Abstract
Despite the fact that all 23S rRNA nucleotides that build the ribosomal peptidyl transferase ribozyme are universally conserved, standard and atomic mutagenesis studies revealed the nucleobase identities being non-critical for catalysis. This indicates that these active site residues are highly conserved for functions distinct from catalysis. To gain insight into potential contributions, we have manipulated the nucleobases via an atomic mutagenesis approach and have utilized these chemically engineered ribosomes for in vitro translation reactions. We show that most of the active site nucleobases could be removed without significant effects on polypeptide production. Our data however highlight the functional importance of the universally conserved non-Watson-Crick base pair at position A2450–C2063. Modifications that disrupt this base pair markedly impair translation activities, while having little effects on peptide bond formation, tRNA drop-off and ribosome-dependent EF-G GTPase activity. Thus it seems that disruption of the A2450–C2063 pair inhibits a reaction following transpeptidation and EF-G action during the elongation cycle. Cumulatively our data are compatible with the hypothesis that the integrity of this A-C wobble base pair is essential for effective tRNA translocation through the peptidyl transferase center during protein synthesis.
INTRODUCTION
From an evolutionary point of view, the ribosome is one of the most ancient cellular particles (1). This complex molecular machine, composed of ∼2/3 ribosomal RNA (rRNA) and 1/3 ribosomal proteins (r-proteins), produces proteins by translating the genetic information carried by messenger RNA (mRNA) sequences as the last step of gene expression according to the central dogma of molecular biology. Biochemical, genetic and structural studies firmly established that the ribosome is a ribozyme polymerizing amino acids into a growing peptide within the catalytic core at a site called the peptidyl transferase center (PTC) (2). This active site is located in a cavity on the interface side of the large ribosomal subunit and consists almost entirely of 23S ribosomal RNA (28S rRNA for eukaryotes) nucleotides (3,4). The active site nucleotides that intimately approach the 3′ CCA ends of both A- and P-site tRNA substrates are universally conserved and have been referred to as the inner shell of the PTC (5).
In order to produce a protein, ribosomes progress through the elongation cycle which consists of the initiation, elongation, termination and finally the recycling stages (6). During the elongation phase the peptide chain extension occurs immediately after accommodation of the aminoacyl-tRNA (aa-tRNA) into the PTC A-site. This results in the transfer of the peptidyl moiety from the P-site bound peptidyl-tRNA (pept-tRNA) to the A-site bound aa-tRNA, the first of two catalytic reactions facilitated by the PTC. Subsequently, the two tRNAs translocate to the P- and E-sites, respectively, in a multi-step process promoted by the elongation factor G (EF-G). In contrast to transpeptidation, where many mechanistic details have been revealed (2), the molecular processes underlying the tRNA translocation steps are far from being understood. According to the hybrid state model, translocation starts upon peptidyl transfer with the spontaneous movement of deacylated tRNA and pept-tRNA acceptor ends from the P-site to the E-site and from the A-site to the P-site correspondingly, while their anticodon stem-loops remain at the previous positions resulting in the hybrid P/E and A/P tRNA states (7). Global conformational rearrangements of the ribosomal subunits together with the ratchet-like intersubunit rotation accompany the tRNAs motions (8). Finally, coupled mRNA-tRNAs movement resulting in the population of classical E/E and P/P sites is driven by the action of the GTPase EF-G. GTP hydrolysis occurs after conformational changes on EF-G induced by its interaction with the ribosome, probably triggered by the A2660 exocyclic N6 amino group in the sarcin-ricin-loop of the 23S rRNA (Escherichia coli 23S rRNA nomenclature is used here and throughout the manuscript) (9). On the other hand, under certain in vitro conditions tRNA translocation can occur even in the complete absence of EF-G revealing this reaction inherent to the ribosome itself (10–13). At the end of the open reading frame and in response to an A-site bound class I release factor, the fully translated protein is released from the P-site located tRNA by pept-tRNA hydrolysis, the second catalytic reaction promoted in the PTC (2,4).
Unexpectedly, given the nature of the PTC with its universally conserved nucleotides, standard (5,14–18) and ‘atomic mutagenesis’ studies (19–21) have shown that the nucleobase identities are actually not critical. In fact, rRNA backbone groups have been identified to directly participate in (the ribose 2′-OH at A2451 for amide bond formation) (22) or indirectly trigger (the ribose at A2602 for pept-tRNA hydrolysis) (19) chemical reactions in the PTC. For peptide bond synthesis another critical ribose 2′-OH backbone group has been identified which resides on the terminal residue of P-site tRNA (23,24), whose functional relevance, however, has recently been challenged (25). It is feasible that the rRNA nucleobase identities of the universally conserved active site residues might be crucial for other ribosomal functions distinct from catalysis. In an attempt to elucidate functions of the inner core nucleotides beyond peptidyl transfer and pept-tRNA hydrolysis, we chemically engineered the PTC via the ‘atomic mutagenesis’ approach (20), a technique that allows the manipulation of single functional groups of 23S rRNA residues in the context of the 70S ribosome. Thus far the atomic mutagenesis approach has been applied to study individual 50S subunit-promoted reactions of the elongation cycle in isolation (9,19–22). To circumvent these limitations we improved this technology to study the consequences of PTC nucleobase manipulations in a more physiologically relevant set-up, namely during in vitro translation of either a poly(U) mRNA analog or a genuine mRNA coding for r-protein S8.
MATERIALS AND METHODS
Reconstitution of Thermus aquaticus ribosomes
To investigate nucleotide positions U2585, A2602, U2506, A2451, A2453, A2450 and C2063, we generated four different gapped-circularly permuted (cp)-23S rRNA constructs (Supplementary Figure S1). The cp-23S rRNAs were generated, in vitro reconstituted to 50S particles and reassociated with native T. aquaticus 30S as described previously (19–21). To investigate position U2585, the synthetic RNA oligo was ligated to the 3′-end of the cp2623-2576 (the first mentioned nucleotide always indicates the new 5′-end and the second residue the 3′-end of the cp-23S rRNA transcript) via a splinter ligation approach prior to in vitro reconstitution (19). Synthetic RNA oligos were purchased from Dharmacon (wild-type, purine, C3-linker modifications), CureVac (isoguanosine) and from Microsynth (2-pyridone), while ribose-abasic modifications were synthesized using the 2′-O-TOM-methodology [(20,22) and references therein].
PolyU-dependent poly(Phe) synthesis
For every time point 20 pmol of reconstituted T. aquaticus 50S particles were associated with 2 pmol of native E. coli 30S subunits and precipitated with three volumes of ethanol for 1 h at −80°C. Ribosomal particles were resuspended in 8 µl buffer containing 20 mM Hepes/KOH pH 7.6, 150 mM NH4Cl, 14 mM MgAc2, 1 mM DTT, 2 mM spermidine, 0.05 mM spermine followed by 15 min incubation with 25 µg polyU at 37°C (26). Translation reactions (25 µl) were launched at 42°C upon addition of recombinant his-tagged E. coli elongation factor (see Supplementary Methods) EF-Tu (0.92 µM), EF-G (0.26 µM), EF-Ts (0.33 µM) and PheRS (1.57 µM) together with ATP (1.2 mM), GTP (0.6 mM), acetyl phosphate (3.4 mM), L-[3H]phenylalanine (0.04 mM, specific activity 640 cpm/pmol) and E. coli deacylated tRNAPhe (1.92 µM) (9). Detection of produced poly(Phe) peptides was carried out by precipitation with TCA followed by filtration through glass-fibre filters and scintillation counting according to (26). Alternatively, we used l-[14C]phenylalanine (270 cpm/pmol) and 4.63 µM tRNAPhe with 40 pmol of reconstituted ribosomes in 21.5 µl reactions for subsequent thin layer chromatography detection of poly(Phe) peptides (see below).
