Abstract
Alternative splicing is a prevalent post-transcriptional process, which is not only important to normal cellular function but is also involved in human diseases. The newly developed second generation sequencing technique provides high-throughput data (RNA-seq data) to study alternative splicing events in different types of cells. Here, we present a computational method, SpliceMap, to detect splice junctions from RNA-seq data. This method does not depend on any existing annotation of gene structures and is capable of finding novel splice junctions with high sensitivity and specificity. It can handle long reads (50–100 nt) and can exploit paired-read information to improve mapping accuracy. Several parameters are included in the output to indicate the reliability of the predicted junction and help filter out false predictions. We applied SpliceMap to analyze 23 million paired 50-nt reads from human brain tissue. The results show at this depth of sequencing, RNA-seq can support reliable detection of splice junctions except for those that are present at very low level. Compared to current methods, SpliceMap can achieve 12% higher sensitivity without sacrificing specificity.
INTRODUCTION
RNA splicing is an important post-transcriptional step where one or more segments of the pre-mRNA are spliced out and the remaining segments (exons) are concatenated to form the mature mRNA product. By alternative splicing, it is possible to produce different transcripts (isoforms) from the same genetic locus. This process occurs in over 90% of multi-exon human genes (1,2) and greatly increases the diversity of possible transcripts in the transcriptome. Aberrant RNA splicing has been found to be associated with many human diseases (3,4). For this reason, techniques to identify and quantify splicing events are important to biology and medicine.
The most popular way to study the structure and abundance of spliced transcripts is through sequencing of expressed sequence tags (ESTs) (5). Traditionally, such studies were expensive and inefficient due to the low throughput of the Sanger method which was the main sequencing technology used in EST projects. However, with the recent advent of second generation sequencing technology (SGS), it is now feasible to conduct deep and comprehensive sequencing of transcriptomes in a high throughput and cost effective manner (6–8), making it possible to detect rare alternative splicing events. In such RNA-seq projects, tens or hundreds of millions of short sequences (30–100 nt) are read randomly from the population of transcripts under study. The first step of the analysis is thus the mapping of each short read to a reference genome to determine the genetic loci that may give rise to this read. For reads that are sampled completely within exonic regions, this mapping task can be handled by any existing short-read mapping programs, such as ELAND (Cox, unpublished software) and SeqMap (9).
However, the reads that are of most interest to us for novel isoform discovery are the ones that span across exon-exon junctions. These ‘junction reads’ cannot be mapped directly to the genome. One approach is to map the reads onto the known transcript sequences from the currently annotated exon library. Since the exon library is incomplete, this method cannot find the junctions that involve novel splicing events (10). In another approach, used in the recently developed TopHat (11) program, reads that are mappable on the reference genome are grouped into distinct clusters such that the reads within each cluster are linked together through overlapping regions. Each cluster then defines a putative exonic region. Subsequently, exon-exon junctions can be searched based on these putative exon definitions.
Clustering is a natural approach to find novel junctions in the first RNA-seq experiments (1,6,10,12–17) because the data generated at the early stage of the development of SGS are mostly very short reads (25–36 nt) that are not suitable for direct de novo detection of exon–exon junctions. However, the technology is improving rapidly, and currently the usable length of reads from some SGS instruments like the Illumina Genome Analyzer are typically in the range of 50–100 nt. The increased read length opens up the possibility to directly map the exon-exon junction without any reference to putative or annotated exons. Here we report a novel algorithm, based on the idea of using the mapping of half-reads as a way to identify the approximate location of a junction. Moreover, this method can be adapted to incorporate the extra information contained in paired-end sequencing data, to achieve a much higher level of specificity than attainable by single end sequencing. The method is implemented in a freely available Python program named SpliceMap (http://biogibbs.stanford.edu/∼kinfai/SpliceMap/).
