also done. Using the same TN93 distance matrix, a hierarchical clustering of motifs was performed using the Hclust program (« single » method parameter) as implemented in the R software (16). A tree of motifs was obtained and manually cut in order to obtain the same number of subtrees as clusters, respectively 19, 20 and 13 for SFFIT, SHITT and FLO motifs. The same strategy was then used on the 1000 replicates, to calculate bootstrap values (Supplementary Figures S7–S9). Tree visualizations used the Cytoscape program with the PhyloTree plugin (developed by Chinmoy Bhatiya). Results obtained with this approach are very similar to those obtained with the graph approach.
Estimation of dN and dS values
In order to get information about functional constraints on megasatellite motifs, we also estimated the number of synonymous substitutions per synonymous site (dS), and the number of non-synonymous substitutions per non-synonymous site (dN), using PAML yn00 program with default parameters (9). We used the same calculation for the non-repeated regions of genes or pseudogenes carrying the megasatellites.
RESULTS
Distance measurement and motif clustering
The aim of this work was to measure sequence divergence between all motifs within each megasatellite family (SFFIT or SHITT in C. glabrata, FLO in S. cerevisiae), in order to infer their evolutive history. For C. glabrata, we used the set of megasatellites described in Thierry et al. (1), consisting of MS#205 to MS#237 and MS#105 to MS#235 for, respectively, SFFIT and SHITT families (Table 1). Several of the megasatellite-containing genes are paralogs. The largest paralogous gene family contains 10 members. There is one family with three members and two families with two members, six genes are singletons. Altogether, a total of 82 SFFIT and 126 SHITT motifs were used for pairwise comparisons. For S. cerevisiae, the three megasatellites in the FLO1, FLO5 and FLO9 genes were used (3), for a total of 36 FLO motifs (Table 1).
Pairwise distances were calculated based on nucleotide substitutions, and motifs were clustered according to such distances (Figure 1). This clustering generated 19 SFFIT clusters (labeled A–S) and 20 SHITT clusters (labeled A to W), represented in Supplementary Figures S4 and S5. For SFFIT motifs, 17 clusters (89%) are made of motifs from one single megasatellite, and two clusters (11%) contain motifs coming from two megasatellites. For SHITT motifs, only 12 clusters (60%) are made of motifs from one single megasatellite, the other eight clusters (40%) contain motifs found in two or three distinct megasatellites (Figure 2). Conversely, some megasatellites are entirely made of motifs belonging to only one single cluster (e.g. MS#215 and MS#225), whereas others are mosaics of motifs belonging to up to five clusters (e.g. MS#231 and MS#214). Thirty-three percent of megasatellites from the SFFIT family contain such mosaics, compared to 44% of the megasatellites from the SHITT family. Megasatellites whose motifs are found in only one cluster are suggestive of a coordinated evolution of motifs (intra-genic evolution). However, mosaic megasatellites are representative of an inter-genic model of evolution, suggesting that a given motif may propagate to several megasatellites. \
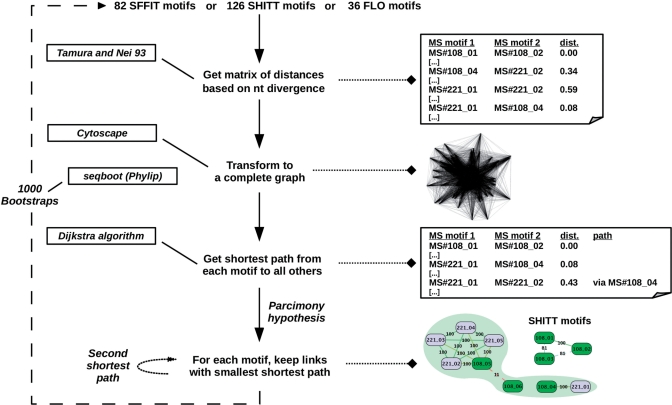
Figure 1.
Schematic representation of the method used to compute distances and clusterize motifs. In order to measure the divergence between megasatellite motifs, we used the transition to transversion TN93 model (10). This detects all nucleotide substitutions among motifs without taking into consideration possible selection on amino-acid conservation. These distances were plotted on a graph. In this complete graph, each node corresponds to a motif and each edge to the pairwise distance between two motifs. Then, the shortest path between two motifs was determined, independently of the megasatellite containing the motif, and in order to deduce relationships between closely related motifs, a minimal evolution model between motifs was favored under a parsimony hypothesis (see ‘Materials and Methods’ section). Center: the mainframe is represented by plain arrows. Left: at each step of the mainframe, the algorithm or program used is depicted (see ‘Materials and methods’ section for exact references). Right: visual output examples of SHITT megasatellites MS#108 and MS#221 are given. The complete graph represents the completely linked 126 SHITT motifs. The bottom graph represents clusters containing MS#108 and MS#221 motifs, and the area shaded in green encompasses the super cluster regrouping V and D clusters. Bootstrap values are indicated on edges.