Thin layer chromatography
In total, 21.5 µl of translation reactions containing [14C]-labeled peptides were stopped after 90 min of incubation upon addition of 2.5 µl of 10% Triton X100 and 10 µl of 0.5 M KOH. After 30 min incubation at 37°C total samples were loaded on an HPTLC F254 Silica gel 60 plate (Merck) in 3 µl aliquots ∼1 cm from the bottom of the plate alongside with the marker di-Phe peptide (1 µg) and penta-Phe (10 µg). The plate was dried with a hair dryer repeatedly after each aliquot application and the vertical TLC was performed (∼90 min) in the running buffer n-butanol:acetic acid:water (4:1:1, v/v) until the buffer front reached 0.5 cm to the top of the plate (27). The plate was dried at 58°C for 10 min. The marker lanes were sprayed with 0.25% ninhydrin solution (in acetone) and the plate was incubated for another 10 min at 58°C to detect the marker spot position. Approximately 300 cpm of [14C]Phe were subsequently applied onto the stained marker spots. The TLC plate was subsequently wrapped in plastic foil and exposed to a phosphoimager screen for 24–72 h.
Peptide bond formation
Peptide bond formation rates were determined using the puromycin assay. Reactions were performed as described before (20) employing 0.3–0.8 pmol N-acetyl-[3H]Phe-tRNAPhe (15 000 cpm/pmol) or f[3H]Met-tRNA (30 000 cpm/pmol) as P-site substrate and 2 mM puromycin as acceptor substrate. Preparation of the pept-tRNA substrates was according to (20,28).
Dipeptide formation and pept-tRNA drop off measurement
To assess the peptidyl transferase activity of reconstituted ribosomes using full-length tRNA substrates 70S ribosomes (associated from 20 pmol reconstituted 50S subunits and 8 pmol native T. aquaticus 30S subunits) were incubated with 80 µg of poly(U) for 15 min at 37°C for P-site binding of 6 pmol unlabeled N-acetyl-Phe-tRNAPhe in a 98 µl reaction containing 20 mM Tris/HCl pH 7.5, 6 mM Hepes/KOH pH 7.5, 7.6 mM MgCl2, 100 mM NH4Cl, 3.6 mM spermidine, 0.04 mM spermine, 5.5 mM ß-mercaptoethanol and 0.05 mM EDTA. The peptidyl transferase reaction was initiated by the addition of ternary complex [3H]Phe-tRNA:EF-Tu:GTP (20 000 cpm/pmol) preformed in the ratio 4 pmol:18.84 pmol:2 nmol at 37°C in the buffer 20 mM Hepes/KOH pH 7.5, 6 mM MgCl2, 150 mM NH4Cl, 2 mM spermidine, 0.05 mM spermine, 4 mM β-mercaptoethanol. After 15 min half of the reaction (50 µl) was stopped by the addition of 7.15 µl of 10 M KOH and incubation at 37°C for 15 min. Subsequently 143 µl of 1 M KH2PO4 and 103 µl of 1 M HCl were added. At this pH value (pH 2.5) the reaction product, N-acetyl-Phe-[3H]Phe, can be extracted into ethyl acetate and was analyzed by liquid scintillation counting (21).
To measure the amount of N-acetyl-Phe-[3H]Phe-tRNAPhe bound to the ribosomes after transpeptidation, the second half of the reaction was diluted on ice with 150 µl of cold buffer containing 60 mM Hepes/KOH pH 7.6, 100 mM NH4Cl, 7.6 mM MgCl2, 9.5 mM β-mercaptoethanol, 1.5 mM spermidine, 0.35 mM spermine. The total reaction was passed through a nitrocellulose filter (Millipore; 0.45 µm pore size) and washed twice with 0.5 ml of the above dilution buffer. Dried filters were soaked in scintillation cocktail and counted.
Uncoupled multiple turnover EF-G GTP hydrolysis
GTP hydrolysis by recombinant his-tagged T. thermophilus EF-G was induced by reassociated 70S ribosomes (reconstituted 50S and native T. aquaticus 30S in a ratio of 10:4). A single time point reaction contained 0.15 µM reconstituted ribosomes, 0.37 µM deacylated tRNAPhe, 46.7 µM γ-[32P]-GTP and 1.13 µM EF-G and was performed in a final volume of 13.4 µl. Reactions were stopped by formic acid addition and the amount of inorganic phosphate released was analyzed by thin layer chromatography (Polygram CEL300 PEI/UV) as described (9).
S8 mRNA in vitro translation
Twenty picomoles of 70S ribosomes (reconstituted T. aquaticus 50S and native E. coli 30S subunits in 10 : 1 ratio) were precipitated for 1 h at −80°C with three volumes of EtOH and dissolved in 18 µl of the buffer containing 44% (v/v) of the S30 premix (Promega), 22% (v/v) of E. coli S100 extract (29), amino acid mixture without Met and Cys (0.11 mM each), 10 µg bulk E. coli tRNA (Sigma), 8 U of RNase inhibitor (Fermentas), 11 mM MgCl2, [35S]Met + [35S]Cys (2.5 mCi/ml, 1000 Ci/mmol). The reaction was pre-incubated (5 min at 37°C) and in vitro translation was initiated upon addition of 1 µg in vitro transcribed mRNA encoding S8 r-protein of M. thermolithotrophicus (30) and simultaneous transfer to 42°C. After 60 min of incubation the protein fraction of the reaction (final volume: 20 µl) was precipitated for 1 h with four volumes of cold acetone at −20°C. Samples were then dissolved in the Laemmli buffer, denaturated at 95°C for 5 min and analyzed by SDS–PAGE. As an S8 marker control we used the protein fraction of E. coli cells containing in vivo synthesized recombinant [35S]Met-labeled S8 of M. thermolithotrophicus. The dried gel was exposed to a phosphoimager screen overnight.
Molecular dynamics simulations
Ribosome crystal structure coordinates have been obtained from Protein Data Bank database [1VQO for the empty ribosome (31); 2WDH and 2WDJ for the PRE state ribosome (32)]. Prior to analysis, the 23S rRNA numbering in the 1VQO structure has been adjusted to follow the E. coli nomenclature. The coordinates of PTC residues in a distance of 32 Å from A2450, A2062 and C2063 have been extracted. These structural PTC fragments have been dissolved with water (TIP3 model) and ionized with 0.5 M NaCl and used for molecular dynamics (MD) simulation with NAMD (33), using all-atom charmm force field parameters. During simulations only residues within a radius of 20 Å from A2450, A2062 and C2063 have been designated as flexible. MD experiment consisted of minimization (0.3 ns) followed by simulation (2 ns) at the initial temperature of 310 K. Result analysis has been performed with the VMD (34) and PyMol software.
RESULTS
Construction of circularly permuted 23S rRNAs (cp-23S rRNAs) for atomic mutagenesis of inner core PTC nucleotides
To implement nucleotide manipulations of PTC residues (Figure 1) on the atomic level, we created a set of four different cp-23S rRNA constructs (Supplementary Figure S1) (19–21). Each cp-23S rRNA transcript has the natural 5′- and 3′-ends covalently linked and new ends established such that a sequence gap is introduced encompassing the PTC nucleotide under investigation. Subsequently the cp-23S rRNA transcript and a synthetic RNA oligo complementing the gap were reconstituted in vitro together with 5S rRNA and the total protein extract of T. aquaticus large ribosomal subunits to assemble functional 50S particles (35). The chemically synthesized RNA oligonucleotides had either the wild-type (wt) sequence or carried single non-natural nucleotide analogs at PTC nucleotide positions C2063, A2450, A2451, A2453, U2506, U2585 or A2602.