MATERIALS AND METHODS
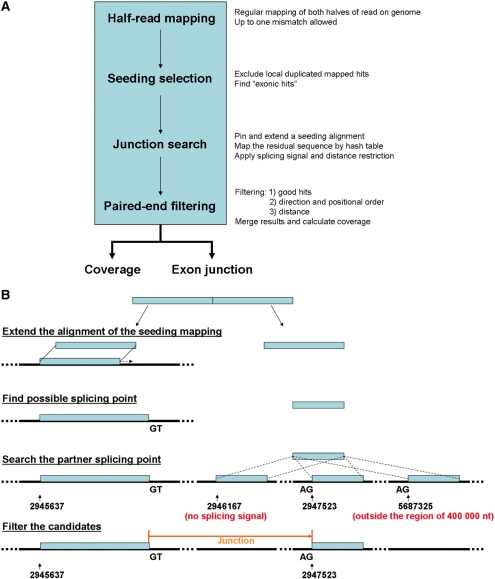
SpliceMap utilizes merely the reference genomic sequence to find the junction independently of existing exon annotation. It is possible to explore all exon splicing events, including known and novel ones, if the sequencing is of sufficient depth. The core notion is to pin down first the junction boundary on one of the two exons that are involved in the splicing event, before the mapping of the full junction. A read that spans a junction must have a match in the reference genome that is not shorter than its half length. Such a match then provides a seeding that can be used to identify a small genomic region for the search of the corresponding junction. There are four main steps in SpliceMap: half-read mapping, seeding selection, junction search and paired-end filtering (Figure 1). The last step is not applied when the data is not paired-end. For reads longer than 50 nt, we extract from them several overlapped 50-nt reads and then apply the standard method. For example, we split a 100-nt read to three segments (1–50, 26–75 and 51–100). An extra filter is added in the post-processing step for the long-read data to check the results with the full length information. In this way, we can find multiply junctions from a single long read.
Figure 1.
Workflow of standard SpliceMap and outline of junction search based on half-read mapping: (a) SpliceMap consists of four steps: half-read mapping, seeding selection, junction search and paired-end filtering. SpliceMap outputs coverage plot and junctions detected. (b) Each half-read is aligned to the genome and extended to obtain the partial alignment. The remaining part of the read, if at least 10 nt, will be used to search for its matches within a neighborhood (400000 nt). The GT–AG splicing signal is also used to filter the matches.
Half-read mapping
Taking advantage of the reasonably long reads (50 nt) offered by the newest models of second-generation sequencers, the half length (25 nt) can be reliably aligned to the reference genomic sequence with high probability. In this step, SpliceMap maps both halves of the read to the reference genome by any currently available short read mapping tools, such as SeqMap (9) and ELAND. The maximum mismatch allowed for the half read mapping can be chosen accordingly, based on the quality of data and read length. After mapping, the following steps are carried out chromosome by chromosome.
Seeding selection
We use the mapped hits of a half-read to narrow the search regions of the junction. These hits are extended base by base in the following step. Thus, we call the half-read mapped hit ‘seeding’. The mapped hits from the above steps are examined for seeding selection. Although the uniquely mapped hits are more reliable as seeding for junction search, one should not simply exclude all multiply mapped reads (i.e. reads mapped to more than one location) because doing so will greatly diminish the chance of detecting junctions with homologous sequences elsewhere, such as those in paralogous genes or pseudogenes. Instead of rejecting all multiply mapped hits, SpliceMap excludes only those hits that are within 400 000 nt of another hit from the same half-read. Because if two regions are identical within a distance of 400 000 nt, false splice predictions tend to form between these two regions which match the reads perfectly.
Junction search
For each seeding identified, the alignment on the reference genome is then extended base by base to find the splicing point (Figure 1). SpliceMap subsequently tries to find the partner splicing point that provides perfect match of the corresponding residual sequence of the original read, within a user-specified distance (set to be 400 000 nt in our examples). When the full reads are 50 nt in size, candidates of splicing point must meet two criteria: first, the alignment extension cannot be longer than 40 nt and the residual length has to be at least 10 nt; and second, the splicing point must be next to the canonical dinucleotides splicing signal GT and AG for donor and acceptor sites, because they appears in 98% known splice sites (18). The mapping of the residual sequence is achieved by searching 10-nt seeding in a pre-computed chromosome-wide hash table and then extending to complete the full alignment. In order to reduce false positive junctions, the results are discarded if the search yields multiple matches of the residual sequence satisfying the above criteria.