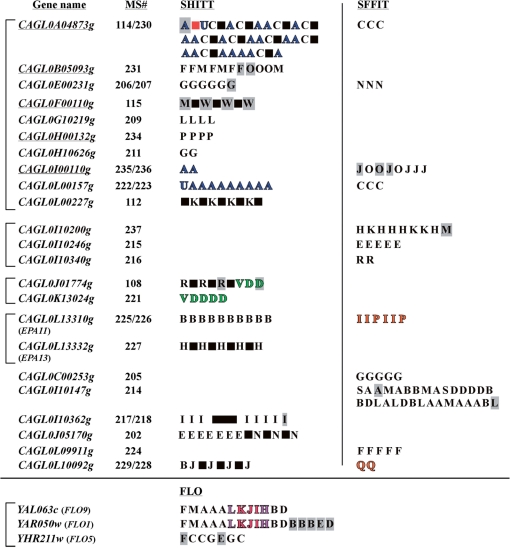
Figure 2.
Summary of motif clusters and super clusters for all C. glabrata and S. cerevisiae megasatellites. Gene names are shown to the left (pseudogenes underlined), brackets indicate paralogous gene families (Table 1). For each gene, megasatellite number(s) is (are) indicated, as in Thierry et al. (1). Each megasatellite motif belongs to a cluster, identified by a letter (Supplementary Figures S1–S3). SHITT megasatellites are shown to the left, SFFIT megasatellites are shown to the right, six genes containing both kinds of megasatellites. Note that CAGL0I10362g contains two SHITT megasatellites, spaced by 1437 bp (large black box). Same color letters indicate a robust super cluster (bootstrap of all edges ≥90%). Small black boxes represent intervening sequences, often found between SHITT motifs (see text). The red box in CAGL0A04873g represents the location of the SFFIT MS#230 within the SHITT MS#114 megasatellite. Grey boxed letters correspond to non robust edges linking a motif to its cluster (bootstrap <90%).
The situation is different in S. cerevisiae, where we found 13 clusters of motifs by applying the same methodology (Supplementary Figure S6). FLO1 and FLO9 motifs are found at precisely the same positions in both genes, whereas five out of the seven FLO5 motifs are specific to this gene (Figure 2). FLO1 and FLO9 genes are respectively located on the right and left subtelomeric arms of chromosome I, whereas FLO5 is located 34-kb away from chromosome VIII right telomere. Although the number of megasatellites in S. cerevisiae is limited, these observations suggest that subtelomeric megasatellite motifs are more conserved than in C. glabrata, where non conservation of subtelomeric megasatellite motifs is the rule (Figure 2).
A hierarchical clustering approach was also used to assess the robustness of the graph approach (see ‘Materials and Methods’ section). This hierarchical clustering is simpler, as the motifs are clustered based on distance information. Motifs are not forced to belong to any cluster. However, the resulting tree had to be manually cut at a given depth in order to obtain the same number of clusters as before. Visual representation of the trees for the three megasatellite families are given in Supplementary Figures S7–S9. For the SFFIT family, five motifs out of 82 (6%) are not included in any cluster previously found, but three out of these five were not supported by the previous bootstrap calculation (Supplementary Figure S7). Only two out of 126 SHITT motifs (1.6%) and one out of 36 FLO motifs (2.8%) were not included in any cluster, but none of these three motifs was previously supported by bootstrap calculation (Supplementary Figures S8 and S9). We concluded that the initial clustering performed by the graph approach gave results almost identical to the hierarchical clustering.
Shortest path between clusters
In order to capture a possible organization of motif clusters into ‘super clusters’, we took into account the second shortest path between motifs (see ‘Materials and methods’ section, Figure 1). We extracted one super cluster supported by bootstraps for SFFIT motifs (regrouping clusters I, P and Q), two super clusters for SHITT motifs (clusters A–U and clusters D–V), and two super clusters for the FLO family (Figure 2 and Supplementary Figures S4–S6). Super clusters tend to regroup clusters of motifs from the same megasatellite (e.g. SFFIT motif clusters I and P from MS#226). This is, however, not always the case. For example, A and C SHITT clusters (in MS#114) are not together in the same super cluster.