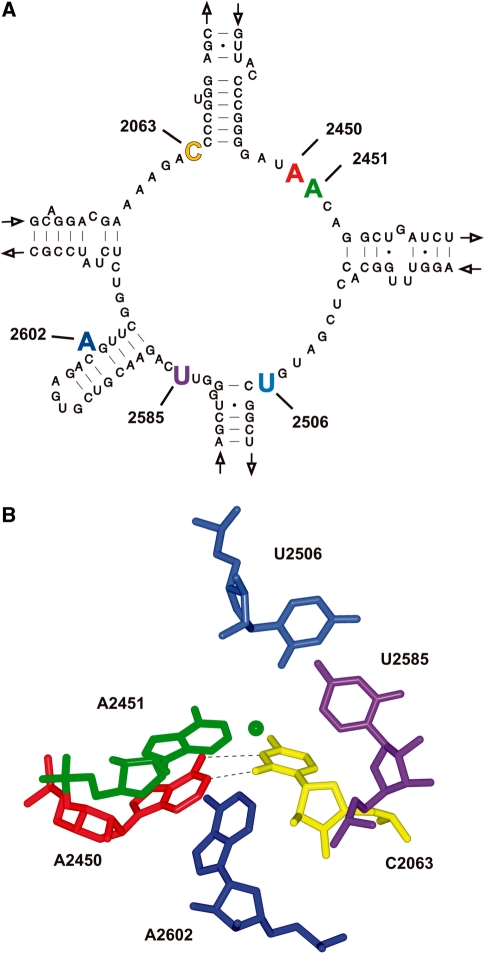
Figure 1.
Structural organization of the inner core PTC nucleotides. (A) Secondary structure of the central loop of domain V of T. aquaticus 23S rRNA (52). The five inner core residues (A2602, U2585, U2506, A2451 and C2063) as well as the ‘second layer’ residue A2450 are highlighted in bold and are colored. (B) The 3D architecture of the PTC structure using the same color code as in (A). The hydrogen bonding interactions of the non-Watson-Crick base pair A2450–C2063 are shown. The position of the nitrogen atom of the attacking α-amino group of the Phe-tRNA in the A-site is depicted as green sphere. The figure was generated from to the T. thermophilus 70S atomic coordinates (pdb files 2WDK and 2WDL) (32).
Removal of active site nucleobases and the effects on in vitro translation activities
Since the removal of nucleobases at the five inner core PTC residues C2063, A2451, U2506, U2585 or A2602 (Figure 1) has been shown previously not to interfere significantly with isolated ribosome-catalyzed reactions (19–21), we now investigated the performance of the chemically engineered ribosomes under biologically more meaningful conditions, namely during poly(U)-directed in vitro translation. In line with previous studies, elimination of the inner core nucleobases by introducing abasic site analogs (at A2451, U2585, U2506) or by deleting the entire nucleotide at A2602 (Δ2602) did not significantly affect the poly(Phe) synthesis (Table 1). Among the PTC nucleotides located in close contact to the CCA ends of both A- and P-site tRNAs, the highly flexible A2602 and U2585 have been proposed to play a pivotal role in the orchestrated movement of the tRNAs (36). Since the individual elimination of the nucleobases did not hamper in vitro translation, we next created the 2585 abasic/Δ2602 double mutant in order to investigate a potential functional redundancy of these residues. This double mutant, however, retained wt-like activities in the poly(Phe) assay. In fact moderately but reproducibly increased translational activities were evident (Table 1). This indicates that neither of these residues is crucial for triggering or assisting the movement of the CCA tRNA ends during in vitro translation. Control poly(Phe) experiments in the presence of thiostrepton or in the absence of EF-G showed, that product formation on chemically engineered ribosomes was still dependent on an authentic tRNA translocation (Supplementary Figure S2). The sole inner core nucleobase whose removal (by introducing an abasic site or a C3-linker nucleotide analog) was not compatible with efficient poly(Phe) synthesis was the cytosine at position 2063 (Figure 2A and C). In this case peptide synthesis was markedly reduced (>4.5-fold), albeit not completely inhibited (Table 1).
Table 1.
In vitro translation activities and peptide bond formation rates of ribosomes carrying nucleotide modifications in the PTC
| Position | Modification | Poly (Phe) synthesisa | Peptide bond formation (krel)b |
|---|---|---|---|
| – | none (wt) | 1.00 | 1.00 |
| A2602 | Δ | 0.74 | 1.09c |
| U2585 | aba | 1.56 | 1.49d |
| 2602/2585 | Δ/aba | 1.78 | 1.32 |
| U2506 | aba | 1.80 | 2.21d |
| A2453 | aba | 1.20 | 0.53e |
| A2451 | aba | 0.72 | 0.53e |
| d-aba | 0.13 | 0.01e | |
| A2450 | aba | 0.07 | 0.61e |
| Pu | 0.03 | 0.70 | |
| isoG | 0.60 | 0.82 | |
| C2063 | aba | 0.22 | 0.98c |
| C3 | 0.34 | 0.71 | |
| 2-pyr | 0.43 | 0.61 |
Incorporated nucleotide analogs (see also Figure 2A): ribose-abasic site (aba), 2′-deoxyribose-abasic (d-aba), C3-linker nucleotide analog (C3); purine (Pu), isoguanosine (isoG), or the 2-pyridone nucleoside analog (2-pyr).
aThe amount of poly(Phe) catalyzed by reconstituted wt ribosomes after 120 min of incubation was taken as 1.00 and compared to the peptide yields produced by ribosomes carrying PTC nucleotide modifications. The values shown represent the mean of at least two independent poly(Phe) reactions.
bPeptide bond formation rates (krel) using the puromycin reaction were determined from experimental points in the linear range of the reaction within the first 15 min of incubation of at least two independent time course experiments. The rate observed with reconstituted ribosomes carrying the wt oligo was taken as 1.00.
cData taken from (19).
dData taken from (21).
eData taken from (20).
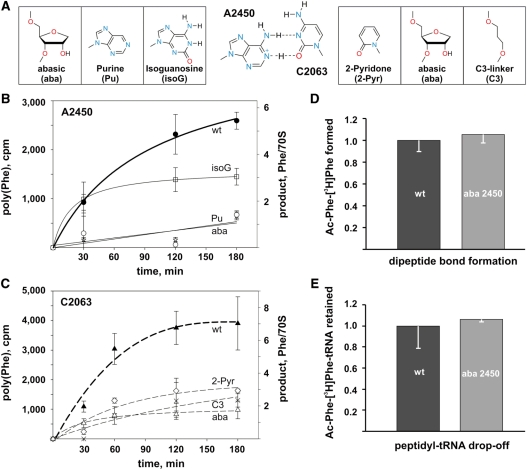
Figure 2.
Effects of A2450–C2063 base pair modification on in vitro translation, peptide bond formation and pept-tRNA drop-off. (A) Schematic illustration of the A–C wobble base pair (middle) and the introduced nucleotide modifications at positions A2450 (left) and C2063 (right). (B) In vitro translation activities of ribosomes with A2450 or (C) with C2063 modifications. The amount of polyphenylalanine produced by reconstituted ribosomes (in cpm or in phenylalanine per 70S ribosome) is plotted as a function of time. (D) The amount of extracted dipeptidyl product (N-acetyl-Phe-[3H]Phe) utilizing N-acetyl-[3H]Phe-tRNAPhe and [3H]Phe-tRNAPhe as reaction substrates formed on reconstituted ribosomes carrying the wt oligo was compared to the product obtained with ribosomes carrying an abasic site at position 2450 (aba). Product formation of wt ribosomes (∼0.1 pmol of N-acetyl-Phe-[3H]Phe per 1 pmol of 70S) was taken as 1.0 (E) The same ribosomal complexes as in (D) were used to quantify the fraction of N-acetyl-Phe-[3H]Phe-tRNAPhe retained on the ribosome after peptide bond formation via filtration through nitrocellulose membranes. The amount of ribosome-bound dipeptidyl-tRNA on reconstituted wt ribosomes was taken as 1.00. Background values of retained [3H]Phe-tRNAPhe on the nitrocellulose membrane in the presence of native 30S subunits alone were subtracted (∼0.025 pmol per 1 pmol of 30S subunits). In (A–E) the values represent mean and standard errors of at least three independent experiments.