Paired-end filtering
When paired-end reads are available, the pairing information is used in this step to improve the specificity of junction detection. First, in the previous steps, three types of hits are identified as ‘good hits’, namely exonic hits, extension hits and junction hits. An exonic hit occurs if the two halves of a full read are mapped to locations that differ by exactly half of the read length. On the other hand, if a half-read hit can be extended maximally to an alignment length that is suitably long but yet shorter than the full read length, then it is regarded as an extension hit. Finally, junction hits are identified as above. To qualify as reliable hits, the hits generated from the two reads from a paired-end reads must satisfy the following conditions (i) both hits are ‘good hits’; (ii) their distance is not longer than 400 000 nt; (iii) the mapping direction and the positional order on the reference genomic sequence of two hits are in the consistent order with the experimental design (Figure 2).
Figure 2.
The direction and positional order of the paired-end reads (R1–R2). If the sequencing sample is the same as the original copy, the read R1 should be mappable in forward direction in 5′-end and R2 in reverse direction in 3′-end. If the sequencing sample is the complementary copy, the read R1 should be mappable in reverse direction in 3′-end and R1 in forward direction in 5′-end.
Parameters for assessment of reliability and optional filtering
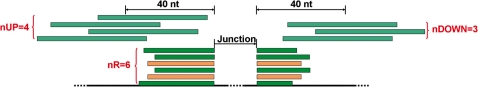
SpliceMap provided several parameters to facilitate the assessment of the reliability and the abundance of the detected junctions. For each junction, SpliceMap computes its nR (number of reads supporting this junction), nNR (number of non-redundant supporting reads), nUM (number of uniquely mapped supporting reads), nUP and nDOWN (number of mapped reads in upstream or downstream adjacent regions respectively) (Figure 3). We examined these parameters on a RNA-seq data set of 23 412 226 paired 50-nt reads provided to us by Illumina. The various requirements of these parameters lead to different optional parametric filterings.
Figure 3.
Schematic of the parameters to assess junctions in SpliceMap. The deep green reads are uniquely mapped supporting reads (nUM = 4) and the wheat reads are multiply mapped supporting reads. Thus, nR of this junction is 6. But some supporting reads are redundant, so nNR = 4. There are four and three uniquely mapped reads (grey green) in upstream and downstream adjacent regions of 40 nt respectively, so nUP = 4 and nDOWN = 3.
We found that for splice junctions near the 3′-end on a gene annotated by RefSeq as having only a single isoform, the number of supporting reads (nR) is significantly correlated with the overall expression (R2 = 0.7173) of the gene as measured by RPKM (number of reads per kb of transcript per million mappable reads) (10). This suggests that when alternative splicing may occur at a junction, we can use nR as an index for its usage. Table 1 shows the distribution of nR among predicted junctions before any optional parametric filtering. While overall only 29% of predicted junctions has nR = 1, this percentage increases to 74% among novel junctions. This is consistent with the notion that junctions not included in current annotations are likely to be relatively rare.
Table 1.
Distributions of nR among all junctions and among novel junctions predicted by SpliceMap from paired-end data (without any optional parametric filtering)
| nR | All junctions | Novel junctionsa |
|---|---|---|
| 1 | 49 888 (28.75%) | 26 744 (74.10%) |
| 2–5 | 49 462 (28.51%) | 7942 (22.00%) |
| 6–20 | 44 577 (25.69%) | 1222 (3.39%) |
| 21 ∼ 50 | 19 074 (10.99%) | 147 (0.41%) |
| 51 ∼ 200 | 10 478 (6.04%) | 34 (0.09%) |
| 201 ∼ 1000 | 1787 (1.03%) | 2 (0.01%) |
| 1000+ | 135 (0.08%) | 0 (0%) |
aNovel junctions are the ones that are not in RefSeq, Ensembl and KnownGene.