We investigated whether motifs found in paralogous genes belong to the same clusters, or super clusters, in other words if megasatellites propagate passively through duplication of the genes that contain them. In the largest paralogous gene family, 11 SHITT clusters and four SFFIT clusters are represented. Out of the 11 SHITT clusters, 9 are not in the same super cluster (Figure 2). For SFFIT motifs, none of the four clusters in this paralogous family assemble into a super cluster. These observations demonstrate an important divergence between motifs contained in paralogous families. A notable exception is the V and D SHITT motifs that are similarly found in CAGL0J01774g and CAGL0K13024g paralogs.
Evolution of megasatellites in paralogous genes and pseudogenes
Seven SHITT and SFFIT megasatellites are contained in seven pseudogenes in C. glabrata (but only five were used in this study, see ‘Materials and Methods’ section). Since these five pseudogenes are indeed under neutral selective pressure, they may be expected to accumulate as many synonymous mutations as non-synonymous mutations per possible site. In order to determine if this holds true for megasatellites, the TN93 motif distances in paralogous genes versus paralogous pseudogenes were compared. Both for SFFIT and SHITT motifs, we observed a significantly higher number of transitions and transversions between motifs carried by pseudogenes, with 1.5- to 2-fold excess of both types of substitutions with respect to motifs carried by genes.
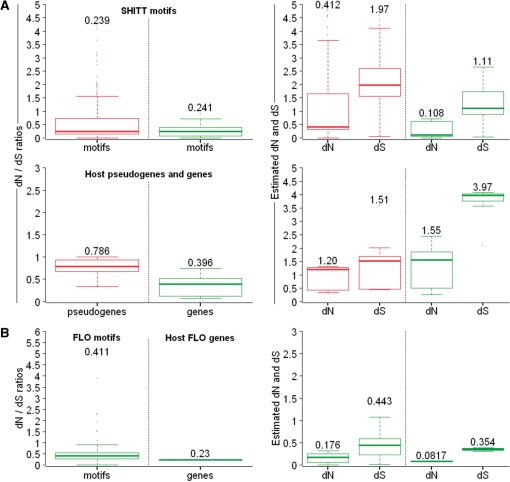
In order to assess selective pressure, we calculated the ratio of non-synonymous to synonymous substitutions (dN/dS, see ‘Materials and Methods’ section) among paralogous gene motifs and paralogous pseudogene motifs. For SHITT motifs, dN/dS median values are significantly under 1, showing that motifs are under strong purifying selection, whether they are located within genes or pseudogenes, although both distributions are significantly different (P = 2 × 10−13, Wilcoxon test, (15)) (Figure 3A). We subsequently measured dN/dS ratios on SHITT-containing genes and pseudogenes, outside of megasatellites. As expected from relaxed selective pressure on pseudogenes, we observed a median dN/dS value of 0.786 for pseudogenes (Figure 3A), and a significantly lower median dN/dS value of 0.396 for genes (P = 6 × 10−4, Wilcoxon test). By comparing dN/dS ratios of genes and pseudogenes to those of megasatellite motifs, we found that genes are a little less constrained than their motifs (median dN/ds = 0.396 for genes, compared to 0.241 for gene motifs, P = 9 × 10−2, Wilcoxon test). Strikingly, this difference is significantly amplified for pseudogenes, in which megasatellite motifs show strong purifying selection (median dN/dS = 0.786 for pseudogenes, compared to 0.239 for pseudogene motifs, P = 6 × 10−3, Wilcoxon test). Therefore, we conclude that genes or pseudogenes and their megasatellites appear to be under very different selective constraints. Remarkably, megasatellite motifs tend to be more conserved through evolution than their containing genes or pseudogenes. Similar calculation could not be performed on SFFIT motifs owing to their smaller number.
Figure 3.
Selection forces acting on megasatellite motifs and their host genes or pseudogenes. (A) Top left: boxplots represent the dN/dS ratios between SHITT motifs contained in paralogous genes (green) or pseudogenes (red) (respectively 1021 and 392 values). Corresponding dN and dS values are on the right. Median values are indicated above distributions. Bottom left: Same representations for the non-repeated regions of the same genes (green) and pseudogenes (red) (respectively 10 and 12 values). Corresponding dN and dS values are on the right. Median values are indicated above distributions. (B) Boxplots on the left represent the dN/dS ratios between FLO motifs and non-repeated regions of FLO genes (respectively 595 and 3 values). Corresponding dN and dS values are on the right. Median values are indicated above distributions.