Disruption of the C2063–A2450 wobble pair inhibits poly(Phe) synthesis
In the catalytic center (Figure 1), C2063 forms an universally conserved non-canonical base pair with A2450 (37). A2450 is a ‘second layer’ PTC nucleotide and is located adjacent to A2451, the key player in peptidyl transfer (22). Removal of the adenine base at A2450 by the incorporation of the analogous abasic site variant at the other side of this A–C base pair resulted in an almost complete inhibition of the synthetic activity of the ribosomal particle (Figure 2B, Table 1). In order to assess the functional importance of the integrity of this non-Watson–Crick pair for in vitro translation, we used the power of the atomic mutagenesis approach and introduced less invasive nucleotide modifications by replacing or altering only single functional groups or atoms at both nucleobases that are expected to either weaken or strengthen the A2450–C2063 interaction. Removing the exocyclic N6 amino group at A2450 by introducing a purine analog, which eliminates a H-bond donor (Figure 2A), also severely interfered with poly(Phe) synthesis (Figure 2B, Table 1). Similarly, when a 2-pyridone nucleoside was incorporated instead of C2063, thereby replacing the N3 by a carbon atom and thus eliminating the potential to serve as a H-bond acceptor for the proton of the A2450 N6 amino group (Figure 2A), the ribosomes were unable to efficiently produce poly(Phe) products (Figure 2C, Table 1). We noted that all A2450 modifications causing base pair disruption (Figure 2A) reproducibly manifested more profound effects on the translation rates than C2063 modifications. In contrast, introducing an isoguanosine (isoG) nucleotide analog at position 2450, which should facilitate hydrogen bonding with C2063 by providing a protonated N1 (Figure 2A), was compatible with in vitro translation. These data are compatible with the idea that the integrity of the A2450–C2063 base pair in the heart of the PTC is of functional importance for protein synthesis. We note that the effects connected with the base pair disruption at A2450–C2063 were unique since disrupting another highly conserved PTC A-C wobble base pair at A2453–C2499 did not inhibit poly(Phe) synthesis (Table 1). This highlights the functional importance of the A2450–C2063 base pair, which likely goes beyond merely stabilizing the productive active site conformation.
Disruption of the A2450–C2063 pair affects a reaction after peptide bond formation
According to the recently proposed catalytic model of peptide bond formation, the most pivotal functional 23S rRNA group is the ribose 2′-OH at A2451 (22). Therefore the almost wt levels of poly(Phe) activity using ribosomes carrying the abasic nucleotide analog at position 2451 could be expected since the ribose moiety was not altered (Table 1). However, the additional removal of the 2′-OH group by placing a deoxyribose-abasic site modification at 2451 severely reduced polypeptide synthesis (Table 1).
Modifications at A2450 and C2063 that were shown to interfere with efficient poly(Phe) synthesis (Figure 2B and C, Table 1) needed to be assessed in the peptidyl transfer reaction. Hence we employed the puromycin reaction under single turnover conditions (20), and showed that the A2450–C2063 modifications that inhibited in vitro translation were efficiently able to form amide bonds (Table 1). Since puromycin is a minimal A-site substrate which has been shown to be hyper-sensitive to active site mutations compared to genuine aa-tRNA substrates (5), we also tested the abasic A2450 ribosome in a dipeptide bond formation assay using two full-length tRNA substrates in the P- and A-sites. Accordingly, the chemically engineered ribosomes formed equal amounts of N-acetyl-Phe-[3H]Phe dipeptidyl product compared to the in vitro reconstituted wt control particles (Figure 2D). It is important to note that subsequent to dipeptidyl-tRNA formation the ribosomal complexes were filtered through nitrocellulose membranes in order to assay for premature dipeptidyl-tRNA drop-off in the abasic 2450 ribosomes. These experiments, however, clearly demonstrated that the modified ribosomes form and retain essentially equal amounts of N-acetyl-[3H]Phe-tRNAPhe compared to the control (Figure 2E) and thus eliminated tRNA drop-off as reason for the observed defects in poly(Phe) production (Figure 2B). Additionally, since abasic A2450 ribosomes can produce N-acetyl-[3H]Phe-tRNAPhe as efficiently as the wt ribosomes (Figure 2D) argues against significant problems during aa-tRNA accommodation into the PTC A-site.
Disruption of the A2450–C2063 pair does not inhibit EF-G GTPase activation
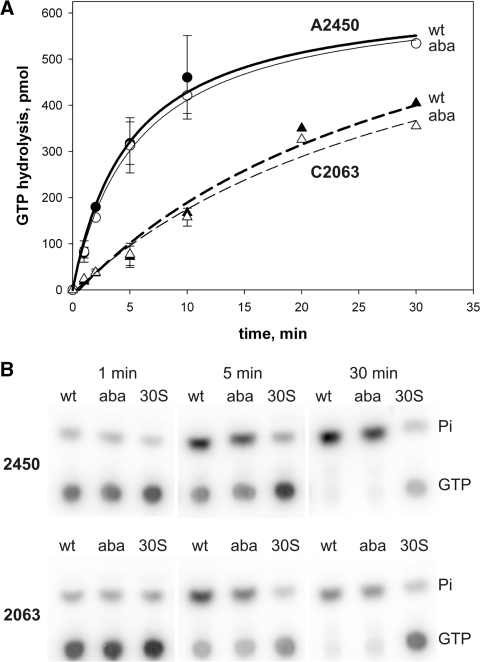
The next step after peptidyl transfer in the ribosomal elongation cycle is the EF-G promoted translocation of pept-tRNA and deacylated tRNA from the A-, and P-sites to the P- and E-sites, respectively. Therefore, we next tested the ribosomes carrying modified 50S subunits in their ability to activate EF-G GTPase. The ribosome induces GTP hydrolysis on EF-G when the elongation factor binds to the intersubunit space near the A-site where it interacts with A2660 of the sarcin-ricin-loop of 23S rRNA which triggers GTPase activation (9). Disruption of the A2450–C2063 base pair, by introducing abasic nucleotide analogs, however, did not affect the EF-G GTPase, since identical GTP hydrolysis rates were observed in the mutant and the respective wt control ribosomes (Figure 3). This indicates that the reconstituted ribosomes harboring abasic site analogs at A2450 or C2063 could productively interact with EF-G and stimulate efficient GTP hydrolysis.
Figure 3.
EF-G GTPase activities triggered by ribosomes containing modifications at A2450 or C2063. (A) EF-G catalyzed GTP hydrolysis induced by reconstituted ribosomes with disrupted (abasic 2450 or 2063, aba) or an intact A2450–C2063 base pair (wt). The amount of GTP hydrolyzed over time is plotted. The GTP input in a single experimental time point was 625 pmol. (B) Products of the ribosome-dependent uncoupled EF-G GTPase reactions were separated via thin-layer chromatography and visualized by phosphor imaging. Values were obtained by quantification of the released inorganic phosphate (Pi) and represent the mean and standard errors of at least three independent time course experiments. Background values (amount of product in the presence of 30S subunits alone) were subtracted from every data point. In the absence of 30S particles, reconstituted 50S subunits do not trigger detectable GTP hydrolysis on EF-G (9).