The nR of a junction is affected by the randomness in the representation or amplification of the RNA fragments in the library to be sequenced. If an RNA fragment is readily captured by the library preparation protocol (random hexamer priming in this case) and then highly amplified then it is likely to generate multiple reads of identical sequence. We call these ‘redundant’ reads. For junctions in low abundance isoforms, if two or more supporting reads are identical in sequence then they are likely to be redundant. In such cases, even if nR > 1 we still may not have a reliable prediction. We can greatly increase the specificity of our prediction by requiring that the junction be supported by more than one ‘non-redundant’ reads (nNR > 1). We can see that the EST validation rate is remarkably high once we require nNR > 1 (114 990/121 718 = 94%). When we restrict to novel junctions, the EST validation of predictions with nNR > 1 is still high (4494/7757 = 58%) (Table 2). Since the EST data is likely to be incomplete in its coverage of novel junctions in rare isoforms, the specificity of predictions with nNR > 1 should be considerably higher than 58%. We will examine this issue further in our discussion of our experimental validation results.
Table 2.
The numbers of detected junctions and the EST validation rates for various optional parametric filters
| SpliceMap |
|||||
|---|---|---|---|---|---|
| Optional filters# | – | nUM | nUP/nDOWN | nNR | nUM + nUP/nDOWN |
| Total junctions | 175 401 | 168 807 | 162 060 | 121 718 | 151 317 |
| Novel junctions | 36 091 | 32 060 | 27 497 | 7757 | 23 020 |
| Junctions with EST validation | 145 517 | 142 610 | 139 880 | 114 990 | 133 010 |
| Novel junctions with EST | 12 053 | 11 549 | 10 562 | 4494 | 9493 |
| EST validation rate | 82.96% | 84.48% | 86.31% | 94.47% | 87.90% |
| EST validation rate (novel) | 33.40% | 36.02% | 38.41% | 57.93% | 41.24% |
# ‘–’ represents no application of any parametric filters; ‘nUM’ filter requires nUM>0; ‘nUP/nDOWN’ filter requires nUP + nDOWN > 0; and ‘nNR’ filter requires nNR>1. For all ‘nUP/nDOWN’ filters, we set K = 40.
Although requiring nNR > 1 was effective in ensuring very high specificity of junction prediction, it reduced the number of predicted junctions by about 31%. Among novel predictions, the potential loss of sensitivity was even greater, i.e. from 36 091 to 7757. This motivated us to design optional filters to discriminate reliable versus unreliable predictions among junctions with nNR = 1. Instead of filtering junction prediction by nR or nNR, we require that the junction must be supported by at least one uniquely mapped reads (nUM > 0). This requirement excludes 3.8% junctions but increases the total specificity from 82.96 to 84.48% (Table 2).
The other optional filter is the requirements of a sufficient number of reads mapped adjacent to the splice site, on each side of the putative junction. To do this, recall that nUP is the numbers of reads (exon or junction reads) falling within a small region (say with a neighborhood size of K nt) upstream of the splice site of interest in the transcript, and nDOWN is the corresponding downstream adjacent reads. Thus our filter is to require each of nUP or nDOWN to be at least 1. Table 3 shows the number of detected junctions and the EST validation rates for neighborhood size K = 40, 80 and 160. We can see that this filtering excludes some junctions but improves the specificity as K decreases. By adjusting K, we can achieve a suitable balance of sensitivity and specificity in our junction predictions. When K = 40, it excludes 7.6% junctions but increases the total specificity from 82.96 to 86.91% (Table 3). The gains and losses of various optional parametric filterings are compared in Table 2.
Table 3.
The numbers of detected junctions and the EST validation rates for various neighborhood sizes used for nUP/nDOWN filtering right after the standard paired-end SpliceMap
| K = 40 | K = 80 | K = 160 | |
|---|---|---|---|
| Total junctions | 156 015 | 162 060 | 165 452 |
| Novel junctions | 25 441 | 27 497 | 29 342 |
| Junctions with EST validation | 135 586 | 139 880 | 141 758 |
| Novel junctions with EST | 9901 | 10 562 | 11 062 |
| EST validation rate | 86.91% | 86.31% | 85.68% |
| EST validation rate (novel) | 38.92% | 38.41% | 37.70% |
RESULTS
Specificity
We tested SpliceMap and TopHat on a RNA-seq data set of 23 412 226 paired 50-nt reads provided to us by Illumina. mRNA was purified from total RNA from human brain tissue with oligo-dT magnetic beads. cDNA was synthesized with random primer priming. ds–cDNA was sequenced using Illumina Genome Analyzers as recommended by the manufacturer. The data is publicly available in database GEO with accession number GSE19166. In this test, SpliceMap requires the reads to cover at least 10 nt on each side of the junction. One mismatch was allowed in seeding mapping based on 25-nt half-reads, but the mapping of the residual sequence is required to be perfect match. The allowance for mismatch on the seeding mapping allows detection of junctions with SNPs in the flanking exon sequences and also accommodates sequencing errors.