The situation is, again, different in S. cerevisiae. The three paralogs, FLO1, FLO5 and FLO9, show lower dN and dS values, both in genes and in motifs, as compared to C. glabrata (Figure 3B). In addition, the dN/dS ratio is lower for genes than for gene motifs, a result opposite to what is observed in C. glabrata (Figure 3). Overall, genes and megasatellite motifs accumulated more synonymous and non-synonymous mutations in C. glabrata than in S. cerevisiae, where purifying selection is stronger on genes than on megasatellite motifs.
DISCUSSION
In the present work, we studied the evolution of C. glabrata and S. cerevisiae megasatellites by using a transition to transversion based model of evolution, in order to estimate distances between megasatellite motifs. Similar studies could not be undertaken before on minisatellites, since their motif length is too short and most of the minisatellites detected in genomes do not belong to conserved families (3). Here, for the first time, the size and number of motifs enabled us to compute evolutionary distances among tandem repeats. All pairwise distances were calculated for the 126 SHITT, 82 SFFIT and 36 FLO motifs, tandemly repeated within 26 different genes, most of them of unknown function [except for the FLO genes and EPA11 and EPA13 (20)]. Note that FLO and EPA genes do not share significant similarity, and although SHITT and FLO motifs have the same size (135 nt), they are not similar in sequence.
Three molecular mechanisms are involved in megasatellite propagation
We show that megasatellite motifs propagate by intra-genic as well as by inter-genic mechanisms. Duplication of a megasatellite-containing gene is one obvious mode of propagation, detected both in C. glabrata and in S. cerevisiae. For example, CAGL0JO1774g and CAGL0K13024g are paralogs that contain closely related megasatellites (Figure 2). Ectopic homologous recombination is a second possible mechanism to propagate megasatellite motifs, in both yeasts. FLO1 is located 10-kb away from the right telomere of chromosome I, whereas FLO9 is located 25-kb away from the left telomere, both genes in the same orientation as compared to the centromere. Although the three FLO genes were apparently duplicated within the same time scale (dN and dS are similar), FLO1 and FLO9 megasatellite motifs are conserved. This is consistent with gene conversion occuring between the two subtelomeric genes (21,22). FLO5 is located 34-kb away from the right end of chromosome VIII, but exhibits very different motifs. This is consistent with the recent observation that chromosome I and VIII arms are located in different subnuclear compartments, reducing the frequency of their interactions (23). Similar examples of subtelomeric motif conservation are also found in C. glabrata (e.g. MS#230 and MS#223 for SFFIT motifs, or MS#236 and MS#223 for SHITT motifs). In S. cerevisiae, it was shown that gene conversion associated to double-strand break repair is a very efficient mechanism to expand or contract minisatellites or large tandem arrays (24,25). Since homologous recombination is functional in C. glabrata (26), and the whole double-strand break repair machinery appears conserved (27), it is likely that this mechanism operates between nearly identical megasatellites of C. glabrata.
Megasatellites (or individual motifs) not originating from the two previous mechanisms are also found in C. glabrata. The eight megasatellites present in six singletons (Figure 2) cannot originate from gene duplications. The second possible mechanism, homologous recombination, is very sensitive to mismatches, and 0.1% sequence divergence is sufficient to dramatically decrease recombination (28). In the present case, motifs belonging to the same cluster exhibit, on average, 4.6% sequence divergence for SFFIT motifs, 5.8% for SHITT motifs, and 3.2% for FLO motifs. This divergence is even higher, as expected, between clusters (0.3–82.4%, mean value: 27.6% for SFFIT motifs, 2.3–82.9%, mean value: 41.5% for SHITT motifs, and 0.7–33.3%, mean value: 19.5% for FLO motifs). Therefore, it is unlikely that homologous recombination between megasatellites explains the propagation of motifs belonging to different clusters. We propose that some motifs are capable to ‘jump’ from a megasatellite to another one, by a new molecular mechanism that remains to be clarified (Figure 4). The first motif of MS#229 (SHITT cluster B) and, to a lesser extent, the last motif of MS#237 (the weakly supported SFFIT cluster M) are representatives of such possible events in C. glabrata. Similarly, MS#226 and MS#228 SFFIT motifs are in the same super cluster and may therefore originate from a similar mechanism. In addition, SHITT megasatellites found within paralogous gene families often contain intervening sequences, of variable sizes, inserted between motifs [e.g. MS#115 or MS#108; (2)]. The structure of such megasatellites cannot be explained either by simple gene duplication or by homologous recombination (Figure 2).