Disruption of the A2450–C2063 pair interferes with productive tRNA translocation
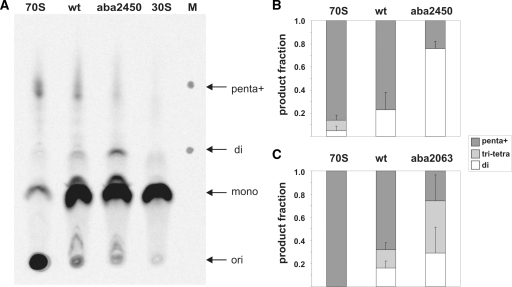
In the standard poly(Phe) assay, the reaction product is analyzed by trichloroacetic acid (TCA) precipitation and subsequent liquid scintillation counting of the radiolabeled poly(Phe) chain. However, it is known that the polypeptide chain has to be larger than 4–7 amino acids in order to be precipitated by TCA (38,39). Thus this detection system does not yield insight into the peptide chain length synthesized on chemically engineered ribosomes. To circumvent these limitations we have used a thin layer chromatography-based detection system (27) of C14-labeled poly(Phe) products synthesized on ribosomes carrying an abasic nucleotide analog either at positions 2450 or 2063. The TLC plates used allow the separation of poly(Phe) peptides based on the increase of hydrophobicity with poly(Phe) chain length extension. The conditions used enable good separation of poly(Phe) length between one and five phenylalanine residues and thus complement the detection gap of the standard TCA precipitation technique. While native ribosomes or reconstituted particles carrying the wt synthetic RNA mainly produced peptides of more than three Phe residues (95–100 and 77–84% for native and reconstituted wt ribosomes, respectively), abasic 2450 ribosomes primarily accumulated only Phe–Phe dipeptides (Figure 4A and B). Seventy-six percent of the product observed during in vitro translation employing abasic A2450 ribosomes was dipeptide and only 24% had chain lengths of three amino acids or longer. The same tendency, albeit again less pronounced, was observed with abasic 2063 ribosomes (Figure 4C). Obviously ribosomes with a disrupted A2450–C2063 can effectively form one peptide bond, which is in good agreement with the unaffected peptidyl transferase activities using puromycin or full-length aa-tRNA substrates (Table 1, Figure 2D), but fail to produce larger polypeptides. Since dipeptidyl-tRNA drop-off from the A-site (Figure 2E) and defects in EF-G GTPase activation (Figure 3) can be excluded as reason for the impaired processivity during the poly(U)-directed poly(Phe) assay, the most likely explanation is a defect in efficient tRNA translocation.
Figure 4.
Effects of A2450–C2063 base pair disruption on the length of the produced poly(Phe) peptides. (A) A representative TLC plate with poly([14C]Phe) peptides synthesized in poly(U)-dependent translation reactions is shown. Lanes show synthesized poly([14C]Phe) peptides and unincorporated [14C]Phe after translation using ribosomes reconstituted with the wt oligo (wt), ribosomes reconstituted with the oligo carrying an abasic site at 2450 (aba2450), native E. coli 70S (70S) as a positive control and E. coli 30S subunits as a negative control. Arrows indicate the loading spots (ori), positions of unincorporated [14C]Phe (mono) as well as the location of di-Phe (di) and penta-Phe peptides (penta) identified on the marker lane (M). Note that peptide products longer than penta-Phe cannot be resolved by this system (indicated by penta+). For quantification the TLC plate was exposed to a phosphoimager screen and analyzed with the Image Quant software. The results of peptide length quantifications using aba2450 (B) or aba2063 (C) ribosomes are shown and compared to native 70S ribosomes as well as to reconstituted wt ribosomes. The produced poly([14C]Phe) peptides were grouped into the length categories ‘di’ (white), ‘tri+tetra’ (light grey) and ‘penta+’ (dark grey) peptides. Radioactivity values measured in reactions containing no 50S ribosomal subunits [30S in (A)] were subtracted as background values from the corresponding category areas. The total amount of poly([14C]Phe) detected on each lane was assigned as 1.0. Graphs represent the results of at least two independent in vitro translation experiments, whereas the error bars indicate standard error.
The most direct assay to assess tRNA translocation on an mRNA template is toeprinting which measures the distance between a pre-hybridized DNA primer on the mRNA and the ribosome via primer extension. Employing the toeprinting assay with ribosomes containing gapped-cp-reconstituted ribosomes turned out to be challenging, mainly due to high background levels of reverse transcriptase stops caused by the known low in vitro assembly efficiencies for functional 50S subunits (21) (see Supplementary Data). Nevertheless, executing the toeprinting analysis with ribosomes carrying an abasic 2450 site repeatedly reduced EF-G-dependent translocation of tRNAs from the PRE to the POST state by 30 ± 5% (determined from nine independent experiments) compared to the reconstituted wt control (Supplementary Figure S3). However, placing an isoG at position 2450, a modification that did not markedly inhibit poly(Phe) production (Table 1), did also not influence translocation in the toeprinting assay (data not shown). Furthermore, the removal of the adenine base at A2453, and thereby disrupting the A2453–C2499 base pair, did not affect translocation efficiencies (Supplementary Figure S3). These data support the conclusion based on the poly(Phe) length measurements (Figure 4), that the disruption of the A2450–C2063 base pair interferes with efficient tRNA movement through the PTC.
Integrity of the A2450–C2063 pair is crucial for the translation of a natural mRNA
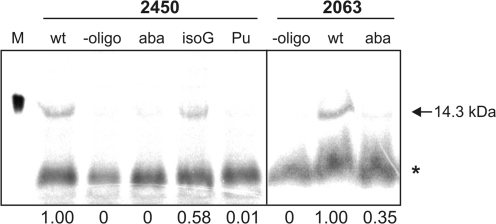
To evaluate the reliability of the poly(Phe) synthesis data on A2450 and C2063 mutant ribosomes under more physiologically relevant conditions, we employed chemically engineered ribosomes for the first time in an in vitro translation assay with a natural mRNA. To this end, the S8 r-protein mRNA from Methanococcus thermolithotrophicus was used as a template for in vitro translation in the presence of labeled [35S]Met and [35S]Cys and the S100 cell extract from E. coli. Successful translation of this 130 codon long mRNA, which requires 129 translocation reactions yielding a protein of 14.3 kDa, was monitored by SDS-PAGE. Ribosomes containing in vitro reconstituted 50S subunits carrying the wt synthetic RNA oligo in the PTC were capable of polymerizing 130 consecutive amino acids to produce clearly detectable amounts of full-length S8 r-protein (Figure 5). Quantification of the produced protein showed that ribosomes carrying modifications that disrupt or weaken the A2450–C2063 base pair (2450 abasic, 2450 purine, 2063 abasic) have lost their ability to translate full-length protein (Figure 5), thus confirming the poly(Phe) synthesis data (Figure 2B and C). Placing the isoG nucleotide analog at position 2450, a modification that was active in the poly(Phe) assay, was also tolerated in the S8 mRNA translation system, although full length protein production was slightly less efficient (Figure 5).
Figure 5.
Natural mRNA in vitro translation is impaired on ribosomes with a disrupted A2450–C2063 base pair. A representative SDS-PAGE of total translation reactions programmed with S8 r-protein mRNA demonstrates full-length [35S]-labeled S8 protein (arrow) produced by reconstituted ribosomes carrying the wt oligo in the PTC. Additional lanes show translation products of ribosomes reconstituted with oligos carrying the abasic site modification (aba) at 2450 or 2063, isoguanosine at 2450 (isoG), purine at 2450 (Pu), or particles reconstituted without synthetic oligo (-oligo). The marker lane (M) shows [35S]-labeled S8 protein produced in vivo in E. coli BL21(DE3) cells. The gel was exposed to a phosphoimager screen and quantified (Image Quant). Background values (radioactivity detected in the areas corresponding to the full-length product in the ‘-oligo’ lanes) were subtracted upon quantification. The asterisk marks a non-specific smear unrelated to translation (see ‘-oligo’ lanes). The quantified relative amounts of the full-length protein (wt was taken as 1.00) is given under the respective lanes.