Here we report the results found by paired-end SpliceMap followed by the nUP/nDOWN filtering (K = 40) and the nUM filtering. In total, SpliceMap found 151 317 exon junctions, including 23 020 novel junctions, which were not reported in RefSeq (19), Ensembl (20) and KnownGene (21). In order to assess SpliceMap’s specificity, the detected junctions are aligned to human ESTs in GenBank (22). Because ESTs are 200–600 nt single reads, they are typically long enough to identify unique transcript fragments with high reliability. Thus if a match can be found in an EST sequence for a junction that was detected by our de novo splice discovery algorithm, then this can be regarded as independent experimental validation of the existence of the junction. As a comparison we also present the corresponding results by TopHat 1.0.12 (11) (the latest version as of 28 October 2009) (Table 4 and Figure 4).
Table 4.
The statistics of the results by TopHat and SpliceMap after nUM and nUP/nDOWN (K = 40) filterings
| SpliceMap | TopHat | |
|---|---|---|
| Total junctions | 151 317 | 133 722 |
| Novel junctionsa | 23 020 | 19 777 |
| Junctions with EST validationb | 133 010 (87.90%) | 117 113 (87.58%) |
| Junctions with nNR > 1 (multiple non-redundant reads) | 119 298 | – |
| Novel junctions with EST | 9493 (41.24%) | 7273 (36.78%) |
| Novel junctions with nNR > 1 | 7187 | – |
| Junctions with EST and with nNR > 1 | 112 962 | – |
| Novel junctions with EST and with nNR > 1 | 4242 | – |
aNovel junctions are the ones that are not in RefSeq, Ensembl and KnownGene.
bThe percentage in the parentheses is the EST validation rate.
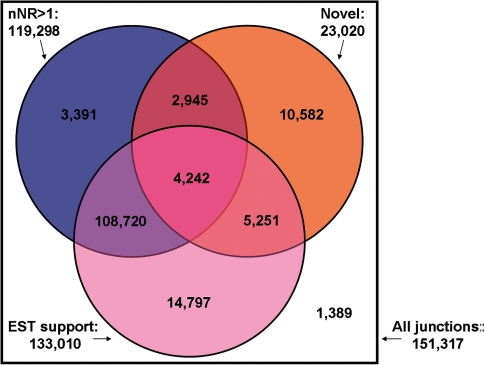
Figure 4.
The Venn diagram of the distribution of redundancy, novelty and EST evidence of the junctions predicted by SpliceMap. Only 1389 known junctions are with single non-redundant read and not supported by EST evidence.
We found that 87.9% (133 010) of the junctions detected by SpliceMap are supported by EST evidence. If we restrict to the 23 020 novel junctions predicted by SpliceMap, the percentage becomes lower (41.2%) (9493 junctions) but still represents a reasonable degree of EST support. We note that EST sequences are not comprehensive representation of the transcriptome and the percentage of junctions supported by EST is only a conservative estimate of specificity. We can see that SpliceMap has the similar EST validation rate but detected more junctions (151 317 versus 133 722) compared with TopHat.
Sensitivity
Table 5 gives the sensitivity of SpliceMap for detecting junctions in the 12 755 genes with a single isoform in RefSeq. The junctions are binned according to the expression level (in RKPM) of the corresponding gene, and the rate of detection is computed for each bin. It is seen that SpliceMap achieves very high sensitivity (>95%) for genes with high expression (RKPM > 20). The sensitivity is still high (90%) for genes with medium expression (5 < RKPM < 20) and remains substantial (40–67%) for genes with relatively low expression (1 < RKPM < 5). However, for genes with very low expression (RKPM < 1) the sensitivity drops to below 7%. Our result suggests that much deeper sequencing than 23 million paired-reads will be necessary for the analysis of such rare transcripts. Finally, Table 5 also shows that at all levels of gene expression, SpliceMap can detect more RefSeq annotated junctions than a representative of current method (TopHat).