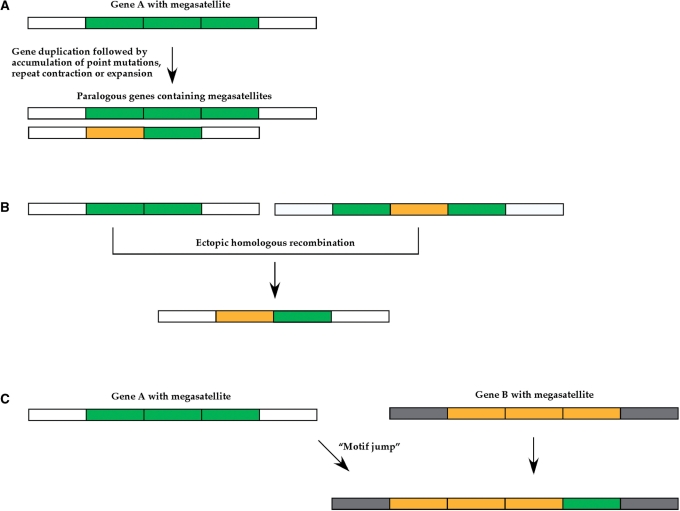
Figure 4.
Three mechanisms propagate megasatellites in yeast genomes. Gene A and gene B are two non-paralogous genes, containing different megasatellites. White and grey boxes represent the non-repeated parts of each gene, outside of megasatellites, green and yellow boxes represent the repeated tandem motifs of each megasatellite. (A) Gene duplication. A megasatellite-containing gene is duplicated, leading to the concomitant duplication of its megasatellite. Tandem repeats may subsequently expand or contract, and accumulate point mutations, leading to sequence divergence. (B) Recombination. Ectopic homologous recombination may occur between two motifs (or two megasatellites) that are not too divergent from each other, and gene conversion tends to homogenize motifs. (C) Motif jump. A motif may ‘jump’ into a gene that contains a megasatellite with different motifs. If the green motif is too divergent from the yellow motifs, gene conversion cannot homogenize the tandem array.
At the present time, we have no experimental data supporting the existence of this new molecular mechanism, tentatively called ‘motif jump’, and we may only speculate about its nature. Based on known mechanisms of DNA transfer, discovered with transposable elements and yeast mitochondrial introns (29), we hypothesize that motifs may ‘jump’ from a megasatellite to another one, either directly by a mechanism relying only on DNA, or using an RNA intermediate. Given that C. glabrata contains only one retrotransposon as a possible source of reverse transcriptase (Tcg3, gene name CAGL0G07183g, The Génolevures Consortium, http://www.genolevures.org/), it is unlikely that reverse transcription is an active phenomenon in this yeast. We cannot however exclude that in a distant past, when C. glabrata may have contained more retrotransposons than now, this mechanism could have been used to propagate megasatellite motifs in the genome of this yeast.
SHITT motifs are under strong purifying selection, both in genes and pseudogenes
Comparison of dN/dS between SHITT motifs and their genes suggests that purifying selection is stronger on motifs than on host genes. Unexpectedly, this is also true for pseudogenes (Figure 3). Nucleotide sequences of the five pseudogenes (CAGL0A04873g, CAGL0B05093g, CAGL0F00110g, CAGL0H00132g and CAGL0I00110g, Figure 2) were verified and confirmed as real pseudogenes (see ‘Materials and Methods’ section). Thus, we may hypothesize that SHITT motifs are transcribed, and confer a selective advantage. It is unlikely that they are translated though, thus we favor a possible role of the transcript in confering this advantage. To the best of our knowledge, there is no RNA interference described in C. glabrata, but it is possible that another mechanism of RNA regulation—relying on the formation of a putative RNA secondary structure—is active in this yeast. By using a dedicated program to look at secondary structures formed by megasatellite motifs, we did not find any evidence for the formation of a recurrent secondary structure common to several motifs (data not shown).