Disruption of the A2450–C2063 pair influences the conformation of A2062
In order to understand the functional defects caused by disrupting the A2450–C2063 base pair and to explore possible structural rearrangements in the active site we run MD simulations. In the vacant ribosome the only gross conformational alterations detected upon disruption of the A2450–C2063 pair were in the dynamics of A2062. In the wt control the adenine base of A2062 is very flexible and it sampled a space of ∼3.8 Å during the time of the simulation. In contrast, A2062 markedly loses its nucleobase flexibility upon disruption of the A2450–C2063 base pair resulting in a restricted motion of only ∼1.2–2 Å (Supplementary Figure S4A). In the presence of bound tRNA substrates, by using the most recently published crystallographic structure of the PTC carrying simultaneously two full length tRNA analogs in A- and P-site (32), additional features of the A2450–C2063 base pair became evident. In the wt situation, the previously highly mobile nucleobase of A2062 seems to have reached and contacted the amino acid side chain of the aa-tRNA analog in the A-site (Figures 6A). In clear contrast, when the A2450–C2063 base pair was disrupted upon removal of either nucleobase at positions 2450 or 2063, A2062 was frozen in a conformation precluding interactions with the amino acid of the A-site tRNA (Figure 6). Significantly, solely removing the N6 exocyclic amino group at A2450 by introducing a purine analog resulted in the loss of this A2062–aa-tRNA interaction in the A-site (Figure 6B). In all the investigated cases where the A2450–C2063 pair was disrupted by A2450 modifications the nucleobase at A2062 lost its inherent flexibility and populated conformations not sampled by this adenine base in the wt control simulation (Supplementary Figure S4C).
Figure 6.
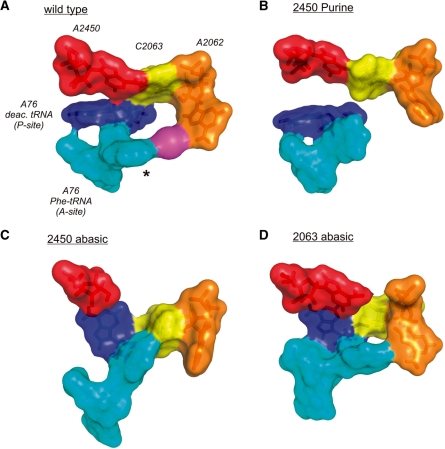
Effects of modulating the A2450–C2063 pair on the PTC architecture. (A) In the surface representation of the structure obtained at the end of the MD simulation of the wt PTC, a continuous ‘interaction ring’ is formed involving the A2450–C2063 pair, A2062, a constrained monovalent ion (magenta), the phenylalanine side chain (asterisk) of A-site bound Phe-tRNAPhe and A76 of deacylated tRNA in the P-site. (B) Removal of the adenine N6-amino group by introducing purine at residue 2450 resulted in the loss of base pairing with C2063 and a complete disconnection of A2062 and the A-site tRNA. (C) Removal of the entire nucleobase at 2450 severely affected the PTC architecture also resulting in the loss of interactions of A2062 with the aa-tRNA in the A-site. Instead the amino acid side chain of Phe-tRNAPhe seems to interact with the nucleobase at C2063. (D) Removal of the base at 2063 showed less dramatic effects since A2062 was still able to reach the aa-tRNA in the A-site. However, the A2450–C2063 base pair is destroyed as well as A76 of P-tRNA is positioned significantly different in respect to A2450 as compared to the wt situation in (A).
DISCUSSION
The PTC is the catalytic heart of the ribosome and its inner core is composed of five universally conserved 23S rRNA residues (Figure 1). Removal or chemical modification of the nucleobases of these nucleotides via an ‘atomic mutagenesis’ approach, however, had only minor consequences on the two chemical reactions promoted by the PTC, namely peptide bond formation and pept-tRNA hydrolysis (2,35). In order to deepen our molecular insight, we have now employed the chemically engineered ribosomes of T. aquaticus in a more physiological relevant set-up, namely during in vitro translation. With this test system it is possible to assess the functional role of the universally conserved PTC nucleobases during multiple rounds of the ribosomal elongation cycle which in short consists of EF-Tu-assisted delivery of aa-tRNAs to the A-site, peptide bond synthesis, and EF-G-driven tRNA movement.
Here, we demonstrate that the individual removal of the inner core PTC nucleobases at 23S rRNA positions A2451, U2506 and U2585 or the complete deletion of residue A2602 did not significantly affect in vitro translation (Figure 2, Table 1). We would like to note that even though the adenine base at position 2451 has previously been suggested to play a direct role in the formation of peptide bonds (40), its complete removal did not significantly affect in vitro translation activities (Table 1) (35). This finding, however, is fully compatible with previous studies pointing at the ribose 2′-OH group of A2451 as the most critical functional group at this pivotal PTC nucleotide for peptide bond synthesis (20–22). The conformationally most flexible inner core residues are A2602 and U2585 (36,41–44). It has been proposed that the N1 of A2602 and the O4 of U2585 are in direct contact with the CCA ends of both A- and P-site tRNAs and thus might play a crucial role in the orchestrated movement of the tRNAs during EF-G promoted translocation (36). Structural flexibility is assumed to be a prerequisite to accompany or even trigger the movement of the tRNAs during translocation. A2602 and U2585 reach into the void lumen of the active site crevice and are the only ones located on and presumably blocking the translocation path for the CCA end of pept-tRNA from the A-to the P-site. Removing the nucleobases at U2585 and A2602 individually or simultaneously, however, did not inhibit poly(Phe) peptide synthesis (Tabel 1), thus arguing against rate limiting functions of driving tRNA movement.
The sole inner core nucleobase that negatively influenced in vitro translation activities upon its ablation was C2063 (Figures 2C and 5, Table 1). In all available crystallographic structures C2063 forms a non-Watson-Crick base pair with A2450 within the PTC (Figure 1). Potential functional importance of this non-conventional A–C base pair is highlighted by its conservation among all three domains of life (37). Nucleobase exchanges at these positions cause lethal phenotypes in both prokaryal and eukaryal organisms (45). However, by using affinity purified ribosomes, in vivo derived A2450–C2063 mutant particles could be tested for PTC functionality (37,46). In both studies the A2450–C2063 pair was mutated to the isosteric G2450:U2063 in the context of E. coli ribosomes, yet yielded quite dissimilar results. While Strobel and colleagues reported an ∼200-fold reduced peptidyl transferase activity (46), Dahlberg and colleagues saw almost wt levels of product formation in an in vitro translation assay (37). Changing the A2450–C2063 wobble pair into a G2450:U2063 base pair simultaneously disrupts several tertiary interactions inside the PTC thus highlighting the limitations of the standard mutagenesis approach in deciphering the functional role of active site residues at the molecular level.