Table 5.
The junction detection sensitivity for genes with different RPKM
| SpliceMap (%) | TopHat (%) | |
|---|---|---|
| 0 < RPKM ≤ 1 (2993) | 6.76 | 5.54 |
| 1 < RPKM ≤ 2 (1199) | 40.86 | 27.74 |
| 2 < RPKM ≤ 5 (2049) | 67.23 | 52.24 |
| 5 < RPKM ≤ 20 (3245) | 89.55 | 80.95 |
| 20 < RPKM ≤ 50 (1340) | 95.55 | 91.10 |
| 50 < RPKM ≤ 100 (522) | 97.18 | 93.87 |
| RPKM > 100 (408) | 95.66 | 88.58 |
We also examine the degree of junction recovery for the genes with single RefSeq isoform. For each gene, we measure its degree of junction discovery by computing the percentage of junctions that have been detected. The results are presented in Table 6. We can see that more genes detected by SpliceMap are of higher degree (80–100%) of completeness in junction discovery. In particular, this completeness may be important in downstream analyses such as isoform reconstruction and abundance estimation.
Table 6.
The distribution of genes with junction recovery
| SpliceMap | TopHat | |
|---|---|---|
| Number of genes detecteda | 8939 | 8777 |
| 1 ≤ Pb < 50 | 1076 | 1729 |
| 51 ≤ P < 80 | 1812 | 2319 |
| 81 ≤ P<100 | 1600 | 1347 |
| P = 100 | 4451 | 3382 |
aA gene is detected if at least one junction of the gene is detected.
bP is the rate (in %) of detection for junctions on the genes within the bin.
PCR validation
In addition to the evidence from EST, our novel junctions are validated experimentally by PCR experiments. Twenty predictions were randomly selected, including 18 novel exon skipping events and two novel exons detections, all without any currently known EST or human mRNA evidence. Eighteen novel skipping events come from five bins with different numbers of non-redundant reads: 1, 2, 3–5, 6–10 and >10. Three to four predictions were randomly selected from each bins. PCR validation showed that 17 (85%) were validated. Of the three false predictions, two occur in the bin of single non-redundant read. Thus, among novel predictions with nNR > 1, the PCR validation rate is 13/14 = 92.86%. This is significantly higher (using Z test, the P-value is 0.004) than the EST validation rate of 58% which we believe is an underestimate of specificity because of incomplete EST coverage for rare isoforms.
Comparison with ERANGE
To see how SpliceMap compares with a simple approach that does use the reference annotation; we also ran an annotation-dependent tool ERANGE (10) on the same data set. ERANGE identified 160 899 junctions, among which 127 043 junctions were also detected by SpliceMap. All results found by ERANGE are not novel but include some junctions that are not detected by SpliceMap, because ERANGE has relaxed requirements of the length of the minor flanking sequence. ERANGE requires at least 4 nt on each flanking sequence, while at least 10 nt is required by SpliceMap. The stricter requirement guarantees the reasonable reliability of the predictions of novel junction by SpliceMap. Novel junction discovery is the major function of SpliceMap, which therefore cannot be replaceable by annotation-independent ERANGE. Among 151 317 junctions found by SpliceMap, 24 274 are not reported by ERANGE, 23 020 of which are novel.
Comparison with BLAT
We also compared SpliceMap with BLAT, a tool commonly used for aligning EST sequences. In order to make a fair comparison, we optimized the parametric setting in BLAT and also filter the results by requiring the presence of canonical splicing signal. We found that when BLAT was run with the most non-stringent parameters and with no filtering, its sensitivity is ∼55% that of SpliceMap. However, in this non-stringent setting the junctions detected by BLAT are mostly false-positives (specificity is only 3%). With more stringent parameters and with the addition of post-alignment filtering steps, we can improve the specificity very substantially but at the expense of further loss of sensitivity. At its best setting, BLAT achieved a similar but still slightly lower level of specificity with a much lower sensitivity (70% lower) as compared to SpliceMap. The details of the comparison between the two methods on the Illumina Brain data set are presented in Table 7.