Selection pressure between orthologs and paralogs is different, and it was previouly shown that dN/dS ratios are lower for duplicated genes than for unique genes (30,31). This rather counterintuitive result was interpreted by proposing that duplicated genes are functionally more constrained because the encoded proteins play important functions in the cell. Megasatellite dN/dS values vary in a large range (from 0 to more than 1, Figure 3), suggesting different times of duplication and divergence. High values may correspond to the substantial relaxed selection observed by Kondrashov et al. (32), acting on recently formed gene duplicates, while lower values may correspond to more ancient duplicates, in which mutations were already fixed. Although the precise timing of duplication events cannot be ascertained, the presence of constrained motifs within ancient duplicates suggests that they play an important function in C. glabrata.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministère de l'Enseignement Supérieur et de la Recherche [Doctoral fellowship to T.R.]. Funding for open access charge: 700000/024310.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to A. Thierry, H. Muller, C. Bouchier and L. Ma for resequencing and reassembling C. glabrata subtelomeric sequences. They are very thankful to Benno Schwikowski and to the members of the Systems Biology group of Institut Pasteur for helpful discussions and strong technical support, as well as to the members of the Unité de Génétique Moléculaire des Levures, especially G. Fischer and I. Lafontaine for helpful comments. B.D. is a member of the Institut Universitaire de France.
REFERENCES
- 1.Thierry A, Bouchier C, Dujon B, Richard G.-F. Megasatellites: a peculiar class of giant minisatellites in genes involved in cell adhesion and pathogenicity in Candida glabrata. Nucleic Acids. Res. 2008;36:5970–5982. doi: 10.1093/nar/gkn594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thierry A, Dujon B, Richard GF. Megasatellites: a new class of large tandem repeats discovered in the pathogenic yeast Candida glabrata. Cell. Mol. Life Sci. 2009;67:671–676. doi: 10.1007/s00018-009-0216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richard G-F, Dujon B. Molecular evolution of minisatellites in hemiascomycetous yeasts. Mol. Biol. Evol. 2006;23:189–202. doi: 10.1093/molbev/msj022. [DOI] [PubMed] [Google Scholar]
- 4.Bowen S, Roberts C, Wheals AE. Patterns of polymorphism and divergence in stress-related yeast proteins. Yeast. 2005;22:659–668. doi: 10.1002/yea.1240. [DOI] [PubMed] [Google Scholar]
- 5.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haber JE, Louis EJ. Minisatellite origins in yeast and humans. Genomics. 1998;48:132–135. doi: 10.1006/geno.1997.5153. [DOI] [PubMed] [Google Scholar]
- 7.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 10.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. Distance methods for inferring phylogenies: A justification. Evolution. 1984;32:16–24. doi: 10.1111/j.1558-5646.1984.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 13.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. III. New York: Academic Press; 1969. pp. 21–123. [Google Scholar]
- 14.Kimura M. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 15.Bauer D. Constructing confidence sets using rank statistics. J. Am. Stat. Assoc. 1972;67:687–690. [Google Scholar]
- 16.R Development Core Team. 2008. R: A language and environment for statistical computing. R Foundation for Statistical Computing ( http://www.R-project.org) [Google Scholar]
- 17.Dijkstra EW. 1959. A note on two problems in connexion with graphs. Numerische Mathematik 1; pp. 269–271. [Google Scholar]
- 18.Felsenstein J. In Department of Genome Sciences. In: UoW, editor. Seattle: 2004. [Google Scholar]
- 19.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castano I, Pan S-J, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 2005;55:1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- 21.Fairhead C, Dujon B. Structure of Kluyveromyces lactis subtelomeres: duplications and gene content. FEMS Yeast Res. 2006;6:428–441. doi: 10.1111/j.1567-1364.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 22.Louis EJ, Naumova ES, Lee A, Naumov G, Haber JE. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics. 1994;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Therizols P, Duong T, Dujon B, Zimmer C, Fabre E. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc. Natl Acad. Sci. USA. 2010;107:2025–2030. doi: 10.1073/pnas.0914187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pâques F, Richard G.-F, Haber JE. Expansions and contractions in 36-bp minisatellites by gene conversion in yeast. Genetics. 2001;158:155–166. doi: 10.1093/genetics/158.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pâques F, Leung W-Y, Haber JE. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cormack BP, Falkow S. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics. 1999;151:979–987. doi: 10.1093/genetics/151.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richard G-F, Kerrest A, Lafontaine I, Dujon B. Comparative genomics of hemiascomycete yeasts: genes involved in DNA replication, repair, and recombination. Mol. Biol. Evol. 2005;22:1011–1023. doi: 10.1093/molbev/msi083. [DOI] [PubMed] [Google Scholar]
- 28.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:318–326. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan IK, Wolf YI, Koonin EV. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol. Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. research0008.0001–0008.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.