Therefore we applied the less invasive and more precisely focused atomic mutagenesis approach, which enables manipulations at the functional group rather than on the entire nucleobase level (35), to investigate the role of the A2450–C2063 interaction during translation of poly(U) or the mRNA coding for the r-protein S8. We show that all modifications that weaken (2450 purine, 2063 2-pyridone) or disrupt (2450 aba, 2063 aba, 2063 C3-linker) this base pair harm in vitro translation activities (Figures 2 and 5, Table 1), while having little effect on peptide bond formation (Table 1, Figure 2D), pept-tRNA drop-off (Figure 2E), and EF-G GTPase activation (Figure 3). Thus it seems that disruption of the A2450–C2063 base pair inhibits a reaction following transpeptidation and EF-G action during the elongation cycle. Obviously manipulation of the A-C pair obstructs the capability of the ribosome in its processivity to form polypeptides. Size analysis of the poly(Phe) chain showed a clear reduction of the peptide length with an overrepresentation of Phe–Phe dipeptides in ribosomes with a disrupted A2450–C2063 pair (Figure 4A and B). These findings suggest that the integrity of the A2450–C2063 base pair might be crucial for effective tRNA translocation through the PTC during protein synthesis. While we can not completely disregard the possibility that the observed defects in translation activity are the result of the introduced modification at either of the individual residues A2450 or C2063, the data presented in Figures 2 and 5 are more compatible with the notion that it is the physical A–C base pair interaction that matters for effective PTC functionality.
What is the molecular basis of these translation defects of ribosomes with a disrupted A2450–C2063 base pair? A2450 stacks on A2451, the central PTC residue whose ribose 2′-OH has been revealed to be directly involved in peptide bond formation (22), and is, furthermore, involved in an A-minor interaction with A76 of P-site tRNA (47). Nevertheless, the loss of these structural duties of A2450 obviously did not markedly influence catalysis in the puromycin reaction as well as in the dipeptide bond formation assay under the applied conditions (Table 1, Figure 2D and E). These findings hint at a more elaborate role of the adenine nucleobase at position 2450 and/or the universally conserved A2450–C2063 base pair. To reveal this potential functional role we have conducted MD simulations starting from published PTC structures in the absence and presence of tRNA substrates in the A- and P-sites (31,32). On the grounds of the MD simulations we assume a connection between the A2450–C2063 base pair integrity and conformational changes at A2062 (Supplementary Figure S4). A2062 is located at the entrance of the nascent peptide exit tunnel and is known as a very dynamic 23S rRNA residue (48), which was also borne out by our MD simulations (Supplementary Figure S4). Disruption of the A-C base pair by introducing an abasic site at position 2450 or 2063 was accompanied by the loss of this inherent flexibility at A2062, while other PTC residues were largely unaffected. Bacteria carrying mutations at A2062 are viable, and mutant ribosomes do not interfere with in vitro translation [(49) and references therein]. In agreement, removal of the adenine base at A2062 also did not inhibit in vitro translation in our reconstituted system (Supplementary Figure S5). A2062 thus appears to have a more elaborate role during protein synthesis. Indeed mutations at A2062 have been shown to be critical for the drug-dependent ribosome stalling that occurs as a consequence of a concerted interaction of the nascent peptide and the bound macrolide antibiotic within the exit tunnel (49). Pyrimidine mutations of A2062 abolished the formation of the stalled ribosomal complex. It has been suggested that drug-dependent ribosome stalling triggers a conformational change in the PTC which results in translation arrest (49). A2062 is the direct neighbor of residue C2063, which in turn is involved in the crucial inner core A2450–C2063 base pair investigated herein. Based on this structural connection to the heart of the PTC it has been suggested that A2062 might be the sensor of the nature of the nascent peptide in the tunnel and might communicate this signal ‘back’ to the catalytic core (49). The requirement of such pathways, which regulatory nascent peptides use to ‘back-talk’ to the PTC and induce translation stalling, arises from recent studies of various nascent peptide-mediated ribosome stalling complexes (50). Our data are in general compatible with this theory and might add another layer of molecular information for this ‘back-talk’ hypothesis. Our MD simulations show that the initially highly dynamic nucleobase at A2062 in the empty PTC (Supplementary Figure S4) interacts in the PRE translocation state ribosome with the amino acid side chain of the aa-tRNA in the A-site (via a monovalent ion) (Figure 6). A similar interaction between A2062 and the amino acid of a nascent polypeptide has very recently been observed via cryo-EM (51). In a surface representation of the PTC it appears that a continuous ‘interaction ring’ is established involving the critical A2450–C2063 pair, the nucleobase of A2062, the amino acid of the aa-tRNA in the A-site and A76 of deacylated tRNA in the P-site (Figure 6A). Modifications of A2450 that disrupt the non-Watson-Crick interaction with C2063 concomitantly destroyed this ‘interaction ring’ which is evident by the inability of the nucleobase at A2062 to interact with the amino acid of the aa-tRNA (Figure 6B and C). Significantly, solely removing the N6 exocyclic amino group at A2450 by introducing a purine analog resulted in the loss of this A2062–aa-tRNA interaction in the A-site (Figure 6B). Consistent with our experimental data, the disruption of the A-C pair by modifying C2063 had less drastic effects on the establishment of this ‘interaction ring’ of the PTC compared to A2450 manipulations, since the A2062–pept-tRNA interaction remains possible (Figure 6D). In summary, our data are compatible with the hypothesis that the universally conserved A2450–C2063 base pair may be the PTC recipient of signals transmitted from the nascent polypeptide in the exit tunnel (via A2062). This signal could subsequently modulate the integrity of the A2450–C2063 which in turn can regulate the efficiency of tRNA translocation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Austrian Science Foundation FWF (Y315 to N.P.) and the Austrian Ministry of Science and Research (GenAU project consortium ‘non-coding RNAs’ D-110420-012-012 to N.P.). M.A. was supported by grants from the Austrian Science Foundation FWF (I317) and the Austrian Ministry of Science and Research (GenAU project consortium ‘non-coding RNAs’ P0726-012-012) to Ronald Micura. Funding for open access charge: Austrian Science Foundation (Y315 to N.P.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We like to thank Ronald Micura, Yuen-Ling Chan, Sean Connell, Dimitri Shcherbakov, Oliver Vesper, Wolfgang Piendl, Gerald Brosch and Alexander Hüttenhofer for valuable comments and suggestions. Our thanks are extended to Tanel Tenson and Aivar Liiv for providing the E. coli EF-G, EF-Tu, EF-Ts and PheRS clones. Martin Taschler is acknowledged for experimental help during a lab rotation.