Table 7.
The comparison of various BLAT setting with SpliceMap
| BLAT | SpliceMap | ||||
|---|---|---|---|---|---|
| Max intron size (nt) | 750 000 | 750 000 | 400 000 | 400 000 | 400 000 |
| Tile size (nt) | 8–12 | 8–12 | 12 | 12 | N/A |
| Minscore | 30 | 30 | 50 | 50 | N/A |
| Mismatch | 1 | 1 | 0 | 0 | 1 |
| No. of splices | – | 1 | 1 | 1 | 1 |
| Min length of flanking seq. | – | 10 | 10 | 10 | 10 |
| Canonical splicing signal | – | – | – | required | required |
| Total junctions | 2 404 632 | 345 195 | 209 673 | 51 325 | 160 076 |
| Valid junctions | 75 147 | 61 671 | 45 714 | 41 173 | 137 965 |
| Specificity | 3.12% | 17.87% | 21.80% | 80.22% | 86.19% |
The first column corresponds to the default setting in BLAT. SpliceMap was run without paired-end information.
BLAT is a well-optimized alignment tool but it was not designed specifically for exon junction detection from short reads. Both SpliceMap and BLAT are based on using fast mapping of segments (seedings) of the reads to narrow the search regions for more detail alignment of the reads. As a splice detection tool specific for short RNA-seq data, SpliceMap uses 25-nt seeding while BLAT usually uses about 8- to 12-nt seeding. SpliceMap generates the seeding by using short-read alignment tools such as ELAND and SeqMap, while BLAT makes use of a hash table. The short seeding allows BLAT to find the splices that span small exons, but leads to many false mappings. As discussed in the above, these false predictions can be detected and removed in post-processing steps based on canonical splicing signal and paired-end information. However the post-processing filters cannot rescue the loss of sensitivity in the BLAT that is evident even when BLAT is run with the most tolerant parameter setting. BLAT has its own ad hoc post-processing steps that are not specially designed for exon junction detection. When the reads are short, these steps may remove many correct splices. Although we expect that BLAT’s performance may improve as the read length increases, for current RNA-seq data, it cannot replace a custom-designed tool for junction detection such as SpliceMap.
Running time
The better performance of SpliceMap is achieved at the cost of longer running time—it took SpliceMap 66 CPU hours to process all the 23 million reads. As a comparison, it took TopHat 12 CPU hours to process the same data set. However, it is worth pointing out that this may be due to the fact that SpliceMap was written in Python, while TopHat was written in C++. Moreover, in our experiments we used a 16-core server so that the overall running time was greatly reduced. The Python part of SpiceMap has been rewritten in C++ and speeds up about 4 times.
DISCUSSION
The use of paired-end sequencing with long reads (50-nt or longer) promises to greatly enhance our ability to characterize transcript isoforms, but this promise can be fulfilled only if suitable computational methods are available to analyze the data. In this paper we reported that 50-nt reads can support an approach of direct de novo detection of splice junctions without the need to first cluster reads to identify putative exons, and that this approach can achieve significantly higher sensitivity in junction detection than current leading methods of RNA-seq analysis. Our results were demonstrated by a systematic analysis of a new data set consisting of 23 million paired 50-nt reads from human brain tissue. Using EST sequences as validation, we found that our method SpliceMap can predict a junction with very high validation rate (95%) if the prediction is supported by at least two non-redundant reads (nNR > 1). However, even predictions supported by only single non-redundant read (nNR = 1) can have high specificity if we implement suitable additional filters to remove unreliable predictions. Including predictions with nNR = 1 that passed these filters greatly increased the sensitivity of detection and still maintained good validation rate (87.9%). Furthermore, EST validation rate is likely to underestimate the specificity of novel predictions, and this was confirmed by our experimental tests of a subset of novel predictions by PCR-based method.