REFERENCES
- 1.Fox GE, Naik AK. The evolutionary history of the ribosome. In: de Pouplana LR, editor. The Genetic Code and the Origin of Life. Texas: Landes Bioscience; 2004. pp. 92–105. [Google Scholar]
- 2.Erlacher MD, Polacek N. Ribosomal catalysis: the evolution of mechanistic concepts for peptide bond formation and peptidyl-tRNA hydrolysis. RNA Biol. 2008;5:5–12. doi: 10.4161/rna.5.1.5922. [DOI] [PubMed] [Google Scholar]
- 3.Maden BE. Historical review: peptidyl transfer, the Monro era. Trends Biochem. Sci. 2003;28:619–624. doi: 10.1016/j.tibs.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Polacek N, Mankin AS. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. 2005;40:285–311. doi: 10.1080/10409230500326334. [DOI] [PubMed] [Google Scholar]
- 5.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 6.Wilson DN, Blaha G, Connell SR, Ivanov PV, Jenke H, Stelzl U, Teraoka Y, Nierhaus KH. Protein synthesis at atomic resolution: mechanistics of translation in the light of highly resolved structures for the ribosome. Curr. Protein Pept. Sci. 2002;3:1–53. doi: 10.2174/1389203023380846. [DOI] [PubMed] [Google Scholar]
- 7.Wilson KS, Noller HF. Molecular movement inside the translational engine. Cell. 1998;92:337–349. doi: 10.1016/s0092-8674(00)80927-7. [DOI] [PubMed] [Google Scholar]
- 8.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr. Op. Chem. Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clementi N, Chirkova A, Puffer B, Micura R, Polacek N. Atomic mutagenesis reveals A2660 of 23S ribosomal RNA as key to EF-G GTPase activation. Nature Chem. Biol. 2010;6 doi: 10.1038/nchembio.341. doi:10.1038/nchembio.341. [DOI] [PubMed] [Google Scholar]
- 10.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 11.Gavrilova LP, Spirin AS. Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Lett. 1971;17:324–326. doi: 10.1016/0014-5793(71)80177-1. [DOI] [PubMed] [Google Scholar]
- 12.Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J. Biol. Chem. 1969;244:1533–1539. [PubMed] [Google Scholar]
- 13.Bergemann K, Nierhaus KH. Spontaneous, elongation factor G independent translocation of Escherichia coli ribosomes. J. Biol. Chem. 1983;258:15105–15113. [PubMed] [Google Scholar]
- 14.Beringer M, Adio S, Wintermeyer W, Rodnina M. The G2447A mutation does not affect ionization of a ribosomal group taking part in peptide bond formation. RNA. 2003;9:919–922. doi: 10.1261/rna.5600503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beringer M, Bruell C, Xiong L, Pfister P, Bieling P, Katunin VI, Mankin AS, Bottger EC, Rodnina MV. Essential mechanisms in the catalysis of peptide bond formation on the ribosome. J. Biol. Chem. 2005;280:36065–36072. doi: 10.1074/jbc.M507961200. [DOI] [PubMed] [Google Scholar]
- 16.Polacek N, Gaynor M, Yassin A, Mankin AS. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature. 2001;411:498–501. doi: 10.1038/35078113. [DOI] [PubMed] [Google Scholar]
- 17.Polacek N, Gomez MG, Ito K, Nakamura Y, Mankin AS. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol. Cell. 2003;11:103–112. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J, Kim DF, O'Connor M, Lieberman KR, Bayfield MA, Gregory ST, Green R, Noller HF, Dahlberg AE. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl Acad. Sci. USA. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amort M, Wotzel B, Bakowska-Zywicka K, Erlacher MD, Micura R, Polacek N. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 2007;35:5130–5140. doi: 10.1093/nar/gkm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erlacher MD, Lang K, Shankaran N, Wotzel B, Huttenhofer A, Micura R, Mankin AS, Polacek N. Chemical engineering of the peptidyl transferase center reveals an important role of the 2′-hydroxyl group of A2451. Nucleic Acids Res. 2005;33:1618–1627. doi: 10.1093/nar/gki308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erlacher MD, Lang K, Wotzel B, Rieder R, Micura R, Polacek N. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2′-OH at A2451 of 23S rRNA. J. Am. Chem. Soc. 2006;128:4453–4459. doi: 10.1021/ja0588454. [DOI] [PubMed] [Google Scholar]
- 22.Lang K, Erlacher M, Wilson DN, Micura R, Polacek N. The role of 23S ribosomal RNA residue A2451 in peptide bond synthesis revealed by atomic mutagenesis. Chem. Biol. 2008;15:485–492. doi: 10.1016/j.chembiol.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Dorner S, Panuschka C, Schmid W, Barta A. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic Acids Res. 2003;31:6536–6542. doi: 10.1093/nar/gkg842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 25.Koch M, Huang Y, Sprinzl M. Peptide-bond synthesis on the ribosome: no free vicinal hydroxy group required on the terminal ribose residue of peptidyl-tRNA. Angew. Chem. Int. Ed. Engl. 2008;47:7242–7245. doi: 10.1002/anie.200801511. [DOI] [PubMed] [Google Scholar]
- 26.Szaflarski W, Vesper O, Teraoka Y, Plitta B, Wilson DN, Nierhaus KH. New features of the ribosome and ribosomal inhibitors: non-enzymatic recycling, misreading and back-translocation. J. Mol. Biol. 2008;380:193–205. doi: 10.1016/j.jmb.2008.04.060. [DOI] [PubMed] [Google Scholar]
- 27.Chan YL, Wool IG. The integrity of the sarcin/ricin domain of 23 S ribosomal RNA is not required for elongation factor-independent peptide synthesis. J. Mol. Biol. 2008;378:12–19. doi: 10.1016/j.jmb.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 28.Triana-Alonso FJ, Spahn CM, Burkhardt N, Rohrdanz B, Nierhaus KH. Experimental prerequisites for determination of tRNA binding to ribosomes from Escherichia coli. Methods Enzymol. 2000;317:261–276. doi: 10.1016/s0076-6879(00)17019-3. [DOI] [PubMed] [Google Scholar]
- 29.Bommer AU, Burkhardt N, Jünemann R, Spahn CM, Triana-Alonso FJ, Nierhaus KH. Ribosomes and polysomes. In: Graham J, Rickwood J, editors. Subcellular Fractionation – A Practical Approach. Washington, DC: IRL Press; 1996. pp. 271–301. [Google Scholar]
- 30.Gruber T, Kohrer C, Lung B, Shcherbakov D, Piendl W. Affinity of ribosomal protein S8 from mesophilic and (hyper)thermophilic archaea and bacteria for 16S rRNA correlates with the growth temperatures of the organisms. FEBS Lett. 2003;549:123–128. doi: 10.1016/s0014-5793(03)00760-9. [DOI] [PubMed] [Google Scholar]
- 31.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Structural insights into the roles of water and the 2' hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.Chirkova A, Erlacher M, Micura R, Polacek N. Chemically engineered ribosomes: a new frontier in synthetic biology. Curr. Org. Chem. 2010;14:148–161. [Google Scholar]
- 36.Agmon I, Auerbach T, Baram D, Bartels H, Bashan A, Berisio R, Fucini P, Hansen HA, Harms J, Kessler M, et al. On peptide bond formation, translocation, nascent protein progression and the regulatory properties of ribosomes. Eur. J. Biochem. 2003;270:2543–2556. doi: 10.1046/j.1432-1033.2003.03634.x. [DOI] [PubMed] [Google Scholar]
- 37.Bayfield MA, Thompson J, Dahlberg AE. The A2453-C2499 wobble base pair in Escherichia coli 23S ribosomal RNA is responsible for pH sensitivity of the peptidyltransferase active site conformation. Nucleic Acids Res. 2004;32:5512–5518. doi: 10.1093/nar/gkh888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg NA, Shipe WF. Comparison of the abilities of trichloroacetic, picric, sulfosalicyclic, and tungstic acids to precipitate protein hydrolysates and proteins. J. Food Sci. 1979;44:735–737. [Google Scholar]
- 39.Yvon M, Chabanet C, Pelissier JP. Solubility of peptides in trichloroacetic acid (TCA) solutions. Hypothesis on the precipitation mechanism. Int. J. Peptide Protein Res. 1989;34:166–176. doi: 10.1111/j.1399-3011.1989.tb00227.x. [DOI] [PubMed] [Google Scholar]
- 40.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 41.Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HA, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 42.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 43.Wilson DN, Schluenzen F, Harms JM, Yoshida T, Ohkubo T, Albrecht R, Buerger J, Kobayashi Y, Fucini P. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 45.Rakauskaite R, Dinman JD. rRNA mutants in the yeast peptidyltransferase center reveal allosteric information networks and mechanisms of drug resistance. Nucleic Acids Res. 2008;36:1497–1507. doi: 10.1093/nar/gkm1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hesslein AE, Katunin VI, Beringer M, Kosek AB, Rodnina MV, Strobel SA. Exploration of the conserved A+C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes. Nucleic Acids Res. 2004;32:3760–3770. doi: 10.1093/nar/gkh672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simonovic M, Steitz TA. A structural view on the mechanism of the ribosome-catalyzed peptide bond formation. Biochim. Biophys. Acta. 2009;1789:612–623. doi: 10.1016/j.bbagrm.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 49.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 50.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 51.Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2010;17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 52.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.