The question of the sensitivity of junction detection from RNA-seq is an important issue that has not be thoroughly examined in the current literature. One way to frame this question is to ask, for a given sequencing depth, what is the C50 which is defined as the lower limit of copy number per cell in order for a given junction to have a 50% chance of being detected. Based on the assumption that one RKPM corresponds to about 0.3 copies per cell for the protocol used in this study (10), we can estimate from Table 5 that for a RNA-seq with a depth of 23 million paired 50-nt reads, the C50 is about one copy per cell.
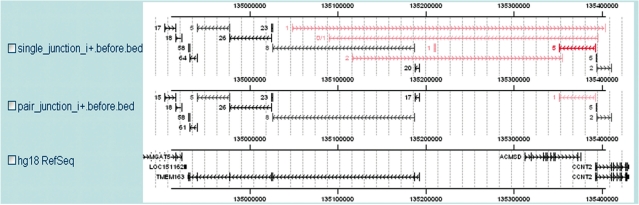
We found that paired-read information can help to reduce false discoveries. For the Illumina protocol used in to produce our data, the distance between two paired-end reads is about 200 nt in the mRNA and there may sometimes exist an intron between the two reads. Since the overwhelming majority of introns are smaller than 400 000 nt in size, SpliceMap will filter out any paired hits that are separated by more than 400 000 nt from all subsequent computations. Figure 5 shows an example of false predictions eliminated by the use of paired-end filtering. Compared to using only single reads (i.e. treating the same data set as 46 million unpaired 50-nt reads), the use of paired-end filtering increases the EST validation rate from 86.19 to 87.9% for all predictions and from 38.53 to 41.24% for novel predictions, without compromising the sensitivity of detection of annotated junctions. Although, single-end SpliceMap still makes better performance than TopHat, we recommend researchers using paired-end sequencing if possible. Since the EST validation rate underestimates the true specificity of novel predictions, the reduction of false predictions should be more substantial than indicated by the above numbers.
Figure 5.
Filtering by paired-end information. The top two tracks are the results from single-end SpliceMap and paired-end SpliceMap before nUP, nDOWN and nUM filtering, respectively. The known junctions detected are in black and the novel ones in red. Single read analysis predicts several junctions that are very long and jump across genes. These are false positive results and the paired-end information helps to remove them.
FUNDING
National Institutes of Health (1R01HG004634 to W.H.W.); Junior faculty grant from the Edward Mallinckrodt Jr. Foundation (to Y.X.) Funding for open access charge: 1R01HG004634.
Conflict of interest statement. None declared.
AUTHOR CONTRIBUTIONS
KFA, HJ and WHW conceived the study, designed the algorithm and carried out data analyses. KFA implemented the algorithm and drafted the manuscript. HJ and WHW revised the manuscript. L.L and XY. performed the experimental validation.
ACKNOWLEDGEMENTS
Shujun Luo and Gary Schroth of Illumina Inc. provided the RNA sequencing data. Gary Schroth also provided helpful comments on the manuscript.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;462:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 3.Nagao K, Togawa N, Fujii K, Uchikawa H, Kohno Y, Yamada M, Miyashita T. Detecting tissue-specific alternative splicing and disease-associated aberrant splicing of the PTCH gene with exon junction microarrays. Hum. Mol. Genet. 2005;14:3379–3388. doi: 10.1093/hmg/ddi369. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Hubbell E, Hu JS, Mei G, Cline M, Lu G, Clark T, Siani-Rose MA, Ares M, Kulp DC, et al. Gene structure-based splice variant deconvolution using a microarray platform. Bioinformatics. 2003;19:315–322. doi: 10.1093/bioinformatics/btg1044. [DOI] [PubMed] [Google Scholar]
- 5.Adams MD, Soares MB, Kerlavage AR, Fields C, Venter JC. Rapid cDNA sequencing (expressed sequence tags) from a directionally cloned human infant brain cDNA library. Nat. Genet. 1993;4:373–380. doi: 10.1038/ng0893-373. [DOI] [PubMed] [Google Scholar]
- 6.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shendure J. The beginning of the end for microarrays? Nat. Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 11.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 14.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 16.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 17.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 18.Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:501–504. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 22.Boguski MS, Lowe TM, Tolstoshev CM. dbEST–database for “expressed sequence tags”. